Abstract
The continuous growth of pathogenic microorganisms and associated biofilms poses severe public health challenges, particularly in food and clinical environments. However, these difficulties have enabled scientists to develop novel and safe methods for combating pathogens. The use of biofilms produced by lactic acid bacteria (LAB) against pathogenic bacteria has recently gained popularity. This review provides an in-depth look at LAB biofilms, their distribution, and mechanisms of action against pathogenic bacteria. More importantly, the bioactive substances produced by LAB-forming biofilm may be active against undesirable microorganisms and their products, which is of great interest in improving human health. Therefore, this review implies that a combination of LAB biofilms and other LAB products like bacteriocins could provide viable alternatives to traditional methods of combating pathogenic microorganisms and their biofilms.
Keywords: Lactic acid bacteria, Biofilms, Extracellular polymeric substances, Microorganisms
1. Introduction
Lactic acid bacteria (LAB) are Gram-positive, catalase-negative, non-spore-forming rods or cocci microorganisms that produce lactic acid as a major metabolic end-product of carbohydrate fermentation [1]. Pediococcus, Lactobacillus, Lactococcus, Leuconostoc, and Streptococcus are common LAB species associated with food fermentation. Due to their remarkable ability to undertake anaerobic fermentation and their good physiological, survival, and probiotic properties, some LAB has a particular interest in improving the stability of raw materials, nutritional values, and sensory properties of food [2,3]. Additionally, these bacteria have been reported to adhere to epithelial surfaces, providing an antagonistic effect against intestinal pathogens by producing antimicrobial substances (e.g., bacteriocins, biosurfactant, H2O2) and forming biofilms, suggesting that this could be a new and exciting field of application in several industries, especially in the food area [4].
Biofilms are surface-adhering communities of microorganisms that produce extracellular polymeric substances (EPS) and can grow on any surface, including metal, plastic, glass, soil particles, stainless steel, wood, and biotic surface [5]. When the term biofilm is mentioned, it is usually associated with relevant risks responsible for human diseases, antibiotic resistance, infections, and difficulty controlling by being resistant to disinfection and cleaning processes [6]. These complications are primarily caused by harmful, detrimental microorganisms such as Listeria monocytogenes, Vibrio parahaemolyticus, Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, Salmonella enterica, and Candida albicans [7,8]. However, it is essential to note that not all biofilms are responsible for the negative challenges observed in the various fields. A certain type of LAB biofilm could be used as a protective biofilm against pathogens and their associated biofilms. Importantly, knowledge about LAB biofilm phenotype could enhance their antimicrobial activities from a fresh perspective. Additionally, LAB biofilms have sparked considerable interest in the biocontrol of pathogenic microorganisms. Scientists worldwide have embraced the LAB biofilm-based techniques instead of the conventional planktonic phase alone [[9], [10], [11]]. At the same time, this approach could be essential for tackling pathogens and is equally crucial for an advanced perspective of combating undesired pathogens.
This review provides a brief overview of LAB biofilm formation, factors influencing biofilm formation, adaptation and interaction of LAB with other microorganisms in biofilms, and the mechanism of action against pathogenic bacteria have been discussed. Furthermore, the use of LAB biofilms against pathogenic microorganisms and their associated biofilms has been paid much attention. Despite the beneficial antimicrobial effects of LAB biofilms, some LAB biofilms may have negative properties, such as being harmful. Some biofilms cause food product misconceptions, and it must be determined whether they are associated with changes in sensory properties. Thus, this paper focused on the common challenges associated with the use of LAB biofilms in controlling pathogenic bacteria in food environments.
2. LAB biofilms properties and their distributions in various environments
2.1. LAB biofilms are ubiquitous in various environments
LAB have been isolated from a variety of sources throughout the global food chain, including fermented food products, milk, soil, silages, bread, sauerkraut, and sausages [[12], [13], [14]], and in the gastrointestinal tract (GIT) of both man and animal [15]. Likewise, LAB biofilms, unlike pathogenic biofilms, have been logically found almost everywhere where LAB exists. This includes food environments (food processing plants, food products, milk, meat and plants), the GIT, human mucosae, vaginal animals, domestic and industrial settings (Table 1). Therefore, LAB biofilms appear to cover a wide range of habitats that are thought to cross-inoculate with each other.
Table 1.
Examples of some LAB biofilms formed in food and domesticated environments.
| Biofilm formed on | Strain involved | Biofilm property | Beneficial/detrimentala and their function | References |
|---|---|---|---|---|
| Grape seed flour | Bifidobacterium strains | Bifidobacterium strains formed strong and weak biofilms, while B. pseudo formed biofilm with a particle diameter (2 mm), and their cell reached 2.04 × 109 CFU/g at 32 h. | B; Biofilms improve cell survival | [16] |
| Daqu | Unculturable Lactobacillus sp. | Increased environmental adaptability and biomass | B; Lactobacillus sp. formed strong biofilms with S. cerevisiae in mixed-species biofilms. | [17] |
| Wine fermentation | Oenococcus oeni | O. oeni biofilm formed on oak can modulate the wood-wine transfer of volatile aromatic compounds during malolactic fermentation | B; The biofilms increase the tolerance to wine stress resulting in the effectiveness of malolactic fermentation. | [18] |
| Man-made environment | Lactococcus sp. Lactobacillus sp. | Biofilms airborne particles, in dust, domestic tools | Biofilm composition in spaces hugely inhabited by humans represents human microbiota | [19,20] |
| Food residue from human gut | L. plantarum, L. acidophilus, L. paracasei | Similar bacteria between biofilms formed on food residues and non-adherent populations | Unique patterns of fermentation product and saccharide utilization between biofilm and non-adhering bacteria | [21] |
| Floor drains | Lactobacillus lactis, Lactobacillus plantarum, and Enterococcus durans | All strains formed strong biofilm on floor drains. | B; Biofilms were able to inhibit the growth of Listeria monocytogenes. | [22] |
| Sausages | Lactobacillus sakei | 43% of L. sakei formed biofilms | B; Improved sensory quality and extended the shelf life. | [23] |
| Ice cream plant | Leuconostoc sp., Streptococcus sp.,Pediococcus sp. | The biofilms were resistant to the cleaning process | D; Biofilms supported the growth of bacterial contamination and low heat transfer exchangers. | [24] |
| Bavarian Wheat Beer | Lactobacillus acetotolerans, Lactobacillus rossiae, L. lactis and Leuconostoc mesenteroides | Do novo biofilm formation in MRS (a medium comparable nutrient composition with wort and most alcohol-free beer types). | B; Biofilm formations of the tested LAB was considered an important hygienic indicator of germs | [25] |
| Mice forestomach | L. reuteri | Biofilms formation are rodent strains- specific only | Bacterial genes involved in biofilm formation are specialized protein transport, adhesion, cell aggregation, environmental sensing, and cell lysis. | [26] |
If relevant; B: beneficial; D: detrimental.
2.2. LAB biofilm components, life cycle, and factors influencing their formation
The compositional differences between LAB biofilms and pathogenic biofilms have not yet been described. However, similar to pathogenic biofilms, it has been hypothesized that LAB biofilms are generally made up of LAB, which produces EPS such as long-chain sugars, lipids, proteins, nucleic acids (RNA and DNA), phospholipids, and other biomolecules [27]. EPS of LAB biofilms play an essential role by acting as a superglue, allowing the LAB to adhere to different surfaces, shielding the LAB from the external environment, and nutrients flow [28,29]. They have been classified into two major types: homopolysaccharides, which contain only one type of sugar monomer, and heteropolysaccharides which contain multiple types of sugar monomers [30]. Water is also an important component in biofilms, accounting for up to 96.8% of the total and being involved in the nutrient movement within biofilms [31].
In general, the composition of the biofilm changes depending on the age of the biofilm, the LAB strain involved, and environmental conditions such as oxygen, temperature, pH, nutrient availability, and desiccation [31,32]. These properties might describe the similarity and differences between LAB biofilms and pathogenic biofilms. Microbial biofilms have been compared to human cities in which people choose a location, select the neighborhood that best meets their needs, build their homes, and eventually leave when living conditions deteriorate. This fascinating behavior has been compared to the planktonic state of LAB, which reside in a specific niche, adheres and colonizes the surface, produces an extracellular matrix resulting in a matured biofilm, and occasionally leaves the biofilm and settles in a new community that suits their needs. Biofilm formation is a sequential process that begins with the attachment of a bacteria to a specific surface. The typical three basic stages of the LAB biofilms lifecycle are as follows: (1) irreversible adhesion to a surface, followed by cell multiplication and production of slimy substances (EPS matrix); (2) the formation of matured 3D biofilms architecture through matrix biosynthesis and cell proliferation. Matured biofilms exhibit increased resistance to the action of antimicrobial agents and host immune defenses; (3) and finally, biofilm dispersal results in cell detachment, allowing for new surface colonization.
External environments, which include physicochemical properties such as bacterial nutritional supply, hydrodynamic properties, coarseness, surface rigidity, topography, humidity, pH, charge, water affinity, material coating properties, material composition, and biotic surfaces, have all influenced the formation and viability of LAB biofilms in food environments [33,34]. Pili (bacterial appendages) have also been shown to contribute to the initial step of LAB biofilm formation. LAB pili have influenced cell adhesion and aggregation, as well as the complexity of biofilms. Lactococcus lactis, for example, has been shown to form a uniform and compact biofilms, which were influenced by the ability of some strains to display pili at their surface, resulting in complex biofilms revealing heterogeneous and aerial structures [32]. Protein Sortase A is also involved in the cell wall adhesion process in Pediococcus pentosaceus MR001 by recognizing the LPxTG motif in the bacterial cell wall protein and catalyzing cell wall sorting reaction and biofilm formation [35]. In addition, a protein known as “modulator of adhesion and biofilm” (MabA) composed of LPxTG signs was discovered to be responsible for adhesion in murine GIT as well as biofilm formation of Lactobacillus rhamnosus GG [36].
Further, intercellular factors such as quorum sensing (QS) (chemical communication between cells) are needed to control cell division, regulate the development of specialized cells, and are responsible for both cell attachment and detachments. By regulating specific genes, QS enables LAB to sense and adapt to their environment based on population densities. Furthermore, two signaling molecules, LuxS-derived autoinducer-2 (AI-2) and post-translationally, which have been reported to affect biofilm adhesion and formation in Lactobacillus spp., have been shown to influence LAB biofilm formation. According to Liu, Wu et al. [37], overexpression of the luxS gene increased the production of AI-2, which promoted the biofilm formation of Lactobacillus paraplantarum L-ZS9. Although various factors have been demonstrated to influence LAB biofilm formation, it is worth noting that biofilm formation is a strain-specific and non-extrapolative property [3].
Understanding the components, life cycle, and factors that influence biofilm formation in LAB is crucial for developing effective strategies for controlling pathogenic bacteria. For example, with knowledge about the composition and structure of LAB biofilm EPS, researchers can develop strategies to disrupt the biofilm and expose pathogenic bacteria to antimicrobial agents. In addition, awareness of the factors that influence biofilm LAB formation can help in identifying LAB strains with superior biofilm-forming abilities and developing strategies to enhance or disrupt biofilm formation. Likewise, each stage of LAB biofilm formation presents a different target for intervention, i.e., influencing attachment can support biofilm formation while targeting dispersal can help to control the spread of pathogenic bacteria.
3. Adaptation and interaction of LAB with other microorganisms in biofilms
In the food industry, the presence and adaptation of LAB allow the bacteria to withstand harsh environmental conditions, and various LAB species can survive several stress conditions by mainly transforming into a physiologically viable but nonculturable (VBNC) state or by forming biofilms [38,39]. Studies have shown that variety of LAB, such as Lactobacillus plantarum [33,40], Lactobacillus acetate [41], when subjected to certain environmental stress during production and application are capable of activating several stress reactions to enter into VBNC state. The VBNC state of LAB is an important survival strategy to cope with several adverse conditions. However, it can have a significant impact on public health and food safety due to their ability to regain culturability after resuscitation; even though they fail to grow and form colonies on the routine bacteriological media, they can remain active and retain their full metabolic activity [39].
On the other hand, the formation of biofilms allows them to withstand harsh environmental conditions, increase biomass and improve the quality of fermented food. For example, a recent study discovered that unculturable Saccharomyces cerevisiae, when co-existed with Lactobacillus sp., displayed a mutualistic relationship to produce biofilms in Daqu, a traditional Chinese fermentation starter. In a mixed biofilm, S. cerevisiae predominated the metabolic activity despite decreased biomass. In contrast, the biomass of Lactobacillus strains increased while metabolic activity was reduced. S. cerevisiae tended to cover the top layer with planktonic cells, while Lactobacillus strains occupied the bottom layer under limited nutrient conditions [17]. Mixed biofilm formation could describe the existence of unculturable microorganisms in other microbial communities in a complex environment. Therefore, research in mixed-species biofilm in the starter community could be important to address the unclear problem about the reconstruction of the starter community, succession and mechanism of fermentation.
In addition, it has to be explored whether biofilm formation by co-cultured LAB reflects symbiosis among the strains or solely firmness. A recent study by Yao et al. [42] determined the growth relationship and biofilm production of Streptococcus thermophilus and four strain of lactobacilli. The results displayed that all four lactobacilli strains significantly promoted the growth of S. thermophilus, while S. thermophilus promoted the growth of Lactobacillus helveticus and Lactobacillus delbrueckii subsp. Bulgaricus showed no effect on Lactobacillus paracasei but inhibited the growth of Lactobacillus kefiranofaciens. In addition, more biofilms were formed in a mixed-species mode than by a single strain, regardless of whether the strains had a symbiotic association. These findings indicate that the formation of biofilms in co-cultured strains is independent of a symbiosis relationship among the strains. However, this review suggests that if there is a symbiotic relationship between strains, molecular analysis is required to analyze the brief mechanism of biofilm formation of co-cultured LAB since their mutualistic or competitive relationship with other microorganisms is not necessarily increase or decrease the biofilm formation.
Moreover, in the clinical environment, the opportunistic yeast Candida albicans and LAB Enterococcus faecalis are often co-isolated from different infection sites on the human body, reflecting a friendly interkingdom, while an antagonism relationship has also been suggested [43,44]. A recent in vitro study aimed to evaluate the phenotypic and transcriptomic biofilm interaction of E. faecalis co-cultured with C. albicans. The results showed that, in the modulation of the pH microenvironment in mixed-species biofilms, these perceived enemies developed into a friendly relationship which influenced the invasion of dental tubules [43]. However, it was also shown that E. faecalis significantly influenced the biofilm formation and morphogenesis of C. albicans which was seen by gene expression levels. In a multi-species biofilm, hyphal-related genes (HWP1 and ECE1) and SAP5 encoding SAP5 enzyme, which are expressed upon hyphal formation, were downregulated in C. albicans, while the adhesion gene ALS3 was upregulated. This effect suggests that E. faecalis affects C. albicans hyphal formation without influencing its surface adhesions. The findings propose that E. faecalis comprises several anti-candida mechanisms despite their common co-existence behavior.
Overall, the ability of LAB to form biofilms and interact with other microorganisms is critical for their survival and function in different environments. Thus, it is important to understand the growth relationship and biofilm production of LAB in order to optimize their use in food processing and improve the quality of fermented foods. Moreover, further research is needed to understand the mechanisms underlying the adaptation and interaction of LAB in biofilms and to explore their potential applications in various fields, including food, medicine, and biotechnology.
4. Common applications of LAB against pathogenic microorganisms
For many years, a variety of LAB culture isolates and their cell-free supernatant (CFS) containing bacteriocins, organic acids, biosurfactants, hydrogen peroxide, reuterin, and diacetyl, have been widely used to inhibit and control the growth of pathogenic bacteria and their associated biofilms [2,45]; N. N [46]. LAB can elicit bacteriostatic and bactericidal effects that disrupt the function of the pathogenic cell membrane, causing membrane permeability, cell lysis, loss of cell contents, and finally, death, depending on the antimicrobial agent, its concentration, and cell density. Table 2, Table 3 summarize the most common approaches to using LAB and their bioactive substances against pathogenic microorganisms and their associated biofilms.
Table 2.
Some common approaches for using the antimicrobial substances of LAB against pathogenic microorganisms.
| Approach used and LAB involved | Pathogenic microorganisms involved | Results summary | References |
|---|---|---|---|
|
Cell-free supernatant (CFS) LAB isolated from Tilapia nicoliticus |
Bacillus cereus, Staphylococcus aureus, Escherichia coli and Salmonella enterica serovar Typhimurium | CFS of LAB incorporated with various spices showed higher antimicrobial activities with inhibition zone ranging from 14 mm to 26 mm at 37 °C for 24 h. | [47] |
| Lactobacillus acidophilus, Lactobacillus plantarum, and Pedioccocus pentosaceus | E. coli | The CFS from the tested LAB inhibited the growth of E. coli in a time-dependent manner | [48] |
| P. pentosaceus and Pediococcus acidilactici | Listeria monocytogenes, Pseudomonas aeruginosa, E. coli, S. aureus, S. Typhimurium and Listeria innocua | The bacteriocins precipitations in CFS showed an increased bacteriocin activity against Listeria monocytogenes and Listeria innocua with arbitrary units ranging from 10240 to 81920 AU/mL | [49] |
| P. pentosaceus | S. aureus and E. coli, | The CFS showed antibacterial activity to the membrane integrity as confirmed by the reduction of cell viability, increased potassium, increased relative electrical conductivity, and reduced absorption at 260 nm | [50] |
| L. plantarum, L. pentosus, and Lactobacillus paracasei | S. aureus, Salmonella enterica, E. coli and Bacillus cereus. | The CFS of eleven strains displayed a strong antibacterial effect ranging from 8.9 to 9.2 mm inhibition zone. | [51] |
| L. paracasei, Lactobacillus fermentum, Lactobacillus parelimentarius and L. plantarum. | Shigella strains | The CFS of L. paracasei Y1-3, L. paracasei M1-1, L. paracasei 18-1, L. plantarum M19-1, L. fermentum Y2-2, and L. parelimentarius M4-3 inhibited the growth of all 12 Shigella strains, with inhibition percent ranging from 58% to 92%. | [52] |
| L. fermentum, L. plantarum, Enterococcus faecium, and Staphylococcus pasteuri | E. coli, Salmonella typhi, P. aeruginosa, L. inocula, E. faecium, and Bacillus Spp. | Thirteen LAB strain produced antibacterial activities against pathogens and foodborne spoilage indicator bacteria, among them E. faecium and S. pasteuri produced bacteriocins | [53] |
|
Cells or/and CFS 6 LAB strains |
S. aureus, B. subtilis, S. Typhimurium and E. coli | Both the cells and supernatants of selected LAB showed good inhibitory activity against the target bacteria. While the supernatant of LAB-HM5 caused complete inhibition of all tested bacteria during 72 h incubation. | [54] |
| Lactobacillus delbrueckii subsp. Bulgaricus Enterococcus durans, Streptococcus thermophilus, Lactobacillus spp., Lactobacillus rhamnosus | E. coli, S. Typhimurium, P. aeruginosa, S. aureus, B. subtilis, Mucor plumbeus, Geotrichum candidum, Fusarium oxysporum, Cladosporium herbarum, Aspergillus flavus, Penicillium aurantioviolaceum, Penicillium spp. and Trichoderma viride | The wide spectra of antibacterial activities were only observed in a simultaneous cultivation of LAB strains while the antifungal Effect of L. rhamnosus in the associations with other LAB strains was partly decreased. | [55] |
| Leuconostoc mesenteroides, L. plantarum, Enteroccocus pseudoavium, L. casei, Lactobacillus curvatus, Lactobacillus farraginis, P. pentosaceus, P. acidilactici, L. paracasei, L. plantarum, Lactobacillus coryniformi, L. brevis, and Lactobacillus uvarum | E. coli, S. aureus, P. aeruginosa, Enterococcus faecalis, and L. monocytogenes | L. uvaruma and L. casei showed the strongest inhibitory activities with inhibition zone ranging from 8 to 45 mm | [56] |
Table 3.
Some common examples of using antimicrobial substances of LAB against biofilms of pathogenic bacteria.
| Approaches and LAB involved | Biofilm formed by: | Results summary | References |
|---|---|---|---|
|
In situ LAB presence Lactobacillus plantarum, Lactobacillus curvatus, Lactobacillus sakei, and Leuconostoc mesenteroides |
Listeria monocytogenes | Biofilm cells were reduced by up to 2.17 log CFU/cm2, 1.62 log CFU/cm2, and 1.09 log CFU/peg on stainless steel (SS), lettuce, and MBEC™, respectively. However, biofilm inhibition on the lettuce surface was lower compared to SS | [57] |
| Lactobacillus kefiri and L. plantarum | Salmonella Enteritidis | About 1 log of S. Enteritidis biofilm was reduced on a polystyrene microtitre plate (PMP)for 48 h at 28 °C | [58] |
| Pediococcus acidilactici | Escherichia coli, Salmonella Typhimurium, Staphylococcus aureus, and L. monocytogene | The sessile form of all target bacteria was reduced by 4 log CFU/coupon under all simulating conditions on SS, polyvinyl chloride, and glass coupons | [59] |
| Lactobacillus paraplantarum | L. monocytogenes | Adhered cells on SS coupons were significantly decreased up to 1.9 log CFU/mL for 72 h. | [60] |
| Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus paracasei, and Lactobacillus rhamnosus | L. monocytogenes | Biofilm cells were reduced (>3 logs) when co-cultured by L. paracasei and L. rhamnosus. | [61] |
|
LAB (Bacteriocins) Enterococcus faecium and Enterococcus faecalis |
L. monocytogenes, Listeria ivanovii, and Listeria innocua | The bacteriocins from the two LAB decreased biomasses and viabilities of both developing and preformed biofilms on 96-well PMP. | [4] |
| E. faecalis (nisin and enterocin B3A-B3B) | L. monocytogenes, Clostridium perfringens, S. aureus, and methicillin resistant S. aureus (MRSA) | The combined Effect of both nisin and B3A-B3B reduced the sessile count of tested bacteria by at least 2 logs after 24 h at 37 °C | [62] |
| P. acidilactici (K10 and HW01) | S. Typhimurium | Both bacteriocins significantly inhibited the biofilm formation of S. Typhimurium in a dose-dependent manner. | [63] |
| Lactobacillus crustorum (BM1157 and BM1300) | L. monocytogenes | SEM and TEM revealed that BM1157 killed L. monocytogenes by biofilm destructions and pore formation | [64] |
|
Cell-free supernatants (CFS) L. curvatus, L. fermentum, Lactobacillus delbrueckii, P. acidilactici, and E. faecium |
L. monocytogenes | In a co-inoculation mode, biofilms of L. monocytogenes strains were significantly inhibited, while the CFS was not able to cause the elimination of preformed biofilms | [65] |
| Lactococcus lactis | L. monocytogenes | Neutralized CFS significantly inhibited biofilm-forming ability of L. monocytogenes | [66] |
| L. lactis subsp. lactis. | L. monocytogenes | Bacterial cell counts were reduced by 4-log while the inhibitory activity was strain-dependent and was influenced by incubation conditions | [67] |
| L. plantarum, Lactobacillus helveticus, P. acidilactici, and E. faecium | S. aureus and E. coli | The neutralized CFSs inhibited the biofilm growth of tested bacteria and 5 LAB strains showed more than 50% inhibitory rate. | [68] |
| Weissella viridescens and Weissella confusa | S. Typhi and of S. Typhimurium. | W. confusa reduced 99.84% of AI-2 signaling interference and showed a maximum biofilm inhibitory activity against S. Typhi while W. viridescens produced a 66.46% biofilm inhibitory activity and 99.99% AI-2 signaling reduction of S. Typhimurium. | [69] |
| L. sakei | E. coli O157:H7 (EHEC) | AI-2 activity and associated virulent were significantly reduced without affecting the cell viability of EHEC | [70] |
| L. helveticus | E. coli, Salmonella, S. aureus, L. monocytogenes, and B. cereus | Biosurfactants significantly inhibited the adhesion of bacteria by nearly 90–100%, while biofilm formation was almost diminished. | [71] |
In situ LAB presence: represent the presence of LAB culture cells directly; LAB (bacteriocins); represent the use of extracted bacteriocins from LAB; Cell-free supernatants: represent the supernatant with either their original acidic or neutralized pH.
Several studies have been conducted to date in order to describe the potential application of LAB and their by-products to combat pathogenic bacteria in the food environment. For example, the antimicrobial and anti-biofilm potential of Leuconostoc mesenteroides and L. plantarum against V. parahaemolyticus, P. aeruginosa, and E. coli on squid and seafood processing surfaces (e.g., rubber and high-density polyethylene plastic) was evaluated [72]. The combined effect of LAB isolates exhibited greater planktonic and biofilm cell inhibition on both abiotic and biotic surfaces. These findings revealed that the ability of LAB to down-regulate the levels of QS signaling molecules and virulence genes involved in biofilm formation inhibited pathogenic biofilm formation [72]. According to Kim et al. [46], bacteriocins produced by Lactobacillus brevis DF01, on the other hand, significantly reduced the biofilm formed by Salmonella Typhimurium KCTC 1925 and E. coli KCTC 1039 on stainless steel. However, the study suggested that while DF01 seems to contain glycoprotein class IV bacteriocins, their mechanisms of action have not yet been determined. In addition, the anti-biofilms activities against the tested foodborne pathogens were opposite to the most commonly used bacteriocins (nisin A), as DF01 lost its biological effect when treated with proteolytic enzymes and α-amylase [46]. While bacteriocins have shown promise in controlling pathogens, there are also concerns associated with their use. One of the concerns is the potential for the emergence of resistance. Bacteria can evolve mechanisms to evade the action of bacteriocins, just as they can with traditional antibiotics. This could limit the effectiveness of bacteriocins over time and may require the development of new bacteriocins or other antimicrobial agents. Therefore, future studies are needed to fully understand their efficacy, safety, and potential impact on a broad range of bacteria.
Moreover, LAB-produced biosurfactant compounds have been reported to have a variety of applications, including antibacterial activities against foodborne bacteria [73]. Cell-bound biosurfactants isolated from L. plantarum and Pediococcus acidilactici strains, for example, have been shown to inhibit S. aureus adhesion and biofilm formation in a dose-dependent manner [74]. The results were confirmed by RT-qPCR assay and a bioluminescence assay, which revealed that the biosurfactants interfered with the expression of genes involved in biofilm formation (sortase A, icaA, dltB, cidA, and agrA) and affected the secretion of AI-2 signalling molecules in QS systems. According to the study, biosurfactant concentrations of 50 mg/mL, 25 mg/mL, and 12.5 mg/mL produced by P. acidilactici significantly reduced the expression of the icaA gene by 3.45-, 3.08-, and 2.19-fold, respectively; whereas, at 50 mg/mL, the expression levels of sarA, agrA, and sortaseA genes decreased by 1.58, 2.05, and 1.65-fold, respectively. On the other hand, the biosurfactant produced by L. plantarum decreased cidA transcription levels in biofilms by 5.68 and 2.87-fold when treated with 50 mg/mL and 25 mg/mL, respectively. It also inhibited the dltB gene by 1.93 and 1.45-fold when compared to the control. At 50 mg/mL, the expression of sarA and agrA genes was reduced by 2.24 and 1.21fold, respectively. The down-regulation of these genes suggests that, at sub-inhibitory concentration, biosurfactants may prevent the growth of biofilms by decreasing expression levels of biofilm-related genes inhibit biofilm growth by lowering the expression levels of biofilm-related genes [74]. Nonetheless, the results of this study provide valuable insights into the mechanisms by which biosurfactants can prevent the growth of biofilms and pave the way for the development of innovative and sustainable solutions for food safety. However, evaluation and determination of the effectiveness of biosurfactants to ensure their safety and regulatory status is mandatory to expand their application.
Moreover, several studies have highlighted the antimicrobial activities of LAB-derived EPS. It has been reported that EPS inhibitory potential depends on autoaggregation and bacterial cell attachment, either by weakening cell-to-cell surface interaction or by differences in monosaccharide compositions and functional groups of exopolysaccharide [75]. For example, differences were observed in exopolysaccharides produced by a probiotic P. pentosaceus M41 [76] but not in exopolysaccharides produced by Lactobacillus kefiranofaciens DN1 [77]. Another reason could be a decrease in cell hydrophobicity interactions in biological adhesion events [78].
Thus, with the increasing demand for natural and sustainable solutions, bioactive compounds from LAB offers an alternative approach for controlling pathogenic bacteria in various industries. In general, there is still a need to fully explore the potential of bioactive compounds from LAB. By improving our understanding of these compounds and optimizing their use, we can develop new and effective strategies for controlling pathogenic microorganisms.
5. Antimicrobial potential of LAB biofilms against pathogenic microorganisms
5.1. LAB biofilms inhibit the growth and surface colonization of foodborne bacterial
Recently, there is an increasing concern about LAB biofilms and their barrier effect against the development of pathogenic microorganisms. However, limited studies have revealed that LAB biofilms could inhibit the growth, adhesion, surface colonization and biofilm formation of pathogenic bacteria. This review has gathered the most recent significant findings on this aspect.
A recent interesting study focused on biofilms formed by L. sakei M129-1 and P. pentosaceus M132-2, which are tolerant to desiccation and can inhibit the growth of foodborne pathogens [79]. The biofilms formed by the tested LAB showed a wide range of antibacterial activity against L. monocytogenes, S. aureus, B. cereus, E. coli, and S. enterica. In 48 h, the initial number of foodborne bacteria was reduced from an average of 7 log CFU/mL to less than 1 log CFU/mL. The results indicated that the antibacterial potential of the two LABs were retained in the biofilm form, despite the fact that their antimicrobial activities were lost after the supernatants were neutralized. Hence the study clearly demonstrated the function of pH in the antagonistic role of Lactobacillus [79]. Another recent study conducted by Cisneros et al. (2021) determined the ability of five sessile LAB strains, L. plantarum CRL 683, P. pentosaceus CRL 908, L. plantarum CRL 1075, L. plantarum CRL 1482, and P. pentosaceus CRL 2145 to inhibit E. coli surface colonization on polystyrene surfaces at a low temperature (10 °C). After 24 h of co-incubation with the five LAB strains tested, pre-formed biofilms of CRL 683, CRL 1075, CRL 1482, and CRL 2145 significantly reduced the colonization of E. coli. In addition, despite the inhibition potential of the four LAB strains, only CRL 1075 showed a special consideration by growing in the presence of the E. coli. Thus, these findings indicated that neither heat-stable bacteriocins nor organic acids were involved in the inhibitory Effect of E. coli. This effect could be explained by an exclusion mechanism of LAB biofilms (Cisneros et al., 2021). Similar findings have also appreciated the use of LAB biofilms against foodborne pathogens and their biofilms [9,10,80,81]. Based on these findings, it has been shown that LAB biofilm can elicit antimicrobial activities in various ways. While both approaches have been proven to have an antagonistic impact against the target pathogens, exclusion mechanisms offer an added advantage when coupled with bioactive substances such as bacteriocins. Therefore, the best way of inhibiting pathogenic growth by using LAB biofilms is when LAB biofilms exert their antimicrobial activities through an exclusion approach in the presence of other bioactive substances. While using LAB biofilms for the biocontrol of pathogenic bacteria is a new and growing research field, their application has shown a promising tool for combating pathogens. The progressive discovery of such beneficial biofilms may pave the way to a novel biotechnological application in the food industry. Therefore, LAB biofilms have proved to be a good candidate for more technological studies to mitigate undesirable bacteria in food processing environments.
5.2. LAB biofilms inhibit biofilm formation of food bacteria
Despite the wide use of LAB and its products in various industries, using LAB biofilm against biofilms formed by foodborne bacteria is still a new field of research, and their applications against foodborne bacteria are still scarce. However, available findings have shown a brighter future in this aspect. For example, E. faecium and P. pentosaceus isolated from fermented chicken and fish could form biofilms on polystyrene surfaces, and their biofilms inhibited the biofilm formed by B. cereus, E. coli, and S. Typhimurium in a co-aggregation process [10]. The results displayed a higher level of LAB autoaggregation ability, favoring their inhibiting potential when co-aggregating with pathogenic bacteria. Also, based on these findings, it seems that the inhibitory potential of the tested LAB biofilms resulted from their bioactive substances present in both neutralized and non-neutralized crude supernatants [10]. On the other hand, Gómez et al. [82] aimed to develop protective biofilms for several LAB strains and test them to exclude the biofilm formation of E. coli, L. monocytogenes, and S. Typhimurium in 12-well polystyrene microtiter plate. A total inhibition of pathogenic biofilms was observed in 24, 48, and 72 h of exposure with L. lactis and Lactobacillus curvatus, which were not influenced by their bacteriocin production. Additionally, the inhibition was time-dependent and varied depending on the target pathogen and the strain involved. In L. monocytogenes, the inhibition was between 4 and 7 log units for 24 and 48 h, which was detected only within the first 24–48 h of pathogenic growth. While in presence of LAB, sessile cells of E. coli and S. Typhimurium were not detected during the first 24 h of incubation. At the same time, during 48 h and 72 h, reductions of S. Typhimurium and E. coli were archived, ranging between 3 and 5 log units [82]. Apart from the effect of LAB biofilms to inhibit bacteria biofilms, it is important to determine whether the surface involved in biofilms formation influences the inhibition process or is it just an effect developed from bioactive content in LAB biofilms. The development of strong LAB biofilms will automatically enhance the design of surface material, which will support their formation and adhesion, and their inhibition might not rely on the presence of the bioactive substance. Therefore, this condition will support the use of LAB biofilms for the LAB which are capable of producing biofilms with no bioactive compounds and pH is excluded in the inhibition process.
5.3. LAB biofilms can inhibit fungi growth
The tremendous benefits of LAB biofilms have not only been noticed against bacterial pathogens but also against fungi, especially in clinical environments. In particular, the formation of LAB biofilm inside the human body is important to health as it can impact functionality and ameliorate microbial persistence [83]. Till now, few studies have investigated and reported some LAB biofilm modes of life, such as Lactobacillus species which can improve the antagonistic effects against microbial pathogens and immunomodulatory and anti-inflammatory properties [16,84,85]. In addition, the antifungal activities of LAB biofilms are also important in order to expand the therapeutical approaches to fungi diseases.
A study performed by Parolin et al. [83] investigated the in vitro ability of 16 vaginal Lactobacillus strains belonging to L. plantarum, L. vaginalis, L. crispatus, and L. gasseri species to form biofilm on polystyrene surface and the culture supernatant obtained from planktonic and biofilm growth was tested against Candida clinical isolates including C. glabrata, C. lusitaniae, C. albicans, C. parapsilosis, C. tropicalis and C. krusei. The results showed that biofilm cell-free supernatant (bf-CFS) from L. crispatus and L. plantarum strains exerted the strongest fungistatic activity against all candida isolates compared to planktonic cell-free supernatant (pk-CFS). To better understand the impact of bf-CFS and their inhibition properties, metabolome assessment by 1H NMR was performed to determine the difference in molecules between bf-CFS and pk-CFS. It was observed that 17 molecules were significantly different; among those, ten amino acids were identified deeply involved in the inhibition mechanism, suggesting that bf-CFS mainly affected nitrogen metabolism, especially amino acidic pathways such as lysine, serine, and alanine. The formation of LAB biofilm in the human body may represent an important factor in host-microbial interaction and microbial physiology, underpinning the progress of new antimycotic approaches using probiotics LAB. This finding, in turn, provides new knowledge in the perspective of using probiotic LAB biofilm to prevent vulvovaginal candidiasis and/or restore the status of vaginal eubiosis. However, the authors suggested further studies on LAB biofilms in the vaginal niche to better understand the LAB-host relationship, its physiological role and anticandidal mechanisms exerted by LAB biofilms.
6. Use of LAB biofilms as a potential approach for detoxification of mycotoxins
Mycotoxins are toxic secondary metabolites mainly produced by some fungi species, including Penicillium, Fusarium and Aspergillus genera and are capable of causing diseases and death in humans and other animals [86]. For years, several detoxification processes such as biological, chemical, and physical approaches have been employed to mitigate mycotoxins in food and feed through modifying, adsorbing or destroying them [13,86]. Even though they have been shown to have some positive outcomes in reducing mycotoxins, however, the chemical and physical process still cannot adsorb them completely, and they contain many drawbacks such as limited implementations, high cost, impact on food nutritional value, safety and insignificant efficacy [87]. Therefore, these approaches are ineffective in mitigating mycotoxins from food and feed.
To date, LAB biofilms have been explored and appreciated as a biological approach of reducing mycotoxins from food and feed. A novel method was developed to eliminate aflatoxin M1 (AFM1) in milk using L. rhamnosus GG biofilm [88]. The detoxification process was carried out by using biofilms developed on polyethylene tubes and polystyrene petri plates for 3 days, which was considered a highly attached and more efficient binding agent. It was shown that L. rhamnosus GG biofilm could bind AFM1 by up to 60.74%. Additionally, there were no significant changes in the quality of milk protein content after AFM1 detoxification, but some changes in total dry matter and fat content were observed. Moreover, the authors have studied the stability of the formed AFM1-biofilm complex using different AFM1 milk concentrations. The findings showed a reversible binding, and some portion of bound AFM1 was liberated after washings. To date, removing mycotoxins by LAB biofilms is a new and modern technique, making it a great challenge and appeal, particularly in removing mycotoxins from foods. Although research on LAB biofilms against mycotoxins is still in its early stages, more findings should be performed (e.g., the mechanism of mycotoxin removal) to support its usefulness to food industries. Thus, in general, using LAB biofilms as a biological adsorbent of mycotoxins is an attractive environmentally friendly approach and may be considered a better substitute for chemical and physical processes presenting a solid foundation for the progress of bio-detoxification methods.
7. Mechanism of antibacterial roles of LAB biofilms
The specific mechanisms of the antibacterial functions of LAB biofilms are not well understood; however, the following approaches are proposed to be involved in inhibiting and preventing bacteria pathogens.
LAB biofilms most likely inhibit the growth of other bacteria through one of two mechanisms; competitive exclusion or by the presence of antimicrobial substances in the biofilms. Once attached to surfaces, the interactions between LAB biofilms and pathogenic cells can evolve in competition for nutrients and space, inhibition of the initial attachment and development of the pathogen biofilms, or reduction in the growth of pathogens in an established biofilm. The Jameson effect, described by Guillier et al. [89], and spatial competitions, described by Habimana et al. [90] are two examples that demonstrate the competitive exclusion model.
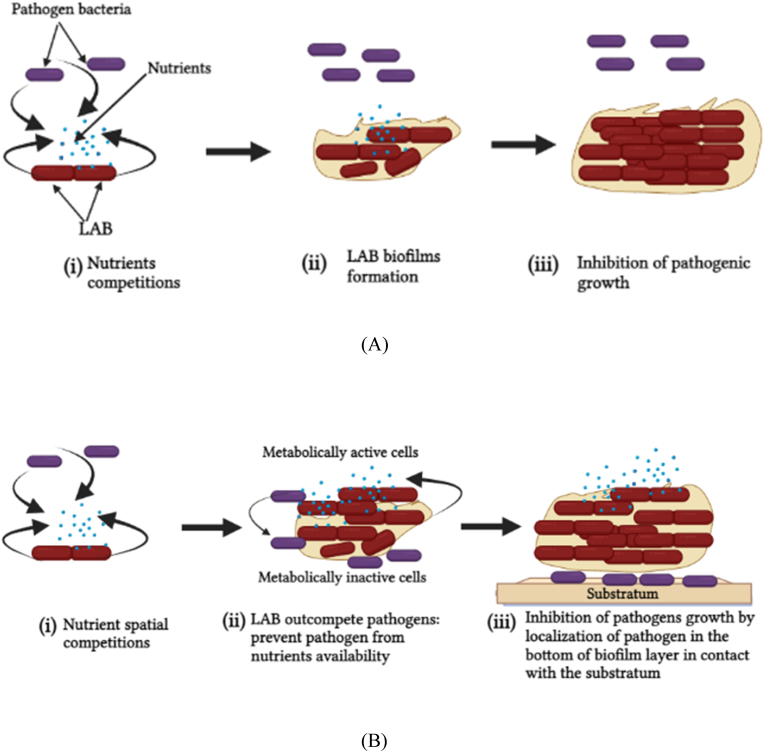
LAB biofilms provide competitive exclusion mechanisms (Jameson effect) (Fig. 1A). Both LAB and pathogens compete for nutrients availability, LAB wins and establishes surface colonization by producing biofilms that inhibit pathogenic nutrients uptake; LAB cells population increases, and the biofilms reach the maturation stage resulting in the inhibition of pathogenic cell growth due to the lack of nutrients and may eventually cause bacterial cell death. The natural biofilm microbiota obtained from cheese-ripening wooden shelves that were used to inhibit the growth of L. monocytogenes was a good example. Despite this, the growth rate of L. monocytogenes in co-culture with these biofilms was not reduced when compared to Listeria alone. When the biofilms reached the stationary phase, L. monocytogenes growth ceased immediately. The exhaustion and nutrient consumption by the bacteria biofilms demonstrated that this scenario reduced the maximum population density of L. monocytogenes [89].
Fig. 1.
Competitive exclusion mechanism of action offered by LAB biofilms against pathogens bacteria: (A) Jameson effect, (i) pathogenic bacteria and LAB are both competing for nutrients, (ii) LAB win the competitions and develop biofilms that prevent pathogenic nutrients consumptions, (iii) eventually pathogenic cell death occurs due to lack of nutrients. (B) Spatial competitions. (i) Nutrients spatial competitions between LAB and pathogenic bacteria (ii) metabolic active LAB cell form biofilms and restrict pathogenic bacteria at the bottom layer of biofilms, inactive pathogenic bacteria cells develop due to insufficient nutrient supply, (iii) inhibition of pathogenic growth occur.
On the other hand, another competitive exclusion approach demonstrated in a simplified individual-based model showed that LAB biofilms can spatially restrict pathogenic cells to the base of the biofilm layers, hence inhibiting their growth [90] (Fig. 1B). For example, in the mixed continuous-flow biofilms of Listeria spp. and L. lactis monitored by confocal laser scanning microscopy, Listeria cells were prevented and entirely covered by L. lactis cells and were localized in the base of biofilm layers in contact with the substratum. Moreover, through the analysis by individual-based modeling, it was suggested that the growth parameters of the two bacteria were governed by the spatial competition occurring during the early stages of biofilm formation. L. monocytogenes showed longer latency and generation time and were outcompeted by L. lactis, which showed a shorter latency time occupying the biofilm surface layer. Moreover, the biofilms developed favoring L. lactis, which initially accessed and utilized the nutrients. At the same time, Listeria cells were spatially exclusively at the bottom of the biofilm with insufficient nutrient supply. Nutrients found in the upper layer of biofilm were highly consumed and utilized compared to the bottom. These spatial races for nutrient consumption could probably explain the growth inhibition of L. monocytogenes cells found in the base layer of mixed-species biofilms [90].
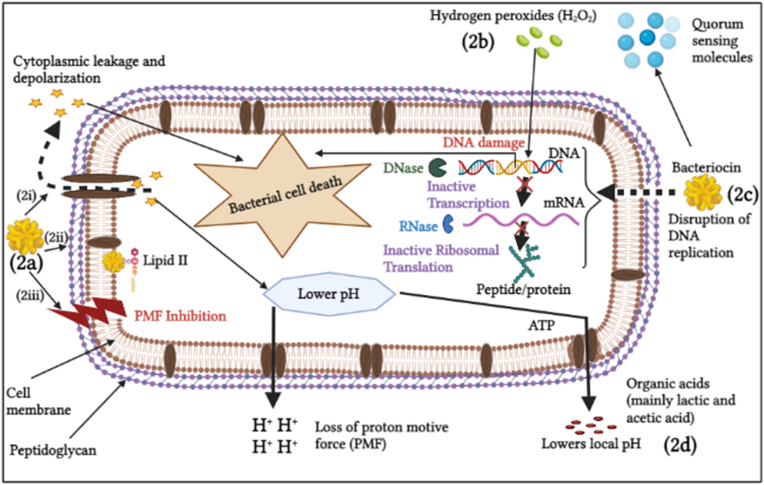
Fig. 2.
Hypothetic mechanisms of action of antimicrobial substances in LAB biofilms against pathogen bacteria: (2a) effect of bacteriocins on the bacteria cell wall, which causes inhibition of cell wall synthesis results into; (2i) pore formation that causes cytoplasmic leakage and depolarization, bacteriocins causes (2ii) inhibition of peptidoglycan synthesis; and (2iii) disruption of the cell membrane. Furthermore, (2b) Hydrogen peroxides disrupt the normal metabolism process of protein, DNA, and RNA, which cause DNA damage and cell death of the targeted bacteria (2c), on the other hand, bacteriocins prevent DNA replication, causing DNA damage, inhibiting DNA gyrase, RNA polymerase, and aspartyle tRNA synthetase, and eventually causing cell death; (2d), the effect of organic acids in the bacteria cell, lactic acid mainly dissociate in the cytoplasm resulting into increasing H+ to be pumped out which uses up cell ATP, as a results bacterial cell loss energy and finally death occur [57].
Another mechanism is through secretion of antimicrobial substances, such as bacteriocin and bacteriocins like substances [91]. Bacteriocin and bacteriocins-like substances function as cytoplasmic membrane perturbators that interact and promote the dissociation of lipid-II molecules. This effect prevents the normal cell cycle and cell wall synthesis of the targeted cell. It causes pore formation in the bacterial cytoplasmic membrane, resulting in cell death through the dissipation of the proton motive force of the bacterial system. Nisin is a good example of the most commonly used bacteriocins as a food preservative. Studies have shown that nisin contains bactericidal effects capable of penetrating even the deepest layer of a biofilm matrix.
Additionally, nisin has been shown to induce the morphological changes of a biofilm structure [92], inhibit the expression of gene-related (fnbA, icaD, rnaIII, and clfB) for biofilm formation of S. aureus [93]. Contrary to that, at sub-inhibitory concentration, nisin promoted the biofilm formation of S. aureus. Based on these results, it has been shown that nisin sub-lethal concentration may trigger the expression of genes associated with biofilm formation, resulting in bacteriocin tolerance. This scenario causes resistance to pathogenic death; even higher concentrations will not kill the bacteria after exposure to sub-lethal doses [94]. While hydrogen peroxide causes cell death of a targeted bacteria by disrupting the normal metabolic process. Lastly, organic acids provide unfriendly acidic environments that prevent the growth of pathogens, causing the dissipation of proton motive force of the bacterial system leading to cell death (Fig. 2) [57].
Despite the fact that the mentioned approaches have shown some effect on the antibacterial mechanism of LAB biofilms, one area that requires further investigation is the mechanism of QS on antibacterial activity. Although QS plays an important role in biofilm formation and antibacterial activity, the exact role of antibacterial activity is still unclear. On the other hand, there is an immediate need to investigate the molecular mechanisms of antibacterial activity exerted by LAB biofilm. Furthermore, this review suggests the need to determine the influence of environmental factors on antibacterial activity. Factors such as temperature, pH, and nutrient availability may affect the formation and structure of LAB biofilms, which in turn may influence their antibacterial properties.
8. Challenges of using LAB biofilms for controlling pathogenic bacteria in food environments
Despite the safety and wide application of LAB in the food industry, LAB are often responsible for food spoilage, especially in raw, under-processed foods and fermented products. Occasional, due to the possibility of several LAB to enter VBNC state under environmental stress conditions can cause false detection during storage, shipment, and marketing, leading to food spoilage resulting in significant profit loss and pose a major concern, particularly in the brewing industry [95]. A good example was shown when VBNC cells of Lactobacillus harbinensis strain was identified in one spoiled beer sample by genome sequencing, and it was observed that the VBNC state was triggered by low-temperature storage and beer subculturing [95]. Therefore, it is important to detect the VBNC state of LAB before biofilm development to avoid any challenge that might come out of it. Likewise, biofilms formed by a certain type of LAB may also have detrimental properties. For instance, some Enterococcus, Streptococcus spp., and psychrotrophic LAB may produce harmful biofilm, which can modify the color or flavor of food [96], and Leuconostoc gasicomitatum may cause spoilage of food products such as meat [97], causing infections that may compromise consumer health. In addition to these challenges, LAB biofilms may reduce heat transfer, subsequent corrosion, and equipment biofouling [98].
Additionally, certain non-starter LAB causes food product misconceptions. For instance, L. curvatus and L. fermentum biofilms can cause cheese defects by producing gas, undesirable flavors or racemase, transforming L (+) –lactic acid into less soluble D (−) lactic acid, producing calcium lactate crystals [99,100]. Although the calcium lactate crystals are not harmful, consumers associate the white haze with mold growth and reject such cheeses. Moreover, the formation of LAB mixed-species biofilms, the most prominent form of biofilms of both LAB and pathogens, can increase food safety problems due to their higher resistance to disinfection than single-species biofilm [5]. Lastly, not all LAB-producing biofilms can inhibit or reduce the growth of foodborne bacteria. Some LAB strains (L. fermentum MP26, Lactobacillus salivarius MP14) could not inhibit L. monocytogenes adhesion, because the viable population remained stable throughout the incubation period [101]. All these challenges may finally result in the rejection of food products. For the aforementioned reasons, investigation and selection of appropriate strains to include in controlled LAB biofilms is demanded before any food application. An in-depth understanding of their side effect will help minimize challenges caused by LAB, hence expanding their application in various industries.
9. Conclusion and future perspective
LAB biofilms have shown tremendous and fantastic effects in preventing undesirable microorganisms, unlike pathogenic biofilms, whose prevention and eradication are severe challenges in a food and clinical environment. LAB biofilms are important candidates to be considered for producing food free of pathogens and treating various infections. Their application opens a door for developing environmentally friendly approaches as potential alternatives to chemical preservatives and antibiotics. The rapid advancement of methods for combating pathogenic microorganisms may help to improve the use of LAB biofilms to enhance their effectiveness in controlling pathogens. Therefore, using LAB biofilms to combat pathogens and their biofilms is more promising than conventional methods. Future studies should look into using LAB biofilms to inhibit/control viruses like Norovirus, Hepatitis A, as well as parasites such as fish-borne trematodes. Furthermore, the application of EPS produced from LAB biofilms should be investigated separately to understand better the underlying mechanism of action of LAB biofilms against undesirable microorganisms. Additionally, we propose that combining LAB biofilms with other LAB products, such as bacteriocins and H2O2, could provide viable alternatives to conventional methods in combating pathogenic bacteria in various industries. Also, this review recommends that more research should be conducted on using LAB in clinical environments, especially on antibiotic-resistant bacteria strains.
Declaration of competing interest
The authors declared that they have no conflicts of interest to this work.
Acknowledgments
This work was written as part of a PhD project in the College of Food Science and Engineering at Yangzhou University, finically supported by the Natural Science Foundation of Jiangsu Province (Grants No BK20210814), the Foundation of China National Key Research & Development Program (2016YFC1300201), Jiangsu Key Research & Development Program (BE2019436-5), and China Postdoctoral Science Foundation (2021TQ0274). The authors wish to thank Lei Yuan and Lusana James for their support in the proofreading and review of the article.
Data availability
Review paper, thus no original data has been included, only published data
References
- 1.Mokoena M.P., Omatola C.A., Olaniran A.O. Applications of lactic acid bacteria and their bacteriocins against food spoilage microorganisms and foodborne pathogens. Molecules. 2021;26(22) doi: 10.3390/molecules26227055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giaouris E. Recent trends in biofilm science and technology. Elsevier Inc; 2020. Application of lactic acid bacteria and their metabolites against foodborne pathogenic bacterial biofilms. [DOI] [Google Scholar]
- 3.Mgomi F.C., Yuan L., Wang Y., Rao S.Q., Yang Z.Q. Physiological properties, survivability and genomic characteristics of Pediococcus pentosaceus for application as a starter culture. Int J Dairy Technol. 2022;0:1–15. doi: 10.1111/1471-0307.12864. [DOI] [Google Scholar]
- 4.Rocha K.R., Perini H.F., de Souza C.M., Schueler J., Tosoni N.F., Furlaneto M.C., Furlaneto-Maia L. Inhibitory effect of bacteriocins from enterococci on developing and preformed biofilms of Listeria monocytogenes, Listeria ivanovii and Listeria innocua. World J Microbiol Biotechnol. 2019;35(7):1–11. doi: 10.1007/s11274-019-2675-0. [DOI] [PubMed] [Google Scholar]
- 5.Mgomi F.C., Yuan L., Chen C., Zhang Y., Yang Z. Bacteriophages: a weapon against mixed‐species biofilms in the food processing environment. J Appl Microbiol. 2021:1–15. doi: 10.1111/jam.15421. 00. [DOI] [PubMed] [Google Scholar]
- 6.Yuan L., Sadiq F.A., Wang N., Yang Z., He G. Recent advances in understanding the control of disinfectant-resistant biofilms by hurdle technology in the food industry. Crit Rev Food Sci Nutr. 2020;0(0):1–16. doi: 10.1080/10408398.2020.1809345. [DOI] [PubMed] [Google Scholar]
- 7.Gonçalves B., Barbosa A., Soares A.R., Henriques M., Silva Só. Sfl1 is required for Candida albicans biofilm formation under acidic conditions. Biochimie. 2023 doi: 10.1016/j.biochi.2023.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Kim U., Kim J.-H., Oh S.-W. Review of multi-species biofilm formation from foodborne pathogens: multi-species biofilms and removal methodology. Crit Rev Food Sci Nutr. 2021;0(0):1–11. doi: 10.1080/10408398.2021.1892585. [DOI] [PubMed] [Google Scholar]
- 9.Speranza B., Liso A., Russo V., Corbo M.R. Evaluation of the potential of biofilm formation of bifidobacterium longum subsp. Infantis and lactobacillus reuteri as competitive biocontrol agents against pathogenic and food spoilage bacteria. Microorganisms. 2020;8(2) doi: 10.3390/microorganisms8020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todhanakasem T., Ketbumrung K. Using potential lactic acid bacteria biofilms and their compounds to control biofilms of foodborne pathogens. Biotechnol Rep. 2020;26 doi: 10.1016/j.btre.2020.e00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanzan M., Achemchem F., Hamadi F., Latrache H., Elmoslih A., Amzil K., Mimouni R. Researchgate.Net; 2020. pp. 18–22. (Antibacterial and anti-adherence activity of Enterococcus spp. against Staphylococcus aureus CECT 976). [Google Scholar]
- 12.Amin M., Adams M.B., Burke C.M., Bolch C.J. Isolation and screening of lactic acid bacteria associated with the gastrointestinal tracts of abalone at various life stages for probiotic candidates. Aquacult Rep. 2020;17(May) doi: 10.1016/j.aqrep.2020.100378. [DOI] [Google Scholar]
- 13.Dao T., Dantigny P. Control of food spoilage fungi by ethanol. Food Control. 2011;22:360–368. doi: 10.1016/j.foodcont.2010.09.019. [DOI] [Google Scholar]
- 14.Reuben R.C., Roy P.C., Sarkar S.L., Alam R.U., Jahid I.K. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019;19(1):1–20. doi: 10.1186/s12866-019-1626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann E., Schmitz‐Esser S., Zebeli Q., Wagner M., Ritzmann M., Metzler-Zebeli B.U. Mucosa‐associated bacterial microbiome of the gastrointestinal tract weaned pigs and dynamics linked to dietary calcium‐phosphorus. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z., Li L., Fang Z.F., Lee Y., Zhao J., Zhang H., Chen W., Li H., Lu W. The biofilm-forming ability of six Bifidobacterium strains on grape seed flour. LWT (Lebensm-Wiss & Technol) 2021;144(February) doi: 10.1016/j.lwt.2021.111205. [DOI] [Google Scholar]
- 17.Fan Y., Huang X., Chen J., Han B. formation of a mixed-species biofilm is a survival strategy for unculturable lactic acid bacteria and Saccharomyces cerevisiae in Daqu, a Chinese traditional fermentation starter. Front Microbiol. 2020;11(February):1–13. doi: 10.3389/fmicb.2020.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastard A., Coelho C., Briandet R., Canette A., Gougeon R., Alexandre H., Guzzo J., Weidmann S. Effect of biofilm formation by Oenococcus oeni on malolactic fermentation and the release of aromatic compounds in wine. Front Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kembel S.W., Meadow J.F., O'Connor T.K., Mhuireach G., Northcutt D., Kline J., Moriyama M., Brown G.Z., Bohannan B.J.M., Green J.L. Architectural design drives the biogeography of indoor bacterial communities. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meadow J.F., Altrichter A.E., Kembel S.W., Maxwell M., O'Connor T.K., Womack A.N., Brown G.Z., Green L.J., Bohannan B.J.M. Bacterial communities on classroom surfaces vary with human contact. Microbiome. 2014;2(7) doi: 10.1186/2049-2618-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macfarlane S., Macfarlane G.T. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl Environ Microbiol. 2006;72:6204–6211. doi: 10.1128/AEM.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao T., Doyle M.P., Zhao P. Control of Listeria monocytogenes in a biofilm by competitive-exclusion microorganisms. Appl Environ Microbiol. 2004;70(7):3996–4003. doi: 10.1128/AEM.70.7.3996-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landeta G., Curiel J.A., Carrascosa A.V., Muñoz R., De las Rivas B. Technological and safety properties of lactic acid bacteria isolated from Spanish dry-cured sausages. Meat Sci. 2013;95(2):272–280. doi: 10.1016/j.meatsci.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 24.Gunduz G.T., Tuncel G. Biofilm formation in an ice cream plant. Antonie van Leeuwenhoek, Int J General Mol Microbiol. 2006;89(3–4):329–336. doi: 10.1007/s10482-005-9035-9. [DOI] [PubMed] [Google Scholar]
- 25.Sun L., Zhang Y., Guo X., Zhang L., Zhang W., Man C., Jiang Y. Characterization and transcriptomic basis of biofilm formation by Lactobacillus plantarum J26 isolated from traditional fermented dairy products. LWT (Lebensm-Wiss & Technol) 2020;125(March) doi: 10.1016/j.lwt.2020.109333. [DOI] [Google Scholar]
- 26.Frese S.A., MacKenzie D.A., Peterson D.A., Schmaltz R., Fangman T., Zhou Y., et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013;9(12) doi: 10.1371/journal.pgen.1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooshdar P., Kermanshahi R.K., Ghadam P., Khosravi-Darani K. A review on production of exopolysaccharide and biofilm in probiotics like lactobacilli and methods of analysis. Biointerf Res Appl Chem. 2020;10(5):6058–6075. doi: 10.33263/BRIAC105.60586075. [DOI] [Google Scholar]
- 28.Kannappan A., Balasubramaniam B., Ranjitha R., Srinivasan R., Packiavathy I.A.S.V., Balamurugan K., Pandian S.K., Ravi A.V. In vitro and in vivo biofilm inhibitory efficacy of geraniol-cefotaxime combination against Staphylococcus spp. Food Chem Toxicol. 2019;125(January):322–332. doi: 10.1016/j.fct.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen P.T., Nguyen T.T., Bui D.C., Hong P.T., Hoang Q.K., Nguyen H.T. Exopolysaccharide production by lactic acid bacteria: the manipulation of environmental stresses for industrial applications. AIMS Microbiol. 2020;6(4):451–469. doi: 10.3934/MICROBIOL.2020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y., Cui Y., Yue F., Liu L., Shan Y., Liu B., Zhou Y., Lü X. Exopolysaccharides produced by lactic acid bacteria and Bifidobacteria: structures, physiochemical functions and applications in the food industry. Food Hydrocolloids. 2019;94:475–499. doi: 10.1016/j.foodhyd.2019.03.032. [DOI] [Google Scholar]
- 31.Nazir R., Zaffar M.R., Amin I. Freshwater microbiology: perspectives of bacterial dynamics in lake ecosystems. Elsevier Inc; 2019. Bacterial biofilms: the remarkable heterogeneous biological communities and nitrogen fixing microorganisms in lakes. [DOI] [Google Scholar]
- 32.Drame I., Lafforgue C., Formosa-Dague C., Chapot-Chartier M.P., Piard J.C., Castelain M., Dague E. Pili and other surface proteins influence the structure and the nanomechanical properties of Lactococcus lactis biofilms. Sci Rep. 2021;11(1):4846. doi: 10.1038/s41598-021-84030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olszewska M.A., Nynca A., Białobrzewski I. Biofilm formation by lactobacilli and resistance to stress treatments. Int J Food Sci Technol. 2019;54(11):3058–3065. doi: 10.1111/ijfs.14219. [DOI] [Google Scholar]
- 34.Pannella G., Lombardi S.J., Coppola F., Vergalito F., Iorizzo M., Succi M., Tremonte P., Iannini C., Sorrentino E., Coppola R. Effect of biofilm formation by lactobacillus plantarum on the malolactic fermentation in model wine. Foods. 2020;9(6) doi: 10.3390/foods9060797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wanna W., Surachat K., Kaitimonchai P., Phongdara A. Evaluation of probiotic characteristics and whole genome analysis of Pediococcus pentosaceus MR001 for use as probiotic bacteria in shrimp aquaculture. Sci Rep. 2021;11(1):1–17. doi: 10.1038/s41598-021-96780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vélez M.P., Petrova M.I., Lebeer S., Verhoeven T.L.A., Claes I., Lambrichts I., Tynkkynen S., Vanderleyden J., De Keersmaecker S.C.J. Characterization of MabA, a modulator of Lactobacillus rhamnosus GG adhesion and biofilm formation. FEMS Immunol Med Microbiol. 2010;59(3):386–398. doi: 10.1111/j.1574-695X.2010.00680.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu L., Wu R., Zhang J., Li P. Overexpression of luxSPromotes stress resistance and biofilm Formation of lactobacillus paraplantarumL-ZS9 by regulating the expression of multiple genes. Front Microbiol. 2018;9(NOV):1–11. doi: 10.3389/fmicb.2018.02628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubota H., Senda S., Nomura N., Tokuda H., Uchiyama H. Biofilm formation by lactic acid bacteria and resistance to environmental stress. J Biosci Bioeng. 2008;106:381–386. doi: 10.1263/jbb.106.381. [DOI] [PubMed] [Google Scholar]
- 39.Liu J., Deng Y., Soteyome T., Li Y., Su J., Li L., Li B., Shirtliff M.E., Xu Z., Peters B.M. Induction and recovery of the viable but Nonculturable state of hop-resistance Lactobacillus brevis. Front Microbiol. 2018;9:2076. doi: 10.3389/fmicb.2018.02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J., Li L., Li B., Peters B.M., Deng Y., Xu Z., Shirtliff M.E. Study on spoilage capability and VBNC state formation and recovery of Lactobacillus plantarum. Microbial Pathol. 2017;110:257–261. doi: 10.1016/j.micpath.2017.06.044. [DOI] [PubMed] [Google Scholar]
- 41.Piao M., Li Y., Wang Y., Wang F., Zhen T., Deng Y. Induction of viable but putatively non-culturable Lactobacillus acetotolerans by thermosonication and its characteristics. LWT-Food Science and Technololgy. 2019;109:313–318. doi: 10.1016/j.lwt.2019.04.046. [DOI] [Google Scholar]
- 42.Yao C.Q., Li J., Jingjing E., Wang R.X., Zhang Q.L., Wang J. The symbiosis among, and the storage stabilities of, starter lactic acid bacterial strains in biofilms. LWT - Food Sci Technol (Lebensmittel-Wissenschaft -Technol) 2022;155 doi: 10.1016/j.lwt.2021.112896. [DOI] [Google Scholar]
- 43.Alshanta O.A., Albashaireh K., McKloud E., Delaney C., Kean R., McLean W., Ramage G. Candida albicans and Enterococcus faecalisbiofilm frenemies: when the relationship sours. Biofilms. 2022;14(4) doi: 10.1016/j.bioflm.2022.100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnamoorthy A.L., Lemus A.A., Solomon A.P., Valm A.M., Neelakantan P. Interactions betweenCandida albicans and Enterococcus faecalis in an organotypic oral epithelial model. Microorganisms. 2020;8(11):1771. doi: 10.3390/microorganisms8111771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aman M., Aneeqha N., Bristi K., Deeksha J., Afza N., Sindhuja V., Shastry R.P. Lactic acid bacteria inhibits quorum sensing and biofilm formation of Pseudomonas aeruginosa strain JUPG01 isolated from rancid butter. Biocatal Agric Biotechnol. 2021;36(April) doi: 10.1016/j.bcab.2021.102115. [DOI] [Google Scholar]
- 46.Kim N.N., Kim W.J., Kang S.S. Anti-biofilm Effect of crude bacteriocin derived from Lactobacillus brevis DF01 on Escherichia coli and Salmonella Typhimurium. Food Control. 2019;98:274–280. doi: 10.1016/j.foodcont.2018.11.004. March 2018. [DOI] [Google Scholar]
- 47.Ismail A., Lani M.N., Zakeri H.A., Hasim N.N., Alias R., Mansor A. Synergistic of antimicrobial activities of lactic acid bacteria in fermented Tilapia nicoliticus incorporated with selected spices. Food Res. 2021;5(3):163–173. doi: 10.26656/fr.2017.5(3).534. [DOI] [Google Scholar]
- 48.Kaewchomphunuch T., Charoenpichitnunt T., Thongbaiyai V., Ngamwongsatit N., Kaeoket K. Cell-free culture supernatants of Lactobacillus spp. and Pediococcus spp. inhibit growth of pathogenic Escherichia coli isolated from pigs in Thailand. BMC Vet Res. 2022;18(1):1–13. doi: 10.1186/s12917-022-03140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bungenstock L., Abdulmawjood A., Reich F. Evaluation of antibacterial properties of lactic acid bacteria from traditionally and industrially produced fermented sausages from Germany. PLoS One. 2020;15(3):1–15. doi: 10.1371/journal.pone.0230345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bajpai V.K., Han J.H., Rather I.A., Park C., Lim J., Paek W.K., Lee J.S., Yoon J.I., Park Y.H. Characterization and antibacterial potential of lactic acid bacterium Pediococcus pentosaceus 4I1 isolated from freshwater fish Zacco koreanus. Front Microbiol. 2016;7(DEC):1–15. doi: 10.3389/fmicb.2016.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren D., Zhu J., Gong S., Liu H., Yu H. Antimicrobial characteristics of lactic acid bacteria isolated from homemade fermented foods. BioMed Res Int. 2018;2018 doi: 10.1155/2018/5416725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirzaei E.Z., Lashani E., Davoodabadi A. Antimicrobial properties of lactic acid bacteria isolated from traditional yogurt and milk against Shigella strains. GMS Hygiene and Infection Control. 2018;13:Doc01. doi: 10.3205/dgkh000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aka-Gbezo S., Konan A.G., N'Cho M., Achi P., Koffi-Nevry R., Koussemon-Camara M., Bonfoh B. Screening of antimicrobial activity of lactic acid bacteria isolated from anango baca slurry, a spontaneously fermented maize product used in Côte d'Ivoire. Int J Brain Cognit Sci. 2018;11(6):2616. doi: 10.4314/ijbcs.v11i6.6. [DOI] [Google Scholar]
- 54.Abubakr M.A.S. Antimicrobial activities of lactic acid bacteria strains isolated from human breast milk against human pathogenic strains. Int J Clin Dev Anat. 2018;4(1):27. doi: 10.11648/j.ijcda.20180401.14. [DOI] [Google Scholar]
- 55.Matevosyan L., Bazukyan I., Trchounian A. Antifungal and antibacterial effects of newly created lactic acid bacteria associations depending on cultivation media and duration of cultivation. BMC Microbiol. 2019;19(1):1–8. doi: 10.1186/s12866-019-1475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartkiene E., Lele V., Sakiene V., Zavistanaviciute P., Ruzauskas M., Bernatoniene J., Jakstas V., Viskelis P., Zadeike D., Juodeikiene G. Improvement of the antimicrobial activity of lactic acid bacteria in combination with berries/fruits and dairy industry by-products. J Sci Food Agric. 2019;99(8):3992–4002. doi: 10.1002/jsfa.9625. [DOI] [PubMed] [Google Scholar]
- 57.Hossain M.I., Mizan M.F.R., Ashrafudoulla M., Nahar S., Joo H.J., Jahid I.K., Park S.H., Kim K.S., Ha S. Do. Inhibitory effects of probiotic potential lactic acid bacteria isolated from kimchi against Listeria monocytogenes biofilm on lettuce, stainless-steel surfaces, and MBECTM biofilm device. LWT (Lebensm-Wiss & Technol) 2020;118(June) doi: 10.1016/j.lwt.2019.108864. [DOI] [Google Scholar]
- 58.Merino L., Trejo F.M., De Antoni G., Golowczyc M.A. Lactobacillus strains inhibit biofilm formation of Salmonella sp. isolates from poultry. Food Res Int. 2019;123:258–265. doi: 10.1016/j.foodres.2019.04.067. [DOI] [PubMed] [Google Scholar]
- 59.Tan X., Han Y., Xiao H., Zhou Z. Pediococcus acidilactici inhibit biofilm formation of foodborne pathogens on abiotic surfaces. Trans Tianjin Univ. 2017;23(1):70–77. doi: 10.1007/s12209-016-0016-z. [DOI] [Google Scholar]
- 60.Winkelströter L.K., Tulini F.L., De Martinis E.C.P. Identification of the bacteriocin produced by cheese isolate Lactobacillus paraplantarum FT259 and its potential influence on Listeria monocytogenes biofilm formation. LWT - Food Sci Technol (Lebensmittel-Wissenschaft -Technol) 2015;64(2):586–592. doi: 10.1016/j.lwt.2015.06.014. [DOI] [Google Scholar]
- 61.Woo J., Ahn J. Probiotic-mediated competition, exclusion and displacement in biofilm formation by foodborne pathogens. Lett Appl Microbiol. 2013;56(4):307–313. doi: 10.1111/lam.12051. [DOI] [PubMed] [Google Scholar]
- 62.Al-Seraih A., Belguesmia Y., Baah J., Szunerits S., Boukherroub R., Drider D. Enterocin B3A-B3B produced by LAB collected from infant faeces: potential utilization in the food industry for Listeria monocytogenes biofilm management. Antonie van Leeuwenhoek, Int J General Mol Microbiol. 2017;110(2):205–219. doi: 10.1007/s10482-016-0791-5. [DOI] [PubMed] [Google Scholar]
- 63.Seo H.J., Kang S.S. Inhibitory effect of bacteriocin produced by Pediococcus acidilactici on the biofilm formation of Salmonella Typhimurium. Food Control. 2020;117 doi: 10.1016/j.foodcont.2020.107361. [DOI] [Google Scholar]
- 64.Yi L., Luo L., Lü X. Heterologous expression of two novel bacteriocins produced by Lactobacillus crustorum MN047 and application of BM1157 in control of Listeria monocytogenes. Food Control. 2018;86:374–382. doi: 10.1016/j.foodcont.2017.11.042. [DOI] [Google Scholar]
- 65.Camargo A.C., de Paula O.A.L., Todorov S.D., Nero L.A. In vitro evaluation of bacteriocins activity against Listeria monocytogenes biofilm formation. Appl Biochem Biotechnol. 2016;178(6):1239–1251. doi: 10.1007/s12010-015-1941-3. [DOI] [PubMed] [Google Scholar]
- 66.Bolocan A.S., Pennone V., O'Connor P.M., Coffey A., Nicolau A.I., McAuliffe O., Jordan K. Inhibition of Listeria monocytogenes biofilms by bacteriocin-producing bacteria isolated from mushroom substrate. J Appl Microbiol. 2017;122(1):279–293. doi: 10.1111/jam.13337. [DOI] [PubMed] [Google Scholar]
- 67.Dygico L.K., O'Connor P.M., Hayes M., Gahan C.G.M., Grogan H., Burgess C.M. Lactococcus lactis subsp. lactis as a natural anti-listerial agent in the mushroom industry. Food Microbiol. 2019;82:30–35. doi: 10.1016/j.fm.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 68.Cui X., Shi Y., Gu S., Yan X., Chen H., Ge J. Antibacterial and antibiofilm activity of lactic acid bacteria isolated from traditional artisanal milk cheese from northeast China against enteropathogenic bacteria. Probiotics and Antimicrobial Prot. 2018;10(4):601–610. doi: 10.1007/s12602-017-9364-9. [DOI] [PubMed] [Google Scholar]
- 69.Pelyuntha W., Chaiyasut C., Kantachote D., Sirilun S. Cell-free supernatants from cultures of lactic acid bacteria isolated from fermented grape as biocontrol against Salmonella Typhi and Salmonella Typhimurium virulence via autoinducer-2 and biofilm interference. PeerJ. 2019;2019(8) doi: 10.7717/peerj.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park H., Yeo S., Ji Y., Lee J., Yang J., Park S., Shin H., Holzapfel W. Autoinducer-2 associated inhibition by Lactobacillus sakei NR28 reduces virulence of enterohaemorrhagic Escherichia coli O157: H7. Food Control. 2014;45:62–69. doi: 10.1016/j.foodcont.2014.04.024. [DOI] [Google Scholar]
- 71.Sharma D., Saharan B.S. Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol Rep. 2016;11:27–35. doi: 10.1016/j.btre.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toushik S.H., Kim K., Ashrafudoulla M., Mizan M.F.R., Roy P.K., Nahar S., Kim Y., Ha S. Do. Korean kimchi-derived lactic acid bacteria inhibit foodborne pathogenic biofilm growth on seafood and food processing surface materials. Food Control. 2021;129(May) doi: 10.1016/j.foodcont.2021.108276. [DOI] [Google Scholar]
- 73.Hippolyte T.M., Alphonse T.S., Augustin M., Robert N., Somashekar D. Biosurfactants from lactic acid bacteria: a critical review on production, extraction, structural characterization and food application. Food Biosci. 2022;46 doi: 10.1016/j.fbio.2022.101598. [DOI] [Google Scholar]
- 74.Yan X., Gu S., Cui X., Shi Y., Wen S., Chen H., Ge J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb Pathog. 2019;127:12–20. doi: 10.1016/j.micpath.2018.11.039. October 2017. [DOI] [PubMed] [Google Scholar]
- 75.Kanmani P., Suganya K., Satish kumar R., Yuvaraj N., Pattukumar V., Paari K.A., Arul V. Synthesis and functional characterization of antibiofilm exopolysaccharide produced by enterococcus faecium mc13 isolated from the gut of fish. Appl Biochem Biotechnol. 2013;169(3):1001–1015. doi: 10.1007/s12010-012-0074-1. [DOI] [PubMed] [Google Scholar]
- 76.Ayyash M., Abu-Jdayil B., Olaimat A., Esposito G., Itsaranuwat P., Osaili T., Obaid R., Kizhakkayil J., Liu S.Q. Physicochemical, bioactive and rheological properties of an exopolysaccharide produced by a probiotic Pediococcus pentosaceus M41. Carbohydr Polym. 2020;229(August) doi: 10.1016/j.carbpol.2019.115462. [DOI] [PubMed] [Google Scholar]
- 77.Jeong D., Kim D.H., Kang I.B., Kim H., Song K.Y., Kim H.S., Seo K.H. Characterization and antibacterial activity of a novel exopolysaccharide produced by Lactobacillus kefiranofaciens DN1 isolated from kefir. Food Control. 2017;78:436–442. doi: 10.1016/j.foodcont.2017.02.033. [DOI] [Google Scholar]
- 78.Mahdhi A., Leban N., Chakroun I., Chaouch M.A., Hafsa J., Fdhila K., Mahdouani K., Majdoub H. Extracellular polysaccharide derived from potential probiotic strain with antioxidant and antibacterial activities as a prebiotic agent to control pathogenic bacterial biofilm formation. Microb Pathog. 2017;109:214–220. doi: 10.1016/j.micpath.2017.05.046. [DOI] [PubMed] [Google Scholar]
- 79.Kim J.H., Lee E.S., Song K.J., Kim B.M., Ham J.S., Oh M.H. Development of desiccation-tolerant probiotic biofilms inhibitory for growth of foodborne pathogens on stainless steel surfaces. Foods. 2022;11(6) doi: 10.3390/foods11060831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jalilsood T., Baradaran A., Song A.A.L., Foo H.L., Mustafa S., Saad W.Z., Yusoff K., Rahim R.A. Inhibition of pathogenic and spoilage bacteria by a novel biofilm-forming Lactobacillus isolate: a potential host for the expression of heterologous proteins. Microb Cell Factories. 2015;14(1):1–14. doi: 10.1186/s12934-015-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turhan E.U., Erginkaya Z., Uney M.H., Ozer E.A. Inactivation effect of probiotic biofilms on growth of Listeria monocytogenes. Kafkas Universitesi Veteriner Fakultesi Dergisi. 2017;23(4):541–546. doi: 10.9775/kvfd.2016.17253. [DOI] [Google Scholar]
- 82.Gómez N.C., Ramiro J.M.P., Quecan B.X.V., de Melo Franco B.D.G. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157: H7 biofilms formation. Front Microbiol. 2016;7(JUN):1–15. doi: 10.3389/fmicb.2016.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parolin C., Croatti V., Laghi L., Giordani B., Tondi M.R., De Gregorio P.R., Foschi C., Vitali B. Lactobacillus biofilms influence anti-Candida activity. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.750368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aoudia N., Rieu A., Briandet R., Deschamps J., Chluba J., Jego G., Garrido C., Guzzo J. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 2016;53:51–59. doi: 10.1016/j.fm.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 85.Rieu A., Aoudia N., Jego G., Chluba J., Yousfi N., Briandet R., et al. The biofilm mode of life boosts the anti-inflammatory properties of Lactobacillus. Cell Microbiol. 2014;16:1836–1853. doi: 10.1111/cmi.12331. [DOI] [PubMed] [Google Scholar]
- 86.Muhialdin B.J., Saari N., Hussin A.S. Review on the biological detoxification of mycotoxins using lactic acid bacteria to enhance the sustainability of foods supply. Molecules. 2020;25 doi: 10.3390/molecules25112655. 10.3390/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li P., Su R., Yin R., Lai D., Wang M., Liu Y., Zhou L. Detoxification of mycotoxins through biotransformation. Toxins. 2020;12:1–37. doi: 10.3390/toxins12020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Assaf J.C., El Khoury A., Chokr A., Louka N., Atoui A. A novel method for elimination of aflatoxin M1 in milk using Lactobacillus rhamnosus GG biofilm. Int J Dairy Technolol. 2019;72:248–256. [Google Scholar]
- 89.Guillier L., Stahl V., Hezard B., Notz E., Briandet R. Modelling the competitive growth between Listeria monocytogenes and biofilm microflora of smear cheese wooden shelves. Int J Food Microbiol. 2008;128(1):51–57. doi: 10.1016/j.ijfoodmicro.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 90.Habimana O., Guillier L., Kulakauskas S., Briandet R. Spatial competition with Lactococcus lactis in mixed-species continuous-flow biofilms inhibits Listeria monocytogenes growth. Biofouling. 2011;27(9):1065–1072. doi: 10.1080/08927014.2011.626124. [DOI] [PubMed] [Google Scholar]
- 91.Pang X., Song X., Chen M., Tian S., Lu Z., Sun J., Li X., Lu Y., Yuk H. Combating biofilms of foodborne pathogens with bacteriocins by lactic acid bacteria in the food industry. Compr Rev Food Sci Food Saf. 2022;21(2):1657–1676. doi: 10.1111/1541-4337.12922. [DOI] [PubMed] [Google Scholar]
- 92.Andre C., de Jesus Pimentel-Filho N., de Almeida Costa P.M., Vanetti M.C.D. Changes in the composition and architecture of staphylococcal biofilm by nisin. Braz J Microbiol. 2019;50(4):1083–1090. doi: 10.1007/s42770-019-00135-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pimentel-Filho N. de J., Martins M.C. de F., Nogueira G.B., Mantovani H.C., Vanetti M.C.D. Bovicin HC5 and nisin reduce Staphylococcus aureus adhesion to polystyrene and change the hydrophobicity profile and Gibbs free energy of adhesion. Int J Food Microbiol. 2014;190:1–8. doi: 10.1016/j.ijfoodmicro.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 94.Shivaee A., Rajabi S., Farahani H.E., Imani Fooladi A.A. Effect of sub-lethal doses of nisin on Staphylococcus aureus toxin production and biofilm formation. Toxicon. 2021;197(March):1–5. doi: 10.1016/j.toxicon.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 95.Liu J., Deng Y., Li L., Li B., Li Y., Zhou S., Shirtliff M.E., Xu Z., Peters B.M. Discovery and control of culturable and viable but non-culturable cells of a distinctive Lactobacillus harbinensis strain from spoiled beer. Scientific Reprod. 2018;8(1) doi: 10.1038/s41598-018-28949-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mende S., Rohm H., Jaros D. Encyclopedia of dairy sciences. Elsevier; 2022. Lactic acid bacteria: exopolysaccharides. [DOI] [Google Scholar]
- 97.Jääskeläinen E., Johansson P., Kostiainen O., Nieminen T., Schmidt G., Somervuo P., Mohsina M., Vanninen P., Auvinen P., Björkroth J. Significance of heme-based respiration in meat spoilage caused by Leuconostoc gasicomitatum. Appl Environ Microbiol. 2013;79(4):1078–1085. doi: 10.1128/AEM.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mozzi F., Raya R.R., Vignolo G.M. second ed. 2016. Biotechnology of lactic acid bacteria: novel applications. (Biotechnology of lactic acid bacteria: novel applications). Second Edition, 1–374. [DOI] [Google Scholar]
- 99.Agarwal S., Sharma K., Swanson B.G., Yüksel G.Ü., Clark S. Non-starter lactic acid bacteria biofilms and calcium lactate crystals in cheddar cheese. J Dairy Sci. 2006;89(5):1452–1466. doi: 10.3168/jds.S0022-0302(06)72213-5. [DOI] [PubMed] [Google Scholar]
- 100.Somers E.B., Johnson M.E., Wong A.C.L. Biofilm formation and contamination of cheese by non-starter lactic acid bacteria in the dairy environment. J Dairy Sci. 2001;84(9):1926–1936. doi: 10.3168/jds.S0022-0302(01)74634-6. [DOI] [PubMed] [Google Scholar]
- 101.Jara J., Pérez-Ramos A., del Solar G., Rodríguez J.M., Fernández L., Orgaz B. Role of Lactobacillus biofilms in Listeria monocytogenes adhesion to glass surfaces. Int J Food Microbiol. 2020;334 doi: 10.1016/j.ijfoodmicro.2020.108804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Review paper, thus no original data has been included, only published data