Abstract
Lloviu virus (LLOV) is a novel filovirus detected in Schreiber's bats in Europe. The isolation of the infectious LLOV from bats has raised public health concerns. However, the virological and molecular characteristics of LLOV remain largely unknown. The nucleoprotein (NP) of LLOV encapsidates the viral genomic RNA to form a helical NP-RNA complex, which acts as a scaffold for nucleocapsid formation and de novo viral RNA synthesis. In this study, using single-particle cryoelectron microscopy, we determined two structures of the LLOV NP–RNA helical complex, comprising a full-length and a C-terminally truncated NP. The two helical structures were identical, demonstrating that the N-terminal region determines the helical arrangement of the NP. The LLOV NP–RNA protomers displayed a structure similar to that in the Ebola and Marburg virus, but the spatial arrangements in the helix differed. Structure-based mutational analysis identified amino acids involved in the helical assembly and viral RNA synthesis. These structures advance our understanding of the filovirus nucleocapsid formation and provide a structural basis for the development of antifiloviral therapeutics.
Keywords: filovirus, Lloviu virus, nucleocapsid, cryo-EM
Significance Statement.
The filovirus helical nucleoprotein (NP)–RNA complex plays a pivotal role in nucleocapsid formation and viral RNA synthesis. Thus, elucidating the molecular structure is essential for understanding the filovirus replication cycle. In this study, we focused on a new filovirus, the Lloviu virus (LLOV), recently isolated in Europe, and are pioneering in establishing the three-dimensional structures of the NP–RNA complex consisting of a full-length NP in filoviruses, using cryoelectron microscopy. Additionally, we determined the residues involved in helical assembly and viral RNA synthesis, based on our structures. Our study reveals that filoviruses of different genera possess a common NP folding mechanism but exhibit different quaternary structures of the NP–RNA helices.
Introduction
Filoviruses such as Ebola virus (EBOV) and Marburg virus (MARV) belong to the Filoviridae family and cause severe hemorrhagic fever in humans and nonhuman primates with a high fatality rate. The genome of Lloviu virus (LLOV), a novel filovirus, was discovered in Schreiber's bats (Miniopterus schreibersii) in Spain, in 2002, as the first filovirus that was not imported from an endemic area in Africa (1). Since LLOV was distinct from EBOV and MARV at the genome level, it was classified as a new species belonging to a new genus Cuevavirus. Although there were no reports on its presence for more than ten years since its initial discovery, the virus genome was again detected in Schreiber's bats in Hungary, in 2016 (2), suggesting its wide distribution and continuous circulation in Europe. Recently, LLOV was successfully isolated from asymptomatic Schreiber's bats (3) as the second filovirus to be isolated from bats, after MARV (4). Although the pathogenicity of LLOV in humans remains unclear, LLOV has been shown to infect and replicate in human- and nonhuman primate-derived cells as well as Schreiber's bat-derived cells in vitro (3), suggesting zoonotic potential.
LLOV possesses a nonsegmented, single-stranded, negative-sense RNA genome. Its viral genomic RNA (vRNA) encodes at least seven proteins: a nucleoprotein (NP), a polymerase cofactor (viral protein 35 [VP35]), a matrix protein (VP40), a glycoprotein (GP), a transcriptional activator (VP30), a nucleocapsid associated protein (VP24), and an RNA-dependent RNA polymerase (L) (1, 5, 6). A morphological analysis of the virus-like particles (VLPs) of the LLOV showed that the virus has a characteristic filamentous structure, like other filoviruses (7). The nucleocapsid, which is located at the center of the filamentous particles, comprises a helical structure of the NP–vRNA complex associated with VP35, VP30, VP24, and L and is responsible for transcribing and replicating the vRNA (6, 8–10).
Recently, the overall structure of the EBOV nucleocapsid in virions was visualized at a nanometer resolution using cryoelectron tomography, demonstrating that the helical NP–RNA complex constitutes the core of the nucleocapsid (11, 12). Single-particle cryoelectron microscopy (cryo-EM) was used to explore high-resolution structures of a C-terminally truncated EBOV NP (1–450 residues)–RNA helix and a C-terminally truncated MARV NP (1–395 residues)–RNA helix (6, 13–15). Studies have shown that both the EBOV and MARV NPs comprise four domains: an N-terminal arm, a core domain responsible for RNA binding, a disordered linker, and a C-terminal tail that is required for the interactions with other nucleocapsid components (6, 13–17). However, the structural details of the LLOV NP–RNA complex remained unknown. In addition, the contribution of C-terminal domains in the NP-RNA helix structure remained unclear.
In this study, to examine the helical assembly of the LLOV NP–RNA structures, we determined two helical LLOV NP–RNA structures comprising a full-length NP and a C-terminally truncated NP, using single-particle cryo-EM. This study advances our understanding of the mechanism of filovirus nucleocapsid formation and can be useful in developing broad-spectrum antifiloviral drugs.
Results
The overall architecture of the LLOV NP–RNA complex
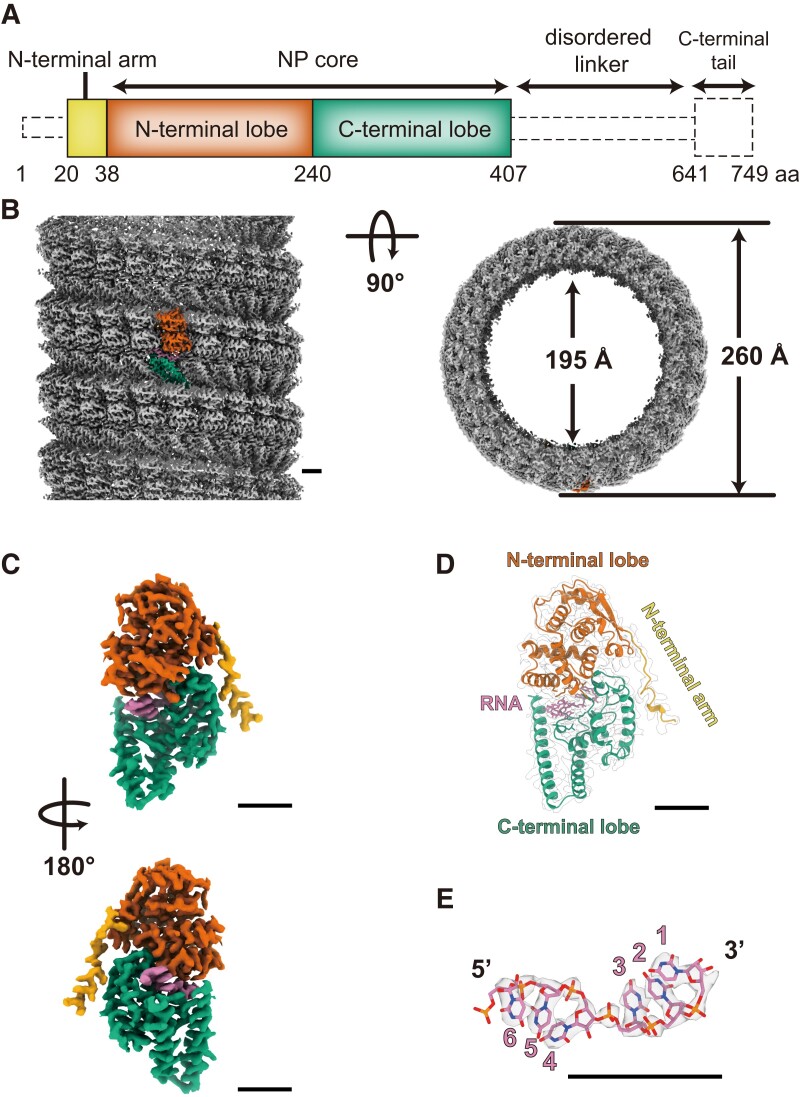
To determine the structure of the LLOV NP–RNA complex, we expressed a full-length and a C-terminally truncated NP comprising its N-terminal 1–450 residues (NP[full] and NP[1–450], respectively) in human embryonic kidney 293T (HEK 293T) cells. The NP–RNA helices were purified using cesium–chloride density–gradient ultracentrifugation (17) and imaged using cryo-EM. Using single-particle analysis, structures of the NP(full)–RNA complex and the NP(1–450)––RNA complex were determined at overall resolutions of 3.0 Å and 3.1 Å, respectively (Figs. S1 and S2). Both the NP(full)–RNA and NP(1–450)–RNA complexes were left-handed helices with 260 Å and 195 Å in outer and inner diameters, respectively, and contained 23.8 subunits per turn (Fig. 1b and Table S1). The high-resolution cryo-EM maps allowed us to generate the atomic models of the respective protomers. The N-terminal 20–407 residues of the LLOV NP could be modeled in both the cryo-EM maps. The N-terminal 19 and C-terminal 342 residues were not modeled because of a lack of interpretable map densities, which is probably a result of the N- and C-termini being composed of disordered regions, and the C-terminal tail being intrinsically flexible or disordered (Fig. 1a). The N-terminal region (20–407 residues) of NP was divided into two domains: the N-terminal arm (20–38 residues) and the core domain (39–407 residues) composed of the N-terminal (39–240 residues) and C-terminal (241–407 residues) lobes (Fig. 1c, d). We observed that NP binds to single-stranded RNA, and the N- and C-terminal lobes of each NP enclose six RNA nucleotides (Fig. 1e). The NP(full)–RNA protomer exhibited a conformation similar to the NP(1–450)–RNA protomer, with only a 0.5 Å backbone root mean square deviation (RMSD), suggesting that the remaining C-terminal region has little impact on the folding of NP (Fig. S3b–d). Similarly, C-terminal truncation had little impact on the overall helical structure (Fig. S3a), because the differences in the helical parameters (NP[full]–RNA, rise = 3.19 Å, twist = −15.11°; NP[1–450]–RNA, rise = 3.16 Å, twist = −15.11°) and the distance between the two NPs in the vertical direction were negligible (Table S2 and Fig. S3e).
Fig. 1.
Overall cryo-EM structure of the LLOV NP(full)–RNA helix and its atomic model. (a) Schematic diagram of the domain architecture of the LLOV NP. Each domain is represented by a unique color, and the unsolved region is represented by a dotted outline. (b) The cryo-EM map of the NP(full)–RNA complex. A single NP–RNA protomer is highlighted with colors: RNA strand in purple and NP in red, green, and yellow, according to the color code shown in (a). (c) The isolated cryo-EM map of the NP–RNA protomer. (d) The atomic model of the NP–RNA complex in ribbon presentation superimposed on the electron microscopy map. (e) The isolated cryo-EM map and atomic model of RNA. RNA bases are modeled as uracil. Scale bar, 20 Å.
Structural comparison of the LLOV NP protomer with that of the EBOV NP (PDB-ID: 5Z9W) and MARV NP (PDB-ID: 7F1M) revealed 1.0 Å and 0.8 Å backbone RMSDs, respectively (Fig. S4), suggesting the absence of drastic conformational differences in the overall structure of the NP protomer. However, the helical structures constituted by each of the three NP protomers exhibited differences in the helical parameters (LLOV NP[full]–RNA, rise = 3.19 Å, twist =−15.11°; EBOV NP[1–450]–RNA, rise = 3.01 Å, twist = −14.73°; and MARV NP[1–395]–RNA, rise = 4.23 Å, twist = −11.81°). These results suggest that filoviruses of different genera exhibit similar NP-folding mechanisms, but their NP–RNA helices have different quaternary structures.
RNA-binding region of the LLOV NP
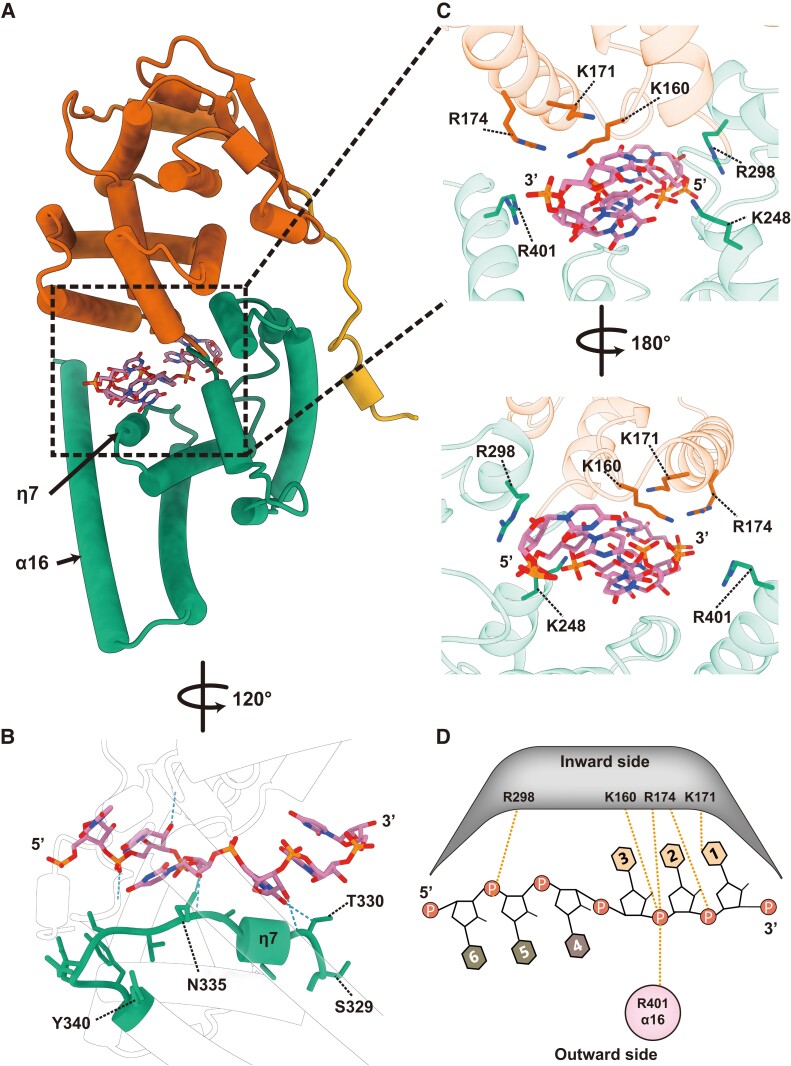
Similar to the C-terminally truncated EBOV and MARV NPs, the LLOV NP(full) formed a positively charged cleft between the N- and C-terminal lobes (Fig. S5). Six RNA nucleotides were embedded in the cleft in a “3-bases-inward, 3-bases-outward” configuration (Figs. 1e and 2d). NP–RNA interactions were mainly composed of hydrogen bonding and electrostatic interactions between the NP and RNA. The RNA strand was likely stabilized by a short helix η7 placed along the RNA backbone from the C-terminal side (Fig. 2b). The side chains of the positively charged amino acids K160, K171, R174, K248, R298, and R401 were located within a distance of <4 Å from the RNA backbone and pointed toward the sugar–phosphate backbone of the RNA (Fig. 2c, d). Some of these positively charged residues (K160, K171) are reportedly important for RNA binding in the EBOV and MARV NPs, while R174 and K248 are dispensable in RNA binding (14, 18–20). Sequence alignment of the EBOV, MARV, and LLOV showed that most positively charged residues were highly conserved (Fig. S6), suggesting that filoviruses of different genera exhibit a similar mechanism for RNA binding. Lastly, NPs interact with the RNA backbone, but not the RNA bases, suggesting that the LLOV NP encapsidates the RNA in a sequence-independent manner, similar to other negative-sense RNA viruses (6, 13–15, 21–25).
Fig. 2.
NP–RNA interactions in the helical NP(full)–RNA complex. (a) The overall structure of the protomeric NP(full)–RNA complex in cylindrical presentation (colored the same as Fig. 1a). (b) Close-up view of the RNA binding site. Hydrogen bonds (distance < 4 Å) were detected by ChimeraX (35) and shown as blue dotted lines. (c) Close-up view of the RNA binding site. Positively charged amino acids are marked. (d) Schematic diagram of the NP–RNA interactions. The RNA bases are numbered in the 3′ to 5′ direction. The RNA bases on the helix, facing inward (light-orange) and outward (light-brown). Secondary structure labels are based on our atomic model. Orange dotted lines represent the potential interactions between the NP residues and RNA backbone.
LLOV NP–NP interactions in the helix
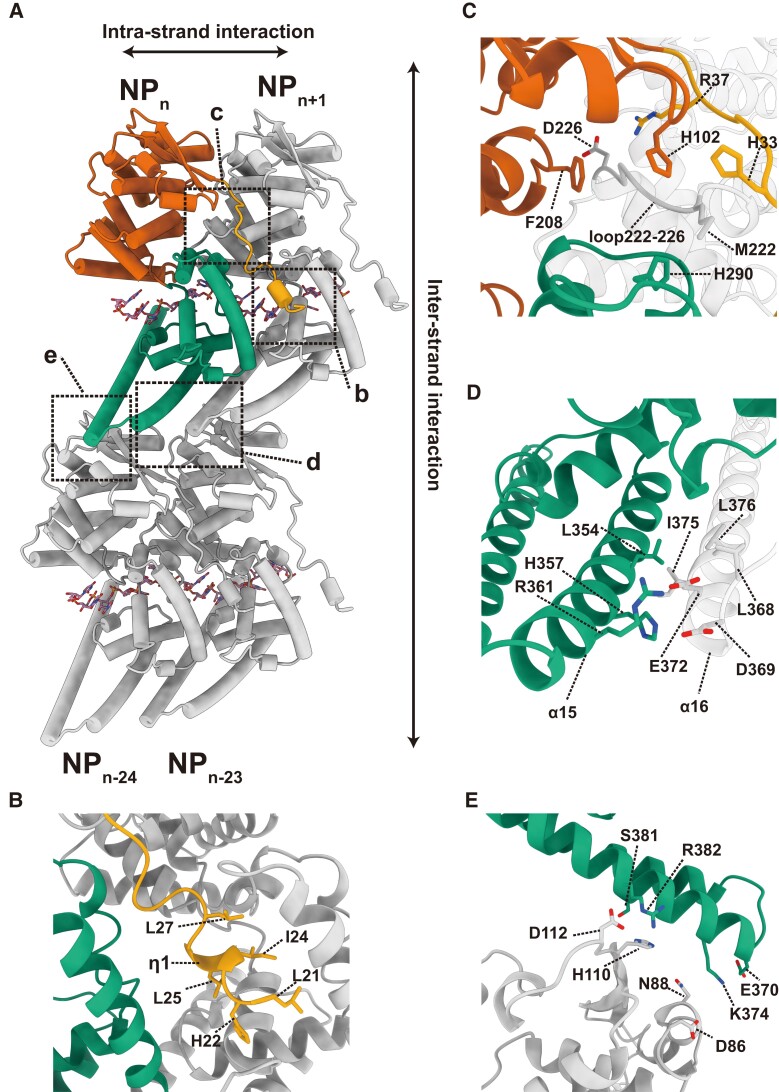
Interactions between NPs are responsible for the helical assembly. Based on the spatial position of the NP–RNA protomers in the NP–RNA helix, NP–NP interactions can be classified into two groups: interactions between neighboring NPs on the strand (intrastrand NP–NP interactions) (Fig. 3a–d) and interactions between helical strands (interstrand NP–NP interactions) (Fig. 3a, e).
Fig. 3.
NP–NP interactions in the helical NP(full)–RNA complex. (a) Four adjacent NP–RNA complexes in a helix. The NPn is colored according to the colors in Fig. 1a. The larger and smaller rectangles with dotted lines highlight (b, c, and d) the close-ups of intrastrand NP–NP interactions between NPn and NPn+1 and (e) interstrand NP–NP interactions between NPn and NPn−24. (b) NP–NP interactions between the N-terminal arm (yellow) of the NPn and the hydrophobic pocket of NPn+1 (gray). (c) NP–NP interactions between the nut structure formed by the N-terminal arm (yellow), N-terminal lobe (red), and C-terminal lobe (green) of the NPn, and the M222 screw structure of the NPn+1 (gray). (d) NP–NP interactions between α15 of NPn (green) and α16 of NPn+1 (gray). (e) Intrastrand NP–NP interactions between the NPn (green) and NPn−24 (gray).
The N-terminal arm is essential for the oligomerization of filovirus NPs (6, 13–15, 17–19). The atomic model developed in this study clearly shows that the helix η1 on the N-terminal arm of the NPn binds to the hydrophobic pocket located in the C-terminal lobe of the NPn+1 (Figs. 3b and S7a, d). Residues in the vicinity of the η1 helix, such as L21, H22, I24, L25, and L27, are likely involved in the interaction between the N-terminal arm and the hydrophobic pocket (Figs. 3b and S7a, d). A loop (residues 222–226) on the N-terminal lobe of the NPn+1 forms a screw-like structure, which fits into a nut-like structure composed of four benzene ring structures of H33 on the N-terminal arm, H102 and F208 on the N-terminal lobe, and H290 on the C-terminal lobe of the NPn. The M222 side chain of the NPn+1 fits into the hole of this nut, possibly stabilizing the contact region between the N-terminal arm and the N-terminal lobe (Figs. 3c and S7c).
Interactions between two C-terminal lobes of adjacent NP molecules were also involved in intrastrand NP–NP interactions (Fig. 3d). Our model shows that these interactions mainly occur between α15 of NPn and α16 of NPn+1, containing several conserved hydrophobic residues, such as L354 of NPn or I375, L368, and L376 of NPn+1 (Figs. 3d and S6). Hydrophobic interactions, along with several electrostatic interactions (H357/R361 on α15 and D369/E372 on α16), might be essential in stabilizing the neighboring NPs in the helical strand, resulting in a stitch-like structure formed by α15 of NPn and α16 of NPn+1 (Fig. 3d).
Unlike the intrastrand interactions, interstrand interactions in a helix were mainly mediated by electrostatic interactions (Fig. 3e). In our model, any arbitrary NPn of the NP(full) formed pairs with NPn−24, but did not exhibit any interaction with NPn−23 or NPn−25 in the helix. The same was true for the helix composed of the LLOV NP(1–450) (Fig. S3e), but not for the helix composed of the C-terminally truncated EBOV NP (Fig. S8), where NPn interacts with both NPn−23 and NPn−24. Given that the LLOV NP-RNA can maintain its compressed helical structure without truncation of the C-terminus, unlike the EBOV NP––RNA helices (12), these paired interstrand NP–NP interactions may strengthen the overall helical structural stability.
Essential residues for the helical assembly and vRNA synthesis
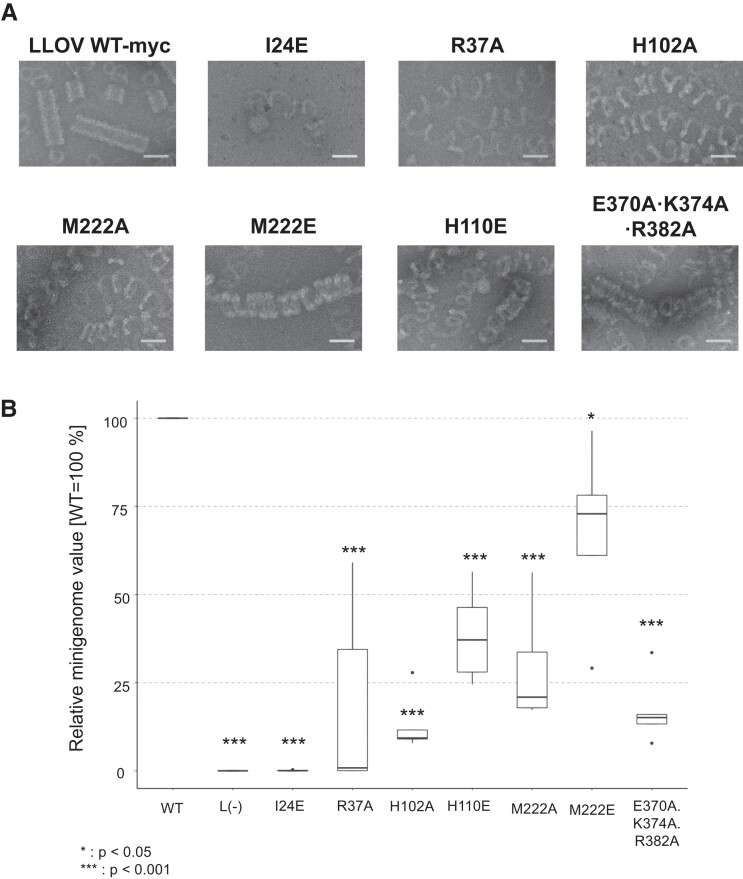
To understand the molecular basis of the helical assembly, we performed structure-based mutational analyses for the residues potentially involved in intrastrand and interstrand NP–NP interactions (Table S2). Western blot analysis showed that the expression levels of all the NP(full) mutants were comparable to those of the wild-type NP(full) (Fig. S9). Negative staining transmission electron microscopy was performed to evaluate their ability to form helical structures. Moreover, a minigenome assay was conducted to assess the impact of the mutation on their transcription and replication (Fig. 4a, b). The NP mutants, except for NP I24E, were able to form the NP–RNA complexes similar to those formed by the wild-type NP. However, none of the NP mutants were able to form rigid NP–RNA helices. The NP I24E mutation on the helix η1 in the N-terminal arm (Fig. 3b) exhibited negligible oligomerization into NP–RNA strands or helices, and the transcription and replication activities were <1% of the wild-type NP, demonstrating the importance of I24 in the interaction with the hydrophobic pocket of a neighboring NP. This finding is consistent with those for the corresponding mutants EBOV I24E and MARV L6E (14). The NPs with the mutations R37A (a mutation close to the nut structure), H102A, or M222A (Fig. 3c) barely formed helical structures, and the helices were irregular and loose. The transcription and replication activities of these mutants were significantly reduced to <50%, indicating that the interactions between the screw and nut structure were involved in helix assembly and viral RNA synthesis (Fig. 3c). The corresponding mutants EBOV R37A and MARV R19A also exhibited significantly reduced transcription and replication activities, but these two mutants formed compressed long helical structures (14). In contrast, the NP with the M222E mutation formed mildly loose but regular helical structures and maintained its transcription and replication activities, suggesting that the M222 side chain can interact with the nut structure H33/H102/F208/H290, through either hydrophobic or electrostatic interactions (Figs. 3c and S7a, c). Since the corresponding residues EBOV V222 and MARV V204 are both hydrophobic residues, the cause of the LLOV NP M222A mutant losing its activity may be its short side chain.
Fig. 4.
Structure-based mutational analysis of the LLOV NP. (a) Transmission electron microscopy images of the negatively stained purified helical structure of the LLOV NP(full) mutant–RNA complex. A myc tag was fused to the C-terminus of the native and mutant LLOV NPs. Scale bar, 50 nm. (b) The assessment of the function of the LLOV NP-myc mutants on genome transcription and replication, evaluated via minigenome assays. The experiments were performed in pentaplicates (n = 5). Significance was set at P < 0.05 (*) and P < 0.001 (***).
The NP H110E and E370A/K374A/R382A mutations, potentially involved in interstrand NP–NP interactions in the helix (Fig. 3e), resulted in barely helical structures, which looked very flexible and irregularly uncoiled (Fig. 4a). The transcription and replication activities of these mutants were reduced to <25% (Fig. 4b), indicating that the interstrand electrostatic interactions are important not only for the maintenance of the helical structure but also for viral RNA synthesis.
Discussion
With the successful isolation of LLOV from bats and the demonstration of a possible zoonotic potential (3), LLOV has emerged as a pathogen of concern. In this study, by using single-particle cryo-EM, we determined high-resolution structures of the LLOV NP(full)–RNA complex and the C-terminally truncated NP(1–450)–RNA complex. The two LLOV NP–RNA protomers have almost identical structures, similar to those of the EBOV and MARV protomers (6, 13–15) yet their helical structures differ from those of EBOV and MARV. We also identified the amino acid residues on the NP that are involved in the intrastrand and interstrand NP–NP interactions in the helix. Furthermore, we experimentally demonstrated the importance of these residues in the helical assembly and viral RNA synthesis. Hence, this study reveals the common structural features of filovirus NP–RNA complexes and will further advance the development of structure-based pan-filovirus antivirals.
Thus far, several cryo-EM structures of helical NP–RNA complexes of mononegaviruses, such as filoviruses and paramyxoviruses, have been reported (13–15, 21, 25). However, these helical structures were determined using C-terminally truncated NPs, and the impact of the C-terminal truncation on the folding of the NP–RNA complex and the helical arrangement of the NP–RNA protomers remained unknown. This study is the first to determine the helical structure of the NP(full)–RNA complex as well as the C-terminally truncated NP–RNA complex, demonstrating that the 450 amino acids of the N-terminal of the NP determine the folding of the NP–RNA complex and its helical arrangement. Considering that the EBOV and MARV NPs cannot maintain their rigid helical structure without the C-terminal truncation (6, 13–15), unlike the LLOV NP, it can be inferred that the C-termini of the respective filovirus NPs may have different structural features in their helical assembly. Since most of the C-terminal regions are disordered and have not been modeled thus far, further structural studies are necessary to reveal the role of the C-terminus of the NP in nucleocapsid formation, along with other nucleocapsid components.
The LLOV NP–RNA binding is maintained by hydrogen bonds and electrostatic interactions between the NP core and the RNA backbone, indicating that the binding of the NP to the RNA is sequence independent (6, 13–15, 21–25). Structural comparison of the LLOV NP, EBOV NP (PDB-ID: 5Z9W), and MARV NP (PDB-ID: 7F1M) revealed that filovirus NPs have similar structures around the RNA-binding cleft, in which several positively charged residues, such as K160, K171, R174, R298, and R401 (K142, K153, R156, and R280 for MARV NP), are conserved (Figs. 2 and S4, 6). Likewise, the sequence of the RNA-binding region of the LLOV NP, which comprises amino acid sequences from 150 to 410, was similar to that of the EBOV and MARV NPs (80.6 and 62.7%, respectively). Thus, it can be inferred that filovirus NPs possibly employ a common RNA recognition and binding mechanism.
The N-terminal arm plays a critical role in the NP oligomerization and the neighboring NP–NP interaction (6, 13–15, 17–19). This is consistent with our finding that a single-residue mutation on a short helix located on the N-terminal arm abolishes the formation of helical structures and even NP–RNA multimers (Fig. 4a). In the case of EBOV and MARV, the hydrophobic pocket in the C-terminal lobe interacts with the N-terminus of VP35, which maintains NP molecules in an RNA-free state at the initial stage of the nucleocapsid formation (18, 19, 26). Since the LLOV NP also possesses a hydrophobic pocket similar to that of the EBOV and MARV NPs and most hydrophobic residues in this region are conserved (Figs. S6, 7a), the mechanism of the NP–VP35 interaction in LLOV is likely similar to that in EBOV and MARV.
In summary, this study presents a cryo-EM structure of the LLOV NP–RNA complex. Since novel filoviruses such as members of the genera Striavirus, Thamnovirus, Dianlovirus, Oblavirus, and the unassigned Tapajós virus (27–30) are continuously being discovered by virtue of advances in sequencing techniques, structural studies on various filovirus NPs will significantly advance our understanding of the filovirus replication mechanisms and contribute to the development of broad-spectrum structure–based antifiloviral therapies.
Materials and methods
Cells
HEK 293T cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin (Nacalai Tesque). The FCS was inactivated at 56°C, for 30 min before use. The cells were incubated at 37°C and supplied with 5% CO2.
Plasmids
All LLOV plasmids (GenBank accession code YP004928135.1) encode wild-type proteins (pCAGGS-NP, pCAGGS-VP35, pCAGGS-VP30, and pCAGGS-L). The pCAGGS-NP(1–450) that expresses the residues 1–450 of the LLOV NP was constructed from the pCAGGS-NP through PCR using primers with digestion enzyme sites. A Kozak sequence was added before the initiation codon. The LLOV NP mutants were generated via recombinant PCR and cloned into the pCAGGS vector. The C-terminal myc-tag was added via PCR. The primers used in this study are listed in Table S3. All constructs were sequenced to confirm the absence of unwanted mutations.
Expression and purification of the NP–RNA complex
For the LLOV NP(full)–RNA complex and its mutant, HEK 293T cells were seeded in two 10-cm dishes (4 × 106 cells/dish) and incubated at 37°C for 24 h. The cells were transfected with 15 µg/dish pCAGGS-LLOV NP(full), using 45 µl/dish polyethylenimine (PEI) MAX (Polysciences) and 1 ml/dish Opti-MEM (Gibco). Two days after transfection, the cells were collected in 1× Dulbecco's phosphate-buffered saline (D-PBS) (−) (Nacalai Tesque), containing KCl 2.68 mm, NaCl 136.89 mm, Na2HPO4 8.1 mm, and KH2PO4 1.46 mm, and further lysed with 0.1% (v/v) Nonidet P-40 (Wako) in Tris-HCl buffer (10 mm Tris-HCl, pH 8.0 [Invitrogen]; 150 mm NaCl [Nacalai Tesque]; 25 mm MgCl2 [Wako]; 5 mm CaCl2 [Wako]; and 1 mm dithiothreitol [Nacalai Tesque]), supplemented with protease inhibitor cocktail tablets (EDTA-free, Roche) and a 10 mm ribonucleoside–vanadyl complex (New England Biolabs, NEB). The lysate was centrifuged at 3200 × g at 4°C for 10 min to remove insoluble compounds. The supernatant was loaded onto a discontinuous 25–40% (w/w) CsCl (Wako) gradient and centrifuged at 191,600 × g at 4°C for 5 h. Fractions containing the NP–RNA complex were collected and centrifuged at 48,500 × g at 4°C for 1 h. The pellets were suspended in the Tris-HCl buffer and stored at 4°C.
For the LLOV NP(1–450)–RNA complex, the HEK 293T cells were seeded in four 15 cm dishes (1 × 107 cells/dish) and incubated at 37°C for 24 h. The cells were transfected with 30 µg/dish pCAGGS-LLOV NP(1–450), using 120 µl/dish PEI MAX and 2 ml/dish Opti-MEM. Three days postransfection, the NP–RNA complexes were purified as per the method described above.
Negative staining
Aliquots of 5 µl purified samples were placed on a carbon-coated copper grid (Thick & Thin Bar Sq 600 mesh Cu, Gilder) and hydrophilized through glow discharge and stained with 2% uranyl acetate. Images were obtained using an HT-7700 transmission electron microscope (Hitachi High-Tech), operated at 80 kV, with an XR81-B CCD camera.
Cryo-EM specimen preparation
For the LLOV NP(full)–RNA complex, an aliquot of a 2.5 µl purified NP–RNA complex solution at a concentration of 1 mg/ml was applied to Quantifoil R1.2/1.3 200 mesh grids and blotted for 7 s, with a humidity of 100% at 4°C, before being rapidly frozen with liquid ethane using the Vitrobot Mark IV system (Thermo Fisher Scientific). Images of the frozen sample stored at insert temperature were acquired as movies on a Titan Krios cryo-TEM (Thermo Fisher Scientific), operated at 300 kV and equipped with a Falcon3EC direct electron detector (Thermo Fisher Scientific), and a Cs corrector (CEOS GmbH) at the Institute for Protein Research, Osaka University.
For the LLOV NP(1–450)–RNA complex, an aliquot of 3 µl purified NP-RNA complex solution at a concentration of 0.3 mg/ml was applied to Quantifoil R1.2/1.3 300 mesh grids and blotted for 6 s, with a humidity of 100% at 4°C, before being rapidly frozen in liquid ethane using the Vitrobot Mark IV system. Images were acquired using a Glacios cryo-TEM (Thermo Fisher Scientific) operated at 200 kV, with a Falcon4 direct electron detector (Thermo Fisher Scientific) in the electron counting mode, at the Institute for Life and Medical Sciences, Kyoto University.
The detailed imaging conditions for both the samples are described in Table S1.
Image processing
Beam-induced motion was corrected using RELION's own implementation, and contrast transfer function parameters were estimated using CTFFIND-4.1.14 (31), for all micrographs. The subsequential helical structure image processing steps for the LLOV NP(full)–RNA and the NP(1–450)–RNA complexes were performed using the software packages RELION3.0 and RELION3.1 (32), respectively. The coordinates of the start- and end-points of the helices on every micrograph were recorded manually. Then, helical segments were extracted from the helices, using a box size of 448 pixels at an unbinned pixel size and an interbox distance of 43 Å; the 2× binned images were subjected to 2D classification.
For the LLOV NP(full)–RNA complex data set, a total of 62,855 segments were selected after two rounds of 2D classification from 3,122 micrographs and further subjected to 3D classification with a featureless cylinder generated by relion_helix_toolbox (33) as the initial model. Then, 3D classification was performed with a symmetry search. The search range for twist is between −14.5 and −15.5°. The search range for rise is between 2.7 and 3.7 Å. After the first 3D classification, one model displayed clear features of the helical structure. The segment stack belonging to this class was selected (60,818 segments) and subjected to the first 3D refinement. Further 3D classification without alignment resulted in a better 3D class average, with 44.1% of the segments displaying clear protein secondary structural features. Then, this subset was subjected to the second 3D refinement with a symmetry search. The search range for twist is between −14.9 and −15.3°. The search range for rise is between 3.0 and 3.5 Å. The helical symmetry parameter converged to a rise = 3.19 Å and twist = −15.11°. The resultant segment set was further processed using CtfRefine, Bayesian polishing, and another CtfRefine, before the third 3D refinement process to obtain the final reconstruction. The resolution estimate of the final reconstruction was based on the Fourier shell correlation (FSC) (34) between independently refined half sets, which yielded a resolution of 3.0 Å.
The LLOV NP(1–450)–RNA complex data set was processed as per the method described above. After the first 3D classification step, CtfRefine and Bayesian polishing were performed, and a mask was created and used in the second 3D classification step. After the second 3D classification without alignment, 90.4% of the segments from their resulting 3D class average displaying the clearest secondary protein structures were reextracted at a refined pixel size of 0.925 Å/pix. The reference map and mask were also rescaled to the same pixel size and subjected to the second 3D refinement along with the reextracted segments. Another CtfRefine and Bayesian polishing were performed before the final 3D refinement step with a helical symmetry local search. The search range for twist is between −14.9 and −15.3°. The search range for rise is between 3.0 and 3.5 Å. The resolution of the final construction was estimated to be 3.1 Å. The helical symmetry parameter converged to a rise and twist of 3.16 Å and −15.11°, respectively.
Model building and refinement
For the LLOV NP(full)–RNA complex structure, the atomic model of the EBOV NP–RNA complex (NP residues: 19–407, PDB-ID: 5Z9W) (13) was used as the initial model and was fitted as a rigid body into the cryo-EM map, using UCSF Chimera (ver 1.13.1) (35). Next, the EBOV residues were replaced with the LLOV residues, and the model was adjusted manually using COOT (36, 37). The stereochemistry of the peptide bonds was monitored using a Ramachandran plot. The N- and C-terminal residues, 1–19 and 407–749, respectively, were not modeled because of a lack of interpretable map features. Then, the atomic models were refined against the corresponding maps in the presence of 8 surrounding models using phenix.real_space_refine within the PHENIX (38) software, in the presence of 8 surrounding subunits, including an extended RNA strand. The resulting models were selected based on clashscore, Ramachandran outliers, and MolProbity score, and the final model was presented using the UCSF Chimera and ChimeraX (35, 39).
For the LLOV NP(1–450)–RNA complex structure, the atomic model of the LLOV NP(full)–RNA complex was used as the initial model, and the model was refined as per the method described above. The detailed model statistics are shown in Table S1.
Minigenome assay
LLOV minigenome assays were performed as described previously (40). Briefly, HEK 293T cells were transfected with the PolII-driven LLOV minigenome (pCAGGS-LLOV-vRNA-hrluc; 125 ng), pCAGGS-LLOV-L (500 ng), pCAGGS-LLOV-VP30 (37.5 ng), and pCAGGS-LLOV-VP35 (62.5 ng), as well as the different myc-tagged NP mutants or NP-WT (62.5 ng), using Transit LT-1, according to manufacturer's instructions. For normalization, the cells were additionally transfected with pCAGGS-luc2 (12.5 ng), and the polymerase was omitted (−L) as the negative control. Two days posttransfection, the cells were lysed in 1× Lysis Juice (PJK) for 10 min and cleared by centrifugation for 3 min at 10,000 × g. Reporter activity was measured by adding 40 µl cleared lysate to either 40 µl Beetle Juice (PJK) or 40 µl Renilla Glo Juice in opaque 96-well plates. Luciferase activity was measured using a Glomax Multi microplate reader (Promega). Reporter activities obtained for Renilla luciferase (minigenome reporter) were normalized against firefly luciferase activity (control luciferase). Only samples that showed NP expression levels between 40 and 250% of NP-myc (+L) were included for reporter activity calculations. Dunnett's posttest was performed for statistical analysis. The experiments were performed in pentaplicates (n = 5). Significance was set at P < 0.05 (*) and P < 0.001 (***).
Western blotting
To determine NP expression levels, 4× SDS-PAGE buffer (125 mm Tris-HCl [pH 6.8], 40% glycerin, 8% SDS, 20% beta-mercaptoethanol, and 10 µg/ml bromophenol blue) was added to the remaining lysates from the luciferase assay and further boiled for 10 min at 99°C, before the samples were subjected to SDS-PAGE and western blotting. Nitrocellulose membranes were blocked with 7% skimmed milk powder in D-PBS (−) and stained with anti-c-myc (rabbit; #PA1-981; Thermo Fisher Scientific) and anti-rabbit (goat anti-rabbit; #111-655-144; Dianova) antibodies for the detection of NP-myc, and anti-GAPDH (mouse; #sc47724; Santa Cruz) and anti-mouse (goat anti-mouse; #926-68070; Li-cor) antibodies were added as loading controls. Western blots were visualized using the Li-cor Odyssey imaging system, and band intensity was determined using the ImageStudio Lite software (Li-cor).
Supplementary Material
Acknowledgments
We thank Ayato Takada (International Institute for Zoonosis Control, Hokkaido University) for providing the plasmid expressing the LLOV NP.
Contributor Information
Shangfan Hu, Laboratory of Ultrastructural Virology, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; Laboratory of Ultrastructural Virology, Graduate School of Biostudies, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; CREST, Japan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan.
Yoko Fujita-Fujiharu, Laboratory of Ultrastructural Virology, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; Laboratory of Ultrastructural Virology, Graduate School of Biostudies, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; CREST, Japan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan.
Yukihiko Sugita, Laboratory of Ultrastructural Virology, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; Laboratory of Ultrastructural Virology, Graduate School of Biostudies, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; Hakubi Center for Advanced Research, Kyoto University, 53 Kawahara-cho, Shogoin, Sakyo-ku, Kyoto 606-8507, Japan.
Lisa Wendt, Laboratory for Integrative Cell and Infection Biology, Institute of Molecular Virology and Cell Biology, Friedrich-Loeffler-Institut, Insel Riems, Greifswald 17493, Germany.
Yukiko Muramoto, Laboratory of Ultrastructural Virology, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; Laboratory of Ultrastructural Virology, Graduate School of Biostudies, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; CREST, Japan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan.
Masahiro Nakano, Laboratory of Ultrastructural Virology, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; Laboratory of Ultrastructural Virology, Graduate School of Biostudies, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; CREST, Japan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan.
Thomas Hoenen, Laboratory for Integrative Cell and Infection Biology, Institute of Molecular Virology and Cell Biology, Friedrich-Loeffler-Institut, Insel Riems, Greifswald 17493, Germany.
Takeshi Noda, Laboratory of Ultrastructural Virology, Institute for Life and Medical Sciences, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; Laboratory of Ultrastructural Virology, Graduate School of Biostudies, Kyoto University, 53 Shogoin Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; CREST, Japan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan.
Supplementary material
Supplementary material is available at PNAS Nexus online.
Funding
This work was supported by a Grant-in-Aid for JSPS Fellows (Number 21J12207) (to Y.F-F.), a Research Grant from the Kazato Research Encouragement Prize, MEXT Grant-in-Aid for Leading Initiative for Excellent Young Researchers, JSPS KAKENHI Grant (Number 21K07052), JST FOREST Program (Number JPMJFR214S, Japan) (to Y.S.), the JSPS Core-to-Core Program A (JPMJCR20HA), the Advanced Research Networks, the AMED (Numbers 21wm0325023j0002, 22wm0325023h9903, JP22fm0208101, and 22KK0115), the JST Core Research for Evolutional Science and Technology (Number JPMJCR20HA) (to T.N.), the Grant for Joint Research Project of the Institute of Medical Science, University of Tokyo, the Joint Usage/Research Center program of Institute for Life and Medical Sciences Kyoto University, and the Takeda Science Foundation (to Y.S. and T.N.). This study was also supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from AMED (Number JP22ama1214032).
Author contributions
S.H., Y.F-F., Y.S., and T.N. designed the study; S.H., Y.F.-F., Y.S., L.W., Y.M., and M.N. performed the experiments; S.H., Y.F-F., Y.S., T. H., and T.N. wrote the manuscript with input from all co-authors. All authors have read and approved the final manuscript.
Data availability
The cryo-EM maps generated in this study have been deposited into the Electron Microscopy Data Bank under accession code EMD-34016 and EMD-34049. The atomic coordinates reported in this paper have been deposited in the Protein Data Bank (PDB) with accession code 7YPW and 7YR8. Raw movies have been deposited in the Electron Microscopy Public Image Archive with accession code EMPIAR-11323. Previously released structural data used in the course of this study: 5Z9W and 7F1M, were obtained from the PDB.
References
- 1. Negredo A, et al. . 2011. Discovery of an Ebolavirus-like Filovirus in Europe. PLoS Pathog. 7:e1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kemenesi G, et al. . 2018. Re-emergence of Lloviu virus in Miniopterus schreibersii bats, Hungary, 2016. Emerg Microbes Infect. 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kemenesi G, et al. . 2022. Isolation of infectious Lloviu virus from Schreiber's Bats in Hungary. Nat Commun. 13:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Towner JS, et al. . 2009. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 5:e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elliott LH, Kiley MP, McCormick JB. 1985. Descriptive analysis of Ebola virus proteins. Virology. 147:169–176. [DOI] [PubMed] [Google Scholar]
- 6. Hu S, Noda T. 2022. Filovirus helical nucleocapsid structures. Microscopy (Oxf). Online ahead of print. doi: 10.1093/jmicro/dfac049 [DOI] [PubMed] [Google Scholar]
- 7. Maruyama J, et al. . 2014. Characterization of the envelope glycoprotein of a novel filovirus, Lloviu virus. J Virol. 88:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruigrok RW, Crépin T, Kolakofsky D. 2011. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 14:504–510. [DOI] [PubMed] [Google Scholar]
- 9. Mühlberger E. 2007. Filovirus replication and transcription. Future Virol. 2:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mühlberger E, Weik M, Volchkov VE, Klenk H-D, Becker S. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J Virol. 73:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wan W, et al. . 2017. Structure and assembly of the Ebola virus nucleocapsid. Nature. 551:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bharat TAM, et al. . 2012. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci U S A. 109:4275–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sugita Y, Matsunami H, Kawaoka Y, Noda T, Wolf M. 2018. Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex at 3.6 Å resolution. Nature. 563:137–140. [DOI] [PubMed] [Google Scholar]
- 14. Fujita-Fujiharu Y, et al. . 2022. Structural insight into Marburg virus nucleoprotein–RNA complex formation. Nat Commun. 13:1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirchdoerfer RN, Saphire EO, Ward AB. 2019. Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex. Acta Cryst F. 75:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Watanabe S, Noda T, Kawaoka Y. 2006. Functional mapping of the nucleoprotein of Ebola virus. J Virol. 80:3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noda T, Hagiwara K, Sagara H, Kawaoka Y. 2010. Characterization of the Ebola virus nucleoprotein–RNA complex. J Gen Virol. 91:1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leung DW, et al. . 2015. An intrinsically disordered peptide from Ebola virus VP35 controls viral RNA synthesis by modulating nucleoprotein-RNA interactions. Cell Rep. 11:376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu B, et al. . 2017. Structural insight into nucleoprotein conformation change chaperoned by VP35 peptide in Marburg virus. J Virol. 91:e00825-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wendt L, et al. . 2022. Evidence for viral mRNA export from Ebola virus inclusion bodies by the nuclear RNA export factor NXF1. J Virol. 96:e0090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Renner M, et al. . 2016. Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein. Elife. 5:e12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gutsche I, et al. . 2015. Structural virology. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 348:704–707. [DOI] [PubMed] [Google Scholar]
- 23. Green TJ, Zhang X, Wertz GW, Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 313:357–360. [DOI] [PubMed] [Google Scholar]
- 24. Hastie KM, et al. . 2011. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci U S A. 108:19365–19370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alayyoubi M, Leser GP, Kors CA, Lamb RA. 2015. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc Natl Acad Sci U S A. 112:E1792–E1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu T, et al. . 2017. Crystal structure of the Marburg virus nucleoprotein core domain chaperoned by a VP35 peptide reveals a conserved drug target for filovirus. J Virol. 91:e00996-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi M, et al. . 2018. The evolutionary history of vertebrate RNA viruses. Nature. 556:197–202. [DOI] [PubMed] [Google Scholar]
- 28. Yang X-L, et al. . 2019. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat Microbiol. 4:390–395. [DOI] [PubMed] [Google Scholar]
- 29. Horie M. 2021. Identification of a novel filovirus in a common lancehead (Bothrops atrox (Linnaeus, 1758)). J Vet Med Sci. 83:1485–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hierweger MM, et al. . 2021. Novel Filoviruses, Hantavirus, and Rhabdovirus in freshwater fish, Switzerland, 2017. Emerg Infect Dis. 27:3082–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zivanov J, et al. . 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He S, Scheres SHW. 2017. Helical reconstruction in RELION. J Struct Biol. 198:163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liao HY, Frank J. 2010. Definition and estimation of resolution in single-particle reconstructions. Structure. 18:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pettersen EF, et al. . 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 36. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 37. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams PD, et al. . 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 39. Goddard TD, et al. . 2018. UCSF Chimerax: meeting modern challenges in visualization and analysis. Protein Sci. 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kämper L, et al. . 2019. Assessment of the function and intergenus-compatibility of Ebola and Lloviu virus proteins. J Gen Virol. 100:760–772. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps generated in this study have been deposited into the Electron Microscopy Data Bank under accession code EMD-34016 and EMD-34049. The atomic coordinates reported in this paper have been deposited in the Protein Data Bank (PDB) with accession code 7YPW and 7YR8. Raw movies have been deposited in the Electron Microscopy Public Image Archive with accession code EMPIAR-11323. Previously released structural data used in the course of this study: 5Z9W and 7F1M, were obtained from the PDB.