Summary
Background
In preclinical models of Type 1 Diabetes (T1D) the integrity of the gut barrier (GB) is instrumental to avoid dysregulated crosstalk between the commensal microbiota and immune cells and to prevent autoimmunity. The GB is composed of the intestinal epithelial barrier (IEB) and of the mucus layer containing mucins and antimicrobial peptides (AMPs) that are crucial to maintain immune tolerance. In preclinical models of T1D the alterations of the GB primarily affect the mucus layer. In human T1D increased gut permeability and IEB damage have been demonstrated but the integrity of the mucus layer was never assessed.
Methods
We evaluated GB integrity by measuring serological markers of IEB damage (serological levels of zonulin) and bacterial translocation such as lipopolysaccharide binding protein (LBP) and myeloid differentiation protein 2 (MD2), and mRNA expression of tight junction proteins, mucins and AMPs in intestinal tissue of T1D patients and healthy controls (HC). Simultaneously, we performed immunological profiling on intestinal tissue and 16S rRNA analysis on the mucus-associated gut microbiota (MAGM).
Findings
Our data show a GB damage with mucus layer alterations and reduced mRNA expression of several mucins (MUC2, MUC12, MUC13, MUC15, MUC20, MUC21) and AMPs (HD4 and HD5) in T1D patients. Mucus layer alterations correlated with reduced relative abundance of short chain fatty acids (SCFA)-producing bacteria such as Bifidobacterium dentium, Clostridium butyricum and Roseburia intestinalis that regulate mucin expression and intestinal immune homeostasis. In T1D patients we also found intestinal immune dysregulation with higher percentages of effector T cells such as T helper (Th) 1, Th17 and TNF-α+ T cells.
Interpretation
Our data show that mucus layer alterations are present in T1D subjects and associated with dysbiosis and immune dysregulation.
Funding
Research Grants from the Juvenile Diabetes Foundation (Grant 1-INO-2018-640-A-N to MF and 2-SRA-2019-680-S-B to JD) and from the Italian Ministry of Health (Grant RF19-12370721 to MF).
Keywords: Mucins, Anti-microbial peptides, Microbiota, Immune dysregulation, Autoimmune diabetess
Research in context.
Evidence before this study
In the recent years a plethora of experimental data suggested that extra-intestinal autoimmune diseases such as Type 1 Diabetes (T1D) are regulated in the gut, possibly by an altered crosstalk between the commensal microbiota and the immune system but the mechanism underlying this effect is largely unknown. We recently demonstrated that loss of gut barrier integrity and mucus layer disruption is responsible for activation of self (islet)-reactive T cells within the gut and occurrence of autoimmune T1D in preclinical models (Sorini C. et al. PNAS 2019). Although signs of increased gut permeability and damage of the intestinal epithelial barrier have been reported in T1D patients early on in disease pathogenesis (Bosi E et al. Diabetologia 2006), the integrity of the mucus layer and its role in modulating gut microbiota composition and mucosal immunity in human T1D have been poorly investigated.
Added value of this study
We had the unique opportunity to collect intestinal tissue samples from T1D patients who performed esophagogastroduodenoscopy for diagnostic purposes and healthy controls for comparison and we assessed: the integrity of physical (IEB) and biological (mucus layer) gut barriers, signs of bacterial translocation, immunological profiling of intestinal immune cells and analysis of the mucus-associated gut microbiota (collected through brushing of the intestinal mucosa). Our report demonstrates for the first time that alterations of the mucus layer composition (mucins and AMPs) are present in T1D subjects and are linked to dysbiosis with reduced representation in the mucus-associated gut microbiota (MAGM) of numerous SCFA-producing species that are crucial to maintain mucus layer integrity and gut immune tolerance like Clostridium butyricum, Roseburia intestinalis and Bifidobacterium dentium. Those alterations were linked to intestinal immune dysregulation with augmented percentages of effector T cells (Th1 cells, Th17 cells and TNFα+ T cells).
Implications of all the available evidence
Our data indicate that modifications of the commensal gut microbiota in human T1D lead to defective function of the mucus barrier with bacterial translocation and immune dysregulation. Further studies are necessary to understand how intestinal immune dysregulation promotes activation of islet-reactive T cells and autoimmune diabetes in humans.
Introduction
Studies in humans and pre-clinical models demonstrated that the gut environment and commensal microbiota regulate the autoimmune pathogenesis of T1D1,2 but the mechanisms underlying this modulatory effect are largely unknown. In physiological conditions, the commensal gut microbiota is contained in the intestinal lumen and its interaction with the host is controlled by physical and biological barriers that maintain tissue homeostasis and prevent translocation of bacterial components into the intestinal and peripheral tissues. These biological and physical components of the GB play a crucial role in regulating the crosstalk between the commensal microbiota and the immune system so that their alteration could lead to abnormal activation of the immune cells within the intestinal mucosa and to autoimmunity in extra-intestinal tissues.3,4 In line with this hypothesis, recent studies in animal models indicates that loss of GB integrity leads to activation of islet-reactive T cells by commensal gut microbiota and occurrence of autoimmune diabetes.5,6 In humans, an increase of gut permeability measured by lactulose/mannitol test is detectable before clinical onset of T1D in individuals with β cell autoimmunity (islet autoantibody-positivity) and no hyperglycemia,7,8 thus suggesting that damage of the GB is directly related to T1D pathogenesis rather than secondary to diabetes-induced metabolic alterations.
The main function of the GB is to control the absorption of nutrients, water and electrolytes while preventing the entrance of pathogens and regulating the crosstalk between the gut microbiota (bacteria, viruses, protozoa, and fungi) and the intestinal immune cells. Hence, the integrity of the GB is fundamental to maintain the equilibrium between tolerance and immunity to food antigens and commensal microbiota but also to self-antigens including those of extra-intestinal tissues such as the pancreatic islets.6,9,10 These key functions of the GB are preserved by a complex structure composed by a mucus layer and a monolayer of epithelial cells interconnected by tight junctions, the IEB. The mucus layer is the first barrier that commensal bacteria encounter before entering in contact with the host and it covers the entire gastrointestinal canal. The mucus is made by specialized Goblet cells (GC) and coat the intestinal epithelial surface with a dense glycocalyx that isolate the gut mucosa from the intestinal lumen content and commensal bacteria. In the small intestine the mucus is thin and composed of a single layer that contains bacterial species. In the large intestine is thicker and has an inner sterile compartment (with no bacteria) and an outer compartment populated by bacterial species in larger number compared to the small intestinal mucus. Mucus is critical for the intestinal health and its damage results in bacterial translocation in the intestinal tissue and, from there, into the systemic circulation. Importantly, the mucus layer is not just a physical barrier but is composed of a complex system of molecules that have crucial immune regulatory and anti-microbial activities such as mucins11 and AMPs.12 Mucins are glycoproteins that can be divided in two categories: secreted and transmembrane. Although in humans there are five secreted mucins (MUC2, MUC5AC, MUC5B, MUC6, and MUC19),13 the mucus layer structure, particularly in the small intestine, is exclusively composed of MUC2 that is produced by GC.14 Transmembrane mucins are crucial components of the glycocalyx on the apical surface of mucosal epithelium and include MUC1, MUC3A, MUC3B, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17, MUC20, and MUC21.15 Transmembrane mucin composition is important to regulate the presence of selected microbial species within the mucus layer because many commensal bacteria utilize those mucin glycans as an energy source.16 Hence, an alteration of transmembrane mucins could be responsible for modification of the bacterial species that resides in the intestinal mucus layer, the so-called mucus-associated gut microbiota (MAGM). Mucins also play key immunoregulatory functions and directly modulate gut mucosal immunity and prevent intestinal inflammation. For example, MUC2 induces differentiation of tolerogenic CD103+ dendritic cells.17 In line with this concept, alterations of the mucus layer and, specifically, of the mucin composition are found in the intestine of individuals affected by inflammatory bowel diseases (IBD).18, 19, 20 Although in preclinical models of T1D alterations of the mucus layer structure and mucin composition have been detected,5 in human T1D the mucus layer integrity was never analyzed.
AMP are concentrated within the firm layer of mucus that is close to the epithelium. Those molecules are fundamental to maintain the multilayered defense against luminal microorganisms by limiting the access of commensal bacteria to the mucosal surface and regulating their interaction with the host immune cells. Virtually all the epithelial cell types in the intestine can produce AMPs, however, the largest amount of these molecules is produced by enterocytes lining the gastrointestinal tract and by Paneth cells (PC) in the small intestine. AMPs include α-defensins (HD), cathelicidins and regenerating islet-derived protein (REG)3 and are fundamental to control commensal microbiota composition, GB integrity, intestinal mucosal immunity and bacterial translocation. AMPs maintain GB integrity by promoting the expression of mucins and tight junction proteins (TJP).21,22 As for mucins, dysregulated expression of AMPs is associated with immune dysregulation and chronic inflammation in IBD.23
In human T1D early signs of intestinal inflammation have been documented,24, 25, 26 possibly due to dysbiosis1,2 and damage of the GB integrity.8 However, while there is clear indication of breakage of the IEB, little is known about the mucus layer and its composition and how it affects bacterial translocation and gut mucosal immunity in human T1D.
Here we analyzed the GB integrity by analyzing serological markers of bacterial translocation as well as mRNA expression levels of components of the IEB and mucus layer (mucins and AMPs) in the small intestine of T1D patients and healthy controls. We confirmed the presence of IEB damage in T1D patients with increased serological levels of zonulin. Moreover, in T1D patients we found signs of bacterial translocation (high serological levels of MD2) and alterations of the mucus layer composition with defective mRNA expression of different structural (MUC2) and transmembrane mucins (MUC12, MUC13, MUC15, MUC20 and MUC21) and AMPs (HD5 and HD6). The alterations of the mucus layer composition in T1D patients were associated with modifications of the MAGM composition and, specifically, with reduced relative abundance of several bacterial species such as Bifidobacterium dentium, Clostridium butyricum and Roseburia intestinalis that release tolerogenic metabolites like short chain fatty acids (SCFA) and enzymes fundamental to promote mucus layer regeneration. Mucus layer alterations and dysbiosis in our human T1D cohort were associated with immune dysregulation and increased percentages of effector Th1, Th17 and TNF-α+ T cells in the intestinal mucosa. Our data suggest that modifications of the physical and biological gut barriers are present in T1D patients and are associated with dysbiosis of mucus-regulating bacteria and immune dysregulation.
Methods
Study population
This is an observational cross-sectional study in which we compared the intestinal environment of T1D patients vs healthy controls. 17 T1D patients and 16 healthy controls (Table S1 of the Supplementary Data for anagraphic data) included in this study were adult male and female individuals who performed esophagogastroduodenoscopy (EGDS) for diagnostic purposes (dyspepsia, gastroesophageal reflux, altered bowel habits, etc.) at the OSR Gastroenterology Unit in the time period between March 2015 and December 2017. None of the T1D patients and healthy controls was diagnosed with gastrointestinal pathology after the EGDS examination. The two cohorts did not show statistically significant difference for age. Diagnosis of T1D was based on the criteria of the American Diabetes Association.27 Exclusion criteria for both T1D and HC were as follows: antibiotics and corticosteroids treatments in the 3 months before EGDS, clinical history of gastroenteritis, gastric ulcer, irritable bowel disease (IBS), IBD and gastric and colorectal cancer. Five out of the 17 T1D patients enrolled in our study were affected by celiac disease and three out of 17 had a recent diagnosis of T1D (<6 months). All patients were caucasian and none of them was obese (BMI between 18 and 30). Clinical information including duration of disease, glucose metabolism indicators (HbA1c), comorbidities including celiac disease, other autoimmune diseases, etc., regarding T1D patients enrolled in this study are presented in Table S2 of the Supplementary Data. At the time of EGDS each T1D patient was asked to fill a 3 day-food questionnaire to verify that their diet was aligned with the Dietary Reference Values for the Italian population (Table S3 of the Supplementary Data).
Samples collection and processing
All EGDS were performed at the Gastroenterology Unit of the San Raffael Hospital (OSR). Intestinal tissue fragments and blood samples were collected simultaneously from each individual at the time of the EGDS procedure. The MAGM was collected by gentle brushing of the gut mucosa at the beginning of the EGDS procedure with a specific tissue brusher (Cytology brush, Cook Medical, USA). Tissue fragments of intestinal mucosa were collected from the descending fragment of the duodenum, immediately placed in complete RPMI 1640 medium (RPMI 1640 containing 10% fetal bovine serum, penicillin/streptomycin (100 μg/ml), 2 mM glutamine, 10 mM Hepes, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, and 0.05 mM β-mercaptoethanol) and rapidly processed. For preparation of RT-qPCR samples, one intestinal tissue fragment was collected in RNA later solution (Invitrogen) and immediately frozen at −80 °C until RNA extraction.
For single cell isolation (FACS analysis), mucus and epithelial cells were removed from the tissue by incubation with 1 mmol/L dithiothreitol (DTT) and 5 mmol/L EDTA in calcium- and magnesium-free Hanks’ balanced salt solution (HBSS) for 15 min at 37 °C. Tissue samples were washed with HBSS, digested with 1 mg/ml collagenase A (Roche Diagnostics Ltd., Indianapolis, IN) in HBSS supplemented with calcium and magnesium, and 5 units/ml DNase I (Roche Diagnostics Ltd.) for 1 h at 37 °C, and mechanically disrupted by gentle pipetting until dissociation was complete. After incubation, intestinal cells released from the tissue samples were passed through a 70 μm cell strainer and washed with complete RPMI 1640 medium. Peripheral blood mononuclear cells were isolated from blood through Ficoll gradient and washing with RPMI 1640 medium.
Serological markers of gut barrier damage and bacterial translocation
Serum samples were obtained from peripheral blood after incubation for 1 h at room temperature and centrifugation at 1600g for 10 min (Multifuge 3SR centrifuge). A quantitative measurement of zonulin, LBP and MD2 in serum was performed with commercial kits that used the ELISA method, following manufacturer's instructions. The following commercial kits were used: human zonulin quantitative ELISA kit (Techno-Genetics); human MD2 quantitative ELISA kit (Sigma-Merck); human LBP quantitative ELISA kit (Biometec GmbH).
Real-time qPCR
RNA was extracted from duodenal biopsies collected during the EGDS by adding 100 μL of chloroform, precipitating the aqueous phase with 300 μL of 70% ethanol, and purifying RNA with RNeasy Mini Kit (QIAGEN). RNA was retrotranscribed with SuperScript III First-Strand Synthesis System following manufacturer's instructions (Life Technologies). Real-time qPCR assay was performed with SYBR Select Master Mix (Life Technologies) using primers specific for different TJP such as zonulin-1 (ZO-1), zonulin-2 (ZO-2), claudin-1 (CLN-1) and occludin (OCLN), mucins (MUC1, MUC2, MUC3A, MUC4, MUC12, MUC13, MUC15, MUC17, MUC20, MUC21) and AMPs (Reg1α, Reg3α, REG3γ, BD1, BD3, LL-37, HD5, and HD6) on a ViiA 7 Real-Time PCR System (Life Technologies). The expression of all genes was normalized to the reference gene GAPDH (the reference gene that showed the lowest variance between different samples) and the results were calculated using the ΔΔCt method. Primers for human intestinal mucins and TJP were purchased from Bio-Rad. Human AMP gene sequences of the specific primers were determined by the online Blast program from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) and compared with the GeneBank database (Table S4 of the Supplementary Data).
FACS analysis
Single cell suspensions from intestinal tissues were stimulated for 4 h in complete RPMI 1640 medium with 50 ng/ml PMA (Sigma) and 1.25 μg/ml ionomycin (Sigma) in the presence of Golgi Plug (BD Biosciences). Cells were harvested in PBS containing 1% FBS and 0.09% NaN3 and stained with Fixable Viability Dye eFlour 506 (eBioscience). Cells were first stained for surface markers (20 min at 4 °C), then fixed and permeabilized using the BD Cytofix/Cytoperm kit (30 min at 4 °C), and finally stained for intracellular cytokines (25 min at 4 °C). The following antibodies were used: APC-Cy7 conjugated anti-human CD3 (RRID: 15829228, BD Biosciences), Pe/Texas Red-conjugated anti-human CD4 (RRID: AB_10371766, Thermo Fisher), eFluor 450-conjugated anti-human interferon (IFN)-γ (RRID: AB_2043866, eBioscience), APC-conjugated anti-human interleukin (IL)-17A (RRID: AB_1724136, eBioscience), PE-conjugated anti-human IL-22 (Catalog Number: IC7821P, R&D Systems), Alexa Fluor-488 conjugated anti-human IL-4 (RRID: AB_493324, BioLegend), PE-cy7 conjugated anti-human tumor necrosis factor (TNF)-α (RRID: AB_1727578, BD Biosciences), and PercP/Cy5.5-conjugated anti-human IL-10 (clone JES3-9D7 BioLegend). Data were acquired using a LSRFortessa cell analyzer (BD Biosciences) and analyzed with FCS Express V4 software (De Novo Software). We identified Th1 cells (CD3+CD4+IFN-γ+), Th17 cells (CD3+CD4+IL-17+), Th22 cells (CD3+CD4+IL-22+), Th2 cells (CD3+CD4+IL-4+), TNF-α+ T cells (CD3+CD4+TNF-α+), and Tr1 cells (CD3+CD4+IL-10+). See Supplementary Figure S1 for gating strategy.
Microbiome sequencing
The human microbiome sequencing was performed by 16S amplicon sequencing on the Illumina Miseq platform (Illumina). Total DNA was purified from brushed material of the duodenum collected during EGDS using the QIAamp DNA Micro kit (Qiagen) following manufacturer's instructions. The V3–V4 region of the 16S rRNA gene was amplified starting from 500 ng of extracted DNA using the FastStart High Fidelity PCR System (Roche), barcoded sample-specific primers: [V3-16S-Fw: CCT ACG GGN GGC WGC AG; V4-16S-rev: GAC TAC HVG GGT ATC TAA TCC]. The following cycling conditions were employed: 94 °C for 2 min, 35 cycles of (94 °C for 30 s, 56 °C for 30 s, and 68 °C for 1 min), and then the samples were stored at 4 °C until usage. Amplicons were purified using the AMPure XP beads (Beckman Coulter, Brea, USA). A second PCR step was performed to add Illumina sequencing adapters to each sample. The Nextera xt Index Kit (Illumina) and the KAPA HiFi HotStart PCR kit (KAPA Biosystem) were used, and the following amplification protocol: 95 °C for 3 min, 8 cycles of (95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 °C, 72 °C for 4 min) and then stored at 4 °C. The purified DNA was quantified using the Qubit Fluorometer (Thermo Fisher) and the 20,100 Bioanalyzer System (Agilent). For metagenomic analysis of 16S rRNA data microbial reads were discriminated against human reads with BMTagger (ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/). To perform the analysis at species/family/order level, reads were mapped to the collection of all available genomes (https://www.ncbi.nlm.nih.gov/genome/) with Kraken2 for exact alignment of k-mers and accurate read classification.28
Statistical analysis
Statistical significance of the differences between two groups (T1D and HC) was calculated using the nonparametric Mann–Whitney U test and expressed as means ± SD. GraphPad Prism, version 8.0 (GraphPad Software, San Diego, CA), was used for statistical analysis, and values of p ≤ 0.05 were considered statistically significant. The power analysis was calculated based on previous data on intestinal immune cell subsets29 and alpha diversity30 in T1D patients vs HC. Group sample sizes of 17 (T1D) and 16 (HC) was expected to achieve 80.00% power to reject the null hypothesis of zero effect size when the population effect size is 1.00 and the significance level (alpha) is 0.050 using a Mann–Whitney U test.
Relative abundance profiling and differential analysis of 16S rRNA data was performed with DESeq2 upon variance-stabilizing transformation,31 where p ≤ 0.05 were considered statistically significant. We used the variance-stabilizing transformation algorithm.28 This function calculates a variance stabilizing transformation from the fitted dispersion-mean relation(s) and then transforms the count data (normalized by division by the size factors or normalization factors), yielding a matrix of values that are now approximately homoskedastic (having constant variance along the range of mean values). The transformation also normalizes with respect to library size. Species alpha diversity was calculated with vegan::diversity (https://cran.rproject.org/web/packages/vegan). The Shannon and Simpson indices extrapolate diversity from the proportional abundance of species within a specific population. Visualization was performed with ggplot232 and Krona.33
Nonparametric multivariate analysis was performed with the ggstatplot R package (https://joss.theoj.org/papers/10.21105/joss.03167) with a significance level = 0.05, confidence level = 0.95, and Bonferroni–Holm method for multiple comparison p-value adjustment.
Ethics
All individuals signed a written informed consent, complied with the study procedure, and were aware that they donated intestinal tissue fragments and blood samples for research purpose. The study was approved by the Institutional Ethical Committee of the IRCCS San Raffaele Scientific Institute on 10/02/2014 (Protocol: T1D-GUT2014), Milan, Italy.
Role of funders
This work was supported by Research Grants from the Juvenile Diabetes Foundation (Grant 1-INO-2018-640-A-N to MF and 2-SRA-2019-680-S-B to JD) and from the Italian Ministry of Health (Grant RF19-12370721 to MF). The funding source had no involvement in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. I was not paid to write this article by a pharmaceutical company or other agency. The authors were not precluded from accessing data in the study, and they accept responsibility to submit for publication.
Results
Alterations of the GB and mucus layer composition in T1D patients
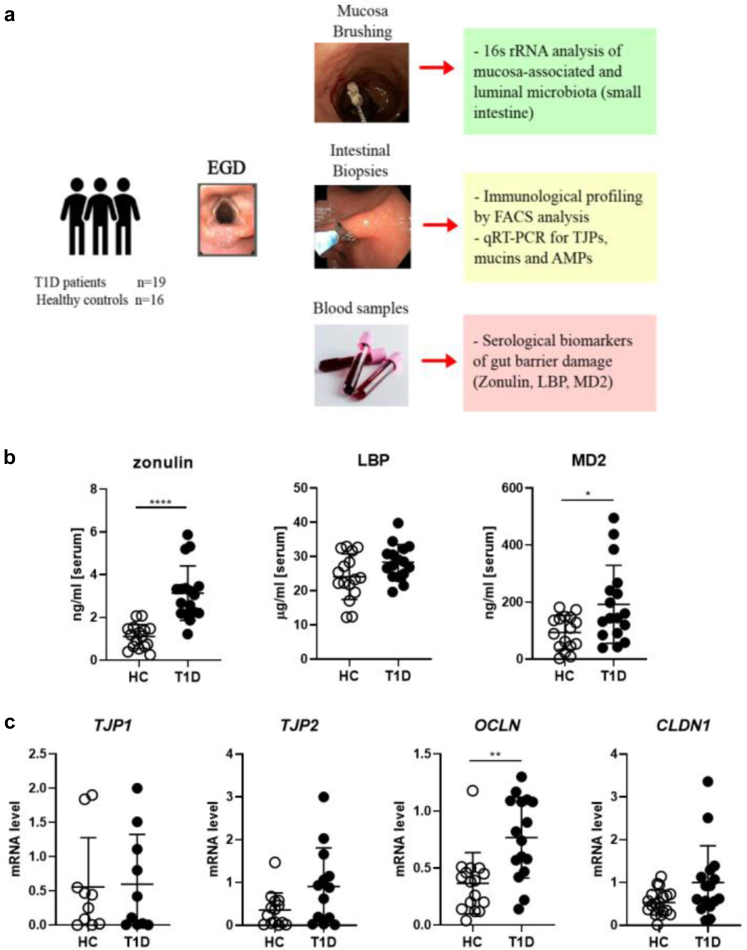
The development of beta cell autoimmunity in patients and pre-clinical models of T1D is preceded by damage of the GB with increased gut permeability.5,7,8 The GB is composed by the IEB, a single layer of epithelial cells held together by a complex junctional system composed of tight junctions, adherent junctions and desmosomes, and by the mucus layer that contains mucins and AMPs, crucial molecules to regulate the crosstalk between the gut microbiota and the immune system. Previous studies in human T1D reported damage of the IEB but the mucus layer integrity was never analyzed. Here we assessed integrity of both components of the GB, the IEB and the mucus layer, in a cohort of T1D patients and healthy controls (Fig. 1a). The IEB integrity was evaluated by measuring serological levels of zonulin as well as mRNA expression of structural TJP in small intestinal tissue samples of T1D patients and healthy controls. Zonulin is an endogenous protein that is released by the IEC upon interaction with pathogens and commensal bacteria such as Escherichia coli and also by dietary protein such as gliadin through the CXCR3 receptor.34 Zonulin presence in the serum is a biological marker of tight junction disassembly and “leaky gut”. Our data confirmed previous findings of increased serological levels of zonulin in T1D subjects compared to healthy controls (∗∗∗∗p ≤ 0.001, Mann–Whitney U test) (Fig. 1b). The IEB damage could lead to uncontrolled passage of bacterial components into the blood circulation. The latter scenario was suggested by the finding that T1D patients had increased serological levels of the myeloid differentiation protein 2 (MD2), a lipopolysaccharide-binding protein that forms an active complex with the TLR4 leading to production of pro-inflammatory cytokines35 (∗p ≤ 0.05, Mann–Whitney U test) (Fig. 1b). We assessed IEB integrity also by measuring by RT-qPCR the differential mRNA expression of a panel of TJP expressed in the small intestine and, specifically, the barrier forming TJPs CLN1, OCLN, ZO-1 and ZO-2. Our data did not reveal alterations of TJPs except for an increased mRNA expression of OCLN in T1D samples that could indicate a compensatory mechanism to restore GB integrity in T1D patients (Fig. 1c).
Fig. 1.
IEB damage and bacterial translocation in T1D subjects. (a) Study design. During EGDS the duodenal mucosa was gently brushed to collect mucus-associated gut microbiota (MAGM). Then, 2–3 intestinal tissue fragments were collected for qRT-PCR for TJPs, mucins and AMP mRNA expression and FACS analysis on intestinal immune cells. An aliquot of peripheral blood (4–5 ml) was also collected from the same individual at time of EGDS for serological analysis of markers of bacterial translocation. (b) Serological level of Zonulin, lipopolysaccharide binding protein (LBP) and myeloid differentiation 2 (MD2) in T1D patients (n = 17) and HC (Ctrl, n = 16) were measured by ELISA assay. (c) Real-time qRT-PCR analysis of TJP gene expression ZO-1, ZO-2, CLN and OCLN in human duodenal samples of T1D patients (n = 17) and HC (n = 16). The Relative Abundance [A.U.] was calculated using the ΔΔCT method. All data are expressed as means ± SD. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗∗p ≤ 0.001 by Mann–Whitney U test.
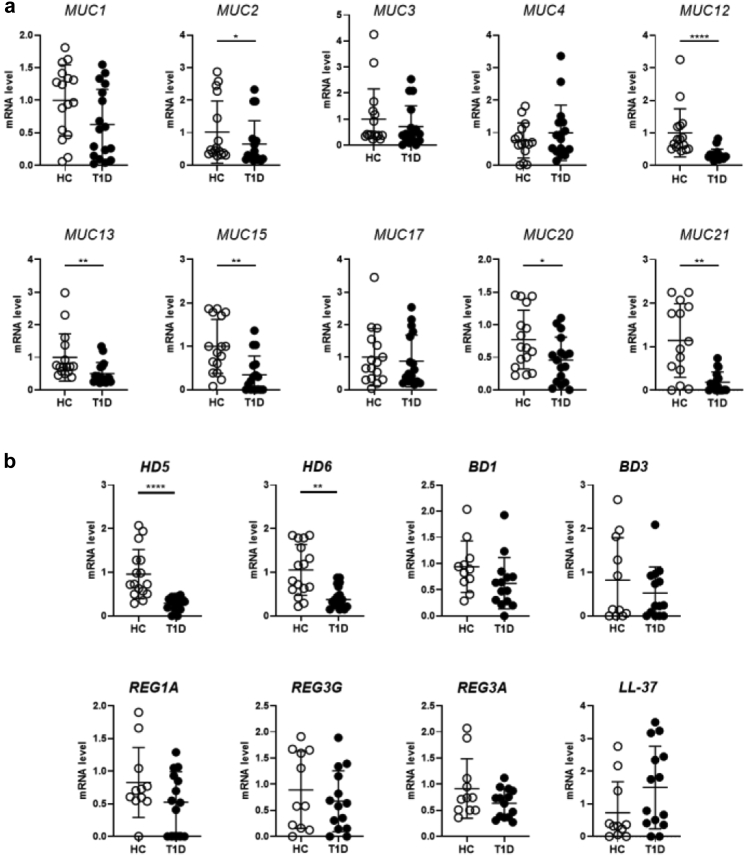
The mucus layer is an important component of the gut barrier that is fundamental to prevent bacterial translocation and protect the gut mucosa. In the spontaneous murine model of T1D, i.e., the non-obese diabetic mouse (NOD), alterations of the mucus structure and composition were recently demonstrated and linked to activation of beta cell autoimmunity and T1D occurrence.5 We assessed the integrity of mucus layer in human T1D patients by measuring mRNA expression of several mucins and AMPs in the small intestinal mucosa. Our RT-qPCR analysis revealed that in T1D subjects compared to healthy controls there was a statistically significant reduction of different mucins including the structural gel-forming mucin MUC2 (∗p ≤ 0.05, Mann–Whitney U test) and several transmembrane mucins such as MUC12, MUC13, MUC15, MUC20 and MUC21 (∗∗∗∗p ≤ 0.001 for MUC12, ∗∗p ≤ 0.01 for MUC13, MUC15 and MUC21 and ∗p ≤ 0.05 for MUC20, Mann–Whitney U test) (Fig. 1a). Importantly, we also detected a significantly decrease in mRNA expression of two important AMPs: the α-defensin 5 (HD5) and α-defensin 6 (HD6) (∗∗∗∗p ≤ 0.001 for HD5 and ∗∗p ≤ 0.01 for HD6, Mann–Whitney U test) in T1D subjects compared to healthy controls, while other AMPs such as BD1, BD3, LL-37 (hCAP18, peptide of the human cationic antimicrobial protein), Reg1α and Reg3α and Reg3γ were not altered (Fig. 2b).
Fig. 2.
Reduced mRNA expression of mucins and AMPs in T1D subjects. (a) Real-time qRT-PCR analysis of gene expression of several mucins (MUC1-4, MUC12-13, MUC15, MUC17, MUC20-21) in human duodenal samples of T1D patients (n = 17) and healthy controls (HC, n = 16). (b) Real-time qRT-PCR analysis of AMP genes for human HD5, HD6, BD, Reg3, and LL-37 (hCAP18, peptide of the human cationic antimicrobial protein) in human duodenal samples of T1D patients and HC. All data are expressed as means ± SD. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗∗p ≤ 0.001 by Mann–Whitney U test.
Mucus layer alteration is associated with reduced relative abundance of butyrate-producing bacteria in the mucosa-associated microbiota of T1D patients
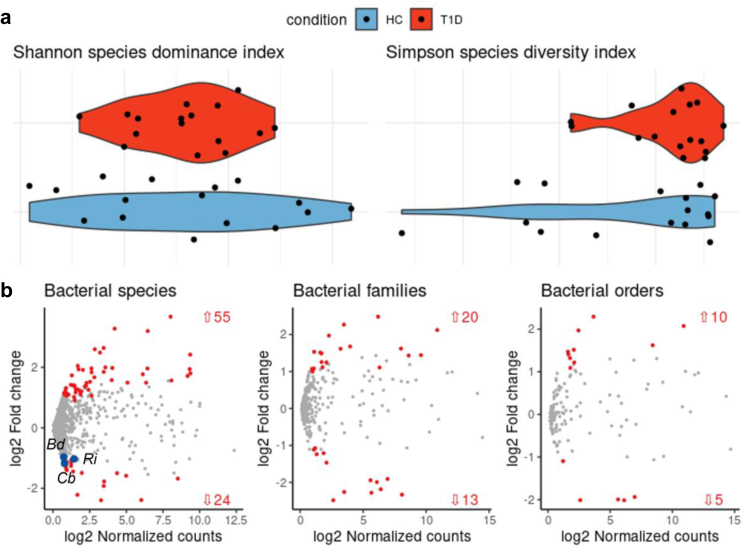
The mucus layer integrity plays an important role in modulating the gut commensal microbiota. For example, transmembrane mucin composition regulates the presence of selected microbial species that use glycans as energy source and reside in the mucus layer in proximity to the gut mucosa (i.e., the mucus-associated gut microbiota: MAGM). Importantly, not only the mucus integrity affects composition of the MAGM, but some microbial species can also regulate mucus production and function. Having found an alteration of the mucus layer in T1D patients we asked whether the MAGM is also dysregulated. To answer this question, we collected bacterial species residing in the mucus layer by gentle brushing the gut mucosa of the duodenum of T1D patients and healthy controls during the EGDS procedure and analyzed the bacterial composition by 16S rRNA analysis. First, we detected a reduced richness with lower bacterial diversity and dominance between T1D patients and healthy controls (Fig. 3a), similarly to what has been detected in the gut commensal microbiota isolated from fecal material of T1D vs HC.30,36, 37, 38 Furthermore, we found 79 bacterial species differentially expressed between the MAGM of T1D patients and healthy controls with 55 species increased and 24 decreased in T1D subjects (Fig. 3b and Table S5 of the Supplementary Data). Notably, 19 out of the 24 bacterial species whose relative abundance was reduced in T1D patients are SCFA-producing (butyrate, propionate or acetate) (Table S5 of the Supplementary Data). Among them, we found statistically significant reduction in the relative abundance of three butyrate-producing obligate anaerobes bacterial species, Bifidobacterium dentium, Clostidrium butyricum and Roseburia intestinalis (∗p ≤ 0.05, DESeq2 statistical test) that are strictly dependent on the mucus layer integrity for their survival and are important for its regeneration (Fig. 3c).
Fig. 3.
Reduced relative abundance of SCFA-producing bacteria in the mucus-associated microbiota of T1D subjects. Gut microbiota profiles on brushed material isolated from the small intestinal mucosa during EGDS were assessed by 16S Amplicon Sequencing of bacterial DNA. (a) Species alpha diversity and dominance in T1D vs HC. (b) 79 bacterial species differentially expressed between the MAGM of T1D and HC with 55 species increased and 24 decreased in T1D vs HC. Three SCFA-producing bacteria, Bifidobacterium dentium, Clostridium butyricum and Roseburia intestinalis with important mucus-regenerating properties were found significantly reduced in T1D patients compared to HC (blue dots). Relative abundance profiling and differential analysis of 16S rRNA data was performed with DESeq2 upon variance-stabilizing transformation,31 where p ≤ 0.05 were considered statistically significant.
Loss of mucus layer integrity in T1D subjects correlates with alterations of intestinal immune homeostasis
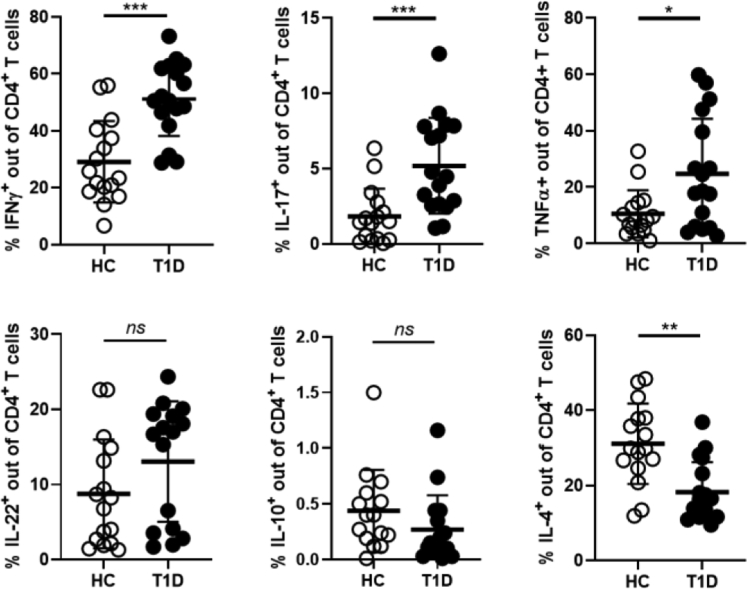
A bi-directional regulation between the mucus layer composition and intestinal mucosal immunity exists. Damage of the mucus layer with passage of bacterial components into the gut mucosa could lead to abnormal activation of immune cells and their acquisition of a pro-inflammatory phenotype. Commensal bacterial species that reside in the mucus layer and depend on transmembrane mucins for their survival contribute to maintenance of immune homeostasis by releasing tolerogenic metabolites such as SCFAs. On the other hand, intestinal immune cells can affect GB integrity by releasing inflammatory cytokines and/or directly modulating the function of intestinal cells that produce mucins and AMPs such as GC and PC. Here we tested whether in our cohort of T1D patients the mucus layer alterations were associated with loss of gut immune homeostasis and with an inflammatory immune profile. To analyze gut mucosal immunity in T1D patients and healthy controls we performed a multiparametric FACS analysis on single cell suspensions isolated from the duodenal biopsies. Our analysis revealed a significant increase in the percentages of several effector T cell subsets including Th1 (CD4+IFN-γ+), Th17 (CD4+IL-17+) and TNF-α+ T cells in the intestine of T1D subjects compared to healthy controls (Fig. 4) (∗∗∗p ≤ 0.005 for Th1 and Th17 cells and ∗p ≤ 0.05 for TNF-α+ T cells, Mann–Whitney U test). In the intestinal mucosa of T1D patients we also detected a decreased in Th2 (CD4+IL-4+) cells, a subset that plays an immune regulatory role and tissue repairing function in the intestinal mucosa39,40 (∗∗p ≤ 0.01, Mann–Whitney U test). We did not detect any difference between T1D patients and healthy controls in the relative percentages of IL-22+ (IL-22+CD4+) and Tr1 (IL-10+CD4+) cells. The increased frequency of T effector cells was observed in the intestinal immune cells but not in the peripheral blood mononuclear cells (PBMC) of T1D patients (data not shown), however we found a decreased in Th2 cells (CD3+CD4+IL-4+) similar to that observed in intestinal immune cells (data not shown). We performed a multivariate analysis to find correlation between the differential presence of gut immune cell subsets, potential confounders (age, sex) and other gut-related parameters altered in T1D patients (zonulin, MD2, HD5, HD6, MUC2, MUC12, MUC20 and MUC21). Our data (Figure S2 of Supplementary Data) show no correlation between age/sex and any of the tested biomarkers of gut alteration in T1D patients.
Fig. 4.
Alterations of the mucus layer composition in T1D subjects correlates with intestinal immune dysregulation. Single cell suspensions isolated from intestinal biopsies of T1D patients and healthy controls were FACS analyzed. Data are expressed as percentages of different T cell subsets out of total CD3+CD4+ T cells. We compared Th1 (CD3+CD4+IFN-γ+), Th17 (CD3+CD4+IL-17+), TNF-α+ (CD3+CD4+TNF-α+), Th22 (CD3+CD4+IL-22+), Tr1 (CD3+CD4+ IL-10+), and Th2 (CD3+CD4+IL-4+) cells in the intestine of T1D subjects compared to healthy controls. In the intestinal mucosa of T1D patients we detected an increased of effector INF-γ+ Th1, IL-17+ Th17, and TNF-α+ T cells while IL-4+ Th2 cells were reduced. All data are expressed as means ± SD. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.005 by Mann–Whitney U test.
Discussion
Growing lines of evidence in humans and pre-clinical models of T1D indicate that the GB integrity is important to maintain immune homeostasis and prevent autoimmune diseases at the intestinal level but also in distal organs such as the pancreatic islets. In line with this idea, increased intestinal permeability precedes the onset of clinical T1D both in humans7 and animal models,5,41,42 thus suggesting that the GB damage is mechanistically linked to the autoimmune pathogenesis of T1D. In human T1D, signs of intestinal inflammation and damage of the GB integrity and, specifically, of the intestinal epithelial barrier have been largely documented.24,43, 44, 45 For example, early studies reported an increased gut permeability measured through the lactulose/mannitol test in T1D patients but also in prediabetic individuals with beta cell autoimmunity but no hyperglycemia.7 Breakage of the GB integrity was confirmed by a study that detected increased zonulin levels in the sera of 50% of T1D patients and 25% of their first-degree relatives.8 Zonulin is an endogenous protein that modulates the intestinal permeability by diassembling the intercellular tight junctions, hence it is considered a biomarker of damage of IEB integrity when found in serum.46 Here, we confirmed damage of the IEB with increased zonulin level in the sera of our T1D cohort, however we did not find a defective mRNA expression of structural IEB proteins such as the barrier forming TJPs CLN1, OCLN, ZO-1 and ZO-2. The mucus layer that is fundamental to maintain immune homeostasis and is altered in preclinical models of autoimmune diabetes was never studied in human T1D. Loss of GB integrity in the NOD mice, the spontaneous preclinical model of T1D, specifically affects the mucus layer more than the IEB.5 Here, in the intestinal tissue of T1D patients we detected a significant reduction of mRNA expression levels of several mucins including the gel forming MUC2 that is fundamental for maintenance of the mucus layer structure as well as transmembrane mucins MUC12, MUC13, MUC15, MUC20 and MUC21 that regulate the colonization of the mucus layer by beneficial commensal microbial species.16 Together with reduction of mucins mRNA expression we also found reduced mRNA expression of AMPs HD5 and HD6 in T1D patients indicating that the overall biological function of the mucus layer including anti-microbial activity is altered in our T1D cohort. Our data are aligned with previous literature showing a reduction of two AMPs, the human cathelicidin AMP (CAMP) and human beta defensin 1 (BD1), in serum of T1D patients.47 We could not validate our RT-qPCR data by qualitative analysis ex-vivo of the mucus layer structure as previously done in preclinical models of T1D5 because collection of intestinal tissue during the EGDS procedure destroys most of the mucus layer and does not allow proper visualization of its structure. However, our data on reduced mucins and AMPs mRNA expression suggest that abnormalities of the mucus layer composition previously found in preclinical models are also present in T1D patients.
The alteration of the mucus layer composition in T1D patients can be related to genetic or environmental factors. In fact, the integrity of the mucus layer and release of mucins and AMP is regulated by gut microbiota as suggested by several observations. For example, germ-free mice have an altered mucus layer that is normalized by colonization with murine commensal microbiota.48 Bacterial glycosidases released by mucus-residing bacteria degrade mucin glycan first and then attack the mucin protein backbone and are fundamental for mucus regeneration and integrity. In parallel, some mucus-residing bacteria such as Clostridium butyricum and Roseburia intestinalis release glycotransferases that are fundamental to modulate the glycosylation profile of the mucus layer. Mucus-residing bacteria also produce key metabolites such as SCFAs that are crucial to promote mucin release and maintain mucus layer integrity.49,50 For example, the SCFA butyrate upregulates MUC2 expression and secretion by GC, increases glycosylation of mucus and induces assembly of tight junctions.51 Hence, alterations in the mucus-residing and, specifically, SCFA-producing bacteria that we found in the gut mucosa of T1D patients and were previously detected also in fecal samples of T1D patients38,52, 53, 54 could explain the defective GC function, reduced expression of MUC2 and transmembrane mucins and disassembly of tight junctions (with increased serological levels of zonulin) that we detected in our T1D patient cohort.
How does the mucus layer alteration affect T1D pathogenesis? The monosaccharides generated by mucin degradation and released are used by the bacterial metabolism to generate SCFAs, metabolites that play important immune regulatory functions. SCFAs that diffuse back to the intestinal mucosa tissue change the metabolic environment and not only affect IEB integrity and regulate mucin expression but also promote immune tolerance at the intestinal level and in organs distal from the intestine such as the pancreatic islets. In fact, previous studies in humans and preclinical models of T1D have shown that a deficiency in SCFA production by the gut microbiota is associated with occurrence of T1D.54,55 Furthermore, oral administration of two SCFA, acetate and butyrate, prevented autoimmune diabetes in NOD mice.56 SCFA induce immune tolerance mechanism and, specifically, FoxP3+ Treg cell differentiation in the gut mucosa.57 Hence, the alterations of the mucus layer and reduced relative abundance of SCFA-producing bacteria in the MAGM of T1D patients could affect immune regulatory mechanism that are crucial to prevent beta cell autoimmunity. In support to the latter hypothesis, a reduction of intestinal differentiation of FoxP3+ Treg cells has been found in T1D patients.29 An alternative mechanism through which mucus barrier alteration could favor T1D pathogenesis is by allowing bacterial translocation and passage of bacterial components in the gut mucosa and systemic circulation. The mucus layer that is mainly composed of MUC2, a mucin whose expression we found dysregulated in T1D patients, normally keeps the commensal bacteria at bay, but when this first defense line fails commensal bacteria come in contact with the epithelium. Once an overwhelming quantity of bacteria reach the immune cells of the lamina propria an overt inflammatory reaction is triggered and is followed by bacterial translocation i.e., uncontrolled passage of bacterial molecules/antigens into the bloodstream and peripheral tissues. These events normally take place in the early pathogenetic events occurring in IBD,16 but our findings of increased serological levels of MD2 in T1D patients suggest that bacterial translocation can also take place in T1D. Although a direct causal link between bacterial translocation and human T1D is yet to be demonstrated, we speculate that passage of bacterial components into the blood circulation can modulate autoimmunity within pancreatic islets with two mechanisms: 1) microbial components, e.g., microbiota-secreted metabolites, can reach the pancreatic tissues and act directly on pancreatic insulin-producing β-cells either inducing protection58 or inflammation and cell damage. 2) The abnormal crosstalk between commensal microbiota and immune cells could lead to activation of islet-reactive T cells and their acquisition of an effector phenotype within the intestinal mucosa. In support to the latter hypothesis, studies in preclinical models of T1D found that commensal bacteria that enter the gut mucosa can stimulate directly islet-reactive T cells through mechanisms of molecular mimicry.5,6,9,10,59 In parallel, bacterial products can activate intestinal dendritic cells within the gut mucosa through pattern recognition receptors60 and drive them towards a pro-inflammatory phenotype that favor differentiation of effector T cells. In line with this view, we found increased effector Th1, Th17 and TNF-α+ T cells in the intestinal mucosa of T1D patients. Although direct evidence of intestinal activation and acquisition of an effector phenotype by islet-reactive T cells exists only in animal models, the observation that T cell infiltrating human pancreatic islets of T1D patients originate in the gut (i.e., express gut homing receptors CCR9 and α4β7) support this pathogenic scenario also in human T1D.61
Together with increased relative percentages of effector T cells in the intestine of T1D patients we also found reduced presence of Th2 cells. Since the release of mucins by intestinal GC is regulated by Th2-type cytokines like IL-4 and IL-13,39,40 we hypothesize that the reduction in the relative percentage of Th2 cells in the intestinal mucosa of T1D patients may contribute to defective GC function and alteration of the mucus layer.
In individuals genetically “at-risk” for T1D several mechanisms including self-antigen presentation by specific HLA alleles, defective thymic selection and failure of peripheral tolerance could lead to the presence of islet-reactive T cells in the peripheral circulation. In most cases those T cells remain quiescent and do not trigger beta cell autoimmunity and T1D unless there are activated in the periphery. Studies in animal models showed that loss of GB integrity and an abnormal crosstalk between the commensal gut microbiota and the immune system leads to peripheral activation of islet-reactive T cells and T1D.5 Our study has several limitations since it was performed in a small cohort of T1D patients exclusively composed of Caucasian individuals living in the Lombardy region and following an italian (i.e., Mediterranean) diet so they only partially represent the overall T1D population. In fact, the gut microbiota and the intestinal environment are largely affected by environmental factors such as diet but also pollution, exercise and other lifestyle and geographic conditions. However, in spite of those limitations and the correlative nature of our data, the results presented here suggest that GB integrity and the function of the mucus barrier maybe important to prevent pathogenic mechanisms leading to autoimmune destruction of pancreatic islets also in humans. Further studies are necessary to confirm these data on different T1D cohorts and to assess whether the alterations of the physical and biological GBs observed in human T1D are directly responsible for activation of beta cell autoimmunity through mechanisms similar to those reported in animal models.5,6 Also, proof-of-concept clinical studies with probiotics and/or dietary interventions in children with islet autoimmunity or genetically at risk for T1D aimed at increasing the relative abundance of SCFA-producing bacteria that we found reduced in T1D patients will be instrumental to link GB integrity with occurrence of T1D in humans.
Contributors
MLC: data curation, formal analysis, investigation, methodology, visualization, writing-original draft. IC: investigation and methodology. RF: data curation, formal analysis, investigation and methodology. MAC: investigation, methodology and validation. AN: data curation, methodology and validation. VP: data curation and formal analysis. LM: data curation, formal analysis, validation and visualization. LAL: data curation, formal analysis and validation. WL: methodology. MR: formal analysis, investigation and methodology. ED: data curation. AMB: investigation. EP: data curation. MS: investigation and supervision. EB: supervision and resources. AF: conceptualisation, methodology and writing-review & editing. FU: conceptualisation and supervision. JD: conceptualisation, resources and supervision. NM: conceptualisation, methodology, supervision and writing-review & editing. MF: conceptualisation, data curation, formal analysis, funding acquisition, project administration, supervision, visualization, writing-original draft.
Data sharing statement
Sequencing data are deposited on the NCBI site (https://www.ncbi.nlm.nih.gov/) with the project's code PRJNA764177. Other data supporting our findings will be made available from the corresponding author on reasonable request.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We thank all the participants (T1D patients and healthy controls) who kindly agreed to donate intestinal mucosa samples for research purposes.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104567.
Appendix A. Supplementary data
References
- 1.Knip M., Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12(3):154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 2.Siljander H., Honkanen J., Knip M. Microbiome and type 1 diabetes. eBioMedicine. 2019;46:512–521. doi: 10.1016/j.ebiom.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaarala O., Atkinson M.A., Neu J. The "perfect storm" for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57(10):2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonini M., Lo Conte M., Sorini C., et al. How the interplay between the commensal microbiota, gut barrier integrity, and mucosal immunity regulates brain autoimmunity. Front Immunol. 2019;10:1937. doi: 10.3389/fimmu.2019.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorini C., Cosorich I., Conte M.L., et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc Natl Acad Sci U S A. 2019;116(30):15140–15149. doi: 10.1073/pnas.1814558116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai N., Peng J., Liu F., et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J Exp Med. 2016;213(10):2129–2146. doi: 10.1084/jem.20160526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosi E., Molteni L., Radaelli M.G., et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49(12):2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 8.Sapone A., de Magistris L., Pietzak M., et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes. 2006;55(5):1443–1449. doi: 10.2337/db05-1593. [DOI] [PubMed] [Google Scholar]
- 9.Hebbandi Nanjundappa R., Ronchi F., Wang J., et al. A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell. 2017;171(3):655–667.e17. doi: 10.1016/j.cell.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Pearson J.A., Kakabadse D., Davies J., et al. Altered gut microbiota activate and expand insulin B15-23-reactive CD8+ T cells. Diabetes. 2019;68(5):1002–1013. doi: 10.2337/db18-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chairatana P., Nolan E.M. Defensins, lectins, mucins, and secretory immunoglobulin A: microbe-binding biomolecules that contribute to mucosal immunity in the human gut. Crit Rev Biochem Mol Biol. 2017;52(1):45–56. doi: 10.1080/10409238.2016.1243654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo R.L., Hooper L.V. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12(7):503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker J., Rossen J.W., Buller H.A., et al. The MUC family: an obituary. Trends Biochem Sci. 2002;27(3):126–131. doi: 10.1016/s0968-0004(01)02052-7. [DOI] [PubMed] [Google Scholar]
- 14.McGuckin M.A., Linden S.K., Sutton P., et al. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9(4):265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 15.Hattrup C.L., Gendler S.J. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 16.Hansson G.C. Mucins and the microbiome. Annu Rev Biochem. 2020;89:769–793. doi: 10.1146/annurev-biochem-011520-105053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan M., Gentile M., Yeiser J.R., et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342(6157):447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raouf A.H., Tsai H.H., Parker N., et al. Sulphation of colonic and rectal mucin in inflammatory bowel disease: reduced sulphation of rectal mucus in ulcerative colitis. Clin Sci (Lond) 1992;83(5):623–626. doi: 10.1042/cs0830623. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y.S., Ho S.B. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12(5):319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto-Furusho J.K., Ascano-Gutierrez I., Furuzawa-Carballeda J., et al. Differential expression of MUC12, MUC16, and MUC20 in patients with active and remission ulcerative colitis. Mediat Inflamm. 2015;2015 doi: 10.1155/2015/659018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson K., Deng Z., Hou Y., et al. Regulation of the intestinal barrier function by host defense peptides. Front Vet Sci. 2015;2:57. doi: 10.3389/fvets.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loonen L.M., Stolte E.H., Jaklofsky M.T., et al. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7(4):939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- 23.Coretti L., Natale A., Cuomo M., et al. The interplay between defensins and microbiota in Crohn's disease. Mediat Inflamm. 2017;2017 doi: 10.1155/2017/8392523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carratu R., Secondulfo M., de Magistris L., et al. Altered intestinal permeability to mannitol in diabetes mellitus type I. J Pediatr Gastroenterol Nutr. 1999;28(3):264–269. doi: 10.1097/00005176-199903000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Westerholm-Ormio M., Vaarala O., Pihkala P., et al. Immunologic activity in the small intestinal mucosa of pediatric patients with type 1 diabetes. Diabetes. 2003;52(9):2287–2295. doi: 10.2337/diabetes.52.9.2287. [DOI] [PubMed] [Google Scholar]
- 26.Secondulfo M., Iafusco D., Carratu R., et al. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis. 2004;36(1):35–45. doi: 10.1016/j.dld.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 27.American Diabetes Association Professional Practice C 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2022. Diabetes Care. 2022;45(Suppl 1):S17–S38. doi: 10.2337/dc22-S002. [DOI] [PubMed] [Google Scholar]
- 28.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badami E., Sorini C., Coccia M., et al. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes. 2011;60(8):2120–2124. doi: 10.2337/db10-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kostic A.D., Gevers D., Siljander H., et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickham H., Hofmann H. Product plots. IEEE Trans Vis Comput Graph. 2011;17(12):2223–2230. doi: 10.1109/TVCG.2011.227. [DOI] [PubMed] [Google Scholar]
- 33.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lammers K.M., Lu R., Brownley J., et al. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor CXCR3. Gastroenterology. 2008;135(1):194–204.e3. doi: 10.1053/j.gastro.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park B.S., Song D.H., Kim H.M., et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 36.Kemppainen K.M., Ardissone A.N., Davis-Richardson A.G., et al. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38(2):329–332. doi: 10.2337/dc14-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi C.J., Zhang Q., Yu M., et al. Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in Chinese children. Chin Med J. 2016;129(11):1298–1304. doi: 10.4103/0366-6999.182841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leiva-Gea I., Sanchez-Alcoholado L., Martin-Tejedor B., et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care. 2018;41(11):2385–2395. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 39.Dabbagh K., Takeyama K., Lee H.M., et al. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J Immunol. 1999;162(10):6233–6237. [PubMed] [Google Scholar]
- 40.Marillier R.G., Michels C., Smith E.M., et al. IL-4/IL-13 independent goblet cell hyperplasia in experimental helminth infections. BMC Immunol. 2008;9:11. doi: 10.1186/1471-2172-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meddings J.B., Jarand J., Urbanski S.J., et al. Increased gastrointestinal permeability is an early lesion in the spontaneously diabetic BB rat. Am J Physiol. 1999;276(4 Pt 1):G951–G957. doi: 10.1152/ajpgi.1999.276.4.G951. [DOI] [PubMed] [Google Scholar]
- 42.Neu J., Reverte C.M., Mackey A.D., et al. Changes in intestinal morphology and permeability in the biobreeding rat before the onset of type 1 diabetes. J Pediatr Gastroenterol Nutr. 2005;40(5):589–595. doi: 10.1097/01.mpg.0000159636.19346.c1. [DOI] [PubMed] [Google Scholar]
- 43.Mooradian A.D., Morley J.E., Levine A.S., et al. Abnormal intestinal permeability to sugars in diabetes mellitus. Diabetologia. 1986;29(4):221–224. doi: 10.1007/BF00454879. [DOI] [PubMed] [Google Scholar]
- 44.Maffeis C., Martina A., Corradi M., et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab Res Rev. 2016;32(7):700–709. doi: 10.1002/dmrr.2790. [DOI] [PubMed] [Google Scholar]
- 45.Gavin P.G., Mullaney J.A., Loo D., et al. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care. 2018;41(10):2178–2186. doi: 10.2337/dc18-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fasano A., Not T., Wang W., et al. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet. 2000;355(9214):1518–1519. doi: 10.1016/S0140-6736(00)02169-3. [DOI] [PubMed] [Google Scholar]
- 47.Brauner H., Luthje P., Grunler J., et al. Markers of innate immune activity in patients with type 1 and type 2 diabetes mellitus and the effect of the anti-oxidant coenzyme Q10 on inflammatory activity. Clin Exp Immunol. 2014;177(2):478–482. doi: 10.1111/cei.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson M.E., Jakobsson H.E., Holmen-Larsson J., et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18(5):582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Da Silva S., Robbe-Masselot C., Ait-Belgnaoui A., et al. Stress disrupts intestinal mucus barrier in rats via mucin O-glycosylation shift: prevention by a probiotic treatment. Am J Physiol Gastrointest Liver Physiol. 2014;307(4):G420–G429. doi: 10.1152/ajpgi.00290.2013. [DOI] [PubMed] [Google Scholar]
- 50.Lili Q., Xiaohui L., Haiguang M., et al. Clostridium butyricum induces the production and glycosylation of mucins in HT-29 cells. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.668766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng L., Li Z.R., Green R.S., et al. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139(9):1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart C.J., Nelson A., Campbell M.D., et al. Gut microbiota of Type 1 diabetes patients with good glycaemic control and high physical fitness is similar to people without diabetes: an observational study. Diabet Med. 2017;34(1):127–134. doi: 10.1111/dme.13140. [DOI] [PubMed] [Google Scholar]
- 53.Cinek O., Kramna L., Mazankova K., et al. The bacteriome at the onset of type 1 diabetes: a study from four geographically distant African and Asian countries. Diabetes Res Clin Pract. 2018;144:51–62. doi: 10.1016/j.diabres.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 54.Vatanen T., Franzosa E.A., Schwager R., et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Groot P.F., Belzer C., Aydin O., et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One. 2017;12(12) doi: 10.1371/journal.pone.0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marino E., Richards J.L., McLeod K.H., et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18(5):552–562. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 57.Arpaia N., Campbell C., Fan X., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun J., Furio L., Mecheri R., et al. Pancreatic beta-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. 2015;43(2):304–317. doi: 10.1016/j.immuni.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 59.Garabatos N., Santamaria P. Gut microbial antigenic mimicry in autoimmunity. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.873607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costa F.R., Francozo M.C., de Oliveira G.G., et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J Exp Med. 2016;213(7):1223–1239. doi: 10.1084/jem.20150744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paronen J., Klemetti P., Kantele J.M., et al. Glutamate decarboxylase-reactive peripheral blood lymphocytes from patients with IDDM express gut-specific homing receptor alpha4beta7-integrin. Diabetes. 1997;46(4):583–588. doi: 10.2337/diab.46.4.583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.