Abstract
Neurodegenerative disorders are characterized by the progressive loss of neuronal structure or function, resulting in memory loss and movement disorders. Although the detailed pathogenic mechanism has not been elucidated, it is thought to be related to the loss of mitochondrial function in the process of aging. Animal models that mimic the pathology of a disease are essential for understanding human diseases. In recent years, small fish have become ideal vertebrate models for human disease due to their high genetic and histological homology to humans, ease of in vivo imaging, and ease of genetic manipulation. In this review, we first outline the impact of mitochondrial dysfunction on the progression of neurodegenerative diseases. Then, we highlight the advantages of small fish as model organisms, and present examples of previous studies regarding mitochondria-related neuronal disorders. Lastly, we discuss the applicability of the turquoise killifish, a unique model for aging research, as a model for neurodegenerative diseases. Small fish models are expected to advance our understanding of the mitochondrial function in vivo, the pathogenesis of neurodegenerative diseases, and be important tools for developing therapies to treat diseases.
Keywords: mitochondria, neurodegenerative disorders, zebrafish, medaka, turquoise killifish
1. Introduction
Neurodegenerative disorders are characterized by the progressive loss of structure or function of neurons and include Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), amyotrophic lateral sclerosis (ALS), spinocerebellar ataxia (SCA), and multiple-system atrophy (MSA). The progressive loss or dysfunction of selectively vulnerable neurons leads to a multitude of symptoms, including memory loss, movement disorders, and behavioral changes [1]. Oxidative stress and inflammation are thought to contribute to progressive neurodegenerative disorders, but the detailed mechanisms of these pathologies remain to be elucidated [2,3,4,5]. In addition, the multifactorial etiology and lack of established biomarkers to predict disease progression contribute to the challenges associated with neurodegenerative disorders. One of the most significant risk factors for neurodegenerative diseases is aging. Although various mechanisms of aging have been proposed, it has long been believed that increased reactive oxygen species (ROS) originating from mitochondria cause oxidative damage, leading to cellular dysfunction and tissue failure (the mitochondrial free radical theory of aging; MFRTA) [6,7,8]. Notably, several studies have shown the link between ROS generation and oxidative stress during aging; however, this theory remains controversial [9].
Mitochondria are unique intracellular organelles that are covered by a double membrane, have their own genome, and can self-replicate independently. They are present in all nucleated cells and perform many functions, including cellular metabolism, energy production, and homeostasis. Mitochondrial damage and dysfunction are caused by mutations in nuclear DNA that encode mitochondrial proteins or mitochondrial DNA (mtDNA), and by cellular stress due to environmental factors. There is a link between pathophysiological changes in several neurodegenerative diseases and mitochondrial dysfunction associated with aging, including oxidative stress and reduced adenosine triphosphate (ATP) production capacity [10,11,12,13,14,15,16]. Notably, the loss of neurons is caused by apoptosis regulated by mitochondria [17]. Therefore, mitochondria could be promising therapeutic targets for preventing age-related diseases.
In this review, we will discuss the roles of mitochondria that affect the process of neurodegenerative disorders. First, we will highlight the connection between mitochondrial dysfunction and neurodegenerative disorders. Then, we will introduce the small fish models (zebrafish, medaka, and turquoise killifish) as beneficial in vivo vertebrate models for studying mitochondrial biology. We will summarize several approaches to studying mitochondrial function using small fish and discuss the advantages and challenges. Additionally, we will discuss the potential of small fish models to contribute to the development of therapeutic strategies for age-related neurodegenerative disorders. It is important to emphasize that mitochondrial dysfunction is not the only factor in aging and neurodegenerative disease, but this review will facilitate understanding of this aspect.
2. Mitochondrial Dysfunctions Affecting the Neurodegenerative Process
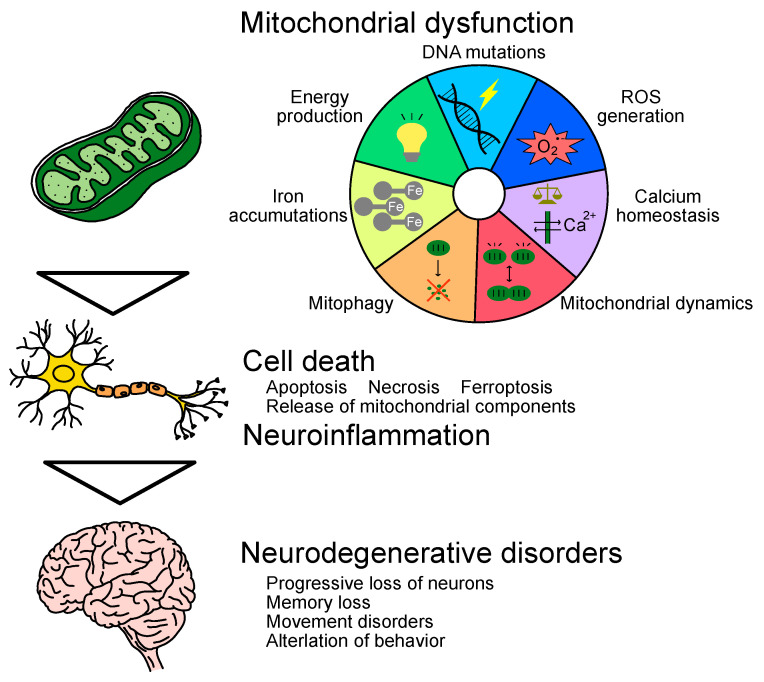
Mitochondrial functions are diverse and complex and essential for cellular homeostasis and survival. Therefore, mitochondrial dysfunction with age leads to cell death and contributes to the progression of neurodegenerative disorders [17] (Figure 1). Mitochondria are estimated to contain 1000–1500 kinds of proteins, of which only 13 are encoded in mtDNA and the rest are encoded in nuclear DNA [18,19]. Proper mitochondrial function depends on the quality control system, such as the transport and translocation of proteins, the turnover of proteins via the ubiquitin–proteasome system, mitochondrial dynamics, as well as the elimination of mitochondria through mitophagy [20]. In the following subsection, we will describe the age-related decline in mitochondrial function and its relation to neurodegenerative disorders.
Figure 1.
Multifactorial effects of mitochondrial dysfunction in the process of neurodegenerative disorders.
2.1. DNA Mutations
Mitochondrial dysfunction is often caused by mutations in nuclear DNA involved in mitochondrial components and maintenance or in mitochondrial DNA. DNA is exposed to both exogenous physical, chemical, and biological stress and endogenous stress from the production of ROS and failed DNA replication. Chromosome aneuploidy caused by abnormal mitosis in the aged brain has been implicated in neurodegenerative diseases [21]. Aneuploidy is also known to lead to mitochondrial dysfunction and increased ROS production, as well as an acceleration of cellular senescence [22]. In addition, mitochondrial DNA is more prone to accumulate mutations than nuclear DNA [23,24,25]. This is due to the lack of histones, ROS generation in the inner membrane, limited repair mechanisms in mitochondrial DNA, and higher replication frequency than nuclear DNA [26,27,28,29]. Moreover, mitochondrial DNA has very few non-coding sequences, with the result that mutations affect functional genes directly [29]. It might be challenging to protect DNA from mutations with age; therefore, the maintenance of its quality control system is essential. Proper regulation of the balance between the removal of damaged mitochondria and the biosynthesis of new mitochondria is important for aging and longevity [30,31].
2.2. Energy Production
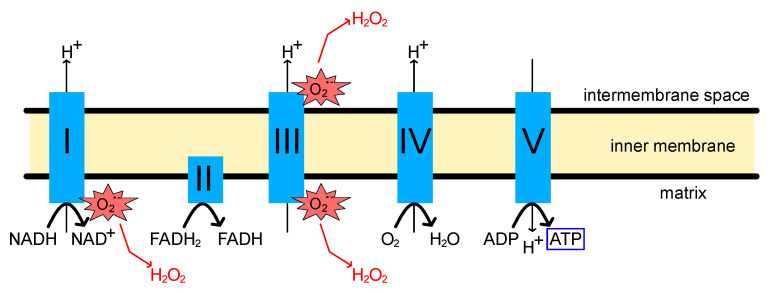
The primary function of mitochondria is to generate ATP via oxidative phosphorylation (OXPHOS). This reaction is carried out by the electron transport chain (ETC) consisting of four respiratory chain (RC) complexes (complexes I-IV) and ATP synthase (complex V), which are present in the mitochondrial inner membrane [32,33,34] (Figure 2). High-energy phosphate production is achieved by coupling electron transfer to proton translocation across the mitochondrial inner membrane, resulting in an electrochemical gradient. It has been suggested that the loss of OXPHOS function may cause various disorders, including non-functional synapses, axonal degeneration, increased ROS production, and cell death [35,36]. Cytochrome c is a small protein tethered to the mitochondrial inner membrane by cardiolipin and functions as an electron donor and receptor during OXPHOS. The release of cytochrome c from mitochondria promotes apoptosis via activation of caspase-9, and the subunit of the RC complex acts as a substrate for caspase [37,38,39,40]. Moreover, age-related decline in ATP levels promotes necrotic inflammation, which may trigger a progression of age-dependent disorders [41].
Figure 2.
Schematic image of oxidative phosphorylation (OXPHOS) process and reactive oxygen species (ROS) production.
There is a link between declines in overall bioenergetic function and the phenotype of aging [42,43]. The brain is particularly vulnerable to a decrease in bioenergetic function due to its high energy demands and relatively high mitochondrial mass. With aging, decreased activity of RC complex I, decreased ATP production capacity, and cytochrome c release have been observed in the brain [44,45,46]. An increased number of cytochrome c oxidase (COX)-deficient neurons with age have also been reported in the substantia nigra and hippocampus in normal human brains [47,48]. Several studies have shown mitochondrial dysfunction and reduced mitochondrial complex I activity in the substantia nigra and frontal cortex of PD patients [49,50,51]. Similarly, mitochondrial complex I dysfunction has been reported in the skeletal muscle and platelets of PD patients [52]. In addition, the induction of a familial PD gene mutation into neuronal cells caused defective complex I activity and synaptic function [53]. Reduced complex IV activity was also observed in postmortem homogenates of AD and PD patients [54,55,56]. Interestingly, a decrease in mitochondrial respiration associated with a decline in the electron transfer rate of complexes I and IV among RC complexes was consistently observed in aging and neurodegenerative diseases. On the other hand, it has been reported that decreased complex II and III activity with increased complex I and IV activity occurred in MSA cerebellar white matter [57]. In addition, decreased mRNA expression of all mitochondrial complexes subunits (I-V) has been observed in the frontal cortical and angular gyrus in PD with dementia [58]. The relationship between these disease-, symptom-, or region-specific alterations of RC complex and neurodegenerative pathogenesis needs to be elucidated in future studies.
2.3. Reactive Oxygen Species/Oxidative Stress
The mitochondrial RC is the primary site of ROS production in the cell [59,60] (Figure 2). ROS produced in the OXPHOS process oxidizes nucleic acids, lipids, and proteins, causing damage, especially within the source origin, mitochondria [61,62,63,64]. Mitochondria possess antioxidant systems to prevent oxidative damage, and properly regulated ROS can trigger various signaling pathways and regulate autophagy [65,66,67]. However, accumulated oxidative damage to mitochondria due to aging and other factors can affect ATP production and other essential functions in mitochondria [68,69,70]. Moreover, ROS themselves also increase mitochondrial membrane permeability, leading to additional ROS release (ROS-induced ROS release, RIRR) [60,71].
The negative cycle associated with ROS production significantly impacts survival in cells with high energy requirements, such as neurons. In addition, the brain is considered vulnerable to oxidative stress due to its high oxygen consumption, an abundance of oxidizable unsaturated fatty acids, and low expression of some antioxidant enzymes [72,73]. Excessive oxidative stress and oxidative changes in mtDNA have been reported in the postmortem brain of AD patients [74,75,76]. Decreased activity of the alpha-ketoglutarate dehydrogenase complex, which is sensitive to oxidants, is a feature found in AD patients’ brains [77,78]. Oxidative damage to the RC complex I was observed in postmortem brain samples from PD patients, as well as oxidative damage to nucleic acids, lipids, and proteins [79,80,81,82]. ALS-associated antioxidant enzyme superoxide dismutase 1 (Sod1) mutant mice showed increased ROS production, decreased expression of NF-E2-related factor 2 (Nrf2), a stress response sensor, and early onset of ALS-like pathology [83,84,85]. Since oxidative damage plays a central role in the common pathophysiology of neurodegenerative diseases, reducing the harmful effects of ROS in the brain may be a promising treatment option to slow the progression of neurodegeneration and alleviate associated symptoms.
2.4. Calcium and Iron Homeostasis
In addition to energy production, mitochondria are the site of critical metabolic and synthetic processes, including fatty acid oxidation, cholesterol synthesis, glucose synthesis, nucleotide synthesis, calcium homeostasis, iron–sulfur clusters (ISC) synthesis, and heme synthesis [86,87]. Here, we focus on the control of calcium and iron levels.
Mitochondrial regulation of calcium levels has a vital role in signaling molecules associated with cell death and cell survival, as well as maintenance of mitochondrial function [88,89]. Mitochondrial regulation of calcium is particularly important in neurons because calcium functions as a second messenger in neurons [90]. To maintain the cytosolic calcium level, the temporary influx of calcium ions that occurs during synaptic activity is taken up by the endoplasmic reticulum and mitochondria and also released to the extracellular space, which requires a large amount of ATP consumption [91,92]. Thus, decreased ATP production capacity affects calcium homeostasis. High cytosolic calcium levels stimulate various Ca2+-dependent catabolic enzymes, such as phospholipases, proteases, and endonucleases, resulting in cell death [93].
In HD patients and mouse models, it has been reported that depolarization at lower calcium loads was caused by mitochondrial calcium abnormalities, which occurred earlier than pathological or behavioral abnormalities [94]. Another study showed that increased cytosolic calcium concentration promoted the degradation of wild-type huntingtin via calcium-dependent proteases, leading to the loss of huntingtin neuroprotective activity [95]. Calcium overload in mitochondria also stimulates ROS generation and releases pro-apoptotic factors such as cytochrome c through the perturbation or rupture of the mitochondrial outer membrane, which triggers calcium-induced cell death [96,97]. To sustain the bioenergetic function of mitochondria, the crosstalk with another calcium storage, the endoplasmic reticulum (ER), is also important. The ε4 allele of apolipoprotein E (APOE4) is considered one of the risk factors of AD. Tambini et al. showed upregulated mitochondria-associated ER membrane (MAM) activity in human fibroblasts or mouse neurons when cultured in APOE4-containing medium, which promotes the transfer of calcium from the ER into the mitochondria [98]. In addition, presenilin (PSEN) mutations in familial AD have been associated with the dysregulation of calcium signaling. PSEN1/2 are abundant in the ER membrane and interact with ER calcium channels such as inositol 1,4,5-trisphosphate receptors (IP3R) and ryanodine receptors (RyR) [99,100,101,102]. It has also been shown that both increased ER-mitochondrial contact sites and the expressions of MAM-related proteins, such as IP3R, RyR, and voltage-dependent anion channel (VDAC1), were found in neurons from sporadic and familial AD patients and AD mouse models [103].
Among the vital metals in the mitochondria, iron plays a central role and is essential for the function of the RC complex. ISC, which is synthesized in the mitochondria, is used for OXPHOS, cellular iron homeostasis, pyrimidine/purine metabolism, tricarboxylic acid cycle (TCA cycle), DNA repair, and heme synthesis [104]. Excessive free iron generates oxidative stress, which is a hallmark of age-related diseases. Iron accumulation within the central nervous system (CNS) was found in AD, PD, HD, and ALS [105,106,107,108,109,110,111,112]. Agrawal et al. demonstrated that human HD and mouse model HD brains accumulated mitochondrial iron and showed increased expression of the iron uptake protein mitoferrin 2 and decreases in the ISC synthesis protein frataxin [113]. Intracellular free iron causes lipid peroxidation and hydroxyl-radical generation, resulting in cell death known as ferroptosis [114]. Lipid peroxidation can transmit from ferroptotic cells to neighboring cells, inducing a chain of further ferroptosis [115]. In ALS, ferroptosis but not necroptosis plays a central role in selective motor neuron death [116]. Therefore, the association of ferroptosis with the pathophysiology of neurodegenerative disorders has gained researchers’ attention [117,118,119,120,121].
2.5. Mitochondrial Dynamics
Mitochondria are dynamic organelles that change their number, size, and DNA copies according to cellular requirements. It has been reported that the copy number of mitochondrial DNA decreases with age [122,123]. Mitochondrial dynamics refers to two opposing phenomena: fission and fusion [124,125]. Both processes are essential for mitochondrial quality control against stress conditions. The rate of mitochondrial fission and fusion depends on metabolic changes and stress intensity. Mitochondrial fission provides a sufficient number of mitochondria to daughter cells during mitosis. Even in non-dividing cells, fission contributes to quality control by isolating damaged mitochondria and targeting them for removal by mitophagy [125,126]. Inhibition of fission in mouse Purkinje cells resulted in morphological abnormalities associated with excess fusion, oxidative damage accumulation, and loss of respiratory function [127]. Excessive mitochondrial fission is an early event in apoptosis and induces apoptosis via permeabilization of the outer membrane [128,129,130]. Mitochondrial fusion can rescue mitochondria with mutations by allowing them to complement each other or mitigate low-level damage by exchanging proteins and lipids with other mitochondria. Therefore, inhibition of fusion leads to the accumulation of mitochondrial damage, resulting in a wide variety of dysfunctions, including heterogeneity of mitochondrial membrane potential, impaired respiratory chain function, disruption of mtDNA integrity, reduced mitochondrial Ca2+ uptake, mitochondrial fragmentation, and apoptosis [131,132,133,134,135].
Disturbances in mitochondrial dynamics have been found to escalate pathogenesis in neurodegenerative disorders [136,137]. Heterogeneous mutations in the mitochondrial fusion gene mitofusin 2 (MFN2) cause the neurodegenerative disease Charcot-Marie-Tooth type 2A (CMT2A) [138,139]. Loss of Mfn2 caused neurodegeneration of Purkinje cells in the cerebellum and dopaminergic neurons [140,141]. In brain tissue from patients with AD and HD, increased expression of fission-related genes such as dynamin related protein 1 (DRP1) and fission protein 1 (FIS1) and decreased expression of fusion-related genes such as MFN1, MFN2, and optic atrophy 1 (OPA1) have been reported, suggesting that excessive fission inducing apoptosis occurs [142,143]. Abnormal interaction of accumulated amyloid-β with DRP1 accelerated mitochondrial fragmentation in AD [142]. Mutant huntingtin also has been reported to interact with DRP1, increasing its enzyme activity and promoting fission [144,145]. Selective inhibition of DRP1 suppressed excessive mitochondrial fragmentation and improved mitochondrial function in cell models of HD and cells derived from HD patients [146]. These disruptions in mitochondrial dynamics potentially have a significant impact on the process of mitophagy.
2.6. Mitophagy
Mitophagy is the removal of dysfunctional mitochondria by autophagy-mediated fusion with lysosomes, which maintain proper cellular homeostasis [147,148]. Mitophagy pathways can occur in response to disturbances such as decreased membrane potential and accumulation of misfolded proteins, and selective mitochondrial fission plays an important role [126,149,150]. Recessive mutations in PTEN-induced putative kinase 1 (Pink1) and Parkin (PARK2) have been identified as genetic causes of familial PD [151,152]. PINK1 is a mitochondria-localized serine-threonine kinase that can phosphorylate ubiquitin to activate Parkin, and Parkin is an E3 ubiquitin ligase in the cytoplasm, and both play central roles in inducing mitophagy [153,154]. In response to mitochondrial damage, such as loss of mitochondrial membrane potential or accumulation of misfolded proteins, PINK1 stabilized on the mitochondrial outer membrane, and Parkin migrated from the cytosol to the damaged mitochondria [153,155]. Disturbed autophagy systems have been reported in other neurodegenerative disorders, such as AD and HD [156]. In HD cellular models, autophagic vacuoles failed to recognize and trap cytosolic cargo, leading to insufficient autophagy and the accumulation of dysfunctional mitochondria [157]. Mitophagy enhancement inhibited amyloid-β and tau pathology in AD models, suggesting mitophagy could be a potential therapeutic target [158].
2.7. Immune System
Mitochondria are thought to have originated as proteobacteria and later became symbiotic in other cells (eukaryotic cells) [159]. Therefore, their components are likely to be recognized as foreign substances by our innate immune system. Mitochondrial DNA is particularly cytotoxic and triggers an innate immune response. In a cultured cell model mimicking Parkinson’s disease, leaked mitochondrial DNA induced an elevated type I interferon response and cell death through the DNA sensor interferon-gamma inducible protein 16 (IFI-16) [160]. In another study, transfection of oxidant-initiated degraded mitochondrial polynucleotides into primary mouse astrocytes stimulated the expression of interleukin 1β (Il-1b), Il-6, monocyte chemotactic protein 1 (Mcp1), and tumor necrosis factor α (Tnfa) [161]. In addition to mitochondrial DNA, mitochondrial components such as oxidized cardiolipin, cytochrome c, ATP, N-formyl peptides, and high mobility group box 1 have been reported to induce inflammatory responses [162,163,164,165,166,167,168,169]. Mitochondrial lysates yielded the expression of Tnfa and Il-8 in a mouse microglial cell line [170]. Interestingly, they also upregulated the expression of amyloid-β precursor protein (App), a precursor of amyloid-β that accumulates in Alzheimer’s disease brains [170]. The microglia of AD patients express cytokines/chemokines such as TNFA, IL-1B, major histocompatibility complex (MHC) class II, cyclooxygenase 2, and MCP1 [171,172]. Similarly, elevated levels of TNFA, interferon γ, IL-2,4,6, and 10 were found in the serum of PD patients [173]. The release of mitochondrial components associated with cell death may induce an immune response and contribute to the progression of neurodegenerative disease with neuroinflammation.
3. Small Fish Models to Study Mitochondrial Function/Dysfunction
Small fish (e.g., zebrafish and medaka) are widely used vertebrate models in developmental genetics and embryology due to the presence of numerous mutants, ease of genetic modification and embryo manipulation, and ease of imaging using transparent embryos and larvae. These have been recognized as human disease models in the last decades because they share a high similarity in genes, organ structures, and disease phenotypes [174,175]. For instance, both zebrafish and medaka have shown PD-like phenotypes by the administration of neurotoxins such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and 6-hydroxydopamine (6-OHDA) [176,177,178,179,180,181]. In the following subsection, the selected examples of genetic models, imaging techniques, and drug screening illustrate the advantages and challenges of small fish models in studying mitochondrial function/dysfunction (Figure 3).
Figure 3.
Advantages and applications of small fish models.
3.1. Genetic Models
Zebrafish and medaka are suitable model organisms to perform gene editing. There are several efficient genome editing methods used for small fishes, such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9 [182]. Small fish release fertilized eggs outside the body, making it easy to introduce genome editing tools by microinjection. In addition, a morpholino antisense oligonucleotide (MO)-based gene knockdown has been widely performed in small fishes. Currently, the mitochondrial gene dysfunction models are mainly evaluated by MO-based gene knockdown (Table 1). This method is easy to introduce, but the effect is temporal, occurring only during the early developmental stage. Furthermore, it has been reported that many genetic knockout models cannot replicate the MO-induced phenotypes, possibly due to off-target effects [183]. Therefore, it is important to establish knockout models or use spontaneous mutants for analyzing gene function [174]. In addition, tissue-specific promoter and/or site-specific recombinase technology (e.g., Cre-Lox recombination system) are required to study tissue-specific effects. Moreover, an inducible recombination system (e.g., heat-shock promoter, chemical-inducible recombination, and light-inducible recombination) may be necessary to analyze the phenotypes in aged populations or avoid lethality. Detailed strategies for spatiotemporal mutagenesis have been summarized in other reviews [184,185]. Notably, fish have undergone a whole-genome duplication that causes them to possess duplicated genes [186,187]. In most cases, one copy loses its function as a pseudogene (nonfunctionalization). However, other cases involve subfunctionalization in which the two copies split the original function, or neofunctionalization in which one copy generates a new function [188]. It is important to remember this fact when analyzing phenotypes and gene function. Here, we summarize zebrafish and medaka models used to study several genes associated with mitochondrial function and mainly neuronal defects. Other models of neurodegenerative disorders can be found in recent reviews [189,190,191,192].
Table 1.
The characteristics of loss or gain of function studied in zebrafish for mitochondrial-related genes.
| Gene | Related Disease | Model | Phenotypes | Refs |
|---|---|---|---|---|
| Neurodegenerative disease model | ||||
| pink1 (park6) | Parkinson’s disease | MO knockdown | Short tail, small eyes and head. Cardiac edema. Enlarged brain ventricles. Reduced number of dopaminergic neurons. Increased caspase-3 activity and ROS levels. | [193] |
| MO knockdown | No alterations in the number of dopaminergic neurons. Disturbed patterning and projection of neurons. | [194] | ||
| MO knockdown | Decreased tyrosine hydroxylase (Th) + neurons. | [195] | ||
| ENU mutagenesis | Reduced number of dopaminergic neurons. Reduced complex I and III activity. Enlarged mitochondria. Increased microglia activity. | [196] | ||
| CRISPR-mediated knockout | Decreased number of dopaminergic neurons and noradrenergic neurons. | [197] | ||
| CRISPR-mediated knockout | Decreased Th + neurons. | [198] | ||
| Parkin (park2) | Parkinson’s disease | MO knockdown | Decreased Complex I activity. Reduced number of dopaminergic neurons. | [199] |
| parl | Parkinson’s disease | MO knockdown | Increased cell death. Low density or mis-patterned dopaminergic neurons. | [200] |
| DJ-1 (park7) | Parkinson’s disease | MO knockdown | No alterations in the number of dopaminergic neurons. Reduced number of dopaminergic neurons under oxidative stress conditions. Increased Sod1 expression level. Increased apoptosis under proteasome inhibition. | [201] |
| MO knockdown | No alterations in the number of dopaminergic neurons. Reduced number of dopaminergic neurons under oxidative stress conditions. Increased apoptosis under the proteasome inhibition condition. | [202] | ||
| CRISPR-mediated knockout | No anomalies in larval development. Small body size. Reduced complex I activity. Reduced Th level in aged fish. | [203] | ||
| CRISPR-mediated knockout | Decreased Th + neurons. | [198] | ||
| lrrk2 | Parkinson’s disease | MO knockdown | Severe embryonic lethality. Small brain, heart edema. Loss of Th + neurons. Deletion of the WD40 domain: Loss of Th + neurons. |
[204] |
| MO knockdown | No alternation in the number of dopaminergic neurons. | [205] | ||
| MO knockdown | Edema, ocular abnormality, abnormal body axis. Reduced number of dopaminergic neurons. Increased ROS level. Increased Sod1 expression level. | [206] | ||
| ZFN-mediated knockout | A weakened antibacterial response. | [207] | ||
| CRISPR-mediated knockout | Increased apoptosis. Reduced number of microglia/leukocytes in the larval brain. Decreased Th + neurons in the larval brain. Progressive increase in monoamine oxidase-dependent catabolism. | [208] | ||
| CRISPR-mediated knockout | No alterations in the number of dopaminergic neurons. | [209] | ||
| sod1 | Amyotrophic lateral sclerosis | Mutant human SOD1 overexpression (temporal) | Abnormal axonal branching. Short axonal length. | [210] |
| Mutant zebrafish sod1 overexpression (stable) | No effect on motor axon outgrowth. Abnormal neuromuscular junction (NMJ). Progressive deficiency in locomotion. (end-stage) with intermittent paralysis. Decreased number of motor neurons. Vacuolated mitochondria. | [211] | ||
| ENU mutagenesis | Decreased NMJ and motor neurons. | [212] | ||
| Neuronal defect | ||||
| mfn2 | Charcot-Marie-Tooth type 2A | MO knockdown | Irregular somite, small eyes, edema in the brain (mild), and small head with encephalic necrosis (severe). Abnormal axonal projections. Underdeveloped motor neurons. Decreased distribution of AChR clusters. Reduced size of myofibers. | [213] |
| ENU mutagenesis | Age-related alteration of NMJ pathology. Reduced number of motile mitochondria. | [214] | ||
| gdap1 | MO knockdown | Reduced density of sensory neurites. Decreased temperature-related activity. | [215] | |
| MO knockdown | Co-suppression of mfn2 + gdap1: Exacerbated phenotype of motor neuron pathology (failed neuronal extension and innervation of myotome) compared with single suppression. | [216] | ||
| slc25a1 | Congenital myasthenic syndromes/ D-2- and L-2-hydroxyglutaric aciduria |
MO knockdown | Abnormal NMJ. Edema of the hindbrain, heart, yolk sac, and tail. | [217] |
| kbp | Goldberg-Shprintzen syndrome | ENU mutagenesis MO knockdown |
Delayed development of peripheral axons. Defects in axonal outgrowth. Axonal degeneration or retraction. Abnormal myelination, microtubule organization, and localization of mitochondria. | [218] |
| actr10 | ENU mutagenesis TALEN-mediated knockout |
Axonal swelling, accumulation of mitochondria. | [219] | |
| PGC-1α (ppargc1a) |
Wallerian degeneration | Laser axotomy + PGC-1α overexpression |
Increased mitochondrial density, attenuated roGFP2 (redox-sensitive sensor) oxidation, delayed degeneration. | [220] |
| SNCA (aSyn) overexpression + PGC-1α overexpression |
Mediated Snca (aSyn) toxicity in axonal neurons. | [221] | ||
| nipsnap1 | CRISPR-mediated knockout | Reduced mitophagy in the head region. Increased ROS production and apoptosis. Loss of dopaminergic neurons. | [222] | |
| Anomaly of brain development | ||||
| tfam | MO knockdown | Decreased mtDNA copy number and OXPHOS activity. Edema, small eyes and brain, non-looped heart, disorganized skeletal muscles. | [223] | |
| opa1 | Optic atrophy | MO knockdown | Abnormal blood circulation, non-looped heart. Small eyes and pectoral fin buds. Obscure midbrain-hindbrain boundary → Enlarged hindbrain ventricle. | [224] |
| MO knockdown | Disturbed mitochondrial network. No effect on sensory neurites and temperature-related activity | [215] | ||
|
surf1
cox5aa cox5ab |
Leigh syndrome | MO knockdown | Impaired COX activity. Shortened rostral-caudal body axis. Abnormal swim bladder, head shape, gut development, jaw formation. Edema, small eyes, and non-looped heart. | [225] |
3.1.1. Neurodegenerative Disease Models
Of the mutated genes that cause familial PD, many encode mitochondria-associated proteins (PINK1, Parkin, PARL, DJ-1, and LRRK2). PINK1 is a protein associated with mitophagy induction through Parkin activation [153]. Knockdown of pink1 in zebrafish reduced the number of dopaminergic neurons [193,195]. Another study of pink1 morphants reported no overall decrease in the number of dopaminergic neurons but disturbed patterning and projection of these neurons [194]. Furthermore, the pink1 null mutant and pink1 knockout model also showed the loss of dopaminergic neurons [196,197,198]. These results suggest that single depletion of pink1 in zebrafish is sufficient to affect dopaminergic neurons and a suitable model of PD. PD is also characterized by movement disorders. Motor deficits have also been observed in many of the pink1 deletion models presented here. Hughes et al. developed a classification method in adult zebrafish movement disorders with PD-like phenotypes using high-resolution video capture and machine learning [198]. These zebrafish models and behavioral assessments will provide further insights into understanding human pathology.
DJ-1 (PARK7) has a role in protecting cells from oxidative and ER stress [226]. Zebrafish knockdown of DJ-1 did not alter the number of dopaminergic neurons; however, they were vulnerable to oxidative stress [201,202]. DJ-1 knockout models showed a reduction in dopaminergic neurons with aging [198,203]. Therefore, mutations in DJ-1 may not directly cause neuronal death, but the weak neuronal cell protection system leads to PD through the accumulation of stress with age.
Leucine-rich repeat kinase 2 (LRRK2) is a multidomain protein interacting with parkin [227,228]. The studies of knockdown or knockout of lrrk2 have reported various but conflicting phenotypes in terms of the number of dopaminergic neurons [204,205,206,207,208,209]. Notably, the mechanism underlying the pathogenic effect of PD by LRRK2 mutation remains unknown because point mutations have been found among different domains [228]. The most frequent mutation in LRRK2 is supposed to be a gain-of-function that increases kinase activity [229,230]. Further investigation will be needed to understand the role of LRRK2 in PD progression by using not only loss-of-function models but also by establishing a gain-of-function model.
Several genetic medaka models of PD have been established. Unlike zebrafish, pink1 or Parkin (park2) single mutations screened from the ENU mutagenesis library did not show dopaminergic cell loss [231,232]. The double deficiency of pink1 and Parkin (park2) led to a deterioration of motor function and loss of dopaminergic neurons [232]. DJ-1 knockout medaka was also established by TALEN and CRISPR/Cas9 systems, but the phenotypes were not reported [233,234]. There have been few analyses of mutants in medaka, and further findings should be obtained in future studies.
Gain-of-function mutations in SOD1 cause familial ALS. Mutated SOD1 aggregates in the mitochondrial inner membrane and is thought to be involved in oxidative stress and apoptosis [235]. Lemmens et al. reported abnormal motor neuron branching and short axons in zebrafish, which transiently overexpressed mutated human SOD1 proteins [210]. On the other hand, no such axonal abnormalities were observed in transgenic lines overexpressing mutant zebrafish sod1, but abnormal neuromuscular junctions (NMJs) were observed [211]. This line showed end-stage manifestations, including reduced swimming behavior, partial paralysis, reduced number of motor neurons, and mitochondrial vacuolation. Decreased NMJs and motor neurons have also been reported in zebrafish mutants of Sod1 [212]. These models recapitulate the ALS phenotype and can be used as valuable models for ALS research.
3.1.2. Neuronal Defects
Charcot-Marie-Tooth disease (CMT) is a peripheral neuropathy resulting in weaker muscles. Mutations in the mitochondrial fusion gene MFN2 lead to CMT2A [139]. Zebrafish knockdown of mfn2 showed abnormal motor neurons and myofiber alignments [213]. In addition, zebrafish mfn2 mutants showing age-related alteration of NMJ pathology and reduced motile mitochondria have been identified [214]. Both morphants and mutants showed dull motor responses to physical stimuli, making them a good model for CMT2A. Similar abnormal NMJ phenotypes could be found in the knockdown of slc25a1, the mitochondrial citrate carrier [217]. Mutations in SLC25A1 are associated with neuromuscular transmission disorders (congenital myasthenic syndromes) and neurometabolic disorders (D-2- and L-2-hydroxyglutaric aciduria) [217,236].
It has been reported that mutation in the MFN2 gene impaired mitochondrial axonal transport [237]. Therefore, defective mitochondrial transport along axons may be associated with NMJ pathology and loss of motor function. Zebrafish kbp is an ortholog of human Kif1-binding protein (KBP/KIAA1279) that regulates mitochondria localization. The zebrafish kbp mutant revealed that kbp has an essential role in the development, growth, and maintenance of axons [218]. Notably, mutations in KIF1B are associated with CMT2A as well as MFN2 [238]. Similarly, the zebrafish mutant of actr10, part of the dynactin complex, led to mitochondria failing to attach to the dynein retrograde motor, leading to axon swelling and accumulation of mitochondria [219].
3.1.3. Anomaly of Brain Development
Knockdown of mitochondrial genes often leads to systemic effects during embryogenesis. Mitochondrial transcription factor A (TFAM) is a multifunctional protein that regulates the transcription and translation of essential mitochondrial genes, mtDNA copy number, and DNA packaging [239,240]. OPA1 is involved in mitochondrial fusion and regulation of apoptosis, and its mutation is associated with autosomal dominant optic atrophy [132]. SURF1 is a COX assembly protein, and its mutation is associated with Leigh syndrome [241]. Even though these genes possess different mitochondrial functions, the morphants showed severe developmental defects in the eye, heart, and brain regions [223,224,225]. These defects have also been reported in mfn2 and slc25a morphants [213,217]. Mitochondrial gene mutations often cause early-onset mitochondrial diseases such as Leigh syndrome and mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) [242]. Mitochondrial disease is clinically complex and can affect any tissue or organ: encephalopathy, neuropathy, blindness, deafness, myopathy, cardiomyopathy, enteropathy, renal disease, liver failure, and anemia. However, many children affected by mitochondrial disease exhibit tissue/organ-specific symptoms in the early stages of the disease [242]. The fast-developing small fish may be used as a good model to approach the pathogenesis of these mitochondrial diseases. In medaka, the knockdown of holo-cytochrome c-type synthase (hccs) showed the phenotype of microphthalmia with linear skin lesions (MLS) through ROS overproduction [243]. Further investigation will be needed by generating tissue- or cell type-specific gene knockout models to understand the tissue/organ specificity and variability of clinical symptoms.
3.2. mtDNA Manipulation
The lengths of mitochondrial DNA of zebrafish and humans are 16,596 and 16,569 bp, respectively, and encode 13 protein genes, 22 tRNAs, 2 rRNAs, and a non-coding control region [244,245]. Many mitochondrial DNA variants, such as point mutations and deletions, have been reported as causative genetic defects of various disorders, including PD and AD [246,247,248,249]. Therefore, developing the tools to edit mitochondrial DNA precisely is essential to understand the etiology of mitochondrial diseases. Current major gene editing methods are also applicable to mitochondrial DNA editing: mtZFN [250,251,252], mitoTALLEN [253,254,255,256], and mito-CRISPR/Cas9 [257,258,259,260]. The mito-CRISPR/Cas9 system was also successfully used in the knock-in strategy in the zebrafish model [261]. However, these strategies face difficulty in delivering the editing components into mitochondria [262]. In addition, mitochondria-targeted nucleases selectively reduced mtDNA haplotypes in the germline, eliminating mitochondrial mutations [263,264]. To date, few studies have been successfully established in in vivo models. Recently, Mok et al. engineered a bacterial cytidine deaminase toxin (DddAtox)-based mitochondrial genome editing tool [265]. DddAtox was split into two inactive portions, which were fused with a transcription activator-like effector (TALE) and a uracil glycosylase inhibitor, resulting in DddA-derived cytosine base editors (DdCBEs). DdCBEs were introduced in zebrafish to create a model of mitochondrial disease. This study showed higher efficiency of mitochondrial nd5 gene mutation associated with Leigh syndrome and MELAS than a mouse model utilizing the same strategy [266,267]. Further new methods will continue to be developed and optimized for precise mitochondrial genome editing for understanding mitochondrial disease and developing therapeutic applications. In this process, small fish can represent strong in vivo models.
3.3. Imaging
Imaging mitochondria is useful for monitoring the structural and functional changes during the pathological process, but measuring mitochondrial function in vivo, especially in mammalian models, involves many technical difficulties. Various fluorescent reporters have been developed and used for in vitro live-cell imaging [268]. To understand the mitochondrial dynamics in vivo, a fluorescent protein fused with a mitochondrial localization sequence (e.g., mito-GFP, mito-CFP, and mito-RFP) has been used in mice and zebrafish models [269,270,271,272]. Dukes et al. reported abnormal mitochondrial transport in vivo in a pharmacological PD model using a transgenic zebrafish in which the mitochondria of dopaminergic neurons are labeled with the fluorescent reporter [273]. Recent advances in fluorescent biosensors enable us to observe the behavior of molecules in live cells with high sensitivity. Using pH-sensitive fluorescent protein, Wrighton et al. established a zebrafish model to monitor physiological stress-induced mitophagy [274]. Vicente et al. fused the Ca2+-sensitive photoprotein to GFP and established a zebrafish model to monitor both the cytoplasmic and mitochondrial Ca2+ during skeletal muscle contraction [275]. A FRET-based ATP biosensor was also used to visualize ATP dynamics in in vivo beating hearts [276]. These recent models will contribute to the elucidation of the disease mechanisms. Since body transparency is only seen during the embryonic and larval stages, intravital imaging within the adult body, such as the brain, is a challenge similar to in mammals. However, there is an option to utilize pigmentation mutants which allow us to see the internal structure in the adult stage to some extent [277].
3.4. Drug Screening
Drug screening processes are used to identify compounds of interest. In such processes, zebrafish is a useful model to evaluate toxicity and effectiveness after the in vitro selection. Needless to say, mice or other mammalian models are evolutionarily closer to humans. However, 71% of human genes have at least one zebrafish orthologue [187]. Furthermore, zebrafish provide beneficial features for high throughput drug screening, including small body size, fast development, ease of laboratory management, and production of large numbers of offspring [174]. Zebrafish can be useful models not only for drug screening but also for determining the mechanism of action [278]. Although high throughput drug screening is available for zebrafish, imaging and analysis of a large number of living organisms are still challenging.
As we discussed above, mitochondria have vital roles in cells, and mitochondrial dysfunction contributes to various disorders. Thus, mitochondria are an important drug target for mutations in mitochondrial DNA, mitochondrial component proteins, and restoration of mitochondrial function [279]. A platform of non-invasive and real-time measurements of metabolic changes in zebrafish larvae has been established and used for drug screening of epilepsy [280,281,282]. Zhang et al. conducted drug screening by using pink1 deficient zebrafish as a model of Parkinson’s disease. Based on a phenotypic screening strategy, they identified trifluoperazine that induces a stress-dependent activation of autophagy to rescue Pink1 deficiency [283]. Another study utilized a nitroreductase-metronidazole system, which induces apoptosis through damage of mitochondrial DNA, to ablate dopaminergic neurons in zebrafish. Through in vivo dopaminergic neuron imaging, the Renin-Angiotensin-Aldosterone System (RAAS) inhibitors were identified as neuroprotective [284]. Zebrafish models for drug screening and disease models will expand more and contribute to future therapeutics.
4. Merits and Demerits of CNS Regeneration Capacity in Zebrafish
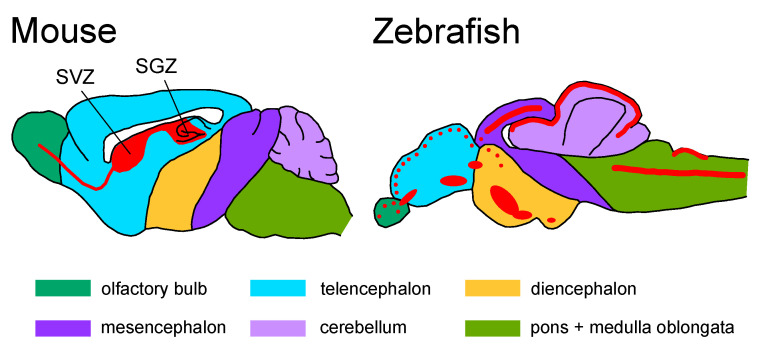
One of the biggest differences between mammals and zebrafish is the ability of neurogenesis. Zebrafish possess pronounced regeneration capacity in various tissues and organs; therefore, they have been widely used as a model to study complex tissue regeneration [285]. They are able to regenerate their injured CNS, such as spinal cord and telencephalon injuries, with functional recoveries [286,287,288,289,290]. In adult mammals, radial glial cells are recognized as the source of new neurons (neural stem/progenitor cells), which are localized in the restricted regions: the subventricular zone (SVZ) and the dentate gyrus subgranular zone (SGZ) [291,292,293,294] (Figure 4).
Figure 4.
Progenitor cells (radial glial cells) distribution in the mouse and zebrafish brain. The red color indicates regions of constitutive proliferation. Based on the data summary from [295]. SVZ, subventricular zone; SGZ, dentate gyrus subgranular zone.
Furthermore, radial glial cells in adult zebrafish brains are widely distributed and form neurogenic niches in the telencephalon, diencephalon, mesencephalon, rhombencephalon, and spinal cord [295,296,297,298,299,300] (Figure 4). These progenitor cells were activated following injury and contributed to regeneration [301,302,303]. In addition, another specified stem cell niche has been identified in the zebrafish cerebellum [304,305,306]. Although these features are important for elucidating the molecular mechanisms that can be translated to therapeutic applications for adult mammals, we should keep their endogenous regeneration ability in mind for understanding the pathological process of neurodegenerative disorders. For example, injection of amyloid-β42-derivates in the zebrafish brain could lead to AD-like phenotypes. However, progenitor cells were activated and processed neurogenesis through the Il-4 signaling pathway [307]. Zebrafish also did not exhibit an age-dependent decline in dopaminergic and noradrenergic neurons, which may be supported by their neurogenesis ability [308]. On the other hand, it has also been reported that the number of newborn neurons and oligodendrocytes decreases with age in the zebrafish telencephalon [309]. Further investigations of regenerative capacity in the fish model will provide knowledge addressing the limited neurogenic capabilities in the mammalian brain.
5. Turquoise Killifish: A New Model for Neurodegenerative Disorders
In 2003, the turquoise killifish was reported to have the shortest lifespan among vertebrates [310]. Since then, the turquoise killifish has attracted attention as a new small fish model for aging research. It shows remarkable aging phenotypes during its short lifespan of only several months, including organ atrophy, scoliosis, and elevated levels of aging-related acidic β-galactosidase [311,312,313,314]. In the body, aging is accompanied by decreased telomere length, mitochondrial copy number, and antibody production capacity, leading to multiple organ failures [315,316,317]. This fish was used to study the relationship between gene expression patterns in youth and longevity. This study identified that mitochondrial RC complex I genes were less active at a young age in long-lived fish. In addition, partial pharmacological inhibition of complex I by the small molecule rotenone extended its lifespan [318]. There is no doubt that mitochondrial function decreases with age, but further investigation will be needed to develop a strategy for improving mitochondrial function in the aged population.
Declines in neuronal regeneration ability with age have also been reported in the optic nerve and telencephalon [319,320]. Interestingly, brain regeneration in young fish was mainly supported by non-glial neural progenitor cells [320], in spite of the presence of radial glia for neurogenesis [321]. Further characterization of neural progenitor cells in young and aged turquoise killifish is necessary. As for neurodegenerative disorders, neurofibrillary degeneration in aged fish was observed in the optic tectum, telencephalon, and brainstem, as indicated by Fluoro-JadeB staining [322]. The turquoise killifish also showed age-related degeneration of dopaminergic and noradrenergic nerves and progression of alpha-synuclein accumulation, similar to pathological phenotypes observed in human Parkinson’s disease [308]. This feature may help to elucidate the mechanism of solitary Parkinson’s disease, which is not dependent on a genetic component. Another recent study reported the decreased expressions of enzymes, transporters, and receptors of brain serotonin (5-HT) that are related to neurodegenerative/neurodevelopmental disorders [323]. This study also revealed the increased monoamine oxidase (MAO) activity in aged fish. Aging-induced increased MAO activity has also been reported in rodents and human brains [324,325]. MAO is localized at mitochondrial outer membranes, and its elevated activity is thought to be associated with age-related diseases, including neurological disorders via increased ROS production and regulation of bioactive amines such as serotonin and catecholamines [326,327,328]. This emerging small fish model is still in its infancy. It is expected that the extremely rapid aging characteristic will be used to advance our understanding of mitochondrial involvement in disease and the mechanisms of neurodegenerative disorders.
6. Conclusions
In this review, we outlined the factors involved in mitochondrial dysfunction in the progression of neurodegenerative disorders and how small fish models can be used to analyze mitochondrial function. Despite many years of research, we do not know much about the mechanisms of neurodegenerative diseases, including how they occur and when they begin. We have not established a therapeutic strategy for their treatment. In addition, whether mitochondrial dysfunction and the progression of neurodegenerative disease are causally associated or correlated is still debatable. The small fish model alone may not be the key tool that unveils everything, and it is important to apply the observed results to other models such as mammals for deeper understanding. However, there are currently various technical difficulties preventing closer investigations, including the analysis of mitochondrial function, which is related to the progression of the disease. Small fish models are undoubtedly useful as vertebrate models for testing new tools that will be developed in the future. Similarly, they are helpful as an entry model for in vivo testing in drug discovery pipelines. These features will facilitate new insights and discoveries to understand human neurological disorders.
Acknowledgments
We acknowledge Shinano Kobayashi, Noriko Matsui and Ai Ito for their technical assistance. Fish images were created by TogoTV (https://togotv.dbcls.jp/en/; accessed on 5 April 2023).
Author Contributions
Writing—original draft preparation, T.O.; writing—review and editing, H.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data and tools described in this manuscript are available upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was funded by JSPS KAKENHI (Grant-in-Aid for Early-Career Scientists, 23K14192) (T.O.), AMED (JP22gm6110028) (H.M.) and JST (Moonshot R&D, Grant Number JPMJMS2024) (H.M.).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Dugger B.N., Dickson D.W. Pathology of Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2017;9:a028035. doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnham K.J., Masters C.L., Bush A.I. Neurodegenerative diseases and oxidatives stress. Nat. Rev. Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 3.Singh A., Kukreti R., Saso L., Kukreti S. Oxidative stress: A key modulator in neurodegenerative diseases. Molecules. 2019;24:1583. doi: 10.3390/molecules24081583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephenson J., Nutma E., van der Valk P., Amor S. Inflammation in CNS neurodegenerative diseases. Immunology. 2018;154:204–219. doi: 10.1111/imm.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon H.S., Koh S.H. Neuroinflammation in neurodegenerative disorders: The roles of microglia and astrocytes. Transl. Neurodegener. 2020;9:42. doi: 10.1186/s40035-020-00221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harman D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 7.Harman D. The Biologic Clock: The Mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 8.Ziada A.S., Smith M.S.R., Côté H.C.F. Updating the Free Radical Theory of Aging. Front. Cell Dev. Biol. 2020;8:908. doi: 10.3389/fcell.2020.575645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava S. The mitochondrial basis of aging and age-related disorders. Genes. 2017;8:398. doi: 10.3390/genes8120398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perier C., Vila M. Mitochondrial biology and Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012;2:a009332. doi: 10.1101/cshperspect.a009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J.L., Lin T.Y., Chen P.L., Guo T.N., Huang S.Y., Chen C.H., Lin C.H., Chan C.C. Mitochondrial Function and Parkinson’s Disease: From the Perspective of the Electron Transport Chain. Front. Mol. Neurosci. 2021;14:315. doi: 10.3389/fnmol.2021.797833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coskun P.E., Wyrembak J., Derbereva O., Melkonian G., Doran E., Lott I.T., Head E., Cotman C.W., Wallace D.C. Systemic mitochondrial dysfunction and the etiology of Alzheimer’s disease and down syndrome dementia. J. Alzheimer’s Dis. 2010;20:S293–S310. doi: 10.3233/JAD-2010-100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desler C., Lillenes M.S., Tønjum T., Rasmussen L.J. The Role of Mitochondrial Dysfunction in the Progression of Alzheimer’s Disease. Curr. Med. Chem. 2017;25:5578–5587. doi: 10.2174/0929867324666170616110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muyderman H., Chen T. Mitochondrial dysfunction in amyotrophic lateral sclerosis–A valid pharmacological target? Br. J. Pharmacol. 2014;171:2191–2205. doi: 10.1111/bph.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J., Wang X., Huo Z., Chen Y., Liu J., Zhao Z., Meng F., Su Q., Bao W., Zhang L., et al. The Impact of Mitochondrial Dysfunction in Amyotrophic Lateral Sclerosis. Cells. 2022;11:2049. doi: 10.3390/cells11132049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monzio Compagnoni G., Di Fonzo A. Understanding the pathogenesis of multiple system atrophy: State of the art and future perspectives. Acta Neuropathol. Commun. 2019;7:113. doi: 10.1186/s40478-019-0730-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y., Chen M., Jiang J. Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion. 2019;49:35–45. doi: 10.1016/j.mito.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Calvo S.E., Mootha V.K. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., Goodman R.P., Grabarek Z., Haas M.E., Hung W.H.W., et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng M.Y.W., Wai T., Simonsen A. Quality control of the mitochondrion. Dev. Cell. 2021;56:881–905. doi: 10.1016/j.devcel.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Faggioli F., Vijg J., Montagna C. Chromosomal aneuploidy in the aging brain. Mech. Ageing Dev. 2011;132:429–436. doi: 10.1016/j.mad.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joy J., Barrio L., Santos-Tapia C., Romão D., Giakoumakis N.N., Clemente-Ruiz M., Milán M. Proteostasis failure and mitochondrial dysfunction leads to aneuploidy-induced senescence. Dev. Cell. 2021;56:2043–2058.e7. doi: 10.1016/j.devcel.2021.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Ballard J.W.O., Whitlock M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294X.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 24.Haag-Liautard C., Coffey N., Houle D., Lynch M., Charlesworth B., Keightley P.D. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6:1706–1714. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X., Xu W., Zhang W., Pan C., Thalacker-Mercer A.E., Zheng H., Gu Z. High-frequency and functional mitochondrial DNA mutations at the single-cell level. Proc. Natl. Acad. Sci. USA. 2023;120:e2201518120. doi: 10.1073/pnas.2201518120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeve A.K., Krishnan K.J., Elson J.L., Morris C.M., Bender A., Lightowlers R.N., Turnbull D.M. Nature of Mitochondrial DNA Deletions in Substantia Nigra Neurons. Am. J. Hum. Genet. 2008;82:228–235. doi: 10.1016/j.ajhg.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogenhagen D., Clayton D.A. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell. 1977;11:719–727. doi: 10.1016/0092-8674(77)90286-0. [DOI] [PubMed] [Google Scholar]
- 28.Lax N.Z., Turnbull D.M., Reeve A.K. Mitochondrial mutations: Newly discovered players in neuronal degeneration. Neuroscientist. 2011;17:645–658. doi: 10.1177/1073858410385469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawless C., Greaves L., Reeve A.K., Turnbull D.M., Vincent A.E. The rise and rise of mitochondrial DNA mutations. Open Biol. 2020;10:200061. doi: 10.1098/rsob.200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Lluch G., Irusta P.M., Navas P., de Cabo R. Mitochondrial biogenesis and healthy aging. Exp. Gerontol. 2008;43:813–819. doi: 10.1016/j.exger.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palikaras K., Lionaki E., Tavernarakis N. Coupling mitogenesis and mitophagy for longevity. Autophagy. 2015;11:1428–1430. doi: 10.1080/15548627.2015.1061172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunnari J., Suomalainen A. Mitochondria: In sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papa S., Martino P.L., Capitanio G., Gaballo A., De Rasmo D., Signorile A., Petruzzella V. The oxidative phosphorylation system in mammalian mitochondria. Adv. Exp. Med. Biol. 2012;942:3–37. doi: 10.1007/978-94-007-2869-1_1. [DOI] [PubMed] [Google Scholar]
- 34.Matsuno-Yagi A., Hatefi Y. Studies on the mechanism of oxidative phosphorylation. Positive cooperativity in ATP synthesis. J. Biol. Chem. 1985;260:11424–11427. doi: 10.1016/S0021-9258(17)38584-8. [DOI] [PubMed] [Google Scholar]
- 35.Breuer M.E., Koopman W.J., Koene S., Nooteboom M., Rodenburg R.J., Willems P.H., Smeitink J.A.M. The role of mitochondrial OXPHOS dysfunction in the development of neurologic diseases. Neurobiol. Dis. 2013;51:27–34. doi: 10.1016/j.nbd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Papa S., De Rasmo D. Complex I deficiencies in neurological disorders. Trends Mol. Med. 2013;19:61–69. doi: 10.1016/j.molmed.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Liu X., Kim C.N., Yang J., Jemmerson R., Wang X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/S0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 38.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M., Mangion J., Jacotot E., Costantini P., Loeffler M., et al. Molecular characterization of mitochodrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 39.Jiang X., Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J. Biol. Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- 40.Ricci J.E., Muñoz-Pinedo C., Fitzgerald P., Bailly-Maitre B., Perkins G.A., Yadava N., Scheffler I.E., Ellisman M.H., Green D.R. Disruption of mitochondrial function during apoptosis is mediated by caspase cleavage of the p75 subunit of complex I of the electron transport chain. Cell. 2004;117:773–786. doi: 10.1016/j.cell.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Miyoshi N., Oubrahim H., Chock P.B., Stadtman E.R. Age-dependent cell death and the role of ATP in hydrogen peroxide-induced apoptosis and necrosis. Proc. Natl. Acad. Sci. USA. 2006;103:1727–1731. doi: 10.1073/pnas.0510346103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gldlöf S., Oldfors A., Wibom R., et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 43.Kujoth C.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 44.Benzi G., Pastoris O., Marzatico F., Villa R.F., Dagani F., Curti D. The mitochondrial electron transfer alteration as a factor involved in the brain aging. Neurobiol. Aging. 1992;13:361–368. doi: 10.1016/0197-4580(92)90109-B. [DOI] [PubMed] [Google Scholar]
- 45.Lenaz G., Bovina C., Castelluccio C., Fato R., Formiggini G., Genova M.L., Marchetti M., Pich M.M., Pallotti F., Castelli G.P., et al. Mitochondrial complex I defects in aging. Mol. Cell. Biochem. 1997;174:329–333. doi: 10.1023/A:1006854619336. [DOI] [PubMed] [Google Scholar]
- 46.Manczak M., Jung Y., Park B.S., Partovi D., Reddy P.H. Time-course of mitochondrial gene expressions in mice brains: Implications for mitochondrial dysfunction, oxidative damage, and cytochrome c in aging. J. Neurochem. 2005;92:494–504. doi: 10.1111/j.1471-4159.2004.02884.x. [DOI] [PubMed] [Google Scholar]
- 47.Itoh K., Weis S., Mehraein P., Müller-Höcker J. Cytochrome c oxidase defects of the human substantia nigra in normal aging. Neurobiol. Aging. 1996;17:843–848. doi: 10.1016/S0197-4580(96)00168-6. [DOI] [PubMed] [Google Scholar]
- 48.Cottrell D.A., Blakely E.L., Johnson M.A., Ince P.G., Borthwick G.M., Turnbull D.M. Cytochrome C oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol. Aging. 2001;22:265–272. doi: 10.1016/S0197-4580(00)00234-7. [DOI] [PubMed] [Google Scholar]
- 49.Schapira A.H.V., Cooper J.M., Dexter D., Clark J.B., Jenner P., Marsden C.D. Mitochondrial Complex I Deficiency in Parkinson’s Disease. J. Neurochem. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 50.Schapira A.H.V., Mann V.M., Cooper J.M., Dexter D., Daniel S.E., Jenner P., Clark J.B., Marsden C.D. Anatomic and Disease Specificity of NADH CoQ1 Reductase (Complex I) Deficiency in Parkinson’s Disease. J. Neurochem. 1990;55:2142–2145. doi: 10.1111/j.1471-4159.1990.tb05809.x. [DOI] [PubMed] [Google Scholar]
- 51.Navarro A., Boveris A., Bández M.J., Sánchez-Pino M.J., Gómez C., Muntané G., Ferrer I. Human brain cortex: Mitochondrial oxidative damage and adaptive response in Parkinson disease and in dementia with Lewy bodies. Free Radic. Biol. Med. 2009;46:1574–1580. doi: 10.1016/j.freeradbiomed.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Mann V.M., Cooper J.M., Krige D., Daniel S.E., Schapira A.H.V., Marsden C.D. Brain, skeletal muscle and platelet homogenate mitochondrial function in parkinson’s disease. Brain. 1992;115:333–342. doi: 10.1093/brain/115.2.333. [DOI] [PubMed] [Google Scholar]
- 53.Morais V.A., Verstreken P., Roethig A., Smet J., Snellinx A., Vanbrabant M., Haddad D., Frezza C., Mandemakers W., Vogt-Weisenhorn D., et al. Parkinson’s disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol. Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chagnon P., Bétard C., Robitaille Y., Cholette A., Gauvreau D. Distribution of brain cytochrome oxidase activity in various neurodegenerative diseases. Neuroreport. 1995;6:711–715. doi: 10.1097/00001756-199503270-00002. [DOI] [PubMed] [Google Scholar]
- 55.Arthur C.R., Morton S.L., Dunham L.D., Keeney P.M., Bennett J.P. Parkinson’s disease brain mitochondria have impaired respirasome assembly, age-related increases in distribution of oxidative damage to mtDNA and no differences in heteroplasmic mtDNA mutation abundance. Mol. Neurodegener. 2009;4:37. doi: 10.1186/1750-1326-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holper L., Ben-Shachar D., Mann J. Multivariate meta-analyses of mitochondrial complex I and IV in major depressive disorder, bipolar disorder, schizophrenia, Alzheimer disease, and Parkinson disease. Neuropsychopharmacology. 2019;44:837–849. doi: 10.1038/s41386-018-0090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foti S.C., Hargreaves I., Carrington S., Kiely A.P., Houlden H., Holton J.L. Cerebral mitochondrial electron transport chain dysfunction in multiple system atrophy and Parkinson’s disease. Sci. Rep. 2019;9:6559. doi: 10.1038/s41598-019-42902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Esparcia P., Koneti A., Rodríguez-Oroz M.C., Gago B., del Rio J.A., Ferrer I. Mitochondrial activity in the frontal cortex area 8 and angular gyrus in Parkinson’s disease and Parkinson’s disease with dementia. Brain Pathol. 2018;28:43–57. doi: 10.1111/bpa.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraga C.G., Shigenaga M.K., Park J.W., Degan P., Ames B.N. Oxidative damage to DNA during aging: 8-Hydroxy-2’-deoxyguanosine in rat organ DNA and urine. Proc. Natl. Acad. Sci. USA. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ugarte N., Petropoulos I., Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid. Redox Signal. 2010;13:539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- 63.Xiao M., Zhong H., Xia L., Tao Y., Yin H. Pathophysiology of mitochondrial lipid oxidation: Role of 4-hydroxynonenal (4-HNE) and other bioactive lipids in mitochondria. Free Radic. Biol. Med. 2017;111:316–327. doi: 10.1016/j.freeradbiomed.2017.04.363. [DOI] [PubMed] [Google Scholar]
- 64.Cui H., Kong Y., Zhang H. Oxidative Stress, Mitochondrial Dysfunction, and Aging. J. Signal Transduct. 2012;2012:646354. doi: 10.1155/2012/646354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Napolitano G., Fasciolo G., Venditti P. Mitochondrial management of reactive oxygen species. Antioxidants. 2021;10:1824. doi: 10.3390/antiox10111824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shadel G.S., Horvath T.L. Mitochondrial ROS Signaling in Organismal Homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y., Azad M.B., Gibson S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 68.Shigenaga M.K., Hagen T.M., Ames B.N. Oxidative damage and mitochondrial decay in aging. Proc. Natl. Acad. Sci. USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.James A.M., Murphy M.P. How mitochondrial damage affects cell function. J. Biomed. Sci. 2002;9:475–487. doi: 10.1007/BF02254975. [DOI] [PubMed] [Google Scholar]
- 70.Stefanatos R., Sanz A. The role of mitochondrial ROS in the aging brain. FEBS Lett. 2018;592:743–758. doi: 10.1002/1873-3468.12902. [DOI] [PubMed] [Google Scholar]
- 71.Zorov D.B., Filburn C.R., Klotz L.O., Zweier J.L., Sollott S.J. Reactive oxygen species (ROS)-induced ROS release: A new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J. Exp. Med. 2000;192:1001–1014. doi: 10.1084/jem.192.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coyle J.T., Puttfarcken P. Oxidative stress, glutamate, and neurodegenerative disorders. Science. 1993;262:689–695. doi: 10.1126/science.7901908. [DOI] [PubMed] [Google Scholar]
- 73.Floyd R.A., Hensley K. Oxidative stress in brain aging: Implications for therapeutics of neurodegenerative diseases. Neurobiol. Aging. 2002;23:795–807. doi: 10.1016/S0197-4580(02)00019-2. [DOI] [PubMed] [Google Scholar]
- 74.Mullaart E., Boerrigter M.E.T.I., Ravid R., Swaab D.F., Vijg J. Increased levels of DNA breaks in cerebral cortex of Alzheimer’s disease patients. Neurobiol. Aging. 1990;11:169–173. doi: 10.1016/0197-4580(90)90542-8. [DOI] [PubMed] [Google Scholar]
- 75.Mecocci P., MacGarvey U., Beal M.F. Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann. Neurol. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 76.Wang J., Xiong S., Xie C., Markesbery W.R., Lovell M.A. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J. Neurochem. 2005;93:953–962. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- 77.Mastrogiacomo F., Lindsay J.G., Bettendorff L., Rice J., Kish S.J. Brain protein and α-ketoglutarate dehydrogenase complex activity in Alzheimer’s disease. Ann. Neurol. 1996;39:592–598. doi: 10.1002/ana.410390508. [DOI] [PubMed] [Google Scholar]
- 78.Gibson G.E., Starkov A., Blass J.P., Ratan R.R., Beal M.F. Cause and consequence: Mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochim. Biophys. Acta-Mol. Basis Dis. 2010;1802:122–134. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keeney P.M., Xie J., Capaldi R.A., Bennett J.P. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bosco D.A., Fowler D.M., Zhang Q., Nieva J., Powers E.T., Wentworth P., Lerner R.A., Kelly J.W. Elevated levels of oxidized cholesterol metabolites in Lewy body disease brains accelerate α-synuclein fibrilization. Nat. Chem. Biol. 2006;2:249–253. doi: 10.1038/nchembio782. [DOI] [PubMed] [Google Scholar]
- 81.Floor E., Wetzel M.G. Increased protein oxidation in human substantia nigra pars compacta in comparison with basal ganglia and prefrontal cortex measured with an improved dinitrophenylhydrazine assay. J. Neurochem. 1998;70:268–275. doi: 10.1046/j.1471-4159.1998.70010268.x. [DOI] [PubMed] [Google Scholar]
- 82.Nakabeppu Y., Tsuchimoto D., Yamaguchi H., Sakumi K. Oxidative damage in nucleic acids and Parkinson’s disease. J. Neurosci. Res. 2007;85:919–934. doi: 10.1002/jnr.21191. [DOI] [PubMed] [Google Scholar]
- 83.Goldsteins G., Keksa-Goldsteine V., Ahtoniemi T., Jaronen M., Arens E., Åkerman K., Chan P.H., Koistinaho J. Deleterious role of superoxide dismutase in the mitochondrial intermembrane space. J. Biol. Chem. 2008;283:8446–8452. doi: 10.1074/jbc.M706111200. [DOI] [PubMed] [Google Scholar]
- 84.Kirby J., Halligan E., Baptista M.J., Allen S., Heath P.R., Holden H., Barber S.C., Loynes C.A., Wood-Allum C.A., Lunec J., et al. Mutant SOD1 alters the motor neuronal transcriptome: Implications for familial ALS. Brain. 2005;128:1686–1706. doi: 10.1093/brain/awh503. [DOI] [PubMed] [Google Scholar]
- 85.Sarlette A., Krampfl K., Grothe C., Neuhoff N.V., Dengler R., Petri S. Nuclear erythroid 2-related factor 2-antioxidative response element signaling pathway in motor cortex and spinal cord in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2008;67:1055–1062. doi: 10.1097/NEN.0b013e31818b4906. [DOI] [PubMed] [Google Scholar]
- 86.Vakifahmetoglu-Norberg H., Ouchida A.T., Norberg E. The role of mitochondria in metabolism and cell death. Biochem. Biophys. Res. Commun. 2017;482:426–431. doi: 10.1016/j.bbrc.2016.11.088. [DOI] [PubMed] [Google Scholar]
- 87.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giorgi C., Agnoletto C., Bononi A., Bonora M., de Marchi E., Marchi S., Missiroli S., Patergnani S., Poletti F., Rimessi A., et al. Mitochondrial calcium homeostasis as potential target for mitochondrial medicine. Mitochondrion. 2012;12:77–85. doi: 10.1016/j.mito.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pivovarova N.B., Andrews S.B. Calcium-dependent mitochondrial function and dysfunction in neurons. FEBS J. 2010;277:3622–3636. doi: 10.1111/j.1742-4658.2010.07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brini M., Calì T., Ottolini D., Carafoli E. Neuronal calcium signaling: Function and dysfunction. Cell. Mol. Life Sci. 2014;71:2787–2814. doi: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gleichmann M., Mattson M.P. Neuronal calcium homeostasis and dysregulation. Antioxid. Redox Signal. 2011;14:1261–1273. doi: 10.1089/ars.2010.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harris J.J., Jolivet R., Attwell D. Synaptic Energy Use and Supply. Neuron. 2012;75:762–777. doi: 10.1016/j.neuron.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 93.Orrenius S., Gogvadze V., Zhivotovsky B. Calcium and mitochondria in the regulation of cell death. Biochem. Biophys. Res. Commun. 2015;460:72–81. doi: 10.1016/j.bbrc.2015.01.137. [DOI] [PubMed] [Google Scholar]
- 94.Panov A.V., Gutekunst C.A., Leavitt B.R., Hayden M.R., Burke J.R., Strittmatter W.J., Greenamyre J.T. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 95.Goffredo D., Rigamonti D., Tartari M., De Micheli A., Verderio C., Matteoli M., Zuccato C., Cattaneo E. Calcium-dependent cleavage of endogenous wild-type huntingtin in primary cortical neurons. J. Biol. Chem. 2002;277:39594–39598. doi: 10.1074/jbc.C200353200. [DOI] [PubMed] [Google Scholar]
- 96.Gandhi S., Wood-Kaczmar A., Yao Z., Plun-Favreau H., Deas E., Klupsch K., Downward J., Latchman D.S., Tabrizi S.J., Wood N.W., et al. PINK1-Associated Parkinson’s Disease Is Caused by Neuronal Vulnerability to Calcium-Induced Cell Death. Mol. Cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giorgi C., Baldassari F., Bononi A., Bonora M., De Marchi E., Marchi S., Missiroli S., Patergnani S., Rimessi A., Suski J.M., et al. Mitochondrial Ca2+ and apoptosis. Cell Calcium. 2012;52:36–43. doi: 10.1016/j.ceca.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tambini M.D., Pera M., Kanter E., Yang H., Guardia-Laguarta C., Holtzman D., Sulzer D., Area-Gomez E., Schon E.A. ApoE4 upregulates the activity of mitochondria-associated ER membranes. EMBO Rep. 2016;17:27–36. doi: 10.15252/embr.201540614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheung K.H., Mei L., Mak D.O.D., Hayashi I., Iwatsubo T., Kang D.E., Foskett J.K. Gain-of-function enhancement of IP3 receptor modal gating by familial Alzheimer’s disease-linked presenilin mutants in human cells and mouse neurons. Sci. Signal. 2010;3:ra22. doi: 10.1126/scisignal.2000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheung K.H., Shineman D., Müller M., Cárdenas C., Mei L., Yang J., Tomita T., Iwatsubo T., Lee V.M.Y., Foskett J.K. Mechanism of Ca2+ Disruption in Alzheimer’s Disease by Presenilin Regulation of InsP3 Receptor Channel Gating. Neuron. 2008;58:871–883. doi: 10.1016/j.neuron.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stutzmann G.E., Smith I., Caccamo A., Oddo S., LaFerla F.M., Parker I. Enhanced ryanodine receptor recruitment contributes to Ca2+ disruptions in young, adult, and aged Alzheimer’s disease mice. J. Neurosci. 2006;26:5180–5189. doi: 10.1523/JNEUROSCI.0739-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chan S.L., Mayne M., Holden C.P., Geiger J.D., Mattson M.P. Presenilin-1 mutations increase levels of ryanodine receptors and calcium release in PC12 cells and cortical neurons. J. Biol. Chem. 2000;275:18195–18200. doi: 10.1074/jbc.M000040200. [DOI] [PubMed] [Google Scholar]
- 103.Hedskog L., Pinho C.M., Filadi R., Rönnbäck A., Hertwig L., Wiehager B., Larssen P., Gellhaar S., Sandebring A., Westerlund M., et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proc. Natl. Acad. Sci. USA. 2013;110:7916–7921. doi: 10.1073/pnas.1300677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheftel A., Stehling O., Lill R. Iron-sulfur proteins in health and disease. Trends Endocrinol. Metab. 2010;21:302–314. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 105.Berg D., Hochstrasser H., Schweitzer K.J., Riess O. Disturbance of iron metabolism in Parkinson’s disease—Ultrasonography as a biomarker. Neurotox. Res. 2006;9:1–13. doi: 10.1007/BF03033302. [DOI] [PubMed] [Google Scholar]
- 106.Connor J.R., Menzies S.L., St. Martin S.M., Mufson E.J. A histochemical study of iron, transferrin, and ferritin in Alzheimer’s diseased brains. J. Neurosci. Res. 1992;31:75–83. doi: 10.1002/jnr.490310111. [DOI] [PubMed] [Google Scholar]
- 107.Connor J.R., Snyder B.S., Beard J.L., Fine R.E., Mufson E.J. Regional distribution of iron and iron-regulatory proteins in the brain in aging and Alzheimer’s disease. J. Neurosci. Res. 1992;31:327–335. doi: 10.1002/jnr.490310214. [DOI] [PubMed] [Google Scholar]
- 108.Rosas H.D., Chen Y.I., Doros G., Salat D.H., Chen N.K., Kwong K.K., Bush A., Fox J., Hersch S.M. Alterations in brain transition metals in Huntington disease: An evolving and intricate story. Arch. Neurol. 2012;69:887–893. doi: 10.1001/archneurol.2011.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kasarskis E.J., Tandon L., Lovell M.A., Ehmann W.D. Aluminum, calcium, and iron in the spinal cord of patients with sporadic amyotrophic lateral sclerosis using laser microprobe mass spectroscopy: A preliminary study. J. Neurol. Sci. 1995;130:203–208. doi: 10.1016/0022-510X(95)00037-3. [DOI] [PubMed] [Google Scholar]
- 110.Suh Y.J., Rathore K.I., Schulz K., Ponka P., Arosio P., David S. Dysregulation of iron homeostasis in the CNS contributes to disease progression in a mouse model of amyotrophic lateral sclerosis. J. Neurosci. 2009;29:610–619. doi: 10.1523/JNEUROSCI.5443-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Altamura S., Muckenthaler M.U. Iron toxicity in diseases of aging: Alzheimer’s disease, Parkinson’s disease and atherosclerosis. J. Alzheimer’s Dis. 2009;16:879–895. doi: 10.3233/JAD-2009-1010. [DOI] [PubMed] [Google Scholar]
- 112.Schneider S.A. Neurodegenerations with Brain Iron Accumulation. Park. Relat. Disord. 2016;22:S21–S25. doi: 10.1016/j.parkreldis.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 113.Agrawal S., Fox J., Thyagarajan B., Fox J.H. Brain mitochondrial iron accumulates in Huntington’s disease, mediates mitochondrial dysfunction, and can be removed pharmacologically. Free Radic. Biol. Med. 2018;120:317–329. doi: 10.1016/j.freeradbiomed.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nishizawa H., Matsumoto M., Chen G., Ishii Y., Tada K., Onodera M., Kato H., Muto A., Tanaka K., Igarashi K. Lipid peroxidation and the subsequent cell death transmitting from ferroptotic cells to neighboring cells. Cell Death Dis. 2021;12:332. doi: 10.1038/s41419-021-03613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang T., Tomas D., Perera N.D., Cuic B., Luikinga S., Viden A., Barton S.K., McLean C.A., Samson A.L., Southon A., et al. Ferroptosis mediates selective motor neuron death in amyotrophic lateral sclerosis. Cell Death Differ. 2022;29:1187–1198. doi: 10.1038/s41418-021-00910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ou M., Jiang Y., Ji Y., Zhou Q., Du Z., Zhu H., Zhou Z. Role and mechanism of ferroptosis in neurological diseases. Mol. Metab. 2022;61:101502. doi: 10.1016/j.molmet.2022.101502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ren J.X., Sun X., Yan X.L., Guo Z.N., Yang Y. Ferroptosis in Neurological Diseases. Front. Cell Neurosci. 2020;14:218. doi: 10.3389/fncel.2020.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yan H., Zou T., Tuo Q.Z., Xu S., Li H., Belaidi A.A., Lei P. Ferroptosis: Mechanisms and links with diseases. Signal Transduct. Target. Ther. 2021;6:49. doi: 10.1038/s41392-020-00428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jakaria M., Belaidi A.A., Bush A.I., Ayton S. Ferroptosis as a mechanism of neurodegeneration in Alzheimer’s disease. J. Neurochem. 2021;159:804–825. doi: 10.1111/jnc.15519. [DOI] [PubMed] [Google Scholar]
- 121.Reichert C.O., de Freitas F.A., Sampaio-Silva J., Rokita-Rosa L., Barros P.d.L., Levy D., Bydlowski S.P. Ferroptosis Mechanisms Involved in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020;21:8765. doi: 10.3390/ijms21228765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barazzoni R., Short K.R., Nair K.S. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J. Biol. Chem. 2000;275:3343–3347. doi: 10.1074/jbc.275.5.3343. [DOI] [PubMed] [Google Scholar]
- 123.Frahm T., Mohamed S.A., Bruse P., Gemünd C., Oehmichen M., Meissner C. Lack of age-related increase of mitochondrial DNA amount in brain, skeletal muscle and human heart. Mech. Ageing Dev. 2005;126:1192–1200. doi: 10.1016/j.mad.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 124.Hales K.G. The machinery of mitochondrial fusion, division, and distribution, and emerging connections to apoptosis. Mitochondrion. 2004;4:285–308. doi: 10.1016/j.mito.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 125.Youle R.J., Van Der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Twig G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kageyama Y., Zhang Z., Roda R., Fukaya M., Wakabayashi J., Wakabayashi N., Kensler T.W., Hemachandra Reddy P., Iijima M., Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J. Cell Biol. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., Youle R.J. The Role of Dynamin-Related Protein 1, a Mediator of Mitochondrial Fission, in Apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/S1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 129.Breckenridge D.G., Stojanovic M., Marcellus R.C., Shore G.C. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Suen D.F., Norris K.L., Youle R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen H., Vermulst M., Wang Y.E., Chomyn A., Prolla T.A., McCaffery J.M., Chan D.C. Mitochondrial fusion is required for mtdna stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 133.Chen Y., Csordás G., Jowdy C., Schneider T.G., Csordás N., Wang W., Liu Y., Kohlhaas M., Meiser M., Bergem S., et al. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca2+ crosstalk. Circ. Res. 2012;111:863–875. doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen H., Chomyn A., Chan D.C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 135.Mourier A., Motori E., Brandt T., Lagouge M., Atanassov I., Galinier A., Rappl G., Brodesser S., Hultenby K., Dieterich C., et al. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J. Cell Biol. 2015;208:429–442. doi: 10.1083/jcb.201411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Burté F., Carelli V., Chinnery P.F., Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 2014;11:11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- 137.Itoh K., Nakamura K., Iijima M., Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reilly M.M., Murphy S.M., Laurá M. Charcot-Marie-Tooth disease. J. Peripher. Nerv. Syst. 2011;16:1–14. doi: 10.1111/j.1529-8027.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- 139.Züchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L., Zappia M., Nelis E., Patitucci A., Senderek J., et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 140.Chen H., McCaffery J.M., Chan D.C. Mitochondrial Fusion Protects against Neurodegeneration in the Cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 141.Pham A.H., Meng S., Chu Q.N., Chan D.C. Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Hum. Mol. Genet. 2012;21:4817–4826. doi: 10.1093/hmg/dds311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Manczak M., Calkins M.J., Reddy P.H. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: Implications for neuronal damage. Hum. Mol. Genet. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shirendeb U., Reddy A.P., Manczak M., Calkins M.J., Mao P., Tagle D.A., Reddy P.H. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: Implications for selective neuronal damage. Hum. Mol. Genet. 2011;20:1438–1455. doi: 10.1093/hmg/ddr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R., et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 2011;17:377–383. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., Mao P., Reddy P.H. Mutant Huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo X., Disatnik M.H., Monbureau M., Shamloo M., Mochly-Rosen D., Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration. J. Clin. Investig. 2013;123:5371–5388. doi: 10.1172/JCI70911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ding W.X., Yin X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Roca-Portoles A., Tait S.W.G. Mitochondrial quality control: From molecule to organelle. Cell. Mol. Life Sci. 2021;78:3853–3866. doi: 10.1007/s00018-021-03775-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yamano K., Fogel A.I., Wang C., van der Bliek A.M., Youle R.J. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Burman J.L., Pickles S., Wang C., Sekine S., Vargas J.N.S., Zhang Z., Youle A.M., Nezich C.L., Wu X., Hammer J.A., et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 2017;216:3231–3247. doi: 10.1083/jcb.201612106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M.K., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 152.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 153.Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., Youle R.J. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pickrell A.M., Youle R.J. The roles of PINK1, Parkin, and mitochondrial fidelity in parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Batlevi Y., La Spada A.R. Mitochondrial autophagy in neural function, neurodegenerative disease, neuron cell death, and aging. Neurobiol. Dis. 2011;43:46–51. doi: 10.1016/j.nbd.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., De Vries R., Arias E., Harris S., Sulzer D., et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fang E.F., Hou Y., Palikaras K., Adriaanse B.A., Kerr J.S., Yang B., Lautrup S., Hasan-Olive M.M., Caponio D., Dan X., et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019;22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sagan L. On the origin of mitosing cells. J. Theor. Biol. 1967;14:225–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 160.Matsui H., Ito J., Matsui N., Uechi T., Onodera O., Kakita A. Cytosolic dsDNA of mitochondrial origin induces cytotoxicity and neurodegeneration in cellular and zebrafish models of Parkinson’s disease. Nat. Commun. 2021;12:3101. doi: 10.1038/s41467-021-23452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mathew A., Lindsley T.A., Sheridan A., Bhoiwala D.L., Hushmendy S.F., Yager E.J., Ruggiero E.A., Crawford D.R. Degraded mitochondrial dna is a newly identified subtype of the damage associated molecular pattern (DAMP) family and possible trigger of neurodegeneration. J. Alzheimer’s Dis. 2012;30:617–627. doi: 10.3233/JAD-2012-120145. [DOI] [PubMed] [Google Scholar]
- 162.Wan M., Hua X., Su J., Thiagarajan D., Frostegård A.G., Haeggström J.Z., Frostegård J. Oxidized but not native cardiolipin has pro-inflammatory effects, which are inhibited by Annexin A5. Atherosclerosis. 2014;235:592–598. doi: 10.1016/j.atherosclerosis.2014.05.913. [DOI] [PubMed] [Google Scholar]
- 163.Codina R., Vanasse A., Kelekar A., Vezys V., Jemmerson R. Cytochrome c-induced lymphocyte death from the outside in: Inhibition by serum leucine-rich alpha-2-glycoprotein-1. Apoptosis. 2010;15:139–152. doi: 10.1007/s10495-009-0412-0. [DOI] [PubMed] [Google Scholar]
- 164.Pullerits R., Bokarewa M., Jonsson I.M., Verdrengh M., Tarkowski A. Extracellular cytochrome c, a mitochondrial apoptosis-related protein, induces arthritis. Rheumatology. 2005;44:32–39. doi: 10.1093/rheumatology/keh406. [DOI] [PubMed] [Google Scholar]
- 165.Kurashima Y., Amiya T., Nochi T., Fujisawa K., Haraguchi T., Iba H., Tsutsui H., Sato S., Nakajima S., Iijima H., et al. Extracellular ATP mediates mast cell-dependent intestinal inflammation through P2X7 purinoceptors. Nat. Commun. 2012;3:1034. doi: 10.1038/ncomms2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Carp H. Mitochondrial n-formylmethionyl proteins as chemoattractants for neutrophils. J. Exp. Med. 1982;155:264–275. doi: 10.1084/jem.155.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Czapiga M., Gao J.L., Kirk A., Lekstrom-Himes J. Human platelets exhibit chemotaxis using functional N-formyl peptide receptors. Exp. Hematol. 2005;33:73–84. doi: 10.1016/j.exphem.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 168.Fiuza C., Bustin M., Talwar S., Tropea M., Gerstenberger E., Shelhamer J.H., Suffredini A.F. Inflammation-promoting activity of HMGB1 on human microvascular endothelial cells. Blood. 2003;101:2652–2660. doi: 10.1182/blood-2002-05-1300. [DOI] [PubMed] [Google Scholar]
- 169.Krysko D.V., Agostinis P., Krysko O., Garg A.D., Bachert C., Lambrecht B.N., Vandenabeele P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32:157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 170.Wilkins H.M., Carl S.M., Weber S.G., Ramanujan S.A., Festoff B.W., Linseman D.A., Swerdlow R.H. Mitochondrial lysates induce inflammation and alzheimer’s disease-relevant changes in microglial and neuronal cells. J. Alzheimer’s Dis. 2015;45:305–318. doi: 10.3233/JAD-142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kitazawa M., Yamasaki T.R., LaFerla F.M. Microglia as a potential bridge between the amyloid β-peptide and tau. Ann. N. Y. Acad. Sci. 2004;1035:85–103. doi: 10.1196/annals.1332.006. [DOI] [PubMed] [Google Scholar]
- 172.Akiyama H., Arai T., Kondo H., Tanno E., Haga C., Ikeda K. Cell mediators of inflammation in the Alzheimer disease brain. Alzheimer Dis. Assoc. Disord. 2000;14:S47–S53. doi: 10.1097/00002093-200000001-00008. [DOI] [PubMed] [Google Scholar]
- 173.Brodacki B., Staszewski J., Toczyłowska B., Kozłowska E., Drela N., Chalimoniuk M., Stepien A. Serum interleukin (IL-2, IL-10, IL-6, IL-4), TNFα, and INFγ concentrations are elevated in patients with atypical and idiopathic parkinsonism. Neurosci. Lett. 2008;441:158–162. doi: 10.1016/j.neulet.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 174.Kodera K., Matsui H. Zebrafish, Medaka and Turquoise Killifish for Understanding Human Neurodegenerative/Neurodevelopmental Disorders. Int. J. Mol. Sci. 2022;23:1399. doi: 10.3390/ijms23031399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bradford Y.M., Toro S., Ramachandran S., Ruzicka L., Howe D.G., Eagle A., Kalita P., Martin R., Moxon S.A.T., Schaper K., et al. Zebrafish models of human disease: Gaining insight into human disease at ZFIN. ILAR J. 2017;58:4–16. doi: 10.1093/ilar/ilw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lam C.S., Korzh V., Strahle U. Zebrafish embryos are susceptible to the dopaminergic neurotoxin MPTP. Eur. J. Neurosci. 2005;21:1758–1762. doi: 10.1111/j.1460-9568.2005.03988.x. [DOI] [PubMed] [Google Scholar]
- 177.Sarath Babu N., Murthy C.L.N., Kakara S., Sharma R., Brahmendra Swamy C.V., Idris M.M. 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine induced Parkinson’s disease in zebrafish. Proteomics. 2016;16:1407–1420. doi: 10.1002/pmic.201500291. [DOI] [PubMed] [Google Scholar]
- 178.Matsui H., Taniguchi Y., Inoue H., Uemura K., Takeda S., Takahashi R. A chemical neurotoxin, MPTP induces Parkinson’s disease like phenotype, movement disorders and persistent loss of dopamine neurons in medaka fish. Neurosci. Res. 2009;65:263–271. doi: 10.1016/j.neures.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 179.Vijayanathan Y., Lim F.T., Lim S.M., Long C.M., Tan M.P., Majeed A.B.A., Ramasamy K. 6-OHDA-Lesioned Adult Zebrafish as a Useful Parkinson’s Disease Model for Dopaminergic Neuroregeneration. Neurotox. Res. 2017;32:496–508. doi: 10.1007/s12640-017-9778-x. [DOI] [PubMed] [Google Scholar]
- 180.Li M., Zhou F., Xu T., Song H., Lu B. Acteoside protects against 6-OHDA-induced dopaminergic neuron damage via Nrf2-ARE signaling pathway. Food Chem. Toxicol. 2018;119:6–13. doi: 10.1016/j.fct.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 181.Matsui H., Ito H., Taniguchi Y., Inoue H., Takeda S., Takahashi R. Proteasome inhibition in medaka brain induces the features of Parkinson’s disease. J. Neurochem. 2010;115:178–187. doi: 10.1111/j.1471-4159.2010.06918.x. [DOI] [PubMed] [Google Scholar]
- 182.Yang Z., Yu Y., Tay Y.X., Yue G.H. Genome editing and its applications in genetic improvement in aquaculture. Rev. Aquac. 2022;14:178–191. doi: 10.1111/raq.12591. [DOI] [Google Scholar]
- 183.Kok F.O., Shin M., Ni C.W., Gupta A., Grosse A.S., VanImpel A., Kirchmaier B.C., Peterson-Maduro J., Kourkoulis G., Male I., et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tian X., Zhou B. Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. J. Biol. Chem. 2021;296:100509. doi: 10.1016/j.jbc.2021.100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kalvaitytė M., Balciunas D. Conditional mutagenesis strategies in zebrafish. Trends Genet. 2022;38:856–868. doi: 10.1016/j.tig.2022.04.007. [DOI] [PubMed] [Google Scholar]
- 186.Meyer A., Schartl M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999;11:699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 187.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L., et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Postlethwait J., Amores A., Cresko W., Singer A., Yan Y.L. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 189.Wang X., Zhang J.B., He K.J., Wang F., Liu C.F. Advances of Zebrafish in Neurodegenerative Disease: From Models to Drug Discovery. Front. Pharmacol. 2021;12:1802. doi: 10.3389/fphar.2021.713963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chia K., Klingseisen A., Sieger D., Priller J. Zebrafish as a model organism for neurodegenerative disease. Front. Mol. Neurosci. 2022;15:376. doi: 10.3389/fnmol.2022.940484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bashirzade A.A., Zabegalov K.N., Volgin A.D., Belova A.S., Demin K.A., de Abreu M.S., Babchenko V.Y., Bashirzade K.A., Yenkoyan K.B., Tikhonova M.A., et al. Modeling neurodegenerative disorders in zebrafish. Neurosci. Biobehav. Rev. 2022;138:104679. doi: 10.1016/j.neubiorev.2022.104679. [DOI] [PubMed] [Google Scholar]
- 192.Wang J., Cao H. Zebrafish and medaka: Important animal models for human neurodegenerative diseases. Int. J. Mol. Sci. 2021;22:10766. doi: 10.3390/ijms221910766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Anichtchik O., Diekmann H., Fleming A., Roach A., Goldsmith P., Rubinsztein D.C. Loss of PINK1 function affects development and results in neurodegeneration in zebrafish. J. Neurosci. 2008;28:8199–8207. doi: 10.1523/JNEUROSCI.0979-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xi Y., Ryan J., Noble S., Yu M., Yilbas A.E., Ekker M. Impaired dopaminergic neuron development and locomotor function in zebrafish with loss of pink1 function. Eur. J. Neurosci. 2010;31:623–633. doi: 10.1111/j.1460-9568.2010.07091.x. [DOI] [PubMed] [Google Scholar]
- 195.Sallinen V., Kolehmainen J., Priyadarshini M., Toleikyte G., Chen Y.C., Panula P. Dopaminergic cell damage and vulnerability to MPTP in Pink1 knockdown zebrafish. Neurobiol. Dis. 2010;40:93–101. doi: 10.1016/j.nbd.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 196.Flinn L.J., Keatinge M., Bretaud S., Mortiboys H., Matsui H., De Felice E., Woodroof H.I., Brown L., McTighe A., Soellner R., et al. TigarB causes mitochondrial dysfunction and neuronal loss in PINK1 deficiency. Ann. Neurol. 2013;74:837–847. doi: 10.1002/ana.23999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Matsui H., Sugie A. An optimized method for counting dopaminergic neurons in zebrafish. PLoS ONE. 2017;12:e0184363. doi: 10.1371/journal.pone.0184363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hughes G.L., Lones M.A., Bedder M., Currie P.D., Smith S.L., Pownall M.E. Machine learning discriminates a movement disorder in a zebrafish model of Parkinson’s disease. DMM Dis. Model. Mech. 2020;13:dmm045815. doi: 10.1242/dmm.045815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Flinn L., Mortiboys H., Volkmann K., Kster R.W., Ingham P.W., Bandmann O. Complex i deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio) Brain. 2009;132:1613–1623. doi: 10.1093/brain/awp108. [DOI] [PubMed] [Google Scholar]
- 200.Noble S., Ismail A., Godoy R., Xi Y., Ekker M. Zebrafish Parla- and Parlb-deficiency affects dopaminergic neuron patterning and embryonic survival. J. Neurochem. 2012;122:196–207. doi: 10.1111/j.1471-4159.2012.07758.x. [DOI] [PubMed] [Google Scholar]
- 201.Bretaud S., Allen C., Ingham P.W., Bandmann O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J. Neurochem. 2007;100:1626–1635. doi: 10.1111/j.1471-4159.2006.04291.x. [DOI] [PubMed] [Google Scholar]
- 202.Baulac S., Lu H., Strahle J., Yang T., Goldberg M.S., Shen J., Schlossmacher M.G., Lemere C.A., Lu Q., Xia W. Increased DJ-1 expression under oxidative stress and in Alzheimer’s disease brains. Mol. Neurodegener. 2009;4:12. doi: 10.1186/1750-1326-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Edson A.J., Hushagen H.A., Frøyset A.K., Elda I., Khan E.A., Di Stefano A., Fladmark K.E. Dysregulation in the Brain Protein Profile of Zebrafish Lacking the Parkinson’s Disease-Related Protein DJ-1. Mol. Neurobiol. 2019;56:8306–8322. doi: 10.1007/s12035-019-01667-w. [DOI] [PubMed] [Google Scholar]
- 204.Sheng D., Qu D., Kwok K.H.H., Ng S.S., Lim A.Y.M., Aw S.S., Lee C.W.H., Sung W.K., Tan E.K., Lufkin T., et al. Deletion of the WD40 domain of LRRK2 in zebrafish causes parkinsonism-like loss of neurons and locomotive defect. PLoS Genet. 2010;6:e1000914. doi: 10.1371/journal.pgen.1000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ren G., Xin S., Li S., Zhong H., Lin S. Disruption of lrrk2 does not cause specific loss of dopaminergic neurons in zebrafish. PLoS ONE. 2011;6:e20630. doi: 10.1371/journal.pone.0020630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Prabhudesai S., Bensabeur F.Z., Abdullah R., Basak I., Baez S., Alves G., Holtzman N.G., Larsen J.P., Møller S.G. LRRK2 knockdown in zebrafish causes developmental defects, neuronal loss, and synuclein aggregation. J. Neurosci. Res. 2016;94:717–735. doi: 10.1002/jnr.23754. [DOI] [PubMed] [Google Scholar]
- 207.Sheng D., See K., Hu X., Yu D., Wang Y., Liu Q., Li F., Lu M., Zhao J., Liu J. Disruption of LRRK2 in Zebrafish leads to hyperactivity and weakened antibacterial response. Biochem. Biophys. Res. Commun. 2018;497:1104–1109. doi: 10.1016/j.bbrc.2018.02.186. [DOI] [PubMed] [Google Scholar]
- 208.Suzzi S., Ahrendt R., Hans S., Semenova S.A., Chekuru A., Wirsching P., Kroehne V., Bilican S., Sayed S., Winkler S., et al. Deletion of lrrk2 causes early developmental abnormalities and age-dependent increase of monoamine catabolism in the zebrafish brain. PLoS Genet. 2021;17:e1009794. doi: 10.1371/journal.pgen.1009794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wint J.M., Sirotkin H.I. Lrrk2 modulation of Wnt signaling during zebrafish development. J. Neurosci. Res. 2020;98:1831–1842. doi: 10.1002/jnr.24687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lemmens R., Van Hoecke A., Hersmus N., Geelen V., D’Hollander I., Thijs V., Van Den Bosch L., Carmeliet P., Robberecht W. Overexpression of mutant superoxide dismutase 1 causes a motor axonopathy in the zebrafish. Hum. Mol. Genet. 2007;16:2359–2365. doi: 10.1093/hmg/ddm193. [DOI] [PubMed] [Google Scholar]
- 211.Ramesh T., Lyon A.N., Pineda R.H., Wang C., Janssen P.M.L., Canan B.D., Burghes A.H.M., Beattie C.E. A genetic model of amyotrophic lateral sclerosis in zebrafish displays phenotypic hallmarks of motoneuron disease. DMM Dis. Model. Mech. 2010;3:652–662. doi: 10.1242/dmm.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Da Costa M.M.J., Allen C.E., Higginbottom A., Ramesh T., Shaw P.J., McDermott C.J. A new zebrafish model produced by TILLING of SOD1-related amyotrophic lateral sclerosis replicates key features of the disease and represents a tool for in vivo therapeutic screening. DMM Dis. Model. Mech. 2014;7:73–81. doi: 10.1242/dmm.012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Vettori A., Bergamin G., Moro E., Vazza G., Polo G., Tiso N., Argenton F., Mostacciuolo M.L. Developmental defects and neuromuscular alterations due to mitofusin 2 gene (MFN2) silencing in zebrafish: A new model for Charcot-Marie-Tooth type 2A neuropathy. Neuromuscul. Disord. 2011;21:58–67. doi: 10.1016/j.nmd.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 214.Chapman A.L., Bennett E.J., Ramesh T.M., De Vos K.J., Grierson A.J. Axonal Transport Defects in a Mitofusin 2 Loss of Function Model of Charcot-Marie-Tooth Disease in Zebrafish. PLoS ONE. 2013;8:e67276. doi: 10.1371/journal.pone.0067276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Eijkenboom I., Vanoevelen J.M., Hoeijmakers J.G.J., Wijnen I., Gerards M., Faber C.G., Smeets H.J.M. A zebrafish model to study small-fiber neuropathy reveals a potential role for GDAP1. Mitochondrion. 2019;47:273–281. doi: 10.1016/j.mito.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 216.Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L.B., Francescatto L., Ozes B., Karaca E., Jhangiani S.N., Bainbridge M.N., et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12:1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chaouch A., Porcelli V., Cox D., Edvardson S., Scarcia P., De Grassi A., Pierri C.L., Cossins J., Laval S.H., Griffin H., et al. Mutations in the mitochondrial citrate carrier SLC25A1 are associated with impaired neuromuscular transmission. J. Neuromuscul. Dis. 2014;1:75–90. doi: 10.3233/JND-140021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lyons D.A., Naylor S.G., Mercurio S., Dominguez C., Talbot W.S. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development. 2008;135:599–608. doi: 10.1242/dev.012377. [DOI] [PubMed] [Google Scholar]
- 219.Drerup C.M., Herbert A.L., Monk K.R., Nechiporuk A.V. Regulation of mitochondria-dynactin interaction and mitochondrial retrograde transport in axons. Elife. 2017;6:e22234. doi: 10.7554/eLife.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.O’Donnell K.C., Vargas M.E., Sagasti A. Wlds and PGC-1α regulate mitochondrial transport and oxidation state after axonal injury. J. Neurosci. 2013;33:14778–14790. doi: 10.1523/JNEUROSCI.1331-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.O’Donnell K.C., Lulla A., Stahl M.C., Wheat N.D., Bronstein J.M., Sagasti A. Axon degeneration and PGC-1α-mediated protection in a zebrafish model of α-synuclein toxicity. Dis. Model. Mech. 2014;7:571–582. doi: 10.1242/dmm.013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Princely Abudu Y., Pankiv S., Mathai B.J., Håkon Lystad A., Bindesbøll C., Brenne H.B., Yoke Wui Ng M., Thiede B., Yamamoto A., Mutugi Nthiga T., et al. NIPSNAP1 and NIPSNAP2 Act as “Eat Me” Signals for Mitophagy. Dev. Cell. 2019;49:509–525.e12. doi: 10.1016/j.devcel.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 223.Otten A.B.C., Kamps R., Lindsey P., Gerards M., Pendeville-Samain H., Muller M., van Tienen F.H.J., Smeets H.J.M. Tfam Knockdown Results in Reduction of mtDNA Copy Number, OXPHOS Deficiency and Abnormalities in Zebrafish Embryos. Front. Cell Dev. Biol. 2020;8:381. doi: 10.3389/fcell.2020.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rahn J.J., Stackley K.D., Chan S.S.L. Opa1 Is Required for Proper Mitochondrial Metabolism in Early Development. PLoS ONE. 2013;8:e59218. doi: 10.1371/journal.pone.0059218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Baden K.N., Murray J., Capaldi R.A., Guillemin K. Early developmental pathology due to cytochrome c oxidase deficiency is revealed by a new zebrafish model. J. Biol. Chem. 2007;282:34839–34849. doi: 10.1074/jbc.M703528200. [DOI] [PubMed] [Google Scholar]
- 226.Yokota T., Sugawara K., Ito K., Takahashi R., Ariga H., Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem. Biophys. Res. Commun. 2003;312:1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 227.Smith W.W., Pei Z., Jiang H., Moore D.J., Liang Y., West A.B., Dawson V.L., Dawson T.M., Ross C.A. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc. Natl. Acad. Sci. USA. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Islam M.S., Moore D.J. Mechanisms of LRRK2-dependent neurodegeneration: Role of enzymatic activity and protein aggregation. Biochem. Soc. Trans. 2017;45:163–172. doi: 10.1042/BST20160264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.West A.B., Moore D.J., Biskup S., Bugayenko A., Smith W.W., Ross C.A., Dawson V.L., Dawson T.M. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Araki M., Ito G., Tomita T. Physiological and pathological functions of LRRK2: Implications from substrate proteins. Neuronal Signal. 2018;2:NS20180005. doi: 10.1042/NS20180005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Matsui H., Taniguchi Y., Inoue H., Kobayashi Y., Sakaki Y., Toyoda A., Uemura K., Kobayashi D., Takeda S., Takahashi R. Loss of PINK1 in medaka fish (Oryzias latipes) causes late-onset decrease in spontaneous movement. Neurosci. Res. 2010;66:151–161. doi: 10.1016/j.neures.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 232.Matsui H., Gavinio R., Asano T., Uemura N., Ito H., Taniguchi Y., Kobayashi Y., Maki T., Shen J., Takeda S., et al. PINK1 and parkin complementarily protect dopaminergic neurons in vertebrates. Hum. Mol. Genet. 2013;22:2423–2434. doi: 10.1093/hmg/ddt095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ansai S., Sakuma T., Yamamoto T., Ariga H., Uemura N., Takahashi R., Kinoshita M. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics. 2013;193:739–749. doi: 10.1534/genetics.112.147645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ansai S., Kinoshita M. Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open. 2014;3:362–371. doi: 10.1242/bio.20148177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Dupuis L., Gonzalez De Aguilar J.L., Oudart H., De Tapia M., Barbeito L., Loeffler J.P. Mitochondria in amyotrophic lateral sclerosis: A trigger and a target. Neurodegener. Dis. 2004;1:245–254. doi: 10.1159/000085063. [DOI] [PubMed] [Google Scholar]
- 236.Nota B., Struys E.A., Pop A., Jansen E.E., Fernandez Ojeda M.R., Kanhai W.A., Kranendijk M., Van Dooren S.J.M., Bevova M.R., Sistermans E.A., et al. Deficiency in SLC25A1, encoding the mitochondrial citrate carrier, causes combined D-2- and L-2-hydroxyglutaric aciduria. Am. J. Hum. Genet. 2013;92:627–631. doi: 10.1016/j.ajhg.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Baloh R.H., Schmidt R.E., Pestronk A., Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J. Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhao C., Takita J., Tanaka Y., Setou M., Nakagawa T., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell. 2001;105:587–597. doi: 10.1016/S0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- 239.Virbasius J.V., Scarpulla R.C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Campbell C.T., Kolesar J.E., Kaufman B.A. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta-Gene Regul. Mech. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 241.Tiranti V., Hoertnagel K., Carrozzo R., Calimberti C., Munaro M., Granatiero M., Zelante L., Gasparini P., Marzella R., Rocchi M., et al. Mutations of SURF-1 in Leigh disease associated with cytochrome C oxidase deficiency. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Rahman S. Mitochondrial disease in children. J. Intern. Med. 2020;287:609–633. doi: 10.1111/joim.13054. [DOI] [PubMed] [Google Scholar]
- 243.Indrieri A., Conte I., Chesi G., Romano A., Quartararo J., Tatè R., Ghezzi D., Zeviani M., Goffrini P., Ferrero I., et al. The impairment of HCCS leads to MLS syndrome by activating a non-canonical cell death pathway in the brain and eyes. EMBO Mol. Med. 2013;5:280–293. doi: 10.1002/emmm.201201739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Anderson S., Bankier A.T., Barrell B.G., De Bruijn M.H.L., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 245.Broughton R.E., Milam J.E., Roe B.A. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001;11:1958–1967. doi: 10.1101/gr.156801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tuppen H.A.L., Blakely E.L., Turnbull D.M., Taylor R.W. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta-Bioenerg. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 247.Lott M.T., Leipzig J.N., Derbeneva O., Michael Xie H., Chalkia D., Sarmady M., Procaccio V., Wallace D.C. MtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinforma. 2013;44:1.23.1–1.23.26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Miller B., Kim S.J., Mehta H.H., Cao K., Kumagai H., Thumaty N., Leelaprachakul N., Jiao H., Vaughan J., Diedrich J., et al. Mitochondrial DNA variation in Alzheimer’s disease reveals a unique microprotein called SHMOOSE. Mol. Psychiatry. 2022:1–14. doi: 10.1038/s41380-022-01769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Müller-Nedebock A.C., Pfaff A.L., Pienaar I.S., Kõks S., van der Westhuizen F.H., Elson J.L., Bardien S. Mitochondrial DNA variation in Parkinson’s disease: Analysis of “out-of-place” population variants as a risk factor. Front. Aging Neurosci. 2022;14:797. doi: 10.3389/fnagi.2022.921412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Minczuk M., Papworth M.A., Kolasinska P., Murphy M.P., Klug A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc. Natl. Acad. Sci. USA. 2006;103:19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Minczuk M., Papworth M.A., Miller J.C., Murphy M.P., Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Gammage P.A., Gaude E., Van Haute L., Rebelo-Guiomar P., Jackson C.B., Rorbach J., Pekalski M.L., Robinson A.J., Charpentier M., Concordet J.P., et al. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucleic Acids Res. 2016;44:7804–7816. doi: 10.1093/nar/gkw676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Bacman S.R., Williams S.L., Pinto M., Peralta S., Moraes C.T. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hashimoto M., Bacman S.R., Peralta S., Falk M.J., Chomyn A., Chan D.C., Williams S.L., Moraes C.T. MitoTALEN: A General Approach to Reduce Mutant mtDNA Loads and Restore Oxidative Phosphorylation Function in Mitochondrial Diseases. Mol. Ther. 2015;23:1592–1599. doi: 10.1038/mt.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yang Y., Wu H., Kang X., Liang Y., Lan T., Li T., Tan T., Peng J., Zhang Q., An G., et al. Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs. Protein Cell. 2018;9:283–297. doi: 10.1007/s13238-017-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Bacman S.R., Kauppila J.H.K., Pereira C.V., Nissanka N., Miranda M., Pinto M., Williams S.L., Larsson N.G., Stewart J.B., Moraes C.T. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat. Med. 2018;24:1696–1700. doi: 10.1038/s41591-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Jo A., Ham S., Lee G.H., Lee Y.I., Kim S., Lee Y.S., Shin J.H., Lee Y. Efficient mitochondrial genome editing by CRISPR/Cas9. Biomed Res. Int. 2015;2015:305716. doi: 10.1155/2015/305716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Loutre R., Heckel A.M., Smirnova A., Entelis N., Tarassov I. Can Mitochondrial DNA be CRISPRized: Pro and Contra. IUBMB Life. 2018;70:1233–1239. doi: 10.1002/iub.1919. [DOI] [PubMed] [Google Scholar]
- 259.Wang B., Lv X., Wang Y., Wang Z., Liu Q., Lu B., Liu Y., Gu F. CRISPR/Cas9-mediated mutagenesis at microhomologous regions of human mitochondrial genome. Sci. China Life Sci. 2021;64:1463–1472. doi: 10.1007/s11427-020-1819-8. [DOI] [PubMed] [Google Scholar]
- 260.Hussain S.R.A., Yalvac M.E., Khoo B., Eckardt S., McLaughlin K.J. Adapting CRISPR/Cas9 System for Targeting Mitochondrial Genome. Front. Genet. 2021;12:402. doi: 10.3389/fgene.2021.627050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bian W.P., Chen Y.L., Luo J.J., Wang C., Xie S.L., Pei D.S. Knock-In Strategy for Editing Human and Zebrafish Mitochondrial DNA Using Mito-CRISPR/Cas9 System. ACS Synth. Biol. 2019;8:621–632. doi: 10.1021/acssynbio.8b00411. [DOI] [PubMed] [Google Scholar]
- 262.Khotina V.A., Bagheri Ekta M., Baig M.S., Wu W.-K., Grechko A.V., Sukhorukov V.N. Challenges of mitochondrial DNA editing in mammalian cells: Focus on treatment of cardiovascular disease. Vessel Plus. 2022;6:65. doi: 10.20517/2574-1209.2022.28. [DOI] [Google Scholar]
- 263.Fan W., Waymire K.G., Narula N., Li P., Rocher C., Coskun P.E., Vannan M.A., Narula J., MacGregor G.R., Wallace D.C. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Reddy P., Ocampo A., Suzuki K., Luo J., Bacman S.R., Williams S.L., Sugawara A., Okamura D., Tsunekawa Y., Wu J., et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell. 2015;161:459–469. doi: 10.1016/j.cell.2015.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mok B.Y., de Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A., Hsu F.S., Radey M.C., Peterson S.B., Mootha V.K., et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Guo J., Zhang X., Chen X., Sun H., Dai Y., Wang J., Qian X., Tan L., Lou X., Shen B. Precision modeling of mitochondrial diseases in zebrafish via DdCBE-mediated mtDNA base editing. Cell Discov. 2021;7:78. doi: 10.1038/s41421-021-00307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lee H., Lee S., Baek G., Kim A., Kang B.C., Seo H., Kim J.S. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat. Commun. 2021;12:1190. doi: 10.1038/s41467-021-21464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Iannetti E.F., Prigione A., Smeitink J.A.M., Koopman W.J.H., Beyrath J., Renkema H. Live-imaging readouts and cell models for phenotypic profiling of mitochondrial function. Front. Genet. 2019;10:131. doi: 10.3389/fgene.2019.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Chandrasekaran K., Hazelton J.L., Wang Y., Fiskum G., Kristian T. Neuron-specific conditional expression of a mitochondrially targeted fluorescent protein in mice. J. Neurosci. 2006;26:13123–13127. doi: 10.1523/JNEUROSCI.4191-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kim M.J., Kang K.H., Kim C.H., Choi S.Y. Real-time imaging of mitochondria in transgenic zebrafish expressing mitochondrially targeted GFP. Biotechniques. 2008;45:331–334. doi: 10.2144/000112909. [DOI] [PubMed] [Google Scholar]
- 271.Plucińska G., Paquet D., Hruscha A., Godinho L., Haass C., Schmid B., Misgeld T. In vivo imaging of disease-related mitochondrial dynamics in a vertebrate model system. J. Neurosci. 2012;32:16203–16212. doi: 10.1523/JNEUROSCI.1327-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Mandal A., Pinter K., Drerup C.M. Analyzing neuronal mitochondria in vivo using fluorescent reporters in zebrafish. Front. Cell Dev. Biol. 2018;6:144. doi: 10.3389/fcell.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Dukes A.A., Bai Q., Van Laar V.S., Zhou Y., Ilin V., David C.N., Agim Z.S., Bonkowsky J.L., Cannon J.R., Watkins S.C., et al. Live imaging of mitochondrial dynamics in CNS dopaminergic neurons in vivo demonstrates early reversal of mitochondrial transport following MPP+ exposure. Neurobiol. Dis. 2016;95:238–249. doi: 10.1016/j.nbd.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wrighton P.J., Shwartz A., Heo J.M., Quenzer E.D., LaBella K.A., Harper J.W., Goessling W. Quantitative intravital imaging in zebrafish reveals in vivo dynamics of physiological-stress-induced mitophagy. J. Cell Sci. 2021;134:jcs256255. doi: 10.1242/jcs.256255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vicente M., Salgado-Almario J., Soriano J., Burgos M., Domingo B., Llopis J. Visualization of mitochondrial Ca2+ signals in skeletal muscle of zebrafish embryos with bioluminescent indicators. Int. J. Mol. Sci. 2019;20:5409. doi: 10.3390/ijms20215409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kioka H., Kato H., Fujita T., Asano Y., Shintani Y., Yamazaki S., Tsukamoto O., Imamura H., Kogo M., Kitakaze M., et al. In vivo real-time ATP imaging in zebrafish hearts reveals G0s2 induces ischemic tolerance. FASEB J. 2020;34:2041–2054. doi: 10.1096/fj.201901686R. [DOI] [PubMed] [Google Scholar]
- 277.White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., et al. Transparent Adult Zebrafish as a Tool for In Vivo Transplantation Analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.MacRae C.A., Peterson R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015;14:721–731. doi: 10.1038/nrd4627. [DOI] [PubMed] [Google Scholar]
- 279.Murphy M.P., Hartley R.C. Mitochondria as a therapeutic target for common pathologies. Nat. Rev. Drug Discov. 2018;17:865–886. doi: 10.1038/nrd.2018.174. [DOI] [PubMed] [Google Scholar]
- 280.Stackley K.D., Beeson C.C., Rahn J.J., Chan S.S.L. Bioenergetic profiling of zebrafish embryonic development. PLoS ONE. 2011;6:e25652. doi: 10.1371/journal.pone.0025652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Kumar M.G., Rowley S., Fulton R., Dinday M.T., Baraban S.C., Patel M. Altered glycolysis and mitochondrial respiration in a zebrafish model of Dravet syndrome. eNeuro. 2016;3:1002–1011. doi: 10.1523/ENEURO.0008-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ibhazehiebo K., Gavrilovici C., De La Hoz C.L., Ma S.C., Rehak R., Kaushik G., Meza Santoscoy P.L., Scott L., Nath N., Kim D.Y., et al. A novel metabolism-based phenotypic drug discovery platform in zebrafish uncovers HDACs 1 and 3 as a potential combined anti-seizure drug target. Brain. 2018;141:744–761. doi: 10.1093/brain/awx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zhang Y., Nguyen D.T., Olzomer E.M., Poon G.P., Cole N.J., Puvanendran A., Phillips B.R., Hesselson D. Rescue of Pink1 Deficiency by Stress-Dependent Activation of Autophagy. Cell Chem. Biol. 2017;24:471–480.e4. doi: 10.1016/j.chembiol.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 284.Kim G.H.J., Mo H., Liu H., Wu Z., Chen S., Zheng J., Zhao X., Nucum D., Shortland J., Peng L., et al. A zebrafish screen reveals Renin-angiotensin system inhibitors as neuroprotective via mitochondrial restoration in dopamine neurons. Elife. 2021;10:e69795. doi: 10.7554/eLife.69795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Gemberling M., Bailey T.J., Hyde D.R., Poss K.D. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29:611–620. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hui S.P., Dutta A., Ghosh S. Cellular response after crush injury in adult zebrafish spinal cord. Dev. Dyn. 2010;239:2962–2979. doi: 10.1002/dvdy.22438. [DOI] [PubMed] [Google Scholar]
- 287.März M., Schmidt R., Rastegar S., Strahle U. Regenerative response following stab injury in the adult zebrafish telencephalon. Dev. Dyn. 2011;240:2221–2231. doi: 10.1002/dvdy.22710. [DOI] [PubMed] [Google Scholar]
- 288.Zambusi A., Ninkovic J. Regeneration of the central nervous system-principles from brain regeneration in adult zebrafish. World J. Stem Cells. 2020;12:8–24. doi: 10.4252/wjsc.v12.i1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ghosh S., Hui S.P. Regeneration of zebrafish CNS: Adult neurogenesis. Neural Plast. 2016;2016:5815439. doi: 10.1155/2016/5815439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Kishimoto N., Shimizu K., Sawamoto K. Neuronal regeneration in a zebrafish model of adult brain injury. DMM Dis. Model. Mech. 2012;5:200–209. doi: 10.1242/dmm.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Alvarez-Buylla A., Lim D.A. For the long run: Maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. doi: 10.1016/S0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 292.Merkle F.T., Mirzadeh Z., Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- 293.Ma D.K., Bonaguidi M.A., Ming G.L., Song H. Adult neural stem cells in the mammalian central nervous system. Cell Res. 2009;19:672–682. doi: 10.1038/cr.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Dimou L., Götz M. Glial cells as progenitors and stem cells: New roles in the healthy and diseased brain. Physiol. Rev. 2014;94:709–737. doi: 10.1152/physrev.00036.2013. [DOI] [PubMed] [Google Scholar]
- 295.Grandel H., Brand M. Comparative aspects of adult neural stem cell activity in vertebrates. Dev. Genes Evol. 2013;223:131–147. doi: 10.1007/s00427-012-0425-5. [DOI] [PubMed] [Google Scholar]
- 296.Than-Trong E., Bally-Cuif L. Radial glia and neural progenitors in the adult zebrafish central nervous system. Glia. 2015;63:1406–1428. doi: 10.1002/glia.22856. [DOI] [PubMed] [Google Scholar]
- 297.Labusch M., Mancini L., Morizet D., Bally-Cuif L. Conserved and Divergent Features of Adult Neurogenesis in Zebrafish. Front. Cell Dev. Biol. 2020;8:525. doi: 10.3389/fcell.2020.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Diotel N., Rodriguez Viales R., Armant O., März M., Ferg M., Rastegar S., Strähle U. Comprehensive expression map of transcription regulators in the adult zebrafish telencephalon reveals distinct neurogenic niches. J. Comp. Neurol. 2015;523:1202–1221. doi: 10.1002/cne.23733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Adolf B., Chapouton P., Lam C.S., Topp S., Tannhäuser B., Strähle U., Götz M., Bally-Cuif L. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev. Biol. 2006;295:278–293. doi: 10.1016/j.ydbio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 300.Grandel H., Kaslin J., Ganz J., Wenzel I., Brand M. Neural stem cells and neurogenesis in the adult zebrafish brain: Origin, proliferation dynamics, migration and cell fate. Dev. Biol. 2006;295:263–277. doi: 10.1016/j.ydbio.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 301.Kroehne V., Freudenreich D., Hans S., Kaslin J., Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138:4831–4841. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- 302.Barbosa J.S., Sanchez-Gonzalez R., Di Giaimo R., Baumgart E.V., Theis F.J., Götz M., Ninkovic J. Live imaging of adult neural stem cell behavior in the intact and injured zebrafish brain. Science. 2015;348:789–793. doi: 10.1126/science.aaa2729. [DOI] [PubMed] [Google Scholar]
- 303.Simon C., Götz M., Dimou L. Progenitors in the adult cerebral cortex: Cell cycle properties and regulation by physiological stimuli and injury. Glia. 2011;59:869–881. doi: 10.1002/glia.21156. [DOI] [PubMed] [Google Scholar]
- 304.Kaslin J., Ganz J., Geffarth M., Grandel H., Hans S., Brand M. Stem cells in the adult zebrafish cerebellum: Initiation and maintenance of a novel stem cell niche. J. Neurosci. 2009;29:6142–6153. doi: 10.1523/JNEUROSCI.0072-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kaslin J., Kroehne V., Ganz J., Hans S., Brand M. Distinct roles of neuroepithelial-like and radial glia-like progenitor cells in cerebellar regeneration. Development. 2017;144:1462–1471. doi: 10.1242/dev.144907. [DOI] [PubMed] [Google Scholar]
- 306.Kaslin J., Kroehne V., Benato F., Argenton F., Brand M. Development and specification of cerebellar stem and progenitor cells in zebrafish: From embryo to adult. Neural Dev. 2013;8:9. doi: 10.1186/1749-8104-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Bhattarai P., Thomas A.K., Cosacak M.I., Papadimitriou C., Mashkaryan V., Froc C., Reinhardt S., Kurth T., Dahl A., Zhang Y., et al. IL4/STAT6 Signaling Activates Neural Stem Cell Proliferation and Neurogenesis upon Amyloid-β42 Aggregation in Adult Zebrafish Brain. Cell Rep. 2016;17:941–948. doi: 10.1016/j.celrep.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 308.Matsui H., Kenmochi N., Namikawa K. Age- and α-Synuclein-Dependent Degeneration of Dopamine and Noradrenaline Neurons in the Annual Killifish Nothobranchius furzeri. Cell Rep. 2019;26:1727–1733.e6. doi: 10.1016/j.celrep.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 309.Edelmann K., Glashauser L., Sprungala S., Hesl B., Fritschle M., Ninkovic J., Godinho L., Chapouton P. Increased radial glia quiescence, decreased reactivation upon injury and unaltered neuroblast behavior underlie decreased neurogenesis in the aging zebrafish telencephalon. J. Comp. Neurol. 2013;521:3099–3115. doi: 10.1002/cne.23347. [DOI] [PubMed] [Google Scholar]
- 310.Valdesalici S., Cellerino A. Extremely short lifespan in the annual fish Nothobranchius furzeri. Proc. R. Soc. B Biol. Sci. 2003;270:S189–S191. doi: 10.1098/rsbl.2003.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Genade T., Benedetti M., Terzibasi E., Roncaglia P., Valenzano D.R., Cattaneo A., Cellerino A. Annual fishes of the genus Nothobarnchius as a model system for aging research. Aging Cell. 2005;4:223–233. doi: 10.1111/j.1474-9726.2005.00165.x. [DOI] [PubMed] [Google Scholar]
- 312.Harel I., Benayoun B.A., Machado B., Singh P.P., Hu C.K., Pech M.F., Valenzano D.R., Zhang E., Sharp S.C., Artandi S.E., et al. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell. 2015;160:1013–1026. doi: 10.1016/j.cell.2015.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Valenzano D.R., Terzibasi E., Cattaneo A., Domenici L., Cellerino A. Temperature affects longevity and age-related locomotor and cognitive decay in the short-lived fish: Nothobranchius furzeri. Aging Cell. 2006;5:275–278. doi: 10.1111/j.1474-9726.2006.00212.x. [DOI] [PubMed] [Google Scholar]
- 314.Hu C.K., Brunet A. The African turquoise killifish: A research organism to study vertebrate aging and diapause. Aging Cell. 2018;17:e12757. doi: 10.1111/acel.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hartmann N., Reichwald K., Lechel A., Graf M., Kirschner J., Dorn A., Terzibasi E., Wellner J., Platzer M., Rudolph K.L., et al. Telomeres shorten while Tert expression increases during ageing of the short-lived fish Nothobranchius furzeri. Mech. Ageing Dev. 2009;130:290–296. doi: 10.1016/j.mad.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 316.Hartmann N., Reichwald K., Wittig I., Dröse S., Schmeisser S., Lück C., Hahn C., Graf M., Gausmann U., Terzibasi E., et al. Mitochondrial DNA copy number and function decrease with age in the short-lived fish Nothobranchius furzeri. Aging Cell. 2011;10:824–831. doi: 10.1111/j.1474-9726.2011.00723.x. [DOI] [PubMed] [Google Scholar]
- 317.Bradshaw W.J., Poeschla M., Placzek A., Kean S., Valenzano D.R. Extensive age-dependent loss of antibody diversity in naturally short-lived turquoise killifish. Elife. 2022;11:e65117. doi: 10.7554/eLife.65117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Baumgart M., Priebe S., Groth M., Hartmann N., Menzel U., Pandolfini L., Koch P., Felder M., Ristow M., Englert C., et al. Longitudinal RNA-seq analysis of vertebrate aging identifies mitochondrial complex i as a small-molecule-sensitive modifier of lifespan. Cell Syst. 2016;2:122–132. doi: 10.1016/j.cels.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 319.Vanhunsel S., Bergmans S., Beckers A., Etienne I., Van Bergen T., De Groef L., Moons L. The age factor in optic nerve regeneration: Intrinsic and extrinsic barriers hinder successful recovery in the short-living killifish. Aging Cell. 2022;21:e13537. doi: 10.1111/acel.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Van Houcke J., Mariën V., Zandecki C., Vanhunsel S., Moons L., Ayana R., Seuntjens E., Arckens L. Aging impairs the essential contributions of non-glial progenitors to neurorepair in the dorsal telencephalon of the Killifish Nothobranchius furzeri. Aging Cell. 2021;20:e13464. doi: 10.1111/acel.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Tozzini E.T., Baumgart M., Battistoni G., Cellerino A. Adult neurogenesis in the short-lived teleost Nothobranchius furzeri: Localization of neurogenic niches, molecular characterization and effects of aging. Aging Cell. 2012;11:241–251. doi: 10.1111/j.1474-9726.2011.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Valenzano D.R., Terzibasi E., Genade T., Cattaneo A., Domenici L., Cellerino A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr. Biol. 2006;16:296–300. doi: 10.1016/j.cub.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 323.Evsiukova V.S., Kulikova E.A., Kulikov A.V. Age-related alterations in the behavior and serotonin-related gene mRNA levels in the brain of males and females of short-lived turquoise killifish (Nothobranchius furzeri) Biomolecules. 2021;11:1421. doi: 10.3390/biom11101421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Banerjee S., Poddar M.K. Carnosine: Effect on aging-induced increase in brain regional monoamine oxidase-A activity. Neurosci. Res. 2015;92:62–70. doi: 10.1016/j.neures.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 325.Saura J., Andrés N., Andrade C., Ojuel J., Eriksson K., Mahy N. Biphasic and region-specific MAO-B response to aging in normal human brain. Neurobiol. Aging. 1997;18:497–507. doi: 10.1016/S0197-4580(97)00113-9. [DOI] [PubMed] [Google Scholar]
- 326.Kumar M.J., Andersen J.K. Perspectives on MAO-B in aging and neurological disease: Where do we go from here? Mol. Neurobiol. 2004;30:77–89. doi: 10.1385/MN:30:1:077. [DOI] [PubMed] [Google Scholar]
- 327.Nicotra A., Pierucci F., Parvez H., Senatori O. Monoamine Oxidase Expression during Development and Aging. Neurotoxicology. 2004;25:155–165. doi: 10.1016/S0161-813X(03)00095-0. [DOI] [PubMed] [Google Scholar]
- 328.Santin Y., Resta J., Parini A., Mialet-Perez J. Monoamine oxidases in age-associated diseases: New perspectives for old enzymes. Ageing Res. Rev. 2021;66:101256. doi: 10.1016/j.arr.2021.101256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and tools described in this manuscript are available upon reasonable request.