Summary
Background
Reprogramming of immunosuppressive tumor-associated macrophages (TAMs) presents an attractive therapeutic strategy in cancer. The aim of this study was to explore the role of macrophage CD5L protein in TAM activity and assess its potential as a therapeutic target.
Methods
Monoclonal antibodies (mAbs) against recombinant CD5L were raised by subcutaneous immunization of BALB/c mice. Peripheral blood monocytes were isolated from healthy donors and stimulated with IFN/LPS, IL4, IL10, and conditioned medium (CM) from different cancer cell lines in the presence of anti-CD5L mAb or controls. Subsequently, phenotypic markers, including CD5L, were quantified by flow cytometry, IF and RT-qPCR. Macrophage CD5L protein expression was studied in 55 human papillary lung adenocarcinoma (PAC) samples by IHC and IF. Anti-CD5L mAb and isotype control were administered intraperitoneally into a syngeneic Lewis Lung Carcinoma mouse model and tumor growth was measured. Tumor microenvironment (TME) changes were determined by flow cytometry, IHC, IF, Luminex, RNAseq and RT-qPCR.
Findings
Cancer cell lines CM induced an immunosuppressive phenotype (increase in CD163, CD206, MERTK, VEGF and CD5L) in cultured macrophages. Accordingly, high TAM expression of CD5L in PAC was associated with poor patient outcome (Log-rank (Mantel–Cox) test p = 0.02). We raised a new anti-CD5L mAb that blocked the immunosuppressive phenotype of macrophages in vitro. Its administration in vivo inhibited tumor progression of lung cancer by altering the intratumoral myeloid cell population profile and CD4+ T-cell exhaustion phenotype, thereby significantly modifying the TME and increasing the inflammatory milieu.
Interpretation
CD5L protein plays a key function in modulating the activity of macrophages and their interactions within the TME, which supports its role as a therapeutic target in cancer immunotherapy.
Funding
For a full list of funding bodies, please see the Acknowledgements.
Keywords: CD5L, Immunotherapy, Lung adenocarcinoma, Macrophage, Monoclonal antibody, Scavenger receptor cysteine rich
Research in context.
Evidence before this study
Immunotherapy is aimed at reactivating the immune system to mount a robust antitumoral response. However, although successful in certain cases, current immunotherapy treatments need improvement. Tumor-associated macrophages (TAMs) are the most abundant immune cells in tumor. Because they are immunosuppressive in most solid tumors, TAMs rise as a remarkably promising cell target in immuno-oncology.
Added value of this study
In this study we report that secreted factors from cancer derived cell lines induced an immunosuppressive profile and enhanced CD5L expression in macrophages. Additionally, TAM expression of the protein CD5L in lung adenocarcinoma human specimens correlated with poor outcomes. We raised a new antibody that specifically binds to CD5L and blocks the immunosuppressive state of macrophages. Administration of this antibody in a mouse model of cancer modified the TME thereby reducing tumor growth.
Implications of all the available evidence
Our results point to CD5L as an immunotherapy target for the treatment of lung cancer that may have an important impact in many solid tumors.
Introduction
The tumor microenvironment (TME) is a complex system comprised of many cell types, including endothelial cells, smooth muscle cells, fibroblasts, and immune cells such as macrophages, dendritic cells (DCs), and T lymphocytes.1 Given the critical role of the TME in modulating tumor progression and response to treatment, therapeutic strategies targeting this environment have recently emerged as a promising approach to tackle cancer.2
Although immune infiltrates vary depending on the type of cancer, numerous studies have demonstrated that tumor-associated macrophages (TAMs) are the major immune component of the tumor stroma.3 Macrophages are plastic cells that can shift their functional phenotypes in response to various microenvironmental signals, including those generated by tumor and stromal cells.4 In this context and depending on their activation states, macrophages can exert pro- and anti-tumor functions. In most established progressing mouse and human tumors, TAMs usually display an immunosuppressive phenotype, geared toward promoting tumor growth and invasiveness directly and via angiogenesis, tissue remodeling, production of specific cytokines/chemokines, and suppression of adaptive immunity.5, 6, 7, 8, 9 The abundance of these cells correlates negatively with survival outcome in several types of solid cancer, including that of the lung10 and liver.8,11
Over recent decades, significant advances in patient treatment options and outcomes have been achieved in oncology.12 Specifically, immunotherapy, including monotherapy and combination therapies, has emerged as the first-line treatment for many types of tumors.13 However, the proportion of cancer patients that can benefit from their clinical application is still limited. Thus, intense research efforts are underway to identify other immune-modulatory strategies for cancer. Selective targeting and re-education of immunosuppressive TAMs in the TME is an attractive option that is currently being intensively studied.14, 15, 16
In recent years, the contribution of Scavenger Receptors (SRs) to macrophage activity in the context of cancer has received increasing attention.17,18 SRs form a large family of proteins predominantly expressed by myeloid cells. They are structurally diverse and participate in a wide range of biological functions, including endocytosis, phagocytosis, adhesion, and signaling, all of which ultimately lead to the elimination of degraded or harmful substances.18 Interestingly, the expression of SRs containing cysteine-rich domains (termed SRCR proteins) such as CD163, SR-A1 and Macrophage Receptor With Collagenous Structure (MARCO) is markedly increased in immunosuppressive macrophages.19 Indeed, CD163 is a widely recognized marker of this phenotype in humans, and its expression is related to poor prognosis in many types of cancer, including breast cancer and lung adenocarcinoma.20,21 In addition, reprogramming TAMs by targeting CD163 and MARCO has been shown to inhibit cancer progression in mouse models of melanoma, glioma, and colon cancer.22, 23, 24
CD5-like (CD5L) is a 40-kDa soluble glycoprotein that belongs to the SRCR superfamily. This protein modulates the activity of macrophages in a wide spectrum of settings, including the pathogenesis of several infectious and inflammatory processes.25,26 Also, we described that CD5L promotes macrophage polarization towards an immunosuppressive phenotype through enhanced autophagy in vitro.17 However, its specific role in the context of TME macrophages remains unknown.
In the present study, we hypothesized that CD5L is a key player in macrophage responses in the context of cancer. We found that CD5L is expressed in TAMs in human specimens of papillary lung adenocarcinoma (PAC) and its increased levels are associated with poor prognosis. We then generated a CD5L-blocking monoclonal antibody (mAb), that when administered in a syngeneic mouse model of lung cancer reduced tumor growth, reprogrammed TAMs, and shifted the anti-inflammatory and pro-tumorigenic TME towards a less tumor-permissive environment. In summary, our results point to CD5L as a target for cancer immunotherapy.
Methods
Ethics approval and consent to participate
All human studies were conducted following the Declaration of Helsinki principles and current legislation on the confidentiality of personal data, and were approved by the Human Ethics Committee of the Germans Trias i Pujol University Hospital (with codes PI-20-049 and PI-20-085). Informed consent from all participants (patients providing tumor tissues) was obtained.
Buffy coats, provided by the Blood and Tissue Bank (Barcelona, Spain), were obtained from healthy blood donors following the institutional standard operating procedures for blood donation and processing, including informed consent.
Animal care and treatment were carried out in accordance with Spanish and EU laws. Protocols requiring animal manipulation were approved by the institutional Animal Experimentation Ethics Committee of the Comparative Medicine and Bioimage Centre of Catalonia (CEEA-CMCiB) (#9455), as well as by animal care and use committees of the Parc Científic de Barcelona (#9672), Universitat de Barcelona (#7088), and the Government of Catalonia (CEA-OH/10300/1).
Human specimens of PAC
Human specimens of PAC were provided by Dr. Masaoki Ito. The samples were obtained at the time of surgery and were examined and characterized by expert pathologists.
Generation and characterization of RImAb monoclonal anti-CD5L antibody
The generation of murine mAbs against human CD5L in BALB/c mice was described recently.27 The CSIC Ethics Committee and the Agriculture Department (Community of Madrid) approved the use of experimental animals PROEX 18/14 by Dr. Leonor Kremer for mAb generation. The 8C4G4D7 mAb was a clone specifically selected from a previous hybridoma collection that we generated by immunizing mice against human CD5L,27 was named RImAb and was produced in culture supernatants and purified by affinity chromatography using Protein G-Sepharose (29-0485-81, GE Healthcare, Chicago, IL, USA). The immunoglobulin subclass was determined by ELISA using specific peroxidase-conjugated antibodies against the heavy (IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM) and light (kappa and lambda) chains of mouse immunoglobulins, following the manufacturer's instructions (37503, Thermo Fisher Scientific, Waltham, MA, USA).
Direct binding enzyme-linked immunosorbent assay (ELISA) was performed by immobilizing 5 μg/ml of recombinant human CD5L (rhCD5L),27 recombinant mouse CD5L (rmCD5L) (2834-CL-050, R&D systems, Minneapolis, MN, USA), human DMBT1 (kindly provided by Dr. Martina Tuttolomondo), and human recombinant CD5 (1636-CD-050, R&D systems) in 96-well microtiter plates, overnight at 4 °C. Plates were blocked with PBS containing 5% Bovine serum albumin (BSA) (A4503, Sigma–Aldrich, Saint Louis, MO, USA) for 1 h at room temperature (RT). The mAb was then added to the wells at the indicated concentrations and incubated for 1 h at RT. Between each step, the plates were washed twice with PBS 0.01% Tween-20 to remove unbound proteins. For bound antibody detection, a 1:5000 dilution of peroxidase (PO)-labeled anti-mouse IgG antibody (A4416, Sigma–Aldrich, RRID: AB_258167) was added at the final step and incubated for 30 min at RT. Unbound antibody was washed three times with PBS 0.01% Tween-20. Yellow color was developed by adding 3,30,5,50-tetramethylbenzidine liquid substrate (T8665, Sigma–Aldrich) for 20 min, after which the reaction was stopped by the addition of 2 M H2SO4, and the optical density was read at 450 nm using a Varioskan Flash microplate reader (Thermo Fisher Scientific). Each assay was repeated three times, with similar results.
Characterization of protein sequence identity and similarity between human CD5L and human DMBT1, human CD5, and mouse CD5L was calculated using the UniProtKB database (NIH, EMBL-EBI), BlastP (NIH) and SIAS (Sequence Identity and Similarity software, Universidad Complutense de Madrid).
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded (FFPE) 4 μm tissue sections were used for the analysis. IHC staining was performed by standard protocol: the sections were deparaffinized with xylene, followed by rehydration in graded alcohol concentration. Endogenous peroxidase blocking was performed with 0.3% H2O2 in ethanol during the rehydration. Heat-induced antigen retrieval (HIER) was performed with autoclave using Citrate buffer (pH 6, 10×, C-9999, Sigma–Aldrich) at 121 °C for 10 min, followed by permeabilization with 0.2% Tween in PBS for 10 min. The slides were later incubated with primary antibodies dissolved in blocking buffer (3% BSA (A4503-504, Sigma–Aldrich)) +5% donkey/goat serum, (S30 and S26 respectively, Millipore, Merck, Burlington, MA, USA) overnight at 4 °C. For human samples, IHC analysis was performed using anti-CD5L antibody (1:100) (HPA068384, Sigma–Aldrich, RRID: AB_2685979) and goat anti-rabbit IgG coupled to horseradish peroxidase (1:200) (HAF008, R&D Systems, RRID: AB_357235) as secondary antibody. For mouse samples, a panel of primary antibodies, namely anti-F4/80 (1:100) (AB100790, RRID: AB_10675322), anti-TNF-α (1:100) (AB6671, RRID: AB_305641), anti-MPO (1:500) (AB45977, RRID: AB_944318), anti-Arg-1 (1:200) (AB92274, RRID: AB_10563668), anti-iNOS (1:100) (AB15323, RRID: AB_301857), all from Abcam (Cambridge, UK), anti-CD31 (1:100) (NB100-2284, Novus Biologicals, RRID: AB_577761) and anti-BCL2 (1:100) (JF104-8, Thermo Fisher Scientific, RRID: AB_2898964), were used with goat anti-rabbit IgG-HRP (1:200) (AB6721, Abcam, RRID:AB_955447) and donkey anti-goat IgG-HRP (1:500) (AB_2340390, Jackson ImmunoResearch, RRID: AB_2340390) as secondary antibodies (30 min incubation). Chromogenic detection of antibody binding was performed using a DAB substrate kit (34002, Thermo Fisher Scientific) for 20 min, protected from light. Counterstaining was done with hematoxylin (1051740500, HX 86017674, Merck). The slides were later mounted with DPX (DPX 06522, Sigma–Aldrich), dried, and observed under a Leica DMI 6000 B microscope (Leica Microsystems, Wetzlar, Germany).
Microscope Koehler illumination was set up to achieve the best image resolution and image quality, as recommended by Leica microsystems. The focused areas were subjected to contrast and brightness adjustments using Leica software and once the optimum conditions were achieved, the images were acquired. For quantification of human samples, images from five random areas were taken. For statistical analysis, mean of CD5L positive cells was taken for each sample. Inclusion and exclusion criteria for the characterization of human CD5L positive macrophages were designed under careful supervision of a pathologist. For mice samples, the captured images were analyzed using Image J, color deconvolution 2 plugin. H DAB was chosen as the vector with an image output of 8 bits. The DAB channel was selected and the threshold was adjusted against a white background to the original image, considering the background and non-specific staining, followed by measurement of stained area percentage. A double blinded analysis was performed by two individuals and the mean value obtained from both evaluators was considered for final analysis. Statistical analysis was performed using GraphPad Prism V9.
Immunofluorescence (IF) of FFPE tissue sections
For IF analysis, 4 μm FFPE tissue sections were used and processed as per the optimized protocol. Human samples were studied using the primary antibodies anti-CD5L (1:100) (HPA068384, Sigma–Aldrich, RRID: AB_2685979) and anti-CD68 (1:100) (Kp-1, 168M-95, Cell Marque, RRID: AB_1158188). For mouse samples, anti-CD5L (1:100) (AF2834, R&D systems, RRID: AB_2076369) and anti-F4/80 (1:100) (AB100790, Abcam, RRID: AB_10675322) were used. Depending on the primary antibody, donkey anti-rabbit (A-21206, RRID: AB_2535792), mouse (A-31570, RRID: AB_2536180), or goat IgG coupled with Alexa-488 and Alexa-555 (A-21432, RRID: AB_2535853), all from Thermo Fisher Scientific, were used as secondary antibodies at a dilution of 1:100. Finally, nuclei were stained with PBS containing Hoechst 33258 (1:20000, 94403, Sigma–Aldrich) for 10 min at RT and observed under the microscope (Leica DMI 6000 B). For IHC and IF, the samples were visualized under 40× magnification and a minimum of five random areas were chosen per sample. The acquired images were analyzed using Image J. A multichannel composite image was created using channel tools, separating the three channels (red, green and blue) respectively. Color balance for the individual channel was performed to adjust the brightness and contrast, eliminating possible background and non-specific staining. The split composite images were later merged for a uniform image with which further analysis and quantification was performed. Statistical analysis was performed using GraphPad Prism V9.
Cell culture
Peripheral blood mononuclear cells (PBMCs) were isolated from leukocyte concentrates from healthy blood donors, as previously described,28 by Ficoll–Paque (17-1440, GE Healthcare) density gradient centrifugation at 400g for 20 min after CD3+ cell depletion by RosetteSep human CD3 depletion cocktail (15621, StemCell Technologies, Vancouver, BC, Canada). Recovered cells were washed twice in PBS and counted using a mouse anti-human CD14 antibody (555397, BD Bioscience, RRID: AB_395798) and Perfect-Count microspheres (CYT-PCM Cytognos, Salamanca, Spain), following the manufacturer's instructions. 5 × 105 or 1 × 106 CD14+ cells per well were seeded in 24- or 6-well plates, respectively, and peripheral blood monocytes (PB monocytes) were isolated by adherence in RPMI supplemented with 10% human AB serum (H4522, Sigma–Aldrich), 1% of 10000 U/ml penicillin and 10 mg/ml streptomycin (P/S; P0781, Sigma–Aldrich) in a 5% CO2 incubator at 37 °C for 30 min. Non-adherent cells were removed, and adherent cells were washed twice with PBS and incubated in RPMI supplemented with 10% heat-inactivated fetal bovine serum (FBS) (DE14-840E, Lonza, Basel, Switzerland) and 1% P/S for 24 h before the experiments. The percentage of adherent CD14+ cells (PB monocytes) routinely obtained was 94.98% ( ± 3.26%).
In assays performed with human monocyte-derived macrophages (HMDMs), PB monocytes were differentiated by incubation in RPMI supplemented with 10% heat-inactivated FBS and 1% P/S for 7 days before the experiments, as previously described.29
HepG2 (RRID: CVCL_0027) and SNU398 (RRID: CVCL_0077) cells were purchased from ATCC (The American Type Culture Collection; Manassas, VA, USA) and Huh7 (RRID: CVCL_0336) cells were purchased from RIKEN BRC Cell Bank. These cell lines were cultured in EMEM (HepG2), DMEM (Huh7) or RPMI (SNU398), supplemented with 1% P/S and 10% heat-inactivated FBS. A459 and PC9 cells were kindly provided by Dr. Rafael Rosell (ICO, IGTP), and were cultured in RPMI supplemented with 1% P/S and 10% heat-inactivated FBS. All human cell lines have been identified and validated using the AmpFLSTR™ Identifiler™ Plus PCR Amplification Kit (A26364, Thermo Fisher Scientific) and the GeneMapper v3.2 software (RRID:SCR_014290). The Lewis Lung Carcinoma (LLC) cell line, 3LL-R, was kindly provided by Dr. Jo van Ginderachter (VIB, Belgium),30 and cultured in RPMI supplemented with 1% P/S, 10% FBS, and 1% Glutamax (13462629, Thermo Fisher Scientific). Characterization of the cell line was performed in.31 This cell line has been used for tumor growth experiments within 15 passages after thawing. Mycoplasma testing was performed and all cell lines tested negative.
For the collection of cancer cell-conditioned media (CM), cells were grown to 90% confluency, then washed with PBS, and the medium was replaced with medium containing 2% FBS. Twenty-four hours later, the supernatant was collected and centrifuged at 10000 rpm for 10 min at 4 °C to remove cellular debris. It was then aliquoted and stored at −80 °C for subsequent experiments, where it was diluted 1:2 with media containing 10% FBS.
In vitro activation of macrophages
PB monocytes were activated with IFN/LPS, 50 ng/ml endotoxin-free IFNγ (Peprotech, 300-02-A, Rocky Hill, NJ, USA) plus 100 ng/ml LPS from E. coli O111:B4 (14391, Sigma–Aldrich); IL4, 40 ng/ml endotoxin-free IL4 (200-04-A, Peprotech); IL10, 50 ng/ml endotoxin-free IL10 (200-10-A, Peprotech) as previously described17; and CM collected from Huh7, HepG2, SNU398, A459, and PC9 cells. Macrophages cultured in growth medium were referred to as controls. Some of the reference stimuli data have been published.17 To assess the role of CD5L blockade on IL10-induced activation, RImAb was added at a final concentration of 5 μg/ml 45 min before IL10 addition to the culture.
Macrophage activation markers by flow cytometry
Flow cytometry analysis of macrophage activation markers was performed as previously described.17 Briefly, PB monocytes were cultured in 6-well plates (106 cells/well) in RPMI medium containing 5% FBS and the indicated stimuli. After 72 h, macrophages were detached with Accutase (Sigma–Aldrich), washed twice in PBS, and incubated with 50 μl of PBS containing 10% human AB serum, 2% FBS, and 0.02% NaN3 (blocking buffer) for 30 min on ice. Cells were then incubated with a combination of fluorescently conjugated anti-human HLA-DR-FITC (555811, RRID: AB_396145), CD80-PE-Cy7 (561135, RRID: AB_10561688), CD206-PE-CF594 (564063, RRID: AB_2732052), CD23-APC (558690, RRID: AB_1645456), and CD163-BV711 (563889, RRID: AB_2738469) mAbs, all from BD Biosciences, for 20 min in Brilliant stain buffer (563794, BD Biosciences) on ice. They were then rinsed with washing buffer (PBS containing 2% FBS and 0.02% NaN3) and fixed with 1% paraformaldehyde. Flow cytometric acquisition was performed on a BD LSR Fortessa instrument using FACSDiva software (BD Biosciences), with 10000 events acquired for each sample. Data analysis was performed using FlowJoTM 10.8.1 software (BD Biosciences, RRID: SCR_008520).
Fluorescence microscopy studies on activated macrophages
Fluorescence studies were performed as previously described17,28: HMDMs (105 cells/well) were plated and incubated with the indicated stimuli for 72 h on Millicell EZ slides (PEZGS0816, Merck). Cells were fixed with PBS containing 4% paraformaldehyde (141451, Panreac), blocked with PBS containing 0.2% Triton X-100 (X100, Sigma–Aldrich), 5% FBS, and 5% human AB serum (blocking buffer), and incubated with 5 μg/ml RImAb in blocking buffer overnight at 4 °C. Cells were subsequently incubated with FITC-anti-mouse IgG/IgM antibody (555988, BD Biosciences, RRID: AB_396275) for 1 h at RT in blocking buffer. Between steps, unbound antibodies were removed with 3 washes with PBS. Finally, nuclei were stained with PBS containing Hoechst 33258 diluted 1:20000 (94403, Sigma–Aldrich) for 10 min at RT. Cells were then washed 5 times with PBS, and coverslips were mounted in Fluoromount media (F4680, Sigma–Aldrich) and stored at 4 °C.
The slides were examined under an Axio Observer Z1 DUO LSM 710 confocal system (Carl Zeiss Microscopy GmbH, Jena, Germany) and analyzed with ZEN Black software (Carl Zeiss Microscopy GmbH). CD5L fluorescence intensity was quantified using ZenLite software (Carl Zeiss Microscopy GmbH).
Syngeneic mouse model
Wild-type C57BL/6 mice (Envigo, Indianapolis, IN, USA) (RRID:MGI:5658887) were injected with 3 × 106 LLC cells (3LL-R) in PBS subcutaneously in the right dorsal flanks, as described previously.30 Tumors were allowed to grow for 7 days, and then 150 μg of RImAb in PBS was administered intraperitoneally (i.p.) or an equal volume of PBS or IgG2a isotype (BE0085, BioXcell, RRID: AB_1107771) was used as a control (n = 8 mice/group), every three days. The number of animals was calculated as follows: accepting an alpha risk of 0.05 and a beta risk of 0.2 in a two-sided test, 8 subjects are necessary in each group to recognize as statistically significant a difference greater than or equal to 450 mm3. The common standard deviation was assumed to be 300 mm3. A drop-out rate of 10% was anticipated. Mice were randomly allocated to the experimental groups. Tumor size was measured manually with a caliper every day, and tumor volume was calculated using the formula V = π × (w2 × l)/6 (width, w, and length, l).32 Mice were euthanized at day 15. Tumors were excised, and blood was collected for further analyses. Serum was obtained after blood centrifugation at 2000g for 15 min and stored at −80 °C until analyzed.
Characterization of intratumoral immune cell populations by flow cytometry and cell sorting
Tumors were minced into small pieces and dissociated using the GentleMACS™ Dissociator (130-093-235 Miltenyi), following the manufacturer's instructions.
CD45+ cells were isolated by magnetic positive selection by using EasySep™ Mouse CD45 Positive Selection Kit (19945, StemCell). The manufacturer's instructions were followed. The CD45+ cell suspension was diluted to a concentration of 5 × 106 cells/ml in PBS and incubated first with Fc block (rat anti-mouse CD16/CD32, BD Biosciences) (1:100 dilution, 30 min, 4 °C). Cells were then incubated with a combination of fluorescently conjugated anti-mouse mAbs for cell surface phenotyping by flow cytometry. CD45-APC-CY7 (557659, BD Biosciences, RRID: AB_396774) was used to verify the purity of the leukocyte population.
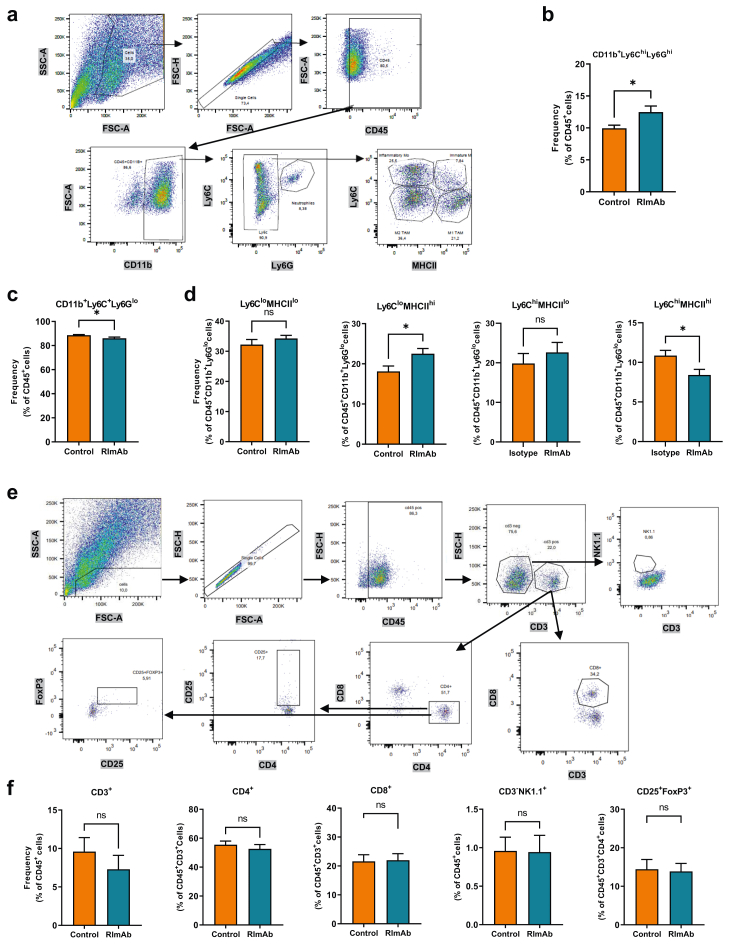
Myeloid cell lineage antibodies comprised CD11b-Alexa647 (557659, RRID: AB_396774), Ly6G-FITC (561105, RRID: AB_10562567), Ly6C-BV421 (562727, RRID: AB_2737748), and I-A/I-E-BV771 (563414, RRID: AB_2738191), all from BD Biosciences, while the following lymphoid cell lineage antibodies were used: anti-mouse CD3-BV650 (564378, RRID: AB_2738779), CD4-BB700 (566407, RRID: AB_2744427), CD8-V500 (560778, RRID: AB_1937329), CD25-PE (558642, RRID: AB_1645250), NK1·1-FITC (561082, RRID: AB_10563221), and FoxP3-Alexa647 (560401, RRID: AB_1645201), all from BD Biosciences. The cells for the lymphoid panel were then permeabilized and fixed using the eBioscience™ FoxP3 staining buffer set (00-5523-00, Invitrogen, Thermo Fisher Scientific), following the manufacturer's specifications. For T-cell memory and exhaustion characterization, the following antibodies were used: CD3-BV650, CD4-BB700, CD8-V500, CD44-FITC (561859, RRID: AB_10894581), CCR7-BV421 (562675, RRID: AB_2737716), LAG3-APC (562346, RRID: AB_11153127), TIGIT-PE (565168, RRID: AB_2739089), from BD Biosciences, and CTLA4-PECy7 (106313, RRID: AB_2564237), PD1-BV785 (135225, RRID: AB_2563680), and TIM3-BV711 (134021, RRID: AB_2890691) from Biolegend. Subsequently, cells were washed by centrifugation at 700g for 5 min, fixed in 100 μl of 1% paraformaldehyde, and stored at 4 °C for a maximum of 24 h. Data were collected using a BD LSR Fortessa instrument and the FACSDiva software (BD Biosciences), with a minimum of 10000 events acquired for each sample (see gating strategy in Fig. 5), and analyzed using FlowJo™ Software (version 10.8.1, BD Biosciences).
Fig. 5.
RImAb modifies the intratumoral myeloid cell compartment in a mouse model of LLC. a) Myeloid cell gating classification using the expression of surface markers CD45, CD11b, Ly6C, Ly6G, and MHCII. b) Percentage of CD45+CD11b+Ly6ChiLy6Ghi neutrophil population. c) Percentage of CD45+CD11b+Ly6C+Ly6Glo myeloid cell subset. d) Percentage of CD45+CD11b+Ly6CloMHCIIlo, CD45+CD11b+Ly6CloMHCIIhi, CD45+CD11b+Ly6ChiMHCIIlo and CD45+CD11b+Ly6ChiMHCIIhi myeloid cell subsets. e) Lymphoid cell gating classification using the expression of the CD45, CD3, CD8, CD4, CD25, NK1.1, and FoxP3 markers. f) Percentage of each lymphocytic subset population analyzed in control (isotype IgG2a) and RImAb-treated mice. Data are presented as the mean of ±SEM (n = 8 per group), and statistical comparisons were performed using the Mann–Whitney t-test.
To sort the CD45+CD11b+Ly6C+Ly6Glo cell population, we repeated the same procedure but cells were incubated only with the CD45-APC-CY7, CD11b-Alexa647, Ly6G-FITC, and Ly6C-BV421 antibodies and were resuspended in PBS-2% FBS after washing. Cells were immediately sorted using the FACSAriaII cell sorter (BD Biosciences) and collected in PBS-2%FBS. Pellets from these cells were frozen at −80 °C.
Analysis of chemokines by Luminex
For tumor sample preparation, 5 mg of each frozen tumor was submerged in 0.5 ml of PBS with the recommended concentration of complete Mini Protease Inhibitor Cocktail (11836153001, Roche) and then homogenized using a Bio-Gen™ PRO200® homogenizer for 30 s and centrifuged at 12000 g for 10 min at 4 °C to remove insoluble materials. The supernatants containing the tumor extracts were then quantified using the Pierce BCA Protein Assay Kit (23227, Thermo Fisher Scientific). Chemokine concentration was measured using the MILLIPLEX® Magnetic Bead Kit (MCYTOMAG-10K, Merck) and following the manufacturer's instructions. For tumor analysis, 10 μg of protein was analyzed, and PBS with protease inhibitor was used as a matrix. For serum analysis, 12.5 μL of serum diluted in assay buffer was added and serum matrix was used. The plate was read on a Luminex® 200™ instrument with xPONENT® software (Merck).
Clinical biochemistry analyses
Analyses for systemic toxicity assessment were performed by the Clinical Analysis Dept. Biochemistry at Hospital Germans Trias i Pujol (HuGTiP), Badalona, Spain. Serum samples obtained at the endpoint of the experiment (n = 4 per treatment group, half of each gender) were analyzed by spectrophotometry using an AU5800 analyzer (Beckman Coulter, Passadena, CA, USA).
Quantification of IgG2a in mouse serum by ELISA
5 μg/ml of capture Ab (anti-mouse IgG2a, 115-005-206, Jackson ImmunoResearch, RRID:AB_2338462) were coated onto a microtiter plate overnight at 4 °C. The plates were blocked for 1 h at RT with PBS containing 5% BSA (A4503, Sigma–Aldrich). Next, 50 μL of mouse serum diluted 1:2000 in PBS-5% BSA was added and incubated for 90 min at RT. Finally, 50 μL of HRP conjugated anti-mouse IgG (A3673, Sigma–Aldrich, RRID: AB_258099) diluted 1:5000 in PBS −5% BSA was added for 1 h at RT. Color was developed as explained above.
RNA extraction and RT-qPCR
RNA extraction from PB monocytes was performed as previously described.17 Briefly, 5 × 105 cells/well cultured in 24-well plates were incubated for 24 h in RPMI medium containing 5% FBS and the indicated stimuli. Cells were then washed with PBS and disrupted with TRIzol reagent (15596026, Thermo Fisher Scientific). Likewise, pellets from cells sorted by flow cytometry (CD45+CD11+Ly6C+Ly6Glo) were also disrupted with TRIzol reagent. For RNA extraction from frozen mouse tumors, tissue was submerged in lysis buffer with β-mercaptoethanol and disrupted mechanically using a PRO 200® homogenizer. In all cases, total RNA was extracted using the RNeasy Mini Kit (74106, Qiagen). The optional on-column DNase digestion using the RNase-Free DNase set (79254, Qiagen, Hilden, Germany) was performed only on the samples that were subsequently sequenced. RNA concentration was measured using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). All RNA samples were kept at −80 °C until the qPCR was performed. Total RNA (0.5–1 μg) was reverse transcribed using the RNA to cDNA EcoDry™ Premix (639549, Clontech, Mountain View, CA, USA). Each RT reaction was then amplified in a LightCycler® 480 PCR system using the KAPA SYBR Fast Master Mix (KAPA Biosystems, 51230-100, Woburn, MA, USA). Samples were incubated for an initial denaturation at 95 °C for 5 min, then 40 PCR cycles were performed using the following conditions: 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 10 s. For CD5L, the RT reaction was amplified using the PrimeTime® Gene Expression Master Mix (1055772, IDT, Newark, NJ, USA) under the following cycling program: polymerase activation at 95 °C for 3 min, and 45 cycles of amplification consisting of denaturation for 15 s at 95 °C, and annealing/extension for 1 min at 60 °C. All primer pairs used in this study are listed in Supp Table S1. Gene expression values were normalized to the expression levels of the housekeeping genes GAPDH (glyceraldehyde 3-phosphate dehydrogenase) for human samples and Actb for mouse samples. Fold induction levels were calculated using the average level of expression of each gene in control samples as a reference.
RNA-sequencing (RNA-seq) and transcriptomic analysis
RNA-seq of CD45+CD11+Ly6C+Ly6Glo cells from mouse tumors sorted by flow cytometry was performed by the Genome Analysis Platform of CIC bioGUNE (Bizkaia Science and Technology Park, Derio, Vizcaya).
The quantity and quality of the isolated RNAs were evaluated using the Qubit RNA HS Assay Kit (Q32855, Thermo Fisher Scientific) and Agilent RNA 6000 Pico Chips (5067-1513, Agilent Technologies, Santa Clara, CA, USA), respectively. Sequencing libraries were prepared following the “TruSeq Stranded mRNA Sample Preparation Guide (Part 15031058 Rev. E)” using the “TruSeq® Stranded mRNA Library Prep” Kit (20020594, Illumina, San Diego, CA, USA) and TruSeq RNA CD Index Plate (96 Indexes, 96 Samples) (20019792, Illumina). Raw reads were filtered using skewer v0.2.233 to remove the low-quality reads and trimming using the Illumina adapter. The STAR program34 against Mus musculus genome (GRCm38) was used to map the reads, followed by gene quantification with the RSEM program35 using GENCODE m15 reference annotation.36 After eliminating genes without an expected value greater than 10, we used the quantile method and limma-voom transformation to normalize non-biological variability. Differential expression was assessed using moderated t-statistics.37
Principal Component Analysis from the expression matrix, heatmaps, and volcano plot were computed and plotted using R statistical software (v-4.2.0). The RNA-seq data are available from the GEO repository of the National Center for Biotechnology Information, U.S. National Library of Medicine (the accession number for these data is GEO: GSE208122). For automated functional annotation and classification of statistically significant genes based on GO Biological process terms, we used DAVID.38 The protein interactome characterization of these genes was done with the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) software (v11.5, ©String Consortium 2022).
Analysis of single-cell gene expression data from GEO127465 dataset
CD5L mRNA expression on lung cancer myeloid cell population was analysed from available GEO dataset, by working the GSE127465_human_counts_normalized_54773x41861.mtx.gz folder, integrated with GSE127465_human_cell_metadata_54773x25.tsv.gz and GSE127465_gene_names_human_41861.tsv.gz from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127465, by using python3 SCANPY.39
Statistical analysis
Statistical analysis and Principal Component Analysis of the in vitro data were performed with GraphPad Prism V.9.3.1 software (La Jolla, CA, USA). Specific statistical tests are indicated in each figure legend. For survival analysis, Kaplan–Meier's method and the log-rank test were performed to compare differences among curves. Values of p ≤ 0.05 were considered significant.
Role of funders
The Funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Cancer cells induce an immunosuppressive macrophage phenotype and stimulate CD5L expression
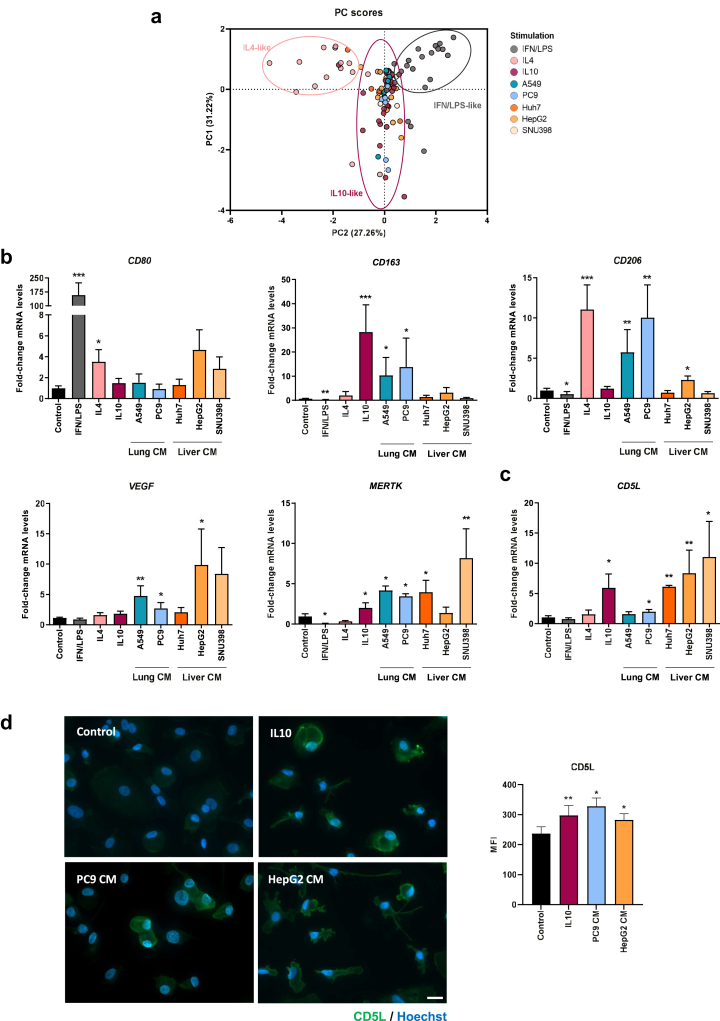
We sought to determine whether cancer cells from two different types of human tumors, namely lung and liver gear macrophage activation towards immunosuppression and whether they influence CD5L expression. To this end, we selected the A549 and PC9 cell lines, derived from human Non-small-cell lung carcinoma (NSCLC), as well as the human liver cancer cell lines Huh7, HepG2, and SNU398. CM of each different cell line were added to PB monocyte cultures, and changes in the expression of HLA-DR and macrophage activation markers CD80, and CD23, CD163, and CD206 were analyzed by flow cytometry. Alterations were compared to those induced by reference activation stimuli, namely IFN/LPS (proinflammatory), and IL4 and IL10 (two types of immunosuppressive cytokines).17 Collectively, the flow cytometry data were examined by Principal Component Analysis (PCA) to reduce dimensionality. PCA analysis revealed that stimulation with cancer cell-CM induced a macrophage receptor expression pattern that predominantly resembled that of IL10-stimulated macrophages (Fig. 1a). Interestingly, examination of the individual receptor expression showed that CM from lung A549 cells significantly increased the expression of CD163. Likewise, lung PC9-CM increased the expression of CD23 and CD163 surface receptors (Supp Fig S1). RT-qPCR analyses reinforced these findings, showing that treatment of PB monocytes with lung cancer cell-CM increased the mRNA expression of CD163, CD206, VEGF and MERTK (Fig. 1b). All these data collectively point to a significant effect of lung cancer cell-CM on macrophages, driving them to an immunosuppressive phenotype. Changes in receptor expression by liver cancer cell-CM provided similar results, both by flow cytometry and RT-qPCR (Supp Fig S1 and Fig. 1b, respectively), thereby highlighting the relevance of the results obtained in lung cancer for other types of solid cancers.
Fig. 1.
Lung and liver cancer cell-conditioned media (CM) induce an IL10-like phenotype and CD5L expression in macrophages.a) Principal Component Analysis (PCA) scatterplot of PB monocytes treated for 72 h with medium alone (control), reference activation stimuli (IFN/LPS, IL4, and IL10), or cancer cell-CM. Projection based on the expression profile of surface markers characterized by multicolor flow cytometry. PC1 and PC2 represented in each axis depict the first and second principal components, respectively. b) Expression of CD80, CD163, CD206, VEGF, MERTK and c)CD5L mRNA was assessed by RT-qPCR in PB monocytes treated for 24 h with the indicated stimuli. mRNA levels normalized to GAPDH, and fold induction levels were calculated using the average expression of each gene in control macrophages as a reference. Data are represented as mean ± SEM (n = 5 to 9). d) CD5L immunofluorescence staining (green) in macrophages treated with the indicated stimuli for 72 h. Nuclei were counterstained with Hoechst 33258 (blue). Scale bar represents 20 μm. CD5L mean fluorescence intensity (MFI) was calculated with ZenLite software and is represented as MFI ± SEM of 50 macrophages scored in random fields (right) (n = 3). Significance was calculated using the Mann–Whitney t-test (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
We next tested whether the CM altered CD5L expression. Very low levels of CD5L mRNA were detected in non-stimulated macrophages, but these were significantly enhanced by all liver cancer-CM, as well as by lung cancer PC9-CM (Fig. 1c). Accordingly, IF staining of CD5L and confocal imaging showed that IL10, lung cancer PC9-CM and liver cancer HepG2-CM induced CD5L protein expression (Fig. 1d, Supp Fig S6a). Therefore, our results reinforce the notion that CD5L mRNA and protein expression is upregulated by tumor-derived stimuli.
CD5L expression by TAMs correlates with poor prognosis in human lung cancer
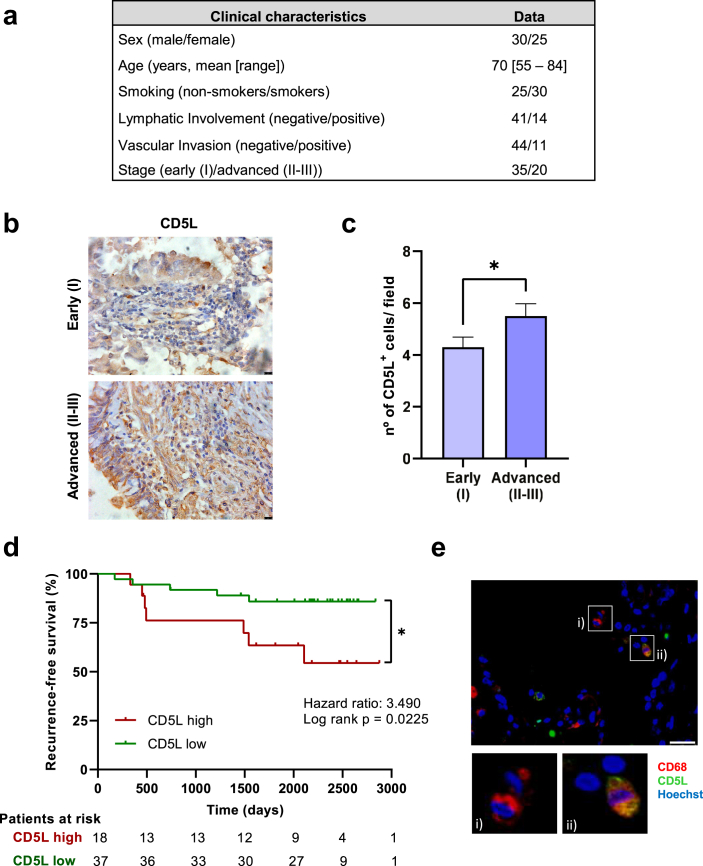
We next investigated whether CD5L is expressed in TAMs in n = 55 specimens of human PAC, a subtype of NSCLC. The main clinical and pathological characteristics of the patients are summarized in Fig. 2a. The IHC analysis revealed the presence of CD5L+ macrophages (Fig. 2b). Moreover, higher levels of CD5L+ macrophages correlated with more advanced stages of the disease (II–III) (median [range] of 6.3 [2.0–8.4]) when compared to early (I) stages (4.0 [1.6–9.6]) (Fig. 2c). Furthermore, cases with higher TAM expression of CD5L showed a 3.5-fold increased likelihood of recurrence when compared to those with lower expression (Fig. 2d). The co-expression of CD5L with CD68, a pan-macrophage marker, was confirmed in six cases by IF analysis (Fig. 2e) (Supp Fig S6b). Reinforcing these data, analysis of CD5L expression data obtained from myeloid single cell transcriptomics in human lung cancers (GSE127465) (Supp Fig S2a,b), suggest that CD5L mRNA is expressed in M2 and M0, but not in M1 macrophages (Supp Fig S2c).40 Taken together, these data indicate that infiltrating macrophages in lung cancer express CD5L, which in turn is associated with a worse prognosis in patients with NSCLC.
Fig. 2.
CD5L expression by TAMs is associated with poor prognosis in papillary lung adenocarcinoma. a) Clinical characteristics of patients diagnosed with Papillary Adenocarcinoma (PAC) who participated in this study. b) Representative immunohistochemistry images showing CD5L expression in early (I) and advanced (II–III) stages of papillary lung adenocarcinoma. Scale bar represents 10 μm. c) Graph shows the number of CD5L+ macrophages per field in early (I, n = 35) and advanced (II–III, n = 20) stages. Data are presented as the mean ± SEM, ∗p < 0.01 determined by the Mann–Whitney t-test. d) Kaplan–Meier analysis of recurrence-free survival in cases with lower and higher TAM CD5L expression. The mean number of CD5L+ macrophages from stage I was taken as the limit value, ∗p < 0.01 determined by the Log-rank (Mantel–Cox) test. e) Representative immunofluorescence image depicting CD68+ (red) (i) and CD5L+ (green) macrophages, and their co-expression (orange), (ii). Scale bar represents 25 μm.
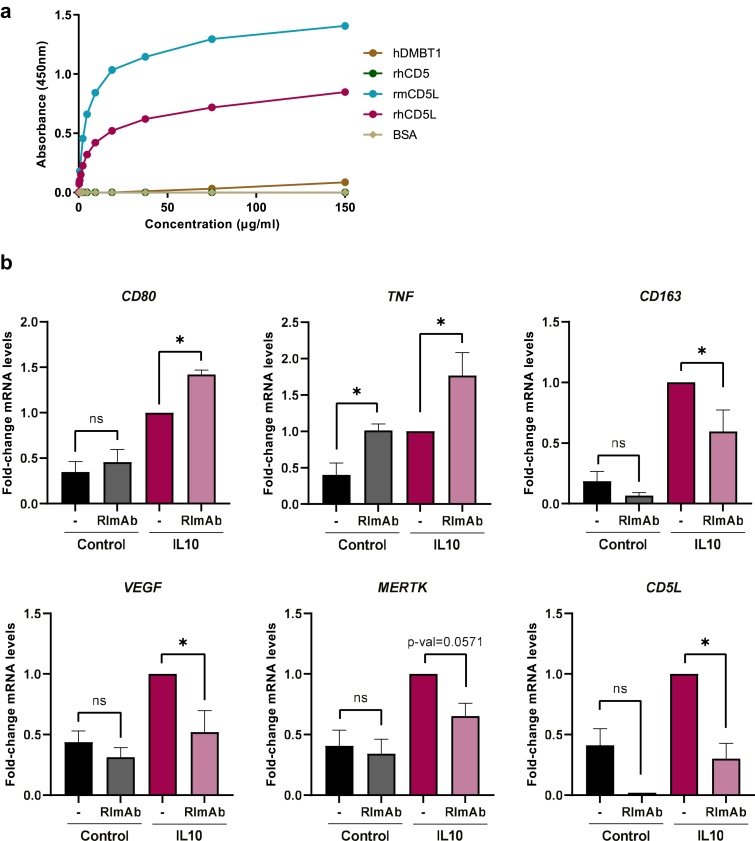
Anti-CD5L antibody RImAb inhibits IL10-induced activation
To explore the therapeutic potential of targeting CD5L, we raised a function-blocking mAb against CD5L, which we named RImAb. The antibody is an IgG2a kappa mAb, and, in a direct binding ELISA, it specifically interacts with recombinant human CD5L (rhCD5L) but not with human DMBT1 (hDMBT1) or human CD5 (hCD5), two structurally related proteins with 44.64% and 36.36% sequence identity and 51.07% and 38.63% similarity, respectively, with CD5L in their SRCR domains (Fig. 3a). Of note, RImAb cross-reacted with the recombinant murine CD5L protein (rmCD5L) (69.29% sequence identity and 73.97% similarity to its human counterpart) (Fig. 3a), thus making it suitable for experimental murine models. We then tested whether RImAb would affect IL10-induced macrophage activation. Interestingly, treatment with 5 μg/ml RImAb before stimulation with IL10 increased the mRNA expression of the co-stimulatory molecule CD80 and the pro-inflammatory cytokine TNFA (Fig. 3b), while inhibiting the induction of the TAM-associated markers CD163, VEGF, and, to a lesser extent, MERTK (Fig. 3b). Accordingly, RImAb also inhibited the IL10-induced increase in CD5L mRNA expression (Fig. 3b), thereby indicating that the IL10-induced phenotype can be prevented by CD5L blockade with RImAb.
Fig. 3.
RImAb specifically binds to human and mouse CD5L and reverts the polarization induced by IL10. a) Direct ELISA of RImAb to hDMBT1, rhCD5, rmCD5L, rhCD5L, or BSA. A representative experiment of three performed is shown. b) RT-qPCR quantification of mRNA expression of CD80, TNFA, CD163, VEGF, MERTK, and CD5L in PB monocytes treated, when indicated, with 5 μg/ml of RImAb for 45 min before the addition of IL10 (50 ng/ml) for 24 h. mRNA levels relative to GAPDH, and fold induction levels were calculated using the expression of each gene in IL10-stimulated macrophages for each donor as a reference. Data are represented as mean ± SEM (n = 5 to 9). Significance was calculated using the Mann–Whitney t-test (∗p ≤ 0.05).
Immunotherapy targeting CD5L (RImAb) inhibits tumor growth, lowers the abundance of tumor macrophages, and shifts the phenotype of this cell population
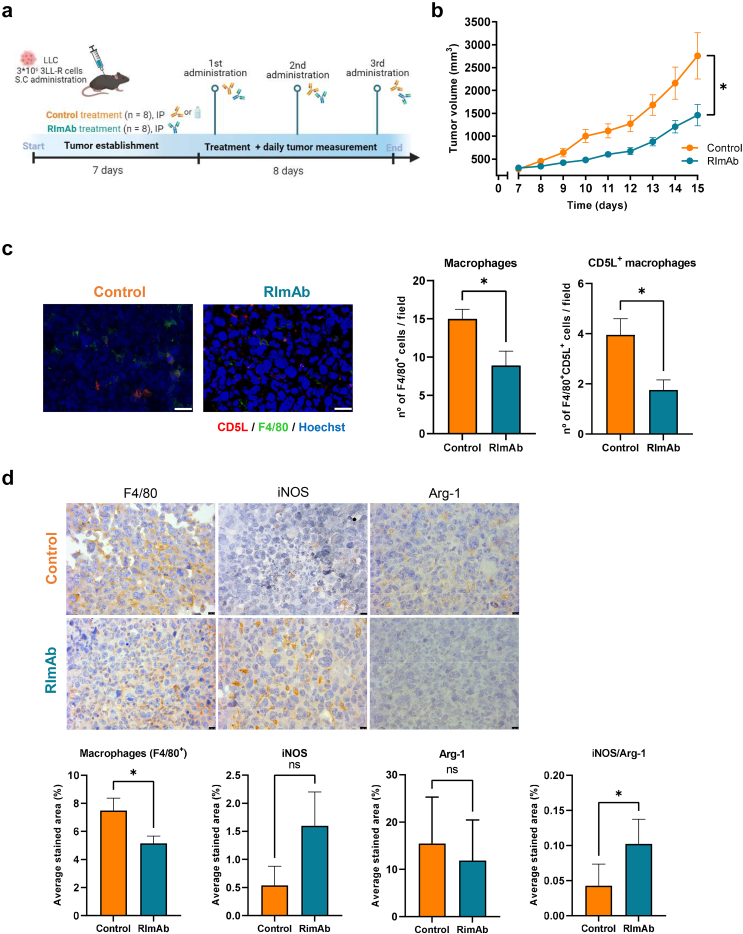
We next assessed CD5L as a target for immunotherapy in a syngeneic mouse model of lung cancer. RImAb, PBS, or the isotype control antibody was administered intraperitoneally (i.p.) every three days after tumor establishment (day 7). A schematic diagram summarizing the protocol is provided in Fig. 4a. RImAb treatment slowed tumor growth over time (Two-way ANOVA, p = 0.019). Consequently, at the end of the study, LLC tumor-bearing mice treated with RImAb presented statistically significant smaller tumors than those treated with PBS or the isotype control antibody (Mann–Whitney t-test; p = 0.0093) (Fig. 4b). This experiment was performed four times with PBS (n = 2) and IgG2a (n = 2) as control of the RImAb arm (n = 4), with lower tumor size in RImAb-treated mice in all cases (Supp Fig S3).
Fig. 4.
Blockade of CD5L slows tumor growth in vivo and reprograms TAMs towards an antitumor profile. a) Study design and timeline for mouse model. b) LLC tumor growth in mm3 in mice treated with control (PBS) or the anti-CD5L RImAb. Data are presented as the mean ± SEM (n = 8 per group). ∗p < 0.01 determined by the two-way repeated measures ANOVA test. c) Left: Immunofluorescence demonstrating CD5L expression (red) by F4/80+ TAMs (green) in control (PBS) and RImAb-treated mice. Nuclei were counterstained with Hoechst 33258 (blue). Scale bars represent 25 μm. Right: graph representing the number of F4/80+ TAMs and F4/80+ TAMs expressing CD5L per field in control and RImAb-treated animals. Data are presented as the mean ± SEM (n = 8 per group). ∗p < 0.01 determined by the Mann–Whitney t-test. d) Immunohistochemistry depicting expression of F4/80, iNOS, and Arg-1 in tumor samples from control (PBS) and RImAb-treated mice. Scale bar represents 10 μm. Average stained area obtained from five random areas was calculated using Image J (color deconvolution) software. Graphs illustrate the average stained area for F4/80, iNOS, Arg-1 and ratio of iNOS/Arg-1 in control (PBS) and RImAb-treated mice. Data are presented as the mean ± SEM (n = 8 per group). ∗p < 0.01 determined by the Mann–Whitney t-test.
Similar circulating levels of both RImAb and isotype control antibodies were observed at the end of treatment (Supp Fig S4a). Regarding potential toxicities, all the animals showed comparable activity, alertness, and reaction to stimuli. Their movement and posture were normal. Their eyes were clear and showed no sign of infection/inflammation. Neither did we observe any differences in weight, or in parameters reflecting liver function, namely alanine and aspartate transaminases (ALT and AST), renal function (creatinine), lipid metabolism, iron concentration, or bilirubin. Of note, all these values were within the standard range, excluding potential functional organ/systemic impairment, as described41,42 (Supp Fig S4b and c).
We next used IF to examine macrophage abundance and phenotype in the tumor tissue and observed a significant reduction in the number of F4/80+ macrophages and of those expressing CD5L in the RImAb group compared to the controls (Mann–Whitney t-test; p = 0.0207 and p = 0.0110, respectively) (Fig. 4c, Supp Fig S6c). IHC using F4/80 confirmed the loss of F4/80 staining, along with higher expression of iNOS (Mann–Whitney t-test; p = 0.0281 and p = 0.054, respectively) (Fig. 4d). Even though Arginase-1 (Arg-1) levels did not show any significant changes, we observed a significant increase in the iNOS/Arg-1 ratio (Mann–Whitney t-test; p = 0.0379) (Fig. 4d), suggesting a shift in TAMs towards the proinflammatory phenotype in the RImAb-treated group.
RImAb treatment reprograms the intratumoral myeloid cell compartment
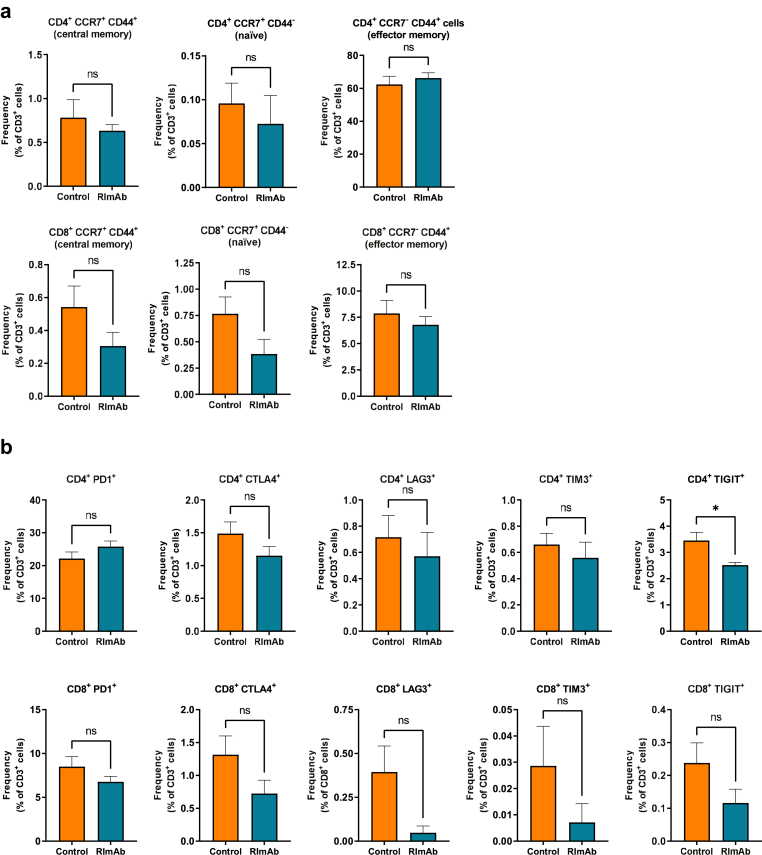
We next used flow cytometry to study whether RImAb administration induced changes in tumor immune cell populations. The gating strategy for myeloid lineage is depicted in Fig. 5a. In these experiments, an imbalance between the CD11b+Ly6ChiLy6Ghi (neutrophil population) and CD11b+Ly6C+Ly6Glo (monocyte/macrophage lineage) populations was observed, with a gain of the former and loss of the latter in RImAb-treated animals (Fig. 5b and c, respectively). Within the monocyte/macrophage populations, four subsets were delineated on the basis of MHCII expression (Fig. 5d). Within Ly6Clow cells, considered TAMs,22,30,43 we observed an increase in MHCIIhi macrophages (Mann–Whitney t-test; p = 0.0379), with no apparent changes in MHCIIlo ones (Mann–Whitney t-test, p = 0.2786) (Fig. 5d). Regarding Ly6Chi populations, considered infiltrating monocytes,22,43 RImAb induced a decrease in those cells with MHCIIhi expression. In contrast, no changes in the number of CD3+CD4+ or CD3+CD8+ T lymphocytes, Tregs (CD3+CD4+CD25+FoxP3+), nor NK T cells (CD3−NK.1.1+) were detected between treatment groups (see gating strategy in Fig. 5e) (Fig. 5f). To further determine the potential differences in the lymphoid population, we analyzed the memory and exhaustion state of intratumoral T-cells. Analysis of CCR7 and CD44 markers revealed no significant differences between the memory state of T-cells (Fig. 6a). In contrast, analysis of exhaustion markers revealed that treatment with RImAb decreased the expression of TIGIT in the CD4+ subpopulation (Fig. 6b), while no significant differences were observed in CTLA4+, PD1+, LAG3+, and TIM3+ cells between treatment groups. Although a decrease in all exhaustion markers was observed in the CD8+ subpopulation, these were not significant (Fig. 6b).
Fig. 6.
RImAb modifies the exhaustion state of intratumoral T-cells in a mouse model of LLC. Flow cytometry analysis of a) CCR7 and CD44 memory markers, and b) CTLA4, PD1, LAG3, TIM3, TIGIT exhaustion markers, in CD4+ (upper panels) or CD8+ (lower panels) T-cells isolated from LLC tumor samples. Data are presented as the mean percentage with respect to CD3+ population ± SEM (n = 8 per group). ∗p < 0.01 determined by the Mann–Whitney t-test.
Taken together, our data suggest that RImAb administration affects mainly the proportion of intratumoral myeloid cell lineages, although it also modulates the exhaustion state of CD4+ T-cells.
RImAb administration induces profound transcriptional changes in intratumoral monocyte/macrophage cells
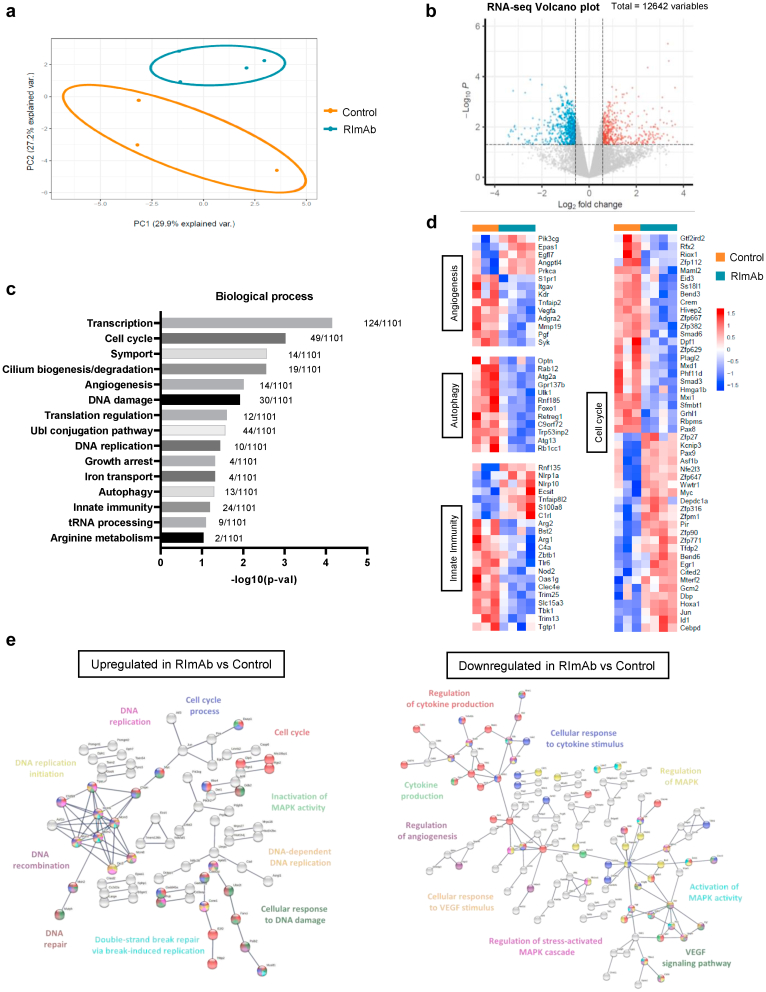
To further dissect how RImAb administration altered tumor populations of monocytes/macrophages, we sorted these cells (CD45+CD11b+Ly6C+Ly6Glo) and analyzed their transcriptomic content by RNA-seq. Sample-level quality control using PCA and clustering methods revealed that the replicates clustered within two distinct groups associated with the distinct treatments (Fig. 7a). In this regard, we observed 1199 differentially expressed genes (FC > ±1.5, p-val < 0.05), of which 512 were downregulated, and 687 upregulated (Fig. 7b). These were classified by biological process through DAVID bioinformatics software, which reported that they were significantly enriched in 15 biological processes (p-val < 0.05) (Fig. 7c), of which our attention was drawn to angiogenesis, autophagy, innate immunity, and cell cycle processes because of their relevance in cancer. Variations in the expression of genes corresponding to these biological processes were then represented in a heatmap, which showed that genes belonging to angiogenesis, autophagy, and innate immunity processes were downregulated mainly in RImAb-treated mice (Fig. 7d). Regarding the genes involved in the cell cycle, we observed a more variable pattern, with several genes upregulated and also downregulated in RImAb-treated animals (Fig. 7d). In addition, using STRING software, we represented the interactome of up- and down-regulated proteins in RImAb-treated animals vs. the control group (Fig. 7e, left and right, respectively). Several GO biological functions were identified as significantly enriched, the most relevant related to DNA replication, repair and damage, and cell cycle for RImAb-upregulated genes, and to cytokine production, MAPK signaling, and angiogenesis for RImAb-downregulated genes (Fig. 7e, Supp Table S2). Taken together, these results suggest that RImAb administration significantly affects the transcriptomic profile of tumor monocyte/macrophages.
Fig. 7.
RImAb induces changes in the gene expression profile of the myeloid population in a mouse model of LLC. a) Principal Component Analysis (PCA) scatterplot of the gene expression profile of CD45+CD11b+Ly6C+Ly6Glo cells in control (isotype IgG2a) (n = 3) and RImAb (n = 4)-treated mice obtained by RNA-seq. Percentages represent variance captured by PCA 1 and 2. b) Volcano-plot representing genes differentially expressed in RImAb-treated vs. control (isotype IgG2a) group as detected by RNA-seq data sets. Red points mark the genes with significantly increased (right, 687 genes) or decreased (left, 512 genes) expression (p < 0.05). Gray line is set to p-value of 0.05. c) Gene Ontology (GO) enrichment analysis obtained from significantly differentially expressed genes (fold-change (FC) > 1.5, p-val < 0.05) between the control (isotype IgG2a) and RImAb-treated groups. Analysis was performed with DAVID database. d) Heatmaps of gene expression of the differentially expressed genes between the control (isotype IgG2a) and RImAb-treated groups for the GO Biological Processes of Angiogenesis, Autophagy, Innate Immunity, and Cell Cycle previously identified by DAVID. e) Interactome analysis of proteins coded by genes upregulated (left) and downregulated (right) in the RImAb-treated vs. control (isotype IgG2a) groups obtained with STRING (v11.5) software.
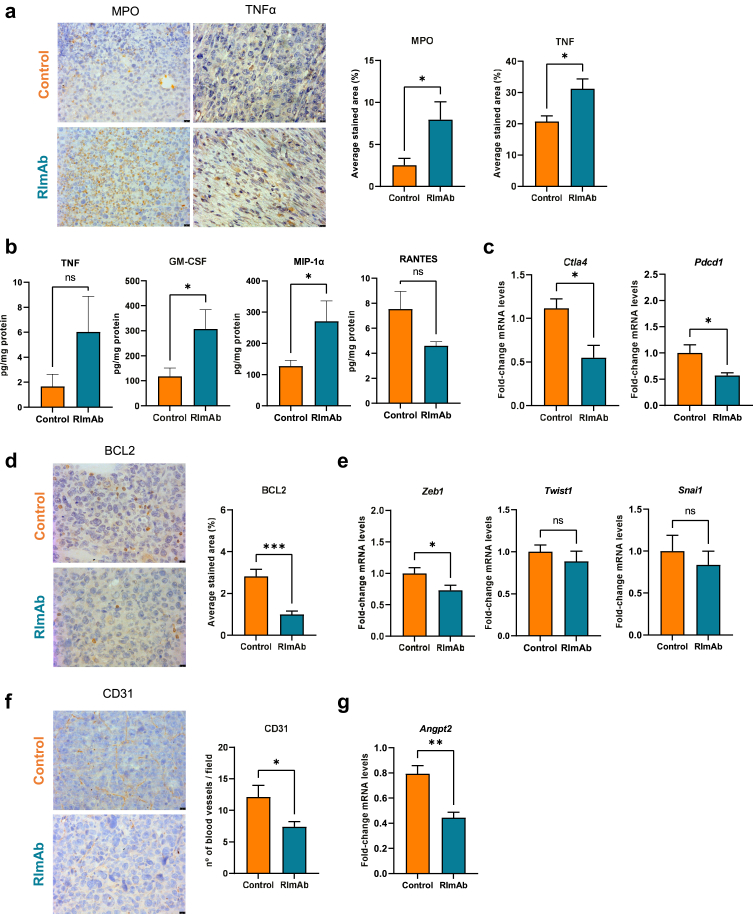
RImAb administration leads to an enhanced antitumor phenotype in the tumor microenvironment
To achieve a deeper understanding of the changes in immune response caused by the treatment, we determined additional proinflammatory markers by IHC. These analyses showed increased myeloperoxidase (MPO) (Mann–Whitney t-test; p = 0.0281) and TNF-α (Mann–Whitney t-test; p = 0.0104) levels in the RImAb treated group (Fig. 8a). Interestingly, MPO expression levels inversely correlated with tumor volume (Mann–Whitney t-test; p = 0.0442), (Supp Fig S5a). In addition, we measured the intratumoral levels of key cytokines/chemokines. TNF-α expression was higher in RImAb-treated mice, although not significant (Mann–Whitney t-test; p = 0.4713). Moreover, these animals showed an increase in Granulocyte Macrophage Colony-Stimulating Factor (GM-CSF), and in Macrophage Inflammatory Protein-1 alpha (MIP-1α or CCL3) (Mann–Whitney t-test; p = 0.0155). In contrast, RImAb-treated animals showed slightly lower expression of the chemokine RANTES ((C–C motif) ligand 5, CCL5) (Mann–Whitney t-test; p = 0.1374) (Fig. 8b). These same cytokines were analyzed in serum, where we observed a significant decrease in GM-CSF in RImAb-treated animals (Mann–Whitney t-test; p = 0.0071) (Supp Fig S4d). We then analyzed the expression of the immune checkpoint Ctla4 and Pdcd1, which were found to be downregulated in the RImAb-treated vs. control mice (Mann–Whitney t-test; p = 0.0293 and p = 0.0350, respectively) (Fig. 8c).
Fig. 8.
RImAb reprograms the tumor microenvironment towards a more inflammatory and antitumoral phenotype in a mouse model of LLC. a)Left: Representative immunohistochemistry images of MPO and TNF-α expression in tumor samples from control (PBS) and RImAb-treated groups. Right: quantification of the average stained area of five random images obtained from each group. b) Levels of TNF-α, GM-CSF, MIP-1α, and RANTES cytokines in tumor homogenates, expressed as pg cytokine/mg protein, in control (isotype IgG2a) and RImAb-treated mice (8 mice per group), measured by the Multiplex Assay. Bar graphs show means with SEM. c) Expression of Ctla4 and Pdcd1 mRNA was assessed by RT-qPCR in total tumor from control (isotype IgG2a) and RImAb-treated mice. d)Left: Representative immunohistochemistry images of BCL2 expression in tumor samples from the control (PBS) and RImAb-treated mice. Right: quantification of the average stained area of 5 random images obtained from each group. e) Expression of EMT markers Zeb1, Snail1, and Twist1 mRNA was assessed by RT-qPCR in total tumor from control (isotype IgG2a) and RImAb-treated mice. f)Left: Representative immunohistochemistry images of CD31 expression in the tumor samples from control (PBS) and RImAb-treated mice. Right: Quantification shows the number of blood vessels per field of five random images obtained from each group. g) Expression of Angpt2 mRNA was assessed by RT-qPCR in total tumor from control (isotype IgG2a) and RImAb-treated mice. For a), d) and f), pictures were quantified using Image J (color deconvolution) software. Scale bars represent 10 μm, and data are presented as the mean ± SEM (n = 8 per group). For c), e), and g), mRNA levels are relative to Actb, and fold induction levels were calculated by using as a reference the average level of expression of each gene in tumors from control (isotype IgG2a) mice, and are represented with a boxplot showing median with whiskers (min to max) (n = 8 per group). For all panels, data are presented as the mean ± SEM, and statistical significance was calculated using the Mann–Whitney t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p ≤ 0.001.
To broaden observations of the consequences of CD5L blockade, we next studied other markers of key aspects of tumorigenesis, namely cancer cell apoptosis, the epithelial–mesenchymal transition (EMT), and tumor vascularity. RImAb administration led to a decrease in the apoptotic regulator BCL2, as analyzed by IHC (Mann–Whitney t-test; p = 0.0003) (Fig. 8d). In terms of the EMT markers, a significant decrease in Zeb1 (Mann–Whitney t-test; p = 0.041) expression was observed, concomitant with a tendency towards a lower expression of Twist1 and Snail1, although the differences were not statistically significant (Mann–Whitney t-test; p = 0.5358, and p = 0.4634, respectively) (Fig. 8e). Importantly, RImAb-treated mice showed decreased tumor vascularity, as reflected by a significant reduction in the number of blood vessels compared to the controls (Mann–Whitney t-test; p = 0.0140) when CD31 was used (Fig. 8f). Moreover, the reduction in the number of blood vessels significantly correlated with the decrease in tumor volume (Mann–Whitney t-test; p = 0.0286) (Supp Fig S5b). In line with these findings, the mRNA expression of the potent angiogenic growth factor angiopoietin-2 (Angpt2) was greatly decreased in RImAb-treated mice (Mann–Whitney t-test; p = 0.0023) (Fig. 8g).
Discussion
Here we show that CD5L is an immune checkpoint in macrophages. Importantly, CD5L protein overexpression in TAMs correlated with a worse patient prognosis in lung cancer. Moreover, through the action of a new specific mAb, we demonstrate that targeting CD5L is a feasible approach to not only reprogram the macrophage transcriptome and phenotype but also reduce the immune-suppressing characteristics of the TME, which together result in reduced tumor growth in vivo.
Previously, we reported that IL10-activated macrophages in vitro overexpressed CD5L.17 Through comprehensive phenotypic profiling of human PB monocytes using up to three complementary techniques, we herein determined that CM from lung and liver cancer cells increase the expression of TAM markers associated with immunosuppression while inducing CD5L levels. Interestingly, IL10, PC9-CM, and HepG2-CM could increase the mRNA level of CD5L in macrophages, in which HepG2-CM exhibited a significantly stronger stimulation of CD5L expression over PC9-CM. However, regarding the protein expression, PC9-CM was associated with the highest MFI compared to IL10 and HepG2-CM. These apparently contradictory results could be explained by previous studies suggesting that CD5L expression is tightly regulated, and its mRNA levels do not always correspond to those of its protein.28,29,44, 45, 46 Additionally, in line with the previous results where CD5L induces an immunosuppressive phenotype,17 we determined that CD5L blockade reverted IL10-induced macrophage activation. Interestingly, blocking CD5L also inhibited CD5L expression, suggesting that this protein undergoes a positive feedback of transcriptional regulation. Together, these observations thus point to a positive feedback loop between CD5L expression and the maintenance of the immunosuppressive phenotype.17 Therefore, we conclude that CD5L is a TAM marker that is induced by factors secreted by cancer cells. Association of high TAM CD5L protein with advanced stages of lung cancer disease and lower event-free survival supports the notion that CD5L contributes to a poor outcome in this pathology. It is also in accordance with the presence of TAMs showing an inverse correlation with prognosis and survival rates in this pathology.47
The generation of a specific antibody, namely RImAb, against CD5L allowed us to explore the therapeutic potential of targeting CD5L in the context of cancer. Of note, using PB monocytes in vitro, the addition of RImAb before stimulation with IL10 reverted the phenotypic changes induced by this cytokine, thereby reinforcing the notion that this mAb blocks macrophage immunosuppression. These data are in agreement with a significant increase in the iNOS/Arg-1 ratio in mice treated with RImAb.48
We sought to further characterize the monocyte/macrophage populations (CD45+CD11b+Ly6C+Ly6Glo) in tumor specimens from mice treated with RImAb. According to recent reports, these populations can be differentiated into four subpopulations by flow cytometry on the basis of Ly6C and MHCII expression, Ly6ChiMHCIIlo being inflammatory monocytes, Ly6ChiMHCIIhi immature macrophages, Ly6CloMHCIIhi TAMs, and Ly6CloMHCIIlo TAMs.22,43 Using this categorization, we did not observe changes in Ly6CloMHCIIlo TAMs, which are described to be immunosuppressive and tumor-promoting in vivo. However, there was an increase in Ly6CloMHCIIhi TAMs, which have a higher antigen-presentation capacity and a more pro-inflammatory gene expression profile.7,30,43 These data reinforced the notion that RImAb administration alters the monocyte and macrophage compartment within the tumor. In agreement, transcriptomic analyses of CD45+CD11b+Ly6C+Ly6Glo cells within the tumor revealed profound changes in mRNA expression between RImAb-treated and control animals. Significant changes were observed in the expression of genes associated with the cell cycle, suggesting that the macrophage cell cycle is altered after treatment with RImAb. Likewise, genes involved in the MAPK pathway were downregulated in RImAb-treated mice, an observation that is consistent with the proliferative role of this pathway and its hyperactivation in cancer.49 Tightly connected to these two activities is the process of autophagy, whose associated genes were downregulated by RImAb. These data are consistent with the participation of autophagy in regulating macrophage responses in cancer.50 Moreover, since CD5L promotes autophagy,17,28 it is consistent that its blockage reduced the expression of autophagy genes.
Furthermore, the RImAb-treated group showed an increase in the CD45+CD11b+Ly6ChiLy6Ghi population, considered neutrophils,51 with respect to the controls. Neutrophils play important and conflicting roles in cancer development.52,53 In our hands, the expression of MPO, a marker of neutrophil activation, negatively correlated with tumor volume, thereby suggesting that neutrophil infiltration contributes to tumor control by RImAb. The RImAb-associated increase in neutrophils is in line with the higher levels of tumor GM-CSF, as this factor is a chemoattractant for neutrophils and may have antitumor activity.54,55 In contrast, we observed diminished levels of GM-CSF in the serum of RImAb-treated animals. Interestingly, increased GM-CSF serum levels are considered a marker of adverse clinical outcome, especially in patients with NSCLC.55
RNAseq also revealed an imbalance in the regulation of cytokine production. Of the many factors involved in this process, we selected macrophage inflammatory protein-1 alpha (MIP-1α/CCL3), and RANTES as two key macrophage-secreted chemokines.56,57 MIP-1α was increased in tumors from the RImAb group. This observation is consistent with its described antitumoral role.57,58 On the other hand, RANTES levels were decreased in tumors from the RImAb-treated group. This observation is relevant since this chemokine promotes macrophage recruitment, immune evasion, and cancer cell invasion in different types of solid tumors, including NSCLC,58,59 and its gene expression is a predictor of survival in stage I NSCLC.60 Collectively, the changes in the expression of these two chemokines indicate that RImAb enhances anti-tumoral immunity.
The hallmarks of cancer proposed by Hanahan and Weinberg summarize the vast complexity of cancer mechanisms in a set of general principles.61,62 Accordingly, here we portray RImAb as a modulator of several hallmarks, namely vascularity, apoptosis and epithelial-to-mesenchymal (EMT) transition factors. RImAb administration led to a notable decrease in tumor vascularity, as reflected by a reduction of VEGF in vitro, CD31 and Angpt2 in tissue, as well as differential expression of Vegfa, Syk, and Kdr in tumor macrophages in the RImAb-treated vs. control mice. Additionally, the RImAb-treated mice showed a significant reduction of BCL2, thereby suggesting a decrease in anti-apoptotic activity, which may lead to increased apoptosis and effective tumor killing. Treatment with RImAb also decreased the expression of Zeb1 but not Snai1 or Twist1. Strikingly, it has been reported that TAMs require ZEB1 for their tumor-promoting functions, and only TAMs that express full levels of ZEB1 accelerate tumor growth.63
The present study reveals that reprogramming macrophages in the TME using an anti-CD5L antibody is safe in mice. Given this finding, this approach may have potential applications for immunotherapy in lung cancer and also in other malignancies.24 Moreover, it would be interesting to study whether RImAb enhances the effect of T-cell-directed immunotherapies, such as those that target the immune checkpoints TIGIT, CTLA4 and PD-L1, as these were found to be downregulated in CD4+ T-cells and whole tumors from RImAb-treated mice, respectively.
In conclusion, here we reveal a key function for CD5L protein in modulating the activity of macrophages and their interactions within the TME. We further propose a new approach against the immunosuppressive TME based on targeting CD5L with a new mAb, which emerges as a promising approach to cancer immunotherapy.
Contributors
LSM, TP, CM, JFD, LS, GA, ET, JC, AFV, LK, and MRS: designed and performed research, analysed data; MI: contributed patient samples and clinical databases; JB: designed research; MT, HD: performed research; GT, CV: designed and performed research and analysed data; MAFS: designed and analysed data; CF, JS, HM, JJL, ARC, and CA: analysed data; LSM, TP, CM, and MRS wrote the paper. All authors read and approved the final version of the manuscript. LSM, TP, CM, and MRS have verified the underlying data.
Data sharing statement
Data reported in this paper will be shared by the corresponding author upon request. This study generated datasets available from the GEO with accession number GEO: GSE208122.
Declaration of interests
A patent protecting a method for the detection of CD5L has been submitted to the European Patent Office (EP3653646A1). Likewise, the RImAb antibody is the object of an EP3476863A1 patent. LK is part of an institutional licensing agreement with SunRock Biopharma, and co-inventor of two patents (EP22382093.7 and 62828195). JB received support from MSD, Grífols and Hipra through institutional grants, and by AlbaJuna Therapeutics S.L. through an Institutional License. He is Founder and CEO of AlbaJuna Therapeutics S.L. from which he owns stock options. He is also consultant for MSD and Nesapor S.L, and received support from Gilead for attending meetings. GT has received honoraria from Takeda for lectures.
Acknowledgements
We thank Gerard Requena (Flow Cytometry Platform, IGTP) and M. Pilar Armengol (Translational Genomics Platform and Microscopy Platform, IGTP) for their valuable support in flow cytometry studies, and RNA and microscopy analysis, respectively. We also wish to acknowledge Maria Doladé Botías (Clinical Analyses and Biochemistry Dept., HUGTiP) for analyzing the mouse serum samples; Dr. Jo van Ginderachter (VIB, Belgium) for providing 3LL-R cells; Ramon Bartolí (IGTP-CIBERehd) for help writing the CMCIB ethical protocol for Animal Experimentation; Ana M Aransay for performing the RNAseq experiments (Genome Analysis Platform, CIC bioGUNE, Spain); Imane Chaib (Cancer Biology and Precision Medicine laboratory of Dr. Rosell ICO-IGTP) for providing the A459 and PC9 lung cancer cell lines used in the study; Elena Ramos for help with antibody purification (Dept. of Immunology and Oncology, CNB-CSIC) and Ismael Varela for his technical assistance with in vivo mice experiments.
This study was supported by the Instituto de Salud Carlos III (ISCIII), and ERDFs from the EU, ‘Una manera de hacer Europa’, grants PI19/00523, 2019 PROD 00118 AGAUR, CI20-00152 and CC22-10181 Caixa Impulse to MRS, and from the Spanish Ministry of Science and Innovation grant PID2020-119875RB-I00 and PID2020-105404RB-I00 to AFV and LK, respectively. This research was also supported by the Consorcio Centro de Investigación Biomédica en Red (CIBERES and CIBEREHD). LSM, TP, JFD, and CM are supported by Juan de la Cierva (FJC2019-041213-I), PFIS (FI20/00115), FPI (PRE2018-085579), and Margarita Salas (MSGD2021-08) fellowships, respectively, all of which are programs run by the Spanish Government and NextGenerationEU, respectively. This study received funding from the EU’s Horizon 2020 Research and Development Program under grant agreement nº 847762. The Innate Immunity lab and the UTE are accredited by the Catalan Agency for Management of University and Research Grants 2017-SGR-490/2021-SGR-01186 and 2017-SGR-500/2021 SGR 00920, respectively. MRS, CV, and CA are researchers at IGTP, which is a member of the CERCA network of institutes supported by the Health Department of the Government of Catalonia.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104555.
Appendix A. Supplementary data
References
- 1.Whiteside T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bejarano L., Jordāo M.J., Joyce J.A., Bejarano L., Jordāo M., Author C. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11(4):933–959. doi: 10.1158/2159-8290.CD-20-1808. [DOI] [PubMed] [Google Scholar]
- 3.Cassetta L., Pollard J.W. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17(12):887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 4.Qian B., Pollard J.W. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casanova-Acebes M., Dalla E., Leader A.M., et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature. 2021;595(7868):578–584. doi: 10.1038/s41586-021-03651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantovani A., Locati M. Tumor-associated macrophages as a paradigm of macrophage plasticity, diversity, and polarization lessons and open questions. Arterioscler Thromb Vasc Biol. 2013;33(7):1478–1483. doi: 10.1161/ATVBAHA.113.300168. [DOI] [PubMed] [Google Scholar]
- 7.Noy R., Pollard J.W. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung O.W.H., Lo C.M., Ling C.C., et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62(3):607–616. doi: 10.1016/j.jhep.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Yuan A., Hsiao Y.J., Chen H.W.H.Y., et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. 2015;5(1):1–12. doi: 10.1038/srep14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtaki Y., Ishii G., Nagai K., et al. Stromal macrophage expressing CD204 is associated with tumor aggressiveness in lung adenocarcinoma. J Thorac Oncol. 2010;5(10):1507–1515. doi: 10.1097/JTO.0b013e3181eba692. [DOI] [PubMed] [Google Scholar]
- 11.Kang F.B., Wang L., Li D., Zhang Y.G., Sun D.X. Hepatocellular carcinomas promote tumor-associated macrophage M2-polarization via increased B7-H3 expression. Oncol Rep. 2015;33(1):274–282. doi: 10.3892/or.2014.3587. [DOI] [PubMed] [Google Scholar]
- 12.Kiyotani K., Toyoshima Y., Nakamura Y. Personalized immunotherapy in cancer precision medicine. Cancer Biol Med. 2021;18(4):955. doi: 10.20892/j.issn.2095-3941.2021.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander M., Kim S.Y., Cheng H. Update 2020: management of non-small cell lung cancer. Lung. 2020;198(6):897–907. doi: 10.1007/s00408-020-00407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martori C., Sanchez-Moral L., Paul T., et al. Macrophages as a therapeutic target in metastatic prostate cancer: a way to overcome immunotherapy resistance? Cancers. 2022;14(2):440. doi: 10.3390/cancers14020440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathria P., Louis T.L., Varner J.A. Targeting tumor-associated macrophages in cancer. Trends Immunol. 2019;40(4):310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Sangaletti S., Ferrara R., Tripodo C., Garassino M.C., Colombo M.P. Myeloid cell heterogeneity in lung cancer: implication for immunotherapy. Cancer Immunol Immunother. 2021;70(9):2429. doi: 10.1007/s00262-021-02916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanjurjo L., Aran G., Téllez É., et al. CD5L promotes M2 macrophage polarization through autophagy-mediated upregulation of ID3. Front Immunol. 2018;9:1–16. doi: 10.3389/fimmu.2018.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.PrabhuDas M.R., Baldwin C.L., Bollyky P.L., et al. A consensus definitive classification of scavenger receptors and their roles in health and disease. J Immunol. 2017;198(10):3775–3789. doi: 10.4049/jimmunol.1700373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarrias M.R., Grønlund J., Padilla O., Madsen J., Holmskov U., Lozano F. The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol. 2004;24:1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- 20.Matsubara E., Komohara Y., Shinchi Y., et al. CD163-positive cancer cells are a predictor of a worse clinical course in lung adenocarcinoma. Pathol Int. 2021;71(10):666–673. doi: 10.1111/pin.13144. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F., Li P., Liu S., et al. β-Catenin-CCL2 feedback loop mediates crosstalk between cancer cells and macrophages that regulates breast cancer stem cells. Oncogene. 2021;40(39):5854–5865. doi: 10.1038/s41388-021-01986-0. [DOI] [PubMed] [Google Scholar]
- 22.Georgoudaki A.M., Prokopec K.E., Boura V.F., et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 2016;15(9):2000–2011. doi: 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- 23.Xing Q., Feng Y., Sun H., et al. Scavenger receptor MARCO contributes to macrophage phagocytosis and clearance of tumor cells. Exp Cell Res. 2021;408(2) doi: 10.1016/j.yexcr.2021.112862. [DOI] [PubMed] [Google Scholar]
- 24.Mantovani A., Allavena P., Marchesi F., Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov. 2022;21:799–820. doi: 10.1038/s41573-022-00520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Moral L., Ràfols N., Martori C., Paul T., Téllez É., Sarrias M.R. Multifaceted roles of cd5l in infectious and sterile inflammation. Int J Mol Sci. 2021;22(8):1–14. doi: 10.3390/ijms22084076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanjurjo L., Aran G., Roher N., Valledor A.F., Sarrias M.R. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J Leukoc Biol. 2015;98(2):173–184. doi: 10.1189/jlb.3RU0215-074R. [DOI] [PubMed] [Google Scholar]
- 27.Bárcena C., Aran G., Perea L., et al. CD5L is a pleiotropic player in liver fibrosis controlling damage, fibrosis and immune cell content. EBioMedicine. 2019;43:513–524. doi: 10.1016/j.ebiom.2019.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjurjo L., Amézaga N., Aran G., et al. The human CD5L/AIM-CD36 axis: a novel autophagy inducer in macrophages that modulates inflammatory responses. Autophagy. 2015;11(3):487–502. doi: 10.1080/15548627.2015.1017183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amézaga N., Sanjurjo L., Julve J., et al. Human scavenger protein AIM increases foam cell formation and CD36-mediated oxLDL uptake. J Leukoc Biol. 2014;95(3):509–520. doi: 10.1189/jlb.1212660. [DOI] [PubMed] [Google Scholar]
- 30.Carbó J.M., León T.E., Font-Díaz J., et al. Pharmacologic activation of LXR alters the expression profile of tumor-associated macrophages and the abundance of regulatory T cells in the tumor microenvironment. Cancer Res. 2021;81(4):968–985. doi: 10.1158/0008-5472.CAN-19-3360. [DOI] [PubMed] [Google Scholar]
- 31.Remels L.M., de Baetselier P.C. Characterization of 3ll-tumor variants generated by in vitro macrophage-mediated selection. Int J Cancer. 1987;39(3):343–352. doi: 10.1002/ijc.2910390313. [DOI] [PubMed] [Google Scholar]
- 32.Casazza A., Laoui D., Wenes M., et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24(6):695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Jiang H., Lei R., Ding S.W., Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A., Gingeras T.R. Mapping RNA-seq reads with STAR. Curr Protoc Bioinforma. 2015;51(1):11–14.1. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frankish A., Diekhans M., Ferreira A.M., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie M.E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman B.T., Hao M., Qiu J., et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50(W1):W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):1–5. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zilionis R., Engblom C., Pfirschke C., et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50(5):1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quimby F.W., Luong R.H. Clinical chemistry of the laboratory mouse. Mouse Biomed Res. 2007;3:171. [Google Scholar]
- 42.Manni I., Di Rocco G., Fusco S., et al. Monitoring the response of hyperbilirubinemia in the mouse brain by in vivo bioluminescence imaging. Int J Mol Sci. 2017;18(1):50. doi: 10.3390/ijms18010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Movahedi K., Laoui D., Gysemans C., et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70(14):5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki T., Hirokami Y., Matsuhashi N., Takatsuka H., Naito M. Increased susceptibility of thymocytes to apoptosis in mice lacking AIM, a novel murine macrophage-derived soluble factor belonging to the scavenger receptor cysteine-rich domain superfamily. J Exp Med. 1999;189:413–422. doi: 10.1084/jem.189.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph S.B., Bradley M.N., Castrillo A., et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119(2):299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Liu X., Zhu K., et al. CD5L-associated gene analyses highlight the dysregulations, prognostic effects, immune associations, and drug-sensitivity predicative potentials of LCAT and CDC20 in hepatocellular carcinoma. Cancer Cell Int. 2022;22(1):1–23. doi: 10.1186/s12935-022-02820-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J.J., Yao P.L., Yuan A., et al. Up-regulation of tumor interleukin-8 expression by infiltrating macrophages: its correlation with tumor angiogenesis and patient survival in non-small cell lung cancer 1. Clin Cancer Res. 2003;9(2):729–737. [PubMed] [Google Scholar]
- 48.Jablonski K.A., Amici S.A., Webb L.M., et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo Y., Pan W., Liu S., Shen Z., Xu Y., Hu L. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med. 2020;19(3):1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deust A., Chobert M.N., Demontant V., et al. Macrophage autophagy protects against hepatocellular carcinogenesis in mice. Sci Rep. 2021;11(1):1–16. doi: 10.1038/s41598-021-98203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cotter M.J., Muruve D.A. Isolation of neutrophils from mouse liver: a novel method to study effector leukocytes during inflammation. J Immunol Methods. 2006;312(1–2):68–78. doi: 10.1016/j.jim.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 52.Rosales C. Neutrophil: a cell with many roles in inflammation or several cell types? Front Physiol. 2018;9(FEB):113. doi: 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coffelt S.B., Wellenstein M.D., De Visser K.E. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16(7):431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 54.Yan W.L., Wu C.C., Shen K.Y., Liu S.J. Activation of GM-CSF and TLR2 signaling synergistically enhances antigen-specific antitumor immunity and modulates the tumor microenvironment. J Immunother Cancer. 2021;9:2758. doi: 10.1136/jitc-2021-002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong I.S. Stimulatory versus suppressive effects of GM-CSF on tumor progression in multiple cancer types. Exp Mol Med. 2016;48(7):e242. doi: 10.1038/emm.2016.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhavsar I., Miller C.S., Al-Sabbagh M. Macrophage inflammatory protein-1 alpha (MIP-1 alpha)/CCL3: as a biomarker. Gen Methods Biomark Res Appl. 2015;1–2:223. [Google Scholar]
- 57.Allen F., Rauhe P., Askew D., et al. CCL3 enhances antitumor immune priming in the lymph node via IFNγ with dependency on natural killer cells. Front Immunol. 2017;8:1390. doi: 10.3389/fimmu.2017.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia L., Zhu X., Zhang L., Xu Y., Chen G., Luo J. EZH2 enhances expression of CCL5 to promote recruitment of macrophages and invasion in lung cancer. Biotechnol Appl Biochem. 2020;67(6):1011–1019. doi: 10.1002/bab.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C., Yao Z., Wang J., et al. Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ. 2019;27(6):1765–1781. doi: 10.1038/s41418-019-0460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moran C.J., Arenberg D.A., Huang C.C., et al. RANTES expression is a predictor of survival in stage I lung adenocarcinoma 1. Clin Cancer Res. 2002;8:3803–3812. [PubMed] [Google Scholar]
- 61.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 62.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 63.Cortés M., Sanchez-Moral L., de Barrios O., et al. Tumor-associated macrophages (TAMs) depend on ZEB1 for their cancer-promoting roles. EMBO J. 2017;36(22) doi: 10.15252/embj.201797345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.