Abstract
Ophirinina is a recently described suborder of jakobid protists (Excavata) with only one described species to date, Ophirina amphinema. Despite the acquisition and analysis of massive transcriptomic and mitogenomic sequence data from O. amphinema, its phylogenetic position among excavates remained inconclusive, branching as sister group either to all Jakobida or to all Discoba. From a morphological perspective, it has not only several typical jakobid features but also unusual traits for this group, including the morphology of mitochondrial cristae (sac-shaped to flattened-curved cristae) and the presence of two flagellar vanes. In this study, we have isolated, morphologically characterized, and sequenced genome and transcriptome data of two new Ophirinina species: Ophirina chinija sp. nov. and Agogonia voluta gen. et sp. nov. Ophirina chinija differs from O. amphinema in having rounded cell ends, subapically emerging flagella and a posterior cell protrusion. The much more distantly related A. voluta has several unique ultrastructural characteristics, including sac-shaped mitochondrial cristae and a complex “B” fiber. Phylogenomic analyses with a large conserved-marker dataset supported the monophyly of Ophirina and Agogonia within the Ophirinina and, more importantly, resolved the conflicting position of ophirinids as the sister clade to all other jakobids. The characterization of the mitochondrial genomes showed that Agogonia differs from all known gene-rich jakobid mitogenomes by the presence of two group II introns and their corresponding maturase protein genes. A phylogenetic analysis of the diversity of known maturases confirmed that the Agogonia proteins are highly divergent from each other and define distant families among the prokaryotic and eukaryotic maturases. This opens the intriguing possibility that, compared to other jakobids, Ophirinina may have retained additional mitochondrial elements that may help to understand the early diversification of eukaryotes and the evolution of mitochondria.
Keywords: Excavata, Jakobida, Ophirinina, mitochondrial genome, phylogenomics
Significance.
Jakobids are famous protists because of their gene-rich mitochondrial genomes, which have prompted several authors to propose that they represent one of the first eukaryotic lineages to have diverged. However, they lack the machinery needed to remove group II introns, an important feature present in the mitochondria of many other protists. We have characterized two new deep-branching jakobids and found this machinery encoded in the mitochondrial genome of one of them, which opens the possibility that it was present since very early in eukaryotic evolution.
Introduction
The supergroup Excavata is a large eukaryotic assemblage composed of Discoba, Malawimonadida, and Metamonada (Hampl et al. 2009; Burki et al. 2020). Within these clades, several species share morphological features that might represent synapomorphies for the Excavata, including a suspension-feeding groove supported by a complex and specific flagellar apparatus as well as a vane-bearing posterior flagellum (Cavalier-Smith 2003; Simpson 2003; Hampl et al. 2009). Recently, Excavata, and more specifically Discoba, have gained relevance due to their possible phylogenetic position at or near the root of the eukaryotic phylogeny (He et al. 2014; Cavalier-Smith 2018). In addition to these groups, a number of enigmatic lineages with excavate morphology exhibit poorly resolved evolutionary affinities. One notorious example is the Malawimonadida, which has most of the “typical excavate” features and branched with some or all other excavates in molecular phylogenies reconstructed with few markers, though usually with modest support (Simpson et al. 2006; Parfrey et al. 2010; Pánek et al. 2015; Zhang et al. 2015), but disconnected from other excavates in most recent phylogenomic analyses based on larger datasets (Hampl et al. 2009; Derelle and Lang 2012; Derelle et al. 2015; Katz and Grant 2015; Burki et al. 2016; Heiss et al. 2018).
The recently described Ophirina amphinema (Suborder Ophirinina) (Yabuki et al. 2018) provides another example of an enigmatic lineage. This species branched either as sister clade to all jakobids in a multigene phylogeny or as sister clade to all Discoba in 18S rDNA phylogenies, in both cases with low support (Yabuki et al. 2018). Not only its phylogenetic position within Discoba remains uncertain but Ophirina has both typical jakobid morphological features and also non-jakobid/discobid characteristics. Among the former are three microtubular roots associated with the posterior basal body, a dorsal fan of microtubules associated with the anterior basal body, and a multi-layered “C” fiber. Among the atypical features are the mitochondrial cristae morphology (discoid-flattened cristae) and the presence of two flagellar vanes (Yabuki et al. 2018). More recently, an unrooted phylogeny reconstructed with a dataset of 14 mitochondrial genes placed Ophirina in an intermediate position between jakobids and other discobids (Ettahi et al. 2021). In this context, the inclusion of sequence data from new Ophirinina species might help to resolve the phylogenetic position of this group in the eukaryotic tree.
Discoba is also a well-known lineage owing to the unique mitogenome features of its members (Gray et al. 2004; Simpson et al. 2006; Hampl et al. 2009). In particular, Jakobida have the most gene-rich and “bacterial-like” mitogenomes known for any eukaryotic group. For example, the mitogenome of Andalucia godoyi contains 100 functionally assignable genes (Burger et al. 2013; Gray et al. 2020). However, despite its gene richness, other elements of alphaproteobacterial origin crucially important during the evolution of the eukaryotic cell, such as group II introns and their associated maturase proteins, have so far never been detected in jakobid mitogenomes. Group II introns are ancient mobile genetic elements that act as retroelements by utilizing both autocatalytic RNAs and enzymatic protein components for their own excision, mobility, and reintegration in genomes (Lambowitz and Belfort 2015). Maturases are the protein components involved in the initial splicing reaction. They bind and promote intron excision, then constituting a retroelement complex that is able to engage in retrotransposition (Wank et al. 1999; Zimmerly et al. 2001; Toor et al. 2008; Haack et al. 2019). Group II introns originated in bacteria and subsequently spread through horizontal gene transfer (HGT) across the entire tree of life, including many mitochondrial and plastid genomes, mostly in plants, fungi, and protists (Zimmerly and Semper 2015; Maciszewski et al. 2022). In the eukaryotic nucleus, group II introns most likely arose from the alphaproteobacterial ancestor of mitochondria, or alternatively, they were transferred from a bacterium to the nuclear genome after the establishment of the mitochondrion. These group II introns most likely gave rise to the more complex spliceosomal introns (Zimmerly and Semper 2015), and their spread through the genome (as considered by eukaryogenetic models) may have been an essential selective force for the maintenance of the nuclear envelope during the evolution of the eukaryotic cell (López-García and Moreira 2006). Documenting the occurrence of group II introns and maturases in mitochondria would help to validate intermediate stages inferred to have occurred during the evolution of mitogenomes and the eukaryotic cell.
In this study, we have isolated, morphologically characterized, and sequenced genomes and transcriptomes of two new ophirinid species, Agogonia voluta gen. et sp. nov. and Ophirina chinija sp. nov., from, respectively, freshwater and marine sediment samples. Our phylogenomic analysis of 350 conserved genes shows that Ophirina and Agogonia are monophyletic and form the sister clade to all other jakobids with full support. All these ophirinids possess gene-rich mitogenomes. In addition, Agogonia exceptionally possesses two group II introns with their corresponding maturase domains, a set of unique molecular traits among all jakobids. These Agogonia maturases are highly divergent from each other and from other eukaryotic clades, suggesting multiple independent origins.
Results
Morphology of the New Ophirinid Species Ophirina chinija and Agogonia voluta
We have isolated two new ophirinid species: Ophirina chinija sp. nov., which defines a new species closely related to the already known Ophirina amphinema (Yabuki et al. 2018), and Agogonia voluta gen. et sp. nov., which is genetically divergent and defines a new genus and species (see the Taxonomic summary below). We characterized their morphology using light microscopy and measurements on 30 cells of each species.
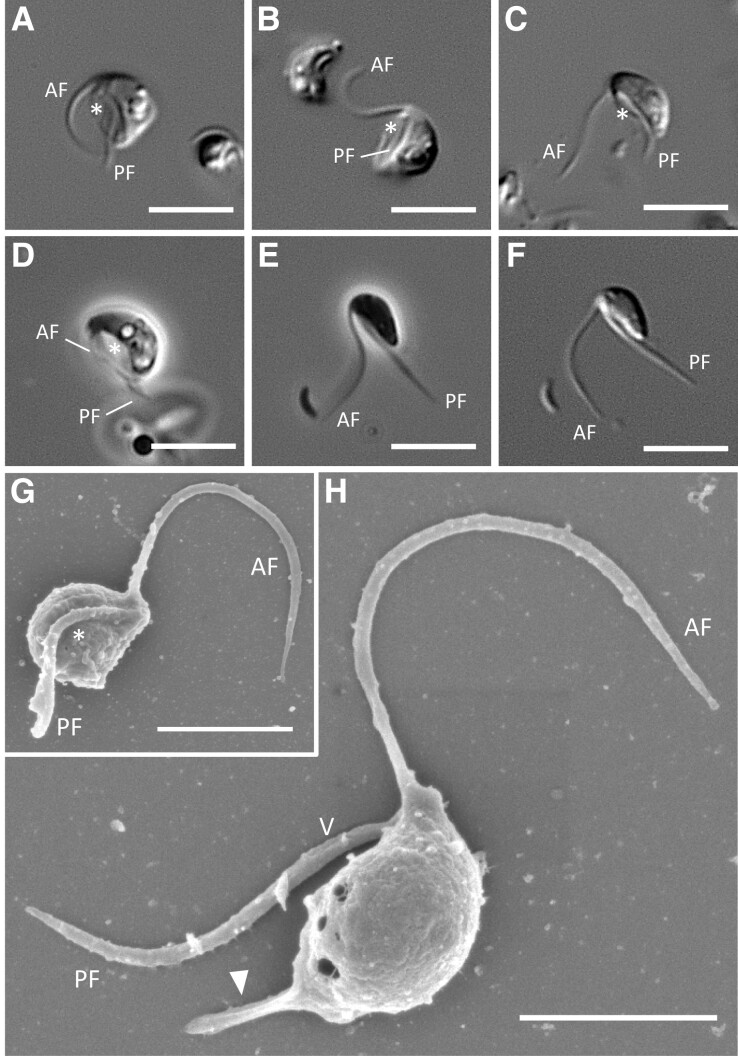
Ophirina chinija is biflagellated, with the anterior (AF) and the posterior (PF) flagella emerging subapically. Both flagella are acronematic (fig. 1). The cells lack any type of covering and are elongated (4.40 ± 0.41 µm length and 2.48 ± 0.4 µm width) with rounded anterior and posterior ends (fig. 1). One or more food vacuoles are observed in the posterior half of the cell, but no contractile vacuole can be identified. We observed a morphological transformation between two cell types: suspension-feeding cells that gather near a substrate and free-swimming cells. Feeding cells (fig. 1A–D) are bean-shaped, with a prominent groove (3.36 ± 0.44 µm in length) that occupies most of the ventral side and that the cells use to ingest bacteria. Swimming cells (fig. 1E and F) are more elongated, with a barely noticeable ventral groove, and are characterized by a mainly rectilinear movement, with rotation of the posterior end of the cell around the longitudinal axis (supplementary movie S1, Supplementary Material online). In feeding cells, the flagella are slightly longer than the cell body (5.91 ± 0.98 and 4.48 ± 0.65 µm for the AF and PF, respectively), whereas the flagella of swimming cells are two times longer than the cell body. The AF is directed above the cell and performs anterior-to-posterior wave-like movements (fig. 1A–C), sometimes vibrating near the ventral side of the cell body (fig. 1D). It periodically pushes off the substrate to help the feeding cells to turn around (supplementary movie S2, Supplementary Material online). The PF is directed backward and lies within the ventral groove in feeding cells (fig. 1A–D) or near the ventral side in swimming cells (fig. 1E and F). Scanning electron microscopy (SEM) observation reveals two poorly visible vanes on the PF (fig. 1H). A well-developed rigid protrusion at the posterior end is also visible in about half of the cells, probably involved in the attachment to substrates (fig. 1H). Cysts are not observed.
Fig. 1.
Light and scanning electron micrographs of Ophirina chinija sp. nov. (A–D) Phase and differential interference contrast (DIC) light micrographs of individual suspension feeding cells of Ophirina chinija strain LJ06. (E and F) Phase and DIC light micrographs of a swimming cell of O. chinija. (G and H) SEM images showing ventral and side views of a cell. Scale bars: 5 µm for A–F; 2 µm for G and H. Asterisk = ventral groove; AF = anterior flagellum; PF = posterior flagellum; V = vane; arrow = posterior protrusion.
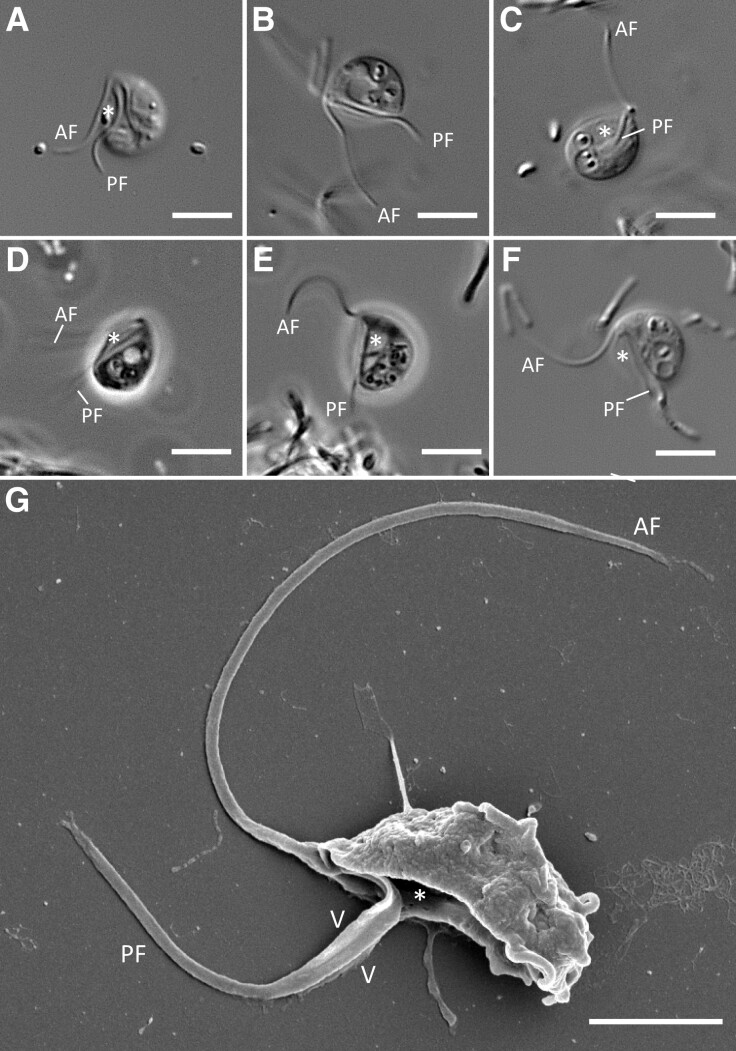
Agogonia voluta cells are naked, without scales, spicules, or any other surface elements (fig. 2). They are bean-shaped, with a convex dorsal side and a slightly pointed anterior end. Their dimensions are 6.8 ± 0.52 µm length and 4.96 ± 0.72 µm width. A well-defined, long (4.46 ± 0.73 µm) ventral groove occupies almost the entire body length (figs. 2G and 3A). Contractile vacuoles are present but not localized in any specific part of the cell (figs. 2B, 2D–F, and 3A). By contrast, food vacuoles, sometimes with visible bacteria inside, are localized mainly in the posterior half of the cells (fig. 2B–F). In contrast with O. chinija, significant morphological differences between feeding and swimming cells are not observed.
Fig. 2.
Light and scanning electron micrographs of Agogonia voluta gen. et sp. nov. (A–E) Phase and differential interference contrast (DIC) light micrographs of individual suspension feeding cells of Agogonia voluta. (F) DIC light micrograph of a swimming cell of A. voluta. (G) SEM image showing ventral and side views of a cell. Scale bars: 5 µm for A–F; 2 µm for G. Asterisk = ventral groove; AF = anterior flagellum; PF = posterior flagellum; V = vane.
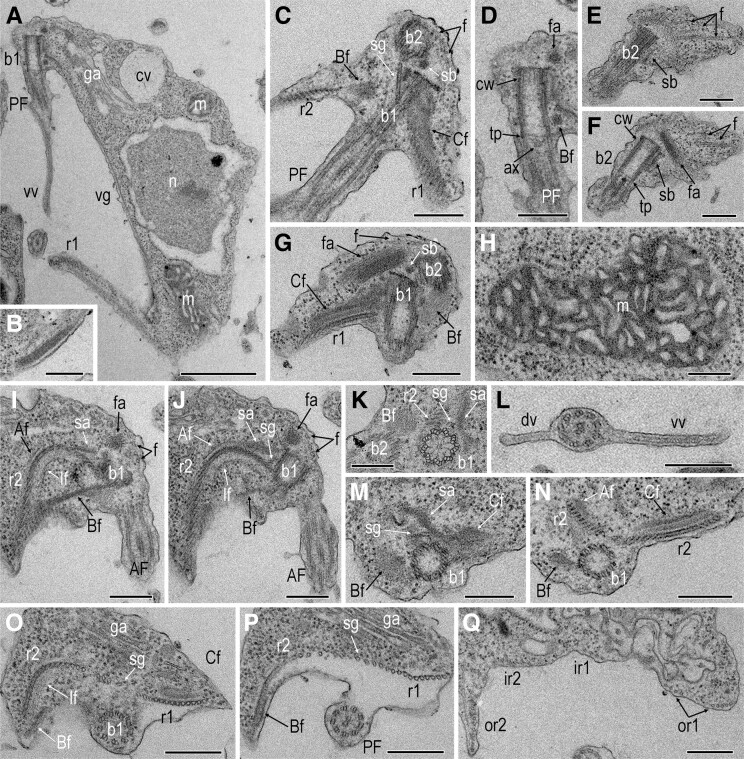
Fig. 3.
Transmission electron micrographs of Agogonia voluta gen. et sp. nov. (A) Longitudinal sagittal section showing the general cell structure. (B) Right posterior edge of the groove with composite fiber with a striated appearance. (C) Longitudinal frontal section of the anterior part of a cell showing the orientation of basal bodies, dorsal fan, and singlet microtubular root. (D) Basal body of posterior flagellum. (E and F) Basal body of anterior flagellum and dorsal fan of microtubules with fan-associated sheet. (G) Oblique sections showing the left side of the cell with basal bodies, left root with associated “C” fiber, and dorsal fan of microtubules with fan-associated sheet. (H) Mitochondrion with sac-shaped cristae. (I and J) Two serial oblique sections showing the right side of the cell with the right root and associated fibers. (K) Cartwheel structure of the posterior basal body. (L) Posterior flagellum with dorsal and ventral vanes. (M and N) Two serial transversal sections showing the origin of roots with associated fibers. (O and P) Two serial transversal sections showing the orientation of roots at the level of posterior flagellum emergence. (Q) Transversal section showing the orientation of the roots at the level of the middle part of the cell. Scale bars: A: 1 µm; С–Q: 300 nm. Af – “A” fiber; AF = anterior flagellum; ax = axosome; b1 = basal body of the posterior flagellum; b2 = basal body of the anterior flagellum; Bf = “B” fiber; Cf = “C” fiber; cv = contractile vacuole; cw = cartwheel structure; dv = dorsal vane of the posterior flagellum; ga = Golgi apparatus; f = dorsal fan of microtubules; fa = fan-associated fiber; If = “I” fiber; m = mitochondrion; n = nucleus; PF = posterior flagellum; r1 = left root; r2 = right root; sa = singlet-root-associated fiber; sg = singlet root; sb = striated band connecting basal bodies; tp = transversal plate; vg = ventral groove; vv = ventral vane of the posterior flagellum.
Agogonia cells exhibit two naked acronematic flagella emerging from the apical part of the cell (fig. 2), with a more prominent acronema on the AF (fig. 2G). The AF is slightly longer (8.06 ± 1.38 µm) than the cell body and describes a bend that directs it ventrally to the posterior end of the cell. In feeding cells, it twitches constantly, making sudden movements back and forth, pushing the cells to turn in opposite directions with jumping movements. In swimming cells, the AF is usually directed forward. On average, the PF has a similar length as the cell body (6.82 ± 2.1 µm) and is directed backward, lying completely in the ventral groove. SEM observations reveal two prominent vanes on the PF (fig. 2G). Usually, feeding cells attach to the substrate and display a vibrating jumping/rolling movement (supplementary movie S3, Supplementary Material online). Swimming cells show spiral movements with rotation around the longitudinal cell axis (supplementary movie S4, Supplementary Material online). Cysts are not observed.
Ultrastructure of Agogonia voluta gen. et sp. nov.
Since the new species Agogonia voluta exhibits important morphological differences with the already known ophirinid O. amphinema (Yabuki et al. 2018) and with our new O. chinija, we decided to determine the extent of that divergence also at the ultrastructural level, following the nomenclature by Yubuki et al. (2013). Transmission electron microscopy (TEM) confirms that the cells of A. voluta are naked (fig. 3A). They contain a single, slightly elongated nucleus (1.5–1.9 µm in diameter), usually located centrally or somewhat posteriorly, and are not directly connected to the flagellar basal bodies (fig. 3A). The nucleolus has a central position. Cells contain a single oblong mitochondrion adjacent to the nucleus, extending from the anterior to the posterior end of the cell (fig. 3A). Mitochondria contain an electron-dense matrix with sac-shaped and (on some sections) flattened-curved cristae (fig. 3H). A single large Golgi apparatus with three to five cisternae is located in the anterior part of the cell, between the nucleus and the basal bodies (fig. 3A, O, and P). No cytostome, microbodies, paranuclear bodies, or extrusomes are observed. One or several food vacuoles are present, usually at the posterior two-thirds of the cell (not shown). A large ventral groove supported by three microtubular roots extends from the posterior basal body (see below).
Both flagella are naked, though the PF bears two prominent vanes, as mentioned above (fig. 3A and L). These vanes are supported internally by approximately 35-nm-thick striated lamella that are not associated with axonemal microtubules (fig. 3A and L, supplementary fig. S1I–M, Supplementary Material online). The ventral vane, directed to the right, is the longest and widest, reaching up to 0.6 µm in width and a length of 2/3 of the flagellum (fig. 2G). The dorsal vane is less prominent, appearing only in the middle of the flagellum, and has an opposite left orientation (fig. 3L). Both flagella exhibit a standard 9 + 2 axoneme structure (fig. 3L and P, supplementary fig. S1F–M, Supplementary Material online). The flagellar basal bodies are located at the anterior apex of the cell and oriented orthogonally or at a slightly acute angle to each other (fig. 3C, G, I, and J). They are approximately 0.5 µm long and 0.2 µm wide and have a longitudinally segmented cartwheel structure in the basal part (fig. 3K) of about 130 nm long (fig. 3A, D, and F). The segmented electron-dense core material of basal bodies is barely visible. The anterior basal body (b2) is turned slightly to the left. The transition zone has a thin transversal plate with an adjacent axosome (fig. 3A, D, and F). An electron-dense striated band connects the two basal bodies (fig. 3C and E–G). The proximal part of b2 is associated with a fan of at least 16 microtubules evenly distributed along the entire dorsal side of the cell (fig. 3C, E–G, I, and J). A wide, electron-dense fan-associated sheet is located dorsally and to the left from b2 but not directly connected to it (fig. 3C, D, F, G, I, and J) and seems not to be associated with the microtubule origin.
Three microtubule roots (left-, right-, and singlet-root) originate near the posterior basal body (b1) and run along the entire surface of the ventral groove, strengthening it. There are four non-microtubular fibers, “A,” “B,” “I,” and “C,” associated with the left (r1) and right (r2) roots in their anterior parts. A dense and striated composite fiber (fig. 3B) supports the right posterior edge of the groove. The r1 consists of a row of 14 microtubules starting on the left side of the posterior basal body (fig. 3C, G, and M–Q; supplementary figs. S1 and S2, Supplementary Material online). Its proximal dorsal part is supported by a well-defined typical jakobid “C” fiber with a conspicuous multi-layered structure (fig. 3C, G, and M–O; supplementary figs. S1B–Eand S2A–D, Supplementary Material online). This “C” fiber starts at the dorsal side of b2 and runs along the dorsal side of r1. Near its origin, the “C” fiber is connected with the singlet-associated fiber and the “A” fiber, which runs toward the opposite direction along the dorsal side of the proximal part of r2 (fig. 3I, J, M, and N; supplementary figs. S1A–Dand S2C–G, Supplementary Material online).
The singlet root consists of a single microtubule originating in the space between b1, r2, and the singlet-associated fiber (fig. 3C, I–K, and M; supplementary figs. S1Aand S2A, Supplementary Material online) and is directed toward the left cell side to join r1 (fig. 3C, O, and P; supplementary figs. S1A–Jand S2A–J, Supplementary Material online). A wide singlet-associated fiber with conspicuous dense material is closely connected to the “A” fiber (fig. 3I, J, M, and N; supplementary figs. S1Aand S2A–C, Supplementary Material online). At the point where the PF separates from the cell body, r1 divides into an outer part (or1), which consists of eight microtubules, and an inner part (ir1), which consists of seven microtubules (supplementary figs. S1I–Mand S2H–M, Supplementary Material online). Toward the middle part of the cell, or1 divides again into two parts, each of which consists of three to five microtubules (fig. 3Q, supplementary fig. S1K–M, Supplementary Material online). The seven internal microtubules are also divided into two branches of three or four microtubules (fig. 3Q, supplementary fig. S2LandM, Supplementary Material online).
The right root (r2) is composed of 22 microtubules and originates on the right dorsal side of b1 (fig. 3I, J, and N–Q; supplementary figs. S1 and S2, Supplementary Material online). The proximal part of r2 forms an arc with its concave part directed dorsally (fig. 3I, J, O, and P; supplementary figs. S1C–E, S2H–J, Supplementary Material online). The r2 is surrounded on the dorsal side by the “A” fiber, and on the ventral side by the “I” and “B” fibers. The complex and thick two-layered “I” fiber (fig. 3I, J, and O; supplementary figs. S1B–Eand S2C–J, Supplementary Material online) corresponds in structure with what has been observed in other jakobids (Simpson et al. 2016). Two parts can be distinguished in the “B” fiber: a wide and electron-light part connecting to each basal body from the right side (fig. 3G, K, and M; supplementary figs. S1A, S2A and B, Supplementary Material online), and a thin and electron-dense part with lateral striation, which extends from b1 to the right-posterior end of the cell and lies along the ventral side of the cell membrane, connecting directly to the outer portion of r2 (fig. 3C, D, I, J, and N–P; supplementary figs. S1A–Hand S2B–K, Supplementary Material online). At this point, the “B” fiber connects with the “I” fiber and terminates soon after, at 1/3 of the cell length. Almost immediately after the origin of the three roots, the microtubules join together and become a single, even row of microtubules (fig. 3P; supplementary figs. S1F–Gand S2H–J, Supplementary Material online). However, on the ventral vane, the outer part of r2 (or2), which includes 14 closely adjacent microtubules (fig. 3Q; supplementary fig. S1K–M, Supplementary Material online), starts to separate from the inner part of r2 (ir2), containing eight microtubules evenly distributed along the right-bottom part of the groove together with the ir1 (fig. 3Q; supplementary fig. S1K–M, Supplementary Material online).
Phylogenomic Analyses
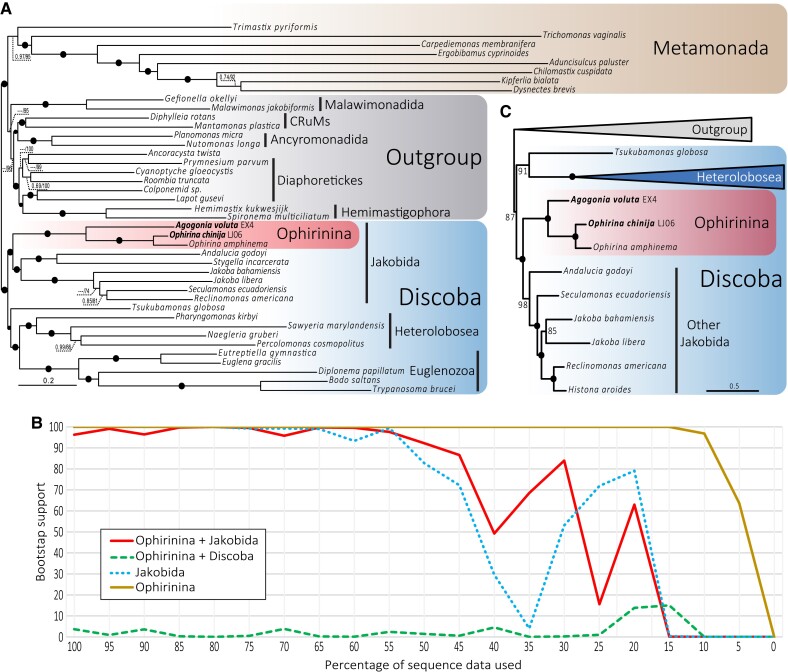
We used a large dataset of 350 conserved markers (112,726 amino acid positions) to reconstruct Bayesian inference (BI) and maximum likelihood (ML) phylogenomic trees with the CAT-GTR and the LG + C60 + F + R7 (PMSF) models, respectively. We obtained an overall identical topology with both methods, only with small differences in terminal branches. In both trees, the monophyly of Discoba, Jakobida, and Ophirinina and the sister relationship of Ophirinina to all other jakobids were recovered with high (≥95%) to full (100%) ML bootstrap support and full BI posterior probability (fig. 4A; supplementary figs. S3 and S4, Supplementary Material online). Ophirina chinija and O. amphinema form a fully supported clade sister to Agogonia voluta. To measure the robustness of this result, we carried out an analysis of the progressive removal of fast-evolving sites of the alignment using ML reconstruction (fig. 4B). First, we analyzed the monophyly of ophirinids and jakobids, recovering a fully (100%) to highly (≥95%) nonparametric bootstrap (bs) support for this clade until we have only 50% of our data left. We then analyzed the alternative hypothesis suggested by the 18S rDNA phylogeny from Yabuki et al. (2018), namely the ophirinids as the sister clade to all Discoba, but never recovered statistical support higher than 14% bs. Thus, this hypothesis was rejected by our analyses.
Fig. 4.
Phylogenomic analysis of Discoba. (A) ML phylogenomic tree based on 350 conserved proteins from Lax et al. (2018). The tree was reconstructed using 41 species and 112,726 amino acid positions with the LG + C60 + F + R7 model using the PMSF approximation for ML and the CAT-GTR model for Bayesian inference (BI). Numbers on branches indicate BI posterior probabilities and ML bootstrap values; bootstrap values <50% are indicated by ---. Branches with support values higher or equal to 0.99 BI posterior probability and 95% ML bootstrap are indicated by black dots. (B) ML bootstrap values for the monophyly of Jakobida (without Ophirinina), Ophirinina, Ophirinina + Jakobida, and the competing hypothesis of Ophirinina sister to all Discoba (Ophirinina + Discoba) as a function of the proportion of fast-evolving sites removed from the phylogenomic dataset. (C) ML phylogenomic tree based on 14 conserved mitochondrion-encoded proteins from Ettahi et al. (2021). The tree was reconstructed using 25 species and 4,683 amino acid positions with the LG + C60 + F + R7 model using the PMSF approximation. Numbers on branches indicate ML bootstrap values; branches with 100% support values are indicated by black dots. The complete tree is provided in supplementary figure S6, Supplementary Material online.
We additionally tested the relationship between ophirinids and the other discobids by using two alternative sequence datasets. First, to avoid possible long branch attraction artifacts (LBA) induced by the very long branches of metamonads, we analyzed a dataset with 34 taxa excluding all metamonads except Trimastix pyriformis, which exhibits the shortest branch among the species of this group. We also recovered ophirinids as the sister lineage to jakobids with high ML bootstrap support (98%) (supplementary fig. S5, Supplementary Material online). Second, we updated the dataset of mitochondrial markers from Ettahi et al. (2021) by incorporating our ophirinid sequences and several outgroup taxa and excluding the very long-branching Euglenozoa. The ML tree based on these markers also supported the sister relation of ophirinids and jakobids with maximum bootstrap value (100%) (fig. 4C; supplementary fig. S6, Supplementary Material online). All datasets and trees are available at https://doi.org/10.6084/m9.figshare.20332842.v3.
Mitochondrial Genomes
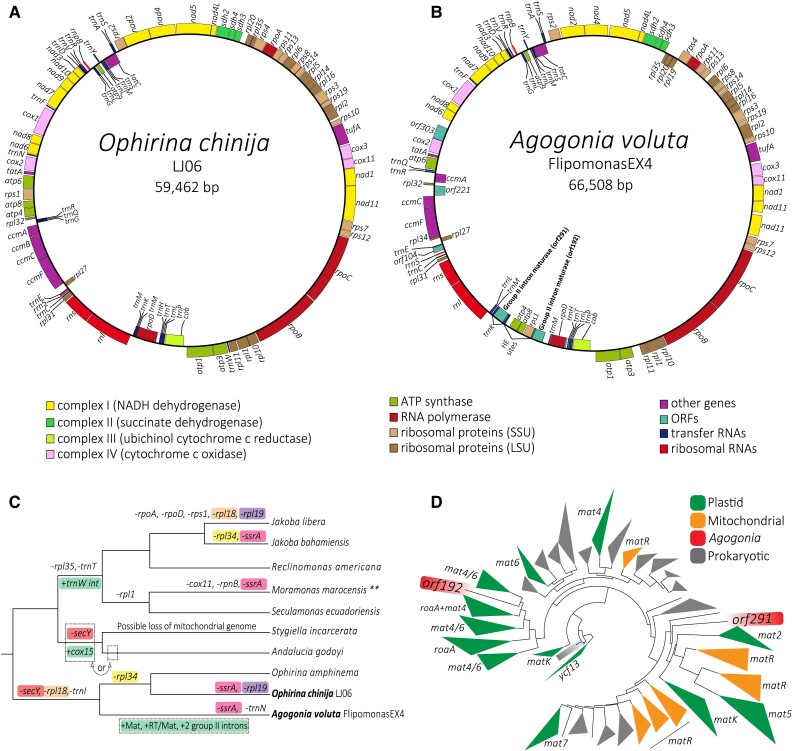
We assembled the complete, circular mitogenomes of both O. chinija and A. voluta. They have sizes of 59,462 and 66,508 bp (fig. 5A and B) and GC content values of 33.7% and 34.3%, respectively, similar to those of O. amphinema (59,094 bp and 39.1%, respectively) (Yabuki et al. 2018). These GC contents are in the upper ranges of GC values known in jakobids (from 26.1% in Reclinomonas americana up to 35.4% in Histiona aroides) (Burger et al. 2013). DELTA-BLAST was used to refine the annotation of genes that were originally identified just as ORFs, including rpoD, ccmB, and two maturase genes of A. voluta (orf291 and orf192). In the end, we were unable to infer the function of only one ORF of A. voluta (orf303). The mitogenome of O. chinija contains 92 genes, including 3 ribosomal RNAs (rnl, rns, and rrn5), 1 RNase-P RNA (rnpB), 26 tRNAs, and 62 protein-coding genes, very similar to the 94 genes of O. amphinema and only differing in the absence of rpl19 in O. chinija. The mitogenome of A. voluta contains 94 genes, including 3 ribosomal RNAs (rnl, rns, and rrn5), 1 RNase-P RNA (rnpB), 25 tRNAs, and 65 protein-coding genes. Coding regions account for 91.6% in O. chinija and 89.8% in A. voluta. We did not detect introns in the mitogenome of O. chinija. However, we detected two group II introns of 1,320 and 1,105 bp in the mitogenome of A. voluta, the first inserted in the trnK gene and the second in an intergenic region between the rps1 and trnM genes (fig. 5B). These introns contained the two maturase genes (orf291 and orf192, respectively) mentioned above. The two annotated mitochondrial genome sequences were deposited in GenBank with accession numbers ON980703 (Ophirina chinija) and ON980704 (Agogonia voluta).
Fig. 5.
Graphical representation of the mitochondrial genome architecture of Ophirina chinija and Agogonia voluta, evolution of gene content in jakobid mitochondria, and phylogeny of maturases. (A and B) Circular-mapping genomes, colors indicate the different types of genes. HE = homing endonuclease. (C) Gene loss and gain events during mitochondrial genome evolution in jakobids. Parallel losses (−) of the same gene are shown with correspondingly colored paired backgrounds, green background represents gains (+). **The complete mitochondrial genome sequence of Moramonas marocensis is not yet released; thus, the diagram shows an incomplete image of its evolution. (D) Schematic ML tree of the diversity of maturases (452 maturase sequences, 1,236 amino acid positions) reconstructed under the VT + F + R8 model (for the detailed tree see supplementary fig. S7; Supplementary Material online).
Phylogenetic Analysis of Maturases
We carried out an ML phylogenetic analysis of a dataset of 452 maturase sequences (1,236 amino acid positions) representing the known diversity of prokaryotic and eukaryotic maturases based on the dataset from Maciszewski et al. (2022), using the VT + F + R8 model (fig. 5D; supplementary fig. S7, Supplementary Material online). We obtained a phylogenetic tree with fully (100%) to highly (≥95% bs) supported monophyly for the mitochondrial maturase genes of Fungi and Streptophyta (matR), and for the matK, mat7, mat2, mat5 roaA, and ycf13 plastid maturases, which suggests that these sequences still contain some ancient phylogenetic signal. The plastid maturases mat4, mat6, roaA, and matK of Rhodophyta appear to have several independent origins within the diversity of Bacteria and Archaea. In the case of mitochondrial maturases (matR), several fungal matR sequences branch with the mat5 sequences from euglenids and several others with the matR sequences of Rhodophyta, suggesting possible cases of eukaryote-to-eukaryote HGT. In general, our tree shows that maturases have been transferred through HGT on several occasions among different prokaryotic and eukaryotic groups, including between organelles. The two maturases found in the Agogonia mitogenome appear to have different origins since they branch in distant parts of the tree, but in both cases associated with other eukaryotic sequences. The Agogonia maturase encoded in orf192 branched with moderate support (92% bs) within the diversity of plastid roaA + mat4 + mat6 maturases (mostly coming from euglenids), indicating that it could have been acquired by HGT from a photosynthetic donor. On the other hand, the maturase encoded in orf291 branched also with moderate support (90% bs) sister to the diversity of mitochondrial maturases from Fungi and plastid maturases matK, mat2, and mat5 (fig. 5D; supplementary fig. S7, Supplementary Material online), suggesting that it may represent a more ancient maturase in Agogonia.
Discussion
Morphology of New Ophirinid Species
In agreement with their phylogenetic proximity, the new species O. chinija has many morphological similarities with the previously described O. amphinema (Yabuki et al. 2018), including the size of the cell body, the presence of two morphologically different types of cells (feeding and swimming cells), two vanes on the posterior flagellum, and the jumping movements of the feeding cells. Although the presence of cysts is widespread in jakobids (Flavin and Nerad 1993; O’Kelly 1993; Lara et al. 2006; Mylnikov and Mylnikov 2014; Strassert et al. 2016), they have never been observed in Ophirina species, including O. chinija. Nevertheless, there are morphological differences between O. chinija and O. amphinema that justify their separation as distinct species. The cells of O. chinija are characterized by the subapical emergence of flagella and more rounded anterior and posterior ends. On SEM images, we also found the presence of a posterior protrusion that has a more rigid structure than pseudopodia and that is probably involved in cell attachment to the substrate, a behavior absent in O. amphinema, which lacks this protrusion. In fact, feeding cells of O. chinija concentrate near the substrate whereas those of O. amphinema remain in the water column.
According to our ultrastructural analysis, Agogonia is characterized by a typical “excavate” appearance, with a conspicuous ventral groove supported by an internal microtubular skeleton and a posterior flagellum that undulates within the groove. Several characteristics support the specific ascription of this new genus within the jakobids. Agogonia cells have a single mitochondrion, which is typical for jakobids (Simpson et al. 2016) except for Moramonas, which seems to have three spherical mitochondria (Strassert et al. 2016). Among the jakobids, a wide variety of types of mitochondrial cristae have been observed, although most species have tubular ones (O’Kelly 1993; Lara et al. 2006; Mylnikov and Mylnikov 2014; Strassert et al. 2016). Exceptions are the discoid cristae found in some undescribed species by O’Kelly (1993), the discoid-flattened cristae observed in the recently described Ophirina amphinema (Yabuki et al. 2018), the flattened cristae of Jakoba libera (O’Kelly 1993), and the absence of cristae in the anaerobic/microaerobic genera Stygiella and Velundella (Simpson et al. 2002; Pánek et al. 2015). In Agogonia, we observed saccular mitochondrial cristae, sometimes with flattened-curved structure, a trait that is new among jakobids, even if, depending on the orientation of the TEM thin sections, it has been noted that the tubular cristae of M. marocensis can show a saccular–vesicular appearance (Strassert et al. 2016). The presence of at least five types of mitochondrial cristae in jakobids is unusual given that cristae generally represent a conserved and consistent feature in most eukaryotic groups (Corliss 1984; Patterson 1988). This is of particular interest since jakobids have been considered to have the most “bacterial-like” mitochondrial genomes of all eukaryotes (Burger et al. 2013). Considering also their possible branching near the root of the eukaryote tree (He et al. 2014; Cavalier-Smith 2018), jakobids may be good models to understand the first mitochondriate eukaryotes. The monophyletic origin of mitochondria is widely accepted (Delihas and Fox 1987; Erdmann et al. 1987; Gray 1989; Wang and Wu 2015; Roger et al. 2017) and mitochondrial cristae probably had a pre-endosymbiotic origin from intracytoplasmic membranes already present in their alphaproteobacterial ancestors, in which they played a similar role to harness energy (Muñoz-Gómez et al. 2017). It remains unclear which form of mitochondrial cristae was ancestral. However, the various forms found in jakobids suggest that morphological diversification of mitochondrial cristae may have occurred at the very beginning of the evolution of early mitochondriate protists (Gray 1992; O’Kelly 1993).
Both the morphology and ultrastructure of the new genus Agogonia show similarities with the recently described genus Ophirina (Yabuki et al. 2018). The most conspicuous feature of both genera is the presence of a second vane on the posterior flagellum, exceptional among jakobids but widespread in other excavates (Simpson 2003). The morphology of these vanes in Agogonia corresponds to that found in O. amphinema, in which the vanes are also reinforced from the inside by a striated lamella. The presence of two vanes in two early branching but not closely related jakobid genera supports the hypothesis that such structure was most likely ancestral. Other similarities of Ophirina and Agogonia include the absence of paranuclear bodies and extrusomes, previously found in other jakobid genera (O’Kelly 1993; Lara et al. 2006; Mylnikov and Mylnikov 2014; Strassert et al. 2016). The two ophirinid genera are also characterized by jumping movements of the feeding cells and the absence of cysts (Yabuki et al. 2018).
Another unique feature of Ophirina and Agogonia is the proximal part of the “B” fiber that originates from both basal bodies and connects them. Such a feature has not been described neither in jakobids nor in any other “typical” excavates, where this fiber usually originates from the posterior basal body or from the left/right root. However, Agogonia (in contrast with Ophirina) has a complex two-parts “B” fiber, with a much wider and electron-lucent proximal part, and a narrow and electron-dense distal part that connects with the outer portion of the right root. Agogonia presents on the dorsal side of its anterior basal body a large, electron-dense fan-associated sheet, with an elongated structure that extends perpendicular to the basal body but not connected with it. Such a prominent structure has not been described in any other jakobid, which usually have a thin sheet tightly appressed to the anterior basal body. We also observed the presence of a large and conspicuous singlet-associated fiber, connected with both the “A” and “C” fibers. This fiber is absent in Ophirina (Yabuki et al. 2018) but a similar structure has been described in the jakobid Stygiella (Simpson and Patterson 2001), though the three fibers are not connected to each other in this species. Another key difference with Ophirina is the presence in Agogonia of twice as many microtubules in the left root and a slightly different orientation of microtubules in all three roots. Basal bodies of Agogonia are connected by a dense striated band, which is quite common in jakobids (Simpson and Patterson 2001; Lara et al. 2006; Mylnikov and Mylnikov 2014; Strassert et al. 2016) but absent in Ophirina (Yabuki et al. 2018). By contrast, the segmented electron-dense core material of basal bodies found in Ophirina (Yabuki et al. 2018) is less pronounced in Agogonia.
Phylogenomics
Both of our ML and BI phylogenomic trees show that O. chinija and O. amphinema form a fully supported clade sister to A. voluta and prove the sister relationship of Ophirinina to all other jakobids. Within Jakobida, our tree shows the same topology as the multigene study by Yabuki et al. (2018) though, thanks to the inclusion of more sequence data, we have significantly improved the overall support of the tree, especially for the position of Ophirinina as the sister clade to all other jakobids, using both nucleus- and mitochondrion-encoded markers and several taxon sampling combinations (fig. 4; supplementary figs. S3–S6, Supplementary Material online).
A fast-evolving sites removal analysis further supports these results, showing that the monophyly of Ophirinina and Jakobida (≥95% bootstrap until 50% of sites left) received in general a support that is even higher than that for the well-accepted monophyly of Jakobida (≥95% bootstrap until 65% of sites left), proving how robust this position is (fig. 4B). Most importantly, we never recovered any strong support (all bootstrap values ≤14%) for the alternative position of ophirinids as sister to all discobids (Yabuki et al. 2018).
Mitochondrial Genomes, Group II Introns, and Maturase Evolution
Our data confirm that the loss of secY, rpl18, and trnI was ancestral to all ophirinids, making the absence of these three genes a defining trait of the whole clade, whereas several losses of rpl34 occurred more recently and in parallel in the genus Ophirina and in Jakoba bahamiensis (fig. 5C). Agogonia voluta has lost trnN, being the first time that the loss of this tRNA is recorded in jakobids (Burger et al. 2013).
As in O. amphinema (Yabuki et al. 2018), no introns were detected in the mitogenome of O. chinija. However, we identified two group II intron-like sequences in the mitogenome of A. voluta, one in an intergenic position between the genes rps1 and trnM (1,105-bp long, containing the orf192) and one in an intragenic position within the gene trnk (1,320-bp long, containing the orf291). The two ORFs contained in these introns encoded two maturase proteins. The presence of these introns in Agogonia is a unique trait among jakobids since only one group II intron had been previously found in the trnW gene of the species belonging to the clade formed by the genera Jakoba, Reclinomonas, Moramonas, and Seculamonas (fig. 5C; Burger et al. 2013). The size of the two Agogonia intron-like sequences and their corresponding maturase genes and homing sites fit the size ranges that have been observed for group II introns in mitogenomes from other eukaryotes (Turmel et al. 2007; Zimmerly and Semper 2015). Another remarkable aspect is that the two maturases encoded in these introns (orf291 and orf192) are highly divergent from each other (fig. 5D), suggesting independent evolutionary origins.
It is widely accepted that group II introns originated within bacterial genomes (Zimmerly and Semper 2015). It has also been hypothesized that intron spreading was important in the evolution and/or maintenance of some key eukaryotic features, including the spliceosome and the nuclear envelope (López-García and Moreira 2006; Lambowitz and Belfort 2015). Several studies have shown that introns have spread through horizontal gene transfer (HGT) across the entire tree of life, including mitochondrial and plastid genomes of several eukaryotic groups (Zimmerly and Semper 2015; Maciszewski et al. 2022). Given the possible position of jakobids near the root of the eukaryote phylogeny (He et al. 2014; Cavalier-Smith 2018), the divergent maturases found in Agogonia can represent interesting models to study the evolution of mitochondrial group II introns, which may have originated within the alphaproteobacterial ancestor of mitochondria or from ancient HGT events. However, at this point, it is difficult to pinpoint if these group II introns and their maturases were integrated into the mitogenome of an ancestor of Agogonia or in that of a more ancient ancestor of ophirinids and then got lost in the genus Ophirina (fig. 5C). The discovery and study of additional ophirinid species may help to elucidate this question.
Taxonomic Summary
Taxonomy: Eukaryota; Excavata; Discoba; Jakobida
suborder Ophirinina Yabuki, Gyaltshen, Heiss, and Kim emend.
Emended diagnosis: Mitochondrion with saccular or flat discoid cristae. Singlet-root-associated fiber can be present or absent. Dorsal fan microtubules project posteriorly and rightward or evenly distributed along the dorsal side of cells.
Agogonidae n. fam. Galindo et al. (2022)
Diagnosis: Unicellular heterotrophic biflagellated protists with a conspicuous ventral groove. The posterior flagellum lies within the ventral groove and bears two vanes. The main and most prominent dextroventral vane extends along 2/3 of the length of the flagellum, and the sinistrodorsal vane appears at the middle of the flagellum length. The basal bodies are interconnected by a dense striated band. The microtubules of the dorsal fan are supported by a wide and electron-dense fan-associated sheet, located dorsally and to the left of the anterior basal body. The ventral groove is supported by three microtubule roots (left, right, and singlet roots) associated with the posterior basal body and with five fibers (“A,” “B,” “C,” “I,” and singlet-associated fiber) supporting these roots. The “B” fiber has a complex structure with a large and electron-lucent part connecting basal bodies with each other from the right, and an electron-dense part connecting the posterior basal body with the outer portion of the right root. A large and conspicuous singlet-associated fiber connects to the “A” fiber and “C” fiber. Mitochondrion with sac-shaped flattened-curved cristae.
Zoobank Registration: urn:lsid:zoobank.org:act:6E3A9690-32A8-4D36-A9E5-A6BED9541469
Type genus: Agogonia.
Agogonia n. gen. Galindo and Prokina (2022)
Diagnosis: Same as family
Etymology: The name Agogonia honors the Spanish chemist and biochemist Antonio González González, who pushed forward science and education in the Canary Islands.
Zoobank registration: urn:lsid:zoobank.org:act:F25EA369-8CD4-4054-8E8D-D65E91D0133D
Type species: Agogonia voluta.
Agogonia voluta n. sp. Galindo and Prokina (2022)
Diagnosis: Cells are bean-shaped, with slightly pointed anterior ends. Cell length is 6.8 ± 0.52 µm; cell width is 4.96 ± 0.72 µm. Two naked acronematic flagella originate from the apical anterior part of the cell. Flagella are slightly longer than the cell length, the anterior flagellum is 8.06 ± 1.38 µm, and the posterior flagellum is 6.82 ± 2.1 µm. Feeding cells attach to a substrate and make vibrating and jumping movements. Swimming cells are characterized by spiral movements and rotation around the longitudinal axis of the cell body. No cysts.
Type material: A block of chemically fixed resin-embedded cells of the type strain, FlipomonasEX4, is deposited in the DEEM culture collection, CNRS and Université Paris-Saclay (France). This constitutes the name-bearing type of the new species (a hapantotype).
Type figure: figure 2.
Gene sequence: The SSU rRNA gene sequence has the GenBank accession number ON997588.
Type culture: A cryopreserved culture is deposited in the DEEM culture collection, CNRS and Université Paris-Saclay (France) as the type strain FlipomonasEX4.
Isolator: Luis Javier Galindo.
Type locality: Specimen isolated from a freshwater pond in the town of Orsay, France.
Etymology: The name voluta refers to the behavior of the cells to roll while feeding.
Zoobank registration: urn:lsid:zoobank.org:act:03EBB812-97D9-4647-8B57-8E555D1ACA05
family Ophirinidae Yabuki et al. (2018)
Ophirina chinija n. sp. Galindo and Prokina (2022)
Diagnosis: Cells are bean-shaped, with rounded anterior and posterior ends; cell length is 4.40 ± 0.41 µm; cell width is 2.48 ± 0.4 µm. Two acronematic flagella emerge subapically; anterior flagellum is 5.91 ± 0.98 µm; posterior flagellum is 4.48 ± 0.65 µm. Two morphologically distinct types of cells: more rounded feeding cells, with a prominent ventral groove and flagella slightly longer than the cell length, and elongated swimming cells, with a poorly distinguishable ventral groove and flagella twice longer than the cell. Feeding cells attach to substrates and perform vibrating and jumping movements, whereas swimming cells show rectilinear motion and rotate around the longitudinal axis of the cell. The posterior end of the cell presents a well-developed posterior protrusion. No contractile vacuole. No cysts.
Type material: A SEM stub of chemically fixed and platinum-coated cells of the type strain, LJ06, is deposited in the DEEM culture collection, CNRS and Université Paris-Saclay (France). This constitutes the name-bearing type of the new species (a hapantotype).
Type figure: figure 1.
Gene sequence: The SSU rRNA gene sequence has the GenBank accession number ON979549.
Type culture: A cryopreserved culture is deposited in the DEEM culture collection, CNRS and Université Paris-Saclay (France) as the type strain LJ06.
Isolator: Luis Javier Galindo.
Type locality: Specimen isolated from an intertidal pool from the island of La Graciosa, Spain.
Etymology: The name chinija refers to the Archipelago of Chinijo (Spain), where the samples were collected, and to the small size of the cells (chinijo is a local colloquial adjective from the area used to describe something small).
Zoobank registration: urn:lsid:zoobank.org:act:7109A458-9FA3-4B5C-8D79-34A087F7E4EA
Materials and Methods
Isolation and Light Microscopy of New Ophirinid Species
Agogonia voluta strain FlipomonasEx4 was isolated from sediment samples of a freshwater pond (48°42′02.0″N 2°10′48.1″E) in Orsay (France). Ophirina chinija strain LJ06 was isolated from sediment samples from a marine intertidal pond (29°13′10.1″N 13°31′23.0″W) in the island of La Graciosa from the Chinijo Archipelago (Canary Islands, Spain). Samples were taken from the upper sediment layer with sterile 15 ml Falcon tubes at a depth of 10 cm below the water surface. A small amount of sediment was inoculated in a Petri dish with 5 ml of sterile Volvic mineral water or seawater supplemented with an equal volume of 1% YT medium (100 mg yeast extract and 200 mg tryptone in 100 ml distilled water as in the protocol from the National Institute for Environmental Studies [NIES, Japan]). After observation of the growth of some ophirinid cells, serial dilution was applied to further enrich the cultures. For that, 250 µl of the initial culture were transferred to a well in a 24-well plate with 1 ml of fresh 1% YT seawater/Volvic medium and then retransferred to the next well, repeating the process 5 times. Single cells of each strain were then isolated from the enriched cultures with an Eppendorf PatchManNP2 micromanipulator using 65 µm VacuTip microcapillaries (Eppendorf) on a Leica Dlll3000 B inverted microscope and inoculated in 24-well plates. After 48 h, we confirmed established monoeukaryotic cultures of Agogonia voluta FlipomonasEx4 and Ophirina chinija LJ06.
Optical microscopy observations were done with a Zeiss Axioplan 2 microscope equipped with oil-immersion differential interference contrast (DIC) and phase contrast objectives. Images were taken with an AxiocamMR camera using the Zeiss AxioVision 4.8.2 SP1 suite. Videos were recorded with a Sony α9 digital camera. Typical morphometric data were obtained at 1,000× magnification on 30 cells. Photos were taken at different planes to visualize the different cell parts.
Electron Microscopy
For SEM, concentrated cell cultures were mixed (1:1) with 4% glutaraldehyde in seawater, deposited on a 9-mm circular coverslip coated with poly-l-lysine, and fixed for 1 h. After fixation, cells were washed with 0.2 M cacodylate buffer (pH 7.2) and dehydrated in a graded series of ethanol baths (30%, 50%, 70%, 80%, 90%, 95%, 100% × 3 times, for 10 min each). Subsequently, coverslips with cells were washed in hexamethyldisiloxane three times for 15 min and dried. Dry coverslips were mounted on aluminum stubs, coated with platinum, and observed using a GeminiSEM 500 (Carl Zeiss, Germany) electron microscope.
For TEM, concentrated cell cultures were fixed on ice for 2 h in 2.5% glutaraldehyde, 0.2 M cacodylate buffer (pH 7.2), centrifuged at 1,000 × g and 10 °C for 20 min, and washed using the same buffer (two times for 10 min). After 1 h of fixation in 1% OsO4, 0.2 M cacodylate buffer (pH 7.2), the cell pellet was washed with distilled water (three times for 5 min), and dehydrated in an ethanol bath series (30%, 50%, 70%, 80%, 90%, 95%, 100% × 2 times, for 10 min each), then in a 1:1 mix of ethanol and acetone (10 min), and finally in pure acetone (twice for 10 min). Cell pellets were then transferred into a 1:1 mix of low-viscosity resin (Agar) and acetone overnight at room temperature, and subsequently into pure resin three times for 2 h. The embedded specimens were included in resin blocks by polymerization overnight in fresh resin at 70 °C. Ultrathin sections (ca. 60 nm) of resin blocks were prepared using an EM UC6 ultramicrotome (Leica Microsystems, Germany), double stained with 2% uranyl acetate and 2% lead citrate, and observed in a JEM-1400 (JEOL, Japan) electron microscope.
RNA and DNA Extraction, Transcriptome and Genome Sequencing and Assembly
The ophirinid strains were grown for 1 to 2 weeks in 75 cm2 cell culture flasks (Falcon) with approximately 10 ml of medium. Fully grown cultures were extracted by gently scratching the bottom of the flask to resuspend the attached cells with a cell scrapper and pooled in 50 ml Falcon tubes to be centrifuged at 10 °C for 15 min at 15,000 × g. Total RNA and DNA were extracted from the cell pellets with the RNeasy mini Kit and the DNeasy PowerMax Soil Kit (QIAGEN), respectively, following the manufacturer’s protocol. Purified RNA/DNA was quantified with a Qubit fluorometer (ThermoFisher Scientific). For each species, one cDNA Illumina library was constructed after polyA mRNA selection. cDNA and DNA libraries were paired-end (2 × 150 bp) sequenced with Illumina HiSeq 2500 chemistry v4 (Eurofins Genomics, Germany). A total of 13,041,150 transcriptome paired reads and 54,201,638 genome paired reads were generated for O. chinija, and 11,300,930 transcriptome paired reads and 42,101,302 genome paired reads were generated for A. voluta. Raw read sequences have been submitted to GenBank under BioProject accession numbers PRJNA858411 and PRJNA858417 for Ophirina chinija and PRJNA858424 and PRJNA858430 for Agogonia voluta.
Paired-end read quality scores were checked with FastQC v0.11.8 (Andrews 2010). Illumina adapters were removed with Trimmomatic v0.32 (Bolger et al. 2014) in Paired-End mode, with the following parameters: ILLUMINACLIP:adapters.fasta:2:30:10LEADING:28 TRAILING:28 SLIDINGWINDOW:4:30. Each genome/transcriptome was de novo assembled using SPAdes v3.13.1 (Bankevich et al. 2012) with -rna mode in the case of the transcriptomes and default parameters. The O. chinija assemblies included 26,290 (transcriptome) and 102,719 (genome) contigs. The A. voluta assemblies included 20,605 (transcriptome) and 77,665 (genome) contigs. All assemblies were then translated into peptides using Transdecoder v2 (http:transdecoder.github.io) with default parameters and multiple identical translated sequences were removed with Cd-hit v4.6 (Li and Godzik 2006) with 100% identity. We then removed possible cross-species contamination using CroCo v1.1 (Simion et al. 2018). To assess genome/transcriptome completeness, we used BUSCO v2.0.1 on the predicted proteomes with the eukaryota_odb9 dataset of 303 near-universal single-copy orthologs (Simão et al. 2015). For O. chinija, we obtained a completeness of 86.9% and 40.6% for its transcriptome and genome, respectively, and for A. voluta, we got a completeness of 81.5% and 23.4% for its transcriptome and genome, respectively. All assemblies and inferred protein sequences can be found at https://doi.org/10.6084/m9.figshare.20332842.v3.
Phylogenomic Analyses
The dataset of 351 conserved proteins from Lax et al. (2018) was updated by Blastp (Altschul et al. 1990) search (with an E-value threshold of 1e-05) against the inferred proteomes of representatives of all known eukaryotic lineages, including our two new ophirinid strains, and allowing to keep more than one hit per species. Sequences of each protein marker were aligned with MAFFTv7 (Katoh and Standley 2013) and trimmed with TrimAl with the automated1 option (Capella-Gutiérrez et al. 2009). Alignments were manually inspected and edited with AliView (Larsson 2014) and Geneious v6.06 (Kearse et al. 2012). Single-protein trees were reconstructed with IQ-TREE v1.6.11 (Nguyen et al. 2015) with the corresponding best-fitting model. Each single-protein tree was manually inspected to discard contaminants, paralogs, and possible cases of horizontal gene transfer or hidden paralogy. At the end of this curation process, we kept a final taxon sampling of 41 species, including 19 species representing the diversity of Discoba, and outgroups from Metamonada, Ancyromonadida, Malawimonadida, Diaphoretickes, Hemimastigophora, and CRuMs (supplementary table S1, Supplementary Material online, available on https://doi.org/10.6084/m9.figshare.20332842.v3), and a supermatrix of 350 markers that were present in at least one of the three Ophirinina species. For this reduced taxon sampling, all proteins were realigned and trimmed as described above, and then concatenated using Alvert.py from the package Barrel-o-Monkeys (http://rogerlab.biochemistryandmolecularbiology.dal.ca/Software/Software.htm), creating a final supermatrix with 112,726 amino acid positions.
Bayesian inference (BI) phylogenetic analyses were done with PhyloBayes-MPI v1.5a (Lartillot et al. 2009) with the CAT-GTR model (Lartillot and Philippe 2004), with four MCMC chains run for 10,000 generations, saving one every 10 trees. Analyses were stopped once convergence thresholds were reached (i.e., maximum discrepancy <0.1 and minimum effective size >100, calculated using bpcomp) and consensus trees constructed after a burn-in of 25%. ML analyses were done with IQ-TREE v1.6.11 (Nguyen et al. 2015) with the best fitting model (LG + C60 + F + R7) as per bayesian information criterion (BIC) using the -TESTNEW option of IQ-TREE and the PMSF approximation (Wang et al. 2018).
We further investigated the phylogenetic position of ophirinids with two alternative datasets. First, a dataset based on the one described above but from which we removed all metamonads with the exception of the short-branched species Trimastix pyriformis (34 species were kept). Second, a dataset of 14 mitochondrion-encoded markers based on that from Ettahi et al. (2021). Both datasets were trimmed using trimAl v1.2 (Capella-Gutiérrez et al. 2009) by removing sites with gaps in more than 80% of the sequences (-gt 0.2) and similarity score below 0.001 (-st 0.001), and restricted minimum resulting length of 20% of the original alignment (-cons 20). ML analyses were carried out with IQ-TREE v1.6.11 (Nguyen et al. 2015) with LG + C60 + F + R7, the best fitting model as per BIC using the -TESTNEW option of IQ-TREE, to generate the input tree for PMSF approximation (Wang et al. 2018).
All trees were visualized and edited with FigTree v1.4.3 (Rambaut 2016).
Phylogenetic Analysis of Maturases
A protein sequence dataset covering the known diversity of maturases was built based on the dataset from Maciszewski et al. (2022), in which the authors carried out Blast (Altschul et al. 1990) and PFAM (Mistry et al. 2020) searches of candidate intron maturase genes focusing on euglenid plastid-encoded maturases, but also including mitochondrial and prokaryotic maturases, for a total of a 234 sequences and 832 amino acid-long alignment. We updated this dataset with the A. voluta maturase sequences (orf291 and orf192), which we used as a query in a Blastp (DELTA-BLAST) search against the NCBI nonredundant protein sequence database (Pruitt et al. 2007) to incorporate the first 100 hits, together with additional opisthokont, streptophyte, archaeal, and bacterial maturase sequences from the NCBI protein database (Clark et al. 2016). MAFFT (Katoh and Standley 2013) was used to incorporate these sequences into the original alignment, and redundant sequences were manually removed. The dataset was trimmed using trimAl v1.2 (Capella-Gutiérrez et al. 2009) by removing sites with gaps in more than 70% of the sequences (-gt 0.3), similarity score below 0.001 (-st 0.001), and restricted minimum resulting length of 30% of the original alignment (-cons 30) (Maciszewski et al. 2022). The final alignment had a total of 452 sequences and 1,236 amino acid positions. An ML tree was reconstructed using IQ-TREE (Nguyen et al. 2015) under the VT + F + R8 model (selected using the -m TEST parameter) with 1,000 ultrafast bootstrap replicates and 1,000 replicates of the SH-like approximate likelihood ratio test. The resulting tree was visualized with FigTree (Rambaut 2016). All alignments, datasets, and trees can be found at https://doi.org/10.6084/m9.figshare.20332842.v3.
Assembly and Annotation of Mitochondrial Genomes
To extract the mitochondrial genome sequences of both ophirinid species from their genome assemblies, we used Blastn with the mitochondrial genome of Ophirina amphinema (GenBank LC369600.1; Yabuki et al. 2018) as a query. In the case of A. voluta, we decided to improve the genome assembly by performing an additional run with SPAdes v3.13.1 set on default parameters using both transcriptomic and genomic reads to reconstruct the best possible mitogenome. The combined genome + transcriptome assembly included 115,042 contigs and increased the genome completeness from 23.4% to 77.7% (according to the BUSCO eukaryota_odb9 dataset). The decision to include transcriptome reads in the assembly came from previous evidence of mitochondrial genomes being obtained from transcriptomes (Yabuki et al. 2018; Forni et al. 2019). We then carried out assemblies of the mitochondrial genomes of both species using the organelle-specialized assemblers GetOrganelle v1.7.6.1 (Jin et al. 2020) and NOVOplasty v3.7 (Dierckxsens et al. 2017) with default parameters, and the O. amphinema mitogenome as a query when needed. Finally, we compared assemblies and selected the most complete one for each species. This resulted in two circular-mapping mitochondrial genomes of 59,462 and 66,508 bp for O. chinija and A. voluta, respectively.
The two mitochondrial genomes were annotated using both AGORA (Jung et al. 2018) and MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) with default parameters and using the mitochondrial genome of O. amphinema as query when needed. After comparing the annotation results of both annotators, the amino acid and/or nucleotide sequences of the open reading frames (ORFs) that remained not annotated by any of them (e.g., orf291 and orf192 of A. voluta) were subjected to DELTA-BLAST (Boratyn et al. 2012) to annotate them based on eventual conserved protein domain sequences. To avoid possible bacterial contamination on the assemblies and to confirm the presence of certain protein domains, an additional examination of all ORFs encoding proteins and genes for both mitochondrial genomes was performed with HMM (hmmscan search) protein searches (Finn et al. 2011) and/or with reciprocal BLAST sequence searches across the GenBank non-redundant database (Pruitt et al. 2007). We generated a graphical representation for the annotation of each mitochondrial genome using OGDRAW v1.3.1 (Greiner et al. 2019). After annotation, we reconstructed the gain and loss patterns of mitochondrial genes through the evolution of Discoba, based on the analysis made by Yabuki et al. (2018).
Supplementary Material
Acknowledgments
We thank Dr. Sergey A. Karpov (Zoological Institute RAS) and Dr. Denis V. Tikhonenkov (IBIW RAS) for their helpful discussions on the ultrastructural observations, and Dr. Hwan Su Yoon (Sungkyunkwan University) for the mitochondrial marker alignments. This work was supported by the European Research Council (ERC) Advanced Grants “Protistworld” and “Plast-Evol” (322669 and 787904, respectively) and the Horizon 2020 research and innovation program under the Marie Skłodowska-Curie ITN project SINGEK H2020-MSCA-ITN-2015-675752 (http://www.singek.eu/). G.T. was supported by the 2019 BP 00208 Beatriu de Pinos-3 Postdoctoral Program (BP3; 801370).
Contributor Information
Luis Javier Galindo, Unité d’Ecologie Systématique et Evolution, CNRS, AgroParisTech, Université Paris-Saclay, Gif-sur-Yvette, France; Department of Biology, University of Oxford, Oxford OX1 3SZ, United Kingdom.
Kristina Prokina, Unité d’Ecologie Systématique et Evolution, CNRS, AgroParisTech, Université Paris-Saclay, Gif-sur-Yvette, France; Laboratory of Microbiology, Papanin Institute for Biology of Inland Waters RAS, Borok, Russia.
Guifré Torruella, Unité d’Ecologie Systématique et Evolution, CNRS, AgroParisTech, Université Paris-Saclay, Gif-sur-Yvette, France; Life Sciences Department, Barcelona Supercomputing Center (BSC-CNS), Barcelona, Catalonia, Spain; Mechanisms of Disease, Institute for Research in Biomedicine (IRB), Barcelona, Catalonia, Spain.
Purificación López-García, Unité d’Ecologie Systématique et Evolution, CNRS, AgroParisTech, Université Paris-Saclay, Gif-sur-Yvette, France.
David Moreira, Unité d’Ecologie Systématique et Evolution, CNRS, AgroParisTech, Université Paris-Saclay, Gif-sur-Yvette, France.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
Genome and transcriptome raw read sequences are available in GenBank under accession numbers PRJNA858411 and PRJNA858417 (Ophirina chinija) and PRJNA858424 and PRJNA858430 (Agogonia voluta). All assemblies, inferred protein sequences, alignments, and phylogenetic datasets and trees can be found at https://doi.org/10.6084/m9.figshare.20332842.v3.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, et al. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct. 7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 5:418–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, et al. 2016. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc R Soc B Biol Sci. 283:20152802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Roger AJ, Brown MW, Simpson AGB. 2020. The new tree of eukaryotes. Trends Ecol Evol. 35:43–55. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2003. The excavate protozoan phyla Metamonada Grassé emend. (Anaeromonadea, Parabasalia, Carpediemonas, Eopharyngia) and Loukozoa emend. (Jakobea, Malawimonas): their evolutionary affinities and new higher taxa. Int J Syst Evol Microbiol. 53:1741–1758. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2018. Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma 255:297–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. Genbank. Nucleic Acids Res. 44:D67–D72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corliss JO. 1984. The kingdom Protista and its 45 phyla. Biosystems 17:87–126. [DOI] [PubMed] [Google Scholar]
- Delihas N, Fox GE. 1987. Origins of the plant chloroplasts and mitochondria based on comparisons of 5S ribosomal RNAs. Ann N Y Acad Sci. 503:92–102. [DOI] [PubMed] [Google Scholar]
- Derelle R, et al. 2015. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci U S A. 112:E693–E699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Lang BF. 2012. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. 29:1277–1289. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann VA, et al. 1987. Evolution of organisms and organelles as studied by comparative computer and biochemical analyses of ribosomal 5S RNA structure. Ann N Y Acad Sci. 503:103–124. [DOI] [PubMed] [Google Scholar]
- Ettahi K, et al. 2021. Evolutionary history of mitochondrial genomes in Discoba, including the extreme halophile Pleurostomum flabellatum (Heterolobosea). Genome Biol Evol. 13:evaa241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin M, Nerad TA. 1993. Reclinomonas americana n. g., n. sp., a new freshwater heterotrophic flagellate. J Eukaryot Microbiol. 40:172–179. [DOI] [PubMed] [Google Scholar]
- Forni G, et al. 2019. Complete mitochondrial genomes from transcriptomes: assessing pros and cons of data mining for assembling new mitogenomes. Sci Rep. 9:14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 1989. The evolutionary origins of organelles. Trends Genet. 5:294–299. [DOI] [PubMed] [Google Scholar]
- Gray MW. 1992. The endosymbiont hypothesis revisited. Int Rev Cytol. 141:233–357. [DOI] [PubMed] [Google Scholar]
- Gray MW, et al. 2020. The draft nuclear genome sequence and predicted mitochondrial proteome of Andalucia godoyi, a protist with the most gene-rich and bacteria-like mitochondrial genome. BMC Biol. 18:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. 2004. Mitochondria of protists. Annu Rev Genet. 38:477–524. [DOI] [PubMed] [Google Scholar]
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47:W59–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack DB, et al. 2019. Cryo-EM structures of a group II intron reverse splicing into DNA. Cell 178:612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, et al. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic ‘supergroups’. Proc Natl Acad Sci U S A. 106:3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Fiz-Palacios O, Fu CJ, Tsai CC, Baldauf SL. 2014. An alternative root for the eukaryote tree of life. Curr Biol. 24:465–470. [DOI] [PubMed] [Google Scholar]
- Heiss AA, et al. 2018. Combined morphological and phylogenomic re-examination of malawimonads, a critical taxon for inferring the evolutionary history of eukaryotes. R Soc Open Sci. 5:171707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-J, et al. 2020. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kim JI, Jeong Y-S, Yi G. 2018. AGORA: organellar genome annotation from the amino acid and nucleotide references. Bioinformatics 34:2661–2663. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, Grant JR. 2015. Taxon-rich phylogenomic analyses resolve the eukaryotic tree of life and reveal the power of subsampling by sites. Syst Biol. 64:406–415. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. 2015. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbiol Spectr. 3.MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara E, Chatzinotas A, Simpson AGB. 2006. Andalucia (n. gen.)–the deepest branch within jakobids (Jakobida; Excavata), based on morphological and molecular study of a new flagellate from soil. J Eukaryot Microbiol. 53:112–120. [DOI] [PubMed] [Google Scholar]
- Larsson A. 2014. Aliview: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. 2009. Phylobayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21:1095–1109. [DOI] [PubMed] [Google Scholar]
- Lax G, et al. 2018. Hemimastigophora is a novel supra-kingdom-level lineage of eukaryotes. Nature 564:410–414. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- López-García P, Moreira D. 2006. Selective forces for the origin of the eukaryotic nucleus. Bioessays 28:525–533. [DOI] [PubMed] [Google Scholar]
- Maciszewski K, Dabbagh N, Preisfeld A, Karnkowska A. 2022. Maturyoshka: a maturase inside a maturase, and other peculiarities of the novel chloroplast genomes of marine euglenophytes. Mol Phylogenet Evol. 170:107441. [DOI] [PubMed] [Google Scholar]
- Mistry J, et al. 2020. Pfam: the protein families database in 2021. Nucleic Acids Res. 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gómez SA, Wideman JG, Roger AJ, Slamovits CH. 2017. The origin of mitochondrial cristae from Alphaproteobacteria. Mol Biol Evol. 34:943–956. [DOI] [PubMed] [Google Scholar]
- Mylnikov AP, Mylnikov AA. 2014. Structure of the flagellar apparatus of the bacterivorous flagellate Histiona aroides Pascher, 1943 (Jakobida, Excavata). Inl Water Biol. 7:331–337. [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kelly CJ. 1993. The jakobid flagellates: structural features of Jakoba, Reclinomonas and Histiona and implications for the early diversification of eukaryotes. J Eukaryot Microbiol. 40:627–636. [Google Scholar]
- Pánek T, et al. 2015. Combined culture-based and culture-independent approaches provide insights into diversity of jakobids, an extremely plesiomorphic eukaryotic lineage. Front Microbiol. 6:1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, et al. 2010. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 59:518–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DJ. 1988. The evolution of protozoa. Mem Inst Oswaldo Cruz. 83(Suppl 1):580–600. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2016. FigTree v1.4.3. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- Roger AJ, Muñoz-Gómez SA, Kamikawa R. 2017. The origin and diversification of mitochondria. Curr Biol. 27:R1177–R1192. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva E V, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simion P, et al. 2018. A software tool ‘CroCo’ detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AGB, et al. 2002. Evolutionary history of ‘early-diverging’ eukaryotes: the excavate taxon Carpediemonas is a close relative of Giardia. Mol Biol Evol. 19:1782–1791. [DOI] [PubMed] [Google Scholar]
- Simpson AGB. 2003. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int J Syst Evol Microbiol. 53:1759–1777. [DOI] [PubMed] [Google Scholar]
- Simpson AGB, et al. 2016. Jakobida. In: Archibald JM, et al., editors. Handbook of the protists. Cham: Springer International Publishing. p. 1–32. [Google Scholar]
- Simpson AGB, Inagaki Y, Roger AJ. 2006. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of ‘primitive’ eukaryotes. Mol Biol Evol. 23:615–625. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Patterson DJ. 2001. On core jakobids and excavate taxa: the ultrastructure of Jakoba incarcerata. J Eukaryot Microbiol. 48:480–492. [DOI] [PubMed] [Google Scholar]
- Strassert JFH, et al. 2016. Moramonas marocensis gen. nov., sp. nov.: a jakobid flagellate isolated from desert soil with a bacteria-like, but bloated mitochondrial genome. Open Biol. 6:150239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. 2007. An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus. BMC Genomics 8:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Minh BQ, Susko E, Roger AJ. 2018. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst Biol. 67:216–235. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu M. 2015. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep. 5:7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM. 1999. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell. 4:239–250. [DOI] [PubMed] [Google Scholar]
- Yabuki A, Gyaltshen Y, Heiss AA, Fujikura K, Kim E. 2018. Ophirina amphinema n. gen., n. sp., a new deeply branching discobid with phylogenetic affinity to jakobids. Sci Rep. 8:16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubuki N, Simpson AGB, Leander BS. 2013. Comprehensive ultrastructure of Kipferlia bialata provides evidence for character evolution within the Fornicata (Excavata). Protist 164:423–439. [DOI] [PubMed] [Google Scholar]
- Zhang Q, et al. 2015. Marine isolates of Trimastix marina form a plesiomorphic deep-branching lineage within Preaxostyla, separate from other known trimastigids (Paratrimastix n. gen.). Protist 166:468–491. [DOI] [PubMed] [Google Scholar]
- Zimmerly S, Hausner G, Wu X. 2001. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 29:1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S, Semper C. 2015. Evolution of group II introns. Mob DNA. 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome and transcriptome raw read sequences are available in GenBank under accession numbers PRJNA858411 and PRJNA858417 (Ophirina chinija) and PRJNA858424 and PRJNA858430 (Agogonia voluta). All assemblies, inferred protein sequences, alignments, and phylogenetic datasets and trees can be found at https://doi.org/10.6084/m9.figshare.20332842.v3.