Abstract
Chimeric antigen receptor (CAR) T cell therapy has made significant strides in the treatment of B-cell malignancies, but its application in treating solid tumors still poses significant challenges. Particularly, the widespread use of viral vectors to deliver CAR transgenes into T cells comes with limitations, including high costs and regulatory restrictions, which hinder the translation of novel genetic engineering concepts into clinical applications. Non-viral methods, such as transposon/transposase and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas systems, offer promising alternatives for stable transgene insertion in CAR-T cells. These methods offer the potential to increase accessibility and efficiency in the development and delivery of CAR-T cell therapies. The main challenge in using non-viral methods, however, is their low knock-in efficiency, which leads to low transgene expression levels. In this review, we discuss recent developments in non-viral approaches for CAR-T cell production, the manufacturing requirements for clinical-grade production of non-viral CAR-T cells, and the adjustments needed in quality control for proper characterization of genomic features and evaluation of potential genotoxicity.
Key words: cell therapy scaling, CRISPR, electroporation, gene delivery, manufacturing, non-viral CAR T cells
Highlights
-
•
High costs and regulatory hurdles associated with viral vectors for CAR T therapies.
-
•
Non-viral gene delivery may overcome limitations of viral vector gene delivery.
-
•
Low knock-in efficiency remains a challenge in non-viral CAR-T cell production.
-
•
Focus on advancements in non-viral vector design and clinical manufacturing.
-
•
Analytical methods to characterize off-target editing and genotoxicity.
Introduction
Chimeric antigen receptor (CAR) T cell therapies, that involve the use of genetically modified T cells expressing CD19-CAR, have transformed the treatment landscape for refractory B-cell and plasma cell malignancies. Several CAR-T cell therapies have received Food and Drug Administration (FDA) approval as second- and third-line treatments, and ongoing clinical trials are evaluating their effectiveness as first-line therapies.1, 2, 3, 4 Current engineering approaches focus on developing the next generation of CAR-T cells to enhance their potency and safety profile.5, 6, 7 These include strategies aimed at improving CAR-T cell fitness (e.g. signaling, killing), overcoming antigen loss (e.g. via dual-targeting CARs), and modifying the tumor microenvironment (e.g. interleukin-18-secreting CARs),5,7, 8, 9 particularly as we progress towards evaluating the efficacy of CAR-T cell therapies in solid tumors.
Despite the great promise of CAR-T cells, the complexity and duration of the current manufacturing process have limited its widespread use. Traditionally, the production of CAR-T cells has relied on viral delivery systems to genetically modify T cells or other immune cells. This method leverages the virus’s inherent ability to deliver genetic material into the host genome efficiently and safely. The high costs of good manufacturing practice (GMP) grade viral vector production, however, reduced lot sizes, and the regulatory burden associated with using a viral vector, present major obstacles to the clinical development of CAR-T cells.10 Thus, there is a pressing need for more flexible, cost-effective approaches to streamline production and logistics related to gene delivery systems.
Non-viral delivery platforms present a promising alternative to address the limitations associated with viral delivery in CAR-T cell therapies.11 Viral vectors exhibit other notable drawbacks such as elevated immunogenic potential, restricted insert size, theoretical possibility of insertional mutagenicity, and substantial production costs. In contrast, non-viral approaches alleviate these limitations through decreased manufacturing expenses, increased cargo capacity, greater design flexibility, and diminished immunogenic profiles. Non-viral gene engineering methods can be classified into those that result in transient gene expression,12,13 and those that result in stable transgene integration. Non-viral stable integration is of particular interest. This can be achieved through either transposon-based systems such as Sleeping Beauty or PiggyBac,14,15 or through directed integration via clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated knock-in.16 The transposase systems can be delivered as proteins, messenger RNAs (mRNAs), or plasmid DNA,15,17 whereas CRISPR/Cas9 system typically uses linearized DNA (double- or single-stranded),16,18, 19, 20 or circular DNA.21,22
This review focuses on the use of non-viral methods for generating CAR-T cells with appropriate purity and yield for clinical use. We examine modifications to the manufacturing process to implement electroporation techniques. Finally, we emphasize strategies to minimize genotoxicity and evaluate its impact in regard to quality control (QC) during non-viral CAR-T cell production.
Challenges in non-viral gene editing for T cell therapy: manufacturing, QC, and safety
Non-viral gene editing systems can be delivered to primary T cells using electroporation, liposome, or nanoparticle transfection methods. Electroporation, being the gold standard, relies on the creation of transient pores in the cell membrane for gene delivery. This method may result in considerable cell loss, however, and the recovery of cell viability requires longer cell expansion times.17,19 To achieve engineered cell yields comparable to viral transduction, a higher number of cells must be activated before electroporation, which necessitates larger starting and expansion media volumes, and alternative cultivation vessels compared with those utilized in viral-based product manufacturing.
Furthermore, non-viral gene integration techniques tend to produce lower CAR delivery and expression levels compared with viral transduction methods. As a result, a final enrichment step is required to increase purity and prevent manufacturing failure.23 The selected enrichment strategy must meet three crucial criteria: lack of immunogenicity, compatibility with GMP grade reagents, and feasibility at a clinical scale. Enrichment methods include those based on surface marker expression,24 as well as drug or mutant enzyme selection systems that confer resistance to specific drugs, resulting in the selection or elimination of non-edited T cells.25 The development of selection tools that enable the enrichment of therapeutic T cells with multiple edits or transgene insertions is necessary. The optimization of non-viral CAR-T cell production processes is thus a pressing matter that needs to be addressed.
The advancements in gene engineering necessitate the development of appropriate QC assessments that evaluate the specific risks associated with the method. Currently, detection of viral load on transduced T cells is done via vector copy number quantification employing techniques like quantitative (q) or digital droplet polymerase chain reaction (ddPCR).26 For transposon/transposase methods, however, transgene copy number quantification is carried out due to their random insertion profile.27 Nonetheless, recent concerns about ‘CAR-T cell lymphoma’ in the CARTELL trial, using PiggyBac transposons, mandate a more thorough analysis of transposon/transposase-based genomic integration,28 as discussed later.
CRISPR/Cas9-based knock-in relies on creating double-stranded DNA (dsDNA) breaks, which raises the possibility of off-target cutting. To address this, thorough testing and optimization of guide RNAs (gRNAs) and identification of their cutting profiles is mandatory before clinical application. Various tools for in silico, in cellula, or in vitro detection of off-target sites have been developed, with next-generation sequencing (NGS) used to provide robust quantification of off-target cutting events.29 Additionally, the inherent potential of dsDNA breaks to cause translocations requires the development of methods to detect or deconvolute these rare translocations, which is a current challenge in the field.30
CAR Transgene delivery into primary T cells for stable integration and persistent expression
The two non-viral methods that enable stable genomic integration of CAR transgenes in primary T cells differ in their pattern of integration, with transposon/transposase-based systems demonstrating an almost random integration pattern and CRISPR/Cas9-based approaches exhibiting targeted integration.31
Transposon/transposase-based transgene insertion
Transposons are ‘jumping’ genetic elements capable of repositioning DNA in the genome. They can be classified into retrotransposons and DNA transposons. Retrotransposons, the most abundant transposons in the genome, consist of long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), and short interspersed nuclear elements (SINEs) and rely on RNA intermediates for transposition.32 DNA transposons, however, also known as class II transposable elements, utilize DNA templates for transposition and are widely used in gene engineering. PiggyBac and Sleeping Beauty are two examples of DNA transposons in use.
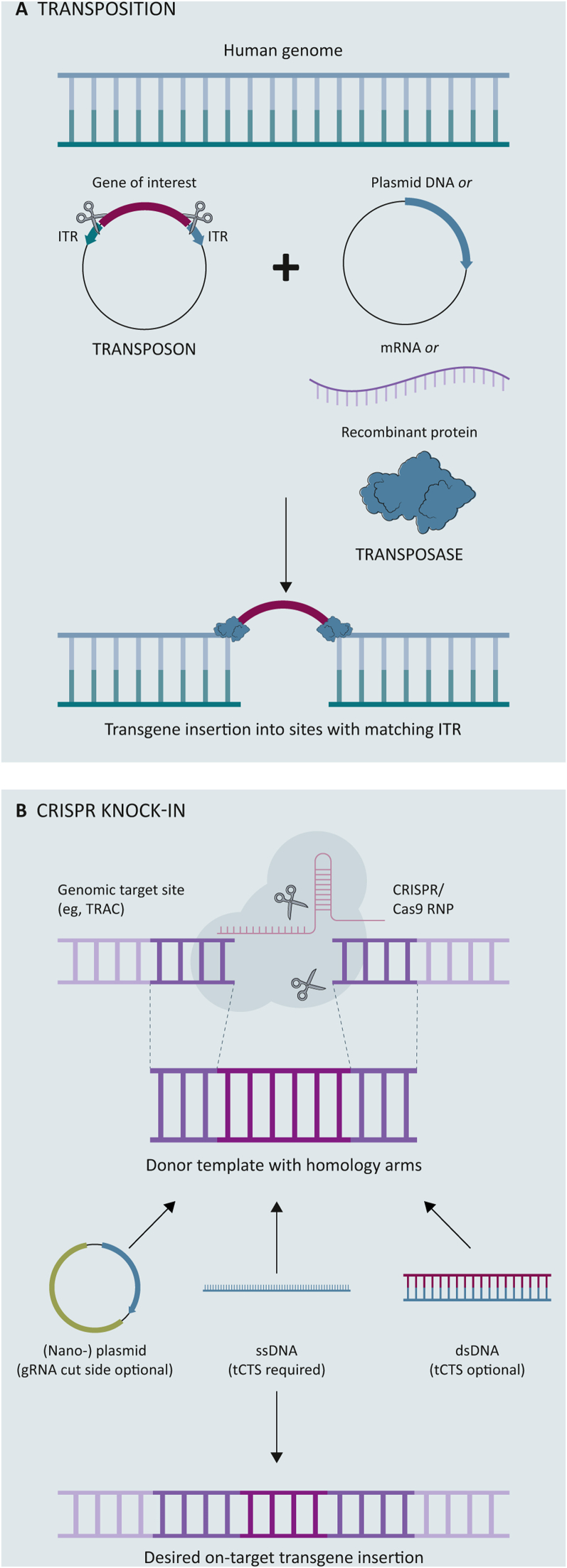
For transposition to occur, the transposon, also known as the gene of interest, must be bounded by inverted terminal repeats (ITRs). These ITRs serve as binding sites for the transposase, enabling excision and separation of the transposon from the donor DNA. The transposase then inserts the transposon into the acceptor DNA at -TA- (Sleeping Beauty) or -TTAA- (PiggyBac) sites31,33 (Figure 1A). The integration may not be specifically targeted, but it is also not completely random, as it is often directed towards preferred insertion sites known as ‘safe harbors’, as observed with Sleeping Beauty.15
Figure 1.
Overview of non-viral transgene insertion strategies. Created with BioRender.com.
CRISPR, clustered regularly interspaced short palindromic repeats; dsDNA, double-stranded DNA; gRNA, guide RNA; ITR, inverted terminal repeat; mRNA, messenger RNA; RNP, ribonucleoprotein; ssDNA, single-stranded DNA; tCTS, truncated Cas9 target sequence.
The transposon, which is responsible for repositioning DNA in the genome, is usually delivered in the form of (nano-)plasmid or minicircle DNA. The transposase enzyme responsible for integrating the transposon is typically delivered in trans as DNA, mRNA, or protein. Research indicates that mRNA-based transposases are preferred over protein-based transposases for their efficiency in cargo insertion, low cytotoxicity, and high level of safety due to their transient expression.15,17 The CARAMBA trial, for example, employed mRNA for Sleeping Beauty and minicircle DNA for the CAR transgene.34 Clinical trials utilizing Sleeping Beauty for the production of CD19-CAR-T cells in B-cell malignancies have been conducted without evidence of genotoxicity or transformation.14,35
The PiggyBac transposase, initially identified in insect cell lines,36 has been applied in the manufacture of CAR-T cells targeting CD19, prostate-specific membrane antigen (PSMA) (Poseida, San Diego, CA, P-PSMA-101), B-cell maturation antigen (BCMA) (P-BCMA-ALL01), and epidermal growth factor receptor (EGFR).14,36, 37, 38 One of the advantages of PiggyBac over Sleeping Beauty is its ability to integrate larger cargo sizes (∼200 kb versus ∼10 kb)39 and its precise excision, which reduces the risk of leaving a DNA footprint and achieve higher transposition activity.32 However, the CARTELL trial using PiggyBac to treat relapsed/refractory B-cell malignancies resulted in lymphoma in 2 out of 10 patients, raising concerns about the safety of PiggyBac and transposon/transposase-based cell therapies. The lymphoma may be related to the manufacturing process or an increase in global copy number changes observed in the products. As such, there is a need for better preclinical genotoxicity models and optimization strategies.28,40 CRISPR-associated transposons, which offer guided transposition and improved genotoxicity profiles, are emerging as a promising alternative.41,42
Targeted transgene delivery using CRISPR/Cas9
Clinical applications of genetic editing have progressed with the advancement of technologies such as transcription activator-like effector nucleases (TALENs), zinc finger endonucleases (ZFNs), and CRISPR/Cas9. The latter has been widely applied for gene editing due to its efficiency (per gene as well as for multiplexed editing) and versatility.43,44 This has paved the way for designing cell therapies, such as CAR-T cells, by utilizing CRISPR/Cas9-mediated transgene insertion.
The CRISPR/Cas9 system involves the use of a gRNA to direct the ribonucleoprotein (RNP) complex to a specific target DNA sequence. Upon nuclease binding, the DNA is broken, and the repair process is initiated, through non-homologous end joining (NHEJ) or by precise homology-directed repair (HDR). NHEJ may result in insertion or deletion mutations which might lead to premature stop codons or protein expression knock-out. HDR is, thus, the preferred gene editing approach as it does not introduce any additional modifications. In the case of CAR-T cell engineering, the delivery of a DNA repair template containing the CAR gene and complementary gene to the insertion site allows specific transgene integration.45 This approach reduces the chance of incorporation near oncogenes or tumor suppressor genes and enables efficient control via an endogenous promoter and simultaneous knock-out of target genes.46
Different delivery platforms have been proposed for the transfer of the DNA template required for CRISPR knock-in, including viral [e.g. adeno-associated virus 6 (AAV6)] and non-viral methods (e.g. linearized DNA, nano-plasmid DNA). Of these, AAV6 has proven to be the most efficient platform to date,16,22,47,48 and several clinical trials are exploring its use in the production of CAR-T cells, such as CTX110 and CTX120.49,50 Its limited payload capacity (<5 kb), however, may not necessarily lead to a reduction in the time and cost required for CAR-T cell production.51 The use of linearized DNA or plasmid DNA as a template for target insertion, however, may address some of the limitations of AAV6-based CRISPR/Cas9 knock-in, but this approach has been associated with high cell toxicity and low knock-in efficiencies.21,22,48 A study evaluating CRISPR/Cas9-mediated knock-in of an anti-CD19 CAR into programmed cell death protein 1 showed improved tumor clearance in a mouse model, suggesting the advantage of targeted CAR insertion. The clinical trial aimed at treating relapsed/refractory B-non-Hodgkin’s lymphoma with this method, however, failed to produce a sufficient dose in three out of eight patients, and the highest number of anti-CD19 CAR-T cells produced for the remaining patients was 2 × 108 CAR-T cells.20
Nguyen et al.52 have demonstrated that incorporating a truncated Cas9 target sequence (tCTS) at both ends of their double-stranded DNA (dsDNA) template and combining it with nuclear localization sequence bound to the RNP, enhances nuclear translocation and results in higher knock-in efficiencies. Subsequently, this approach has been extended to single-stranded DNA (ssDNA) templates, which have a reputation for lower toxicity but also lower knock-in efficiencies for CRISPR/Cas9-based knock-in. Shy et al.16 found that using ssDNA for CRISPR/Cas9-based knock-in of a BCMA CAR in a clinical setting yielded efficiencies of 40% and adequate yields after a 10-day manufacturing process. These outcomes provide optimism for the future of non-viral CRISPR/Cas9-based CAR-T cell production (Figure 1B).
To further enhance HDR efficiency, strategies have been proposed to inhibit cell cycle progression and NHEJ DNA repair in a timely manner. A CRISPR screen identified inhibitors of HDR. For example, targeting CDC7 kinase with XL413 can temporarily slow down S-phase progression, thereby prolonging the period for homology-directed recombination and resulting in a 3.5-fold increase in HDR efficiency.53 Alternatively, blocking NHEJ and promoting DNA repair via HDR increases knock-in frequency.54 Inhibiting DNA-dependent protein kinase with NU7441 and M3814 also resulted in 43% and 46% increases in knock-in efficiency, respectively.16 Interfering with DNA repair mechanisms post double-stranded breaks (DSBs) can cause genomic instability, however, making these strategies less appealing for the therapeutic engineering of non-viral CRISPR CAR-T cells.55
Required modifications for automated and closed manufacturing of non-viral CAR-T cells
The manufacturing of CAR-T cells using non-viral methods in a GMP setting poses unique challenges, including cell recovery and transgene insertion efficiency. Current non-viral manufacturing platforms utilize enriched CD4+/CD8+ T cells from leukapheresis, followed by activation, electroporation, expansion to desired cell numbers, and enrichment of CAR-T cells before harvest. To make non-viral CAR-T cell manufacturing feasible on automated and closed platforms, efforts should focus on improving electroporation, knock-out/knock-in efficiencies, and post-electroporation cell recovery and expansion yields.
Platforms for non-viral manufacturing
Upon receipt of starting material, manufacturing processes begin with the removal of platelets and red blood cells (RBCs) to enable optimal labeling of target cells, such as CD4+/CD8+ T-cell subsets, and subsequent enrichment and activation. Automated cell washing devices, including the Sepax C-Pro (Cytiva, Marlborough, MA), Lovo or Cue (Fresenius Kabi, Bad Homburg, Germany), and Rotea (Thermo Fisher Scientific, Waltham, MA), have been used in clinical trials to carry out this critical step effectively.56,57 The low insertion efficiency and cell loss associated with non-viral manufacturing approaches necessitate the implementation of highly efficient selection mechanisms.19,48 Although immunomagnetic labeling and selection of CD4 and CD8 T-cell subsets are the most widely used methods, the enrichment of CD62L+ (central memory T cells) or depletion of natural killer cells has also been employed.58 For cell enrichment, widely used devices include the CliniMACS Plus and CliniMACS Prodigy system (both from Miltenyi Biotec, Bergisch Gladbach, Germany), and the Robosep-C (STEMCELL Technologies, Vancouver, BC), which rely on immunomagnetic sorting, either column-based (Miltenyi Biotec devices) or non-column-based (Robosep-C). One advantage of the CliniMACS Prodigy and Robosep-C is their ability to integrate selection with automated platelet and RBC wash steps, reducing operator involvement.
To enhance genetic manipulation, a more accessible chromatin landscape is desirable, which can be achieved by activating T cells through CD3/T cell receptor and CD28 signaling pathways.17,48,59 The field has moved away from the traditional, time-consuming Dynabeads method (Thermo Fisher Scientific) to more efficient options such as magnetic beads coated with antibodies, including TransAct (Mitenyi Biotec) and Immunocult (STEMCELL Technologies). Artificial non-viable antigen-presenting cells that co-express tumor-associated antigens or stimulatory molecules are also gaining popularity.58 The CARAMBA trial, which investigated anti-SLAMF7 CAR-T cells for the treatment of multiple myeloma, employed T-cell activation and enrichment of CD4+/CD8+ positive T cells before electroporation on day 2 after activation, resulting in a 14-day production process.34 In contrast, two clinical trials that utilized the transposon/transposase system for transgene insertion relied on peripheral blood mononuclear cells (PBMCs) as starting material and did not activate the T cells before electroporation.14,35 This approach resulted in challenges in target cell population recovery, requiring the application of activated and propagating cells and extended cell cultures.
The primary method of non-viral gene delivery is through electroporation, where electrical fields are applied to cells to generate pores in the cell membrane, allowing the transgene to enter. This technique, however, often results in cell loss and temporary decreased viability due to the stress on the cells.17,22,48 In addition, high DNA concentrations and/or plasmid size can induce further cell death. To optimize the process, the pulse parameters (number and/or length), cell density, DNA concentration, and buffer composition must be carefully considered.48 Using a higher cell number before electroporation can increase the yield of edited cells, but this may be limited by the percentage of cells in the apheresis product.60,61 Electroporation also requires significant volume reduction, which can be achieved through manual cell centrifugation or with automated platforms mentioned earlier. The CliniMACS Prodigy electroporation platform is an exception, and recent studies have shown the feasibility of scaling up electroporation using the CliniMACS Electroporator and the Sleeping Beauty system, leading to a fully automated non-viral manufacturing process for generating anti-CD19 CAR-T cells in 12 days.62
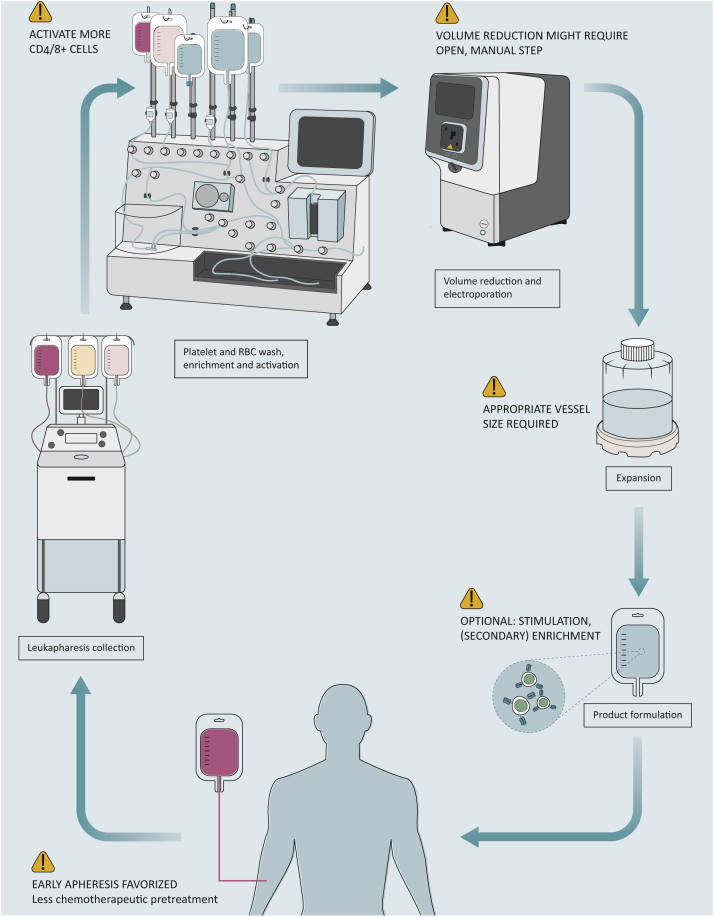
Despite the challenges posed by non-viral gene delivery, clinical trials have seen success in generating CAR-T cells for patients with B-cell leukemia and lymphoma by using electroporation with various devices, including the ExPERT GTx (MaxCyte Inc., Gaithersburg, MD), 4D-Nuclefector LV Unit (Lonza, Basel, Switzerland), CliniMACS Electroporator (Miltenyi Biotec), and the CTS Xenon (Thermo Fisher Scientific).12,14,28,35,62,63 To increase the yield of edited cells, the development of cultivation vessels and closed systems that can handle larger volumes of starting material and cell expansion is crucial.12,15 Miltenyi Biotec is releasing a new tubing set (TS-620), which can accommodate more cells than the previous cultivation chamber (TS-520). Alternative systems include the G-Rex (Wilson Wolf, Saint Paul, MN)64 and Xuri Bioreactor (Cytiva),65 both capable of expanding cultures up to 20 billion cells. The CliniMACS Prodigy is currently the only device that has the capabilities for cell washing, enrichment, electroporation, and expansion. Other devices with specific capabilities, however, can be connected to each other in an aseptic manner to create an automated and/or closed system12,63 (Figure 2, Table 1). Bozza et al.12 recently demonstrated the feasibility of an automated and rapid CAR transgene delivery process using enriched T cells as the starting material, with electroporation on day 4 and harvest on day 5, with a starting number of 1 × 109 cells.
Figure 2.
Requirements needed for automated and closed manufacturing of non-viral CAR-T cell. Created with BioRender.com.
CAR, chimeric antigen receptor; RBC, red blood cell.
Table 1.
Overview of fully non-viral CAR-T cell trials with published manufacturing process
| Trial | Target | Disease | Insertion strategy | Electroporator and manufacturing platform |
|---|---|---|---|---|
| CARTELL (ACTRN12617001579381) | CD19 | B-NHL | PiggyBac | Neon, G-Rex |
| NCT03182816 | EGFR | NSCLC | PiggyBac | Lonza, NA |
| P-BCMA-ALLO1 (NCT04960579) | BCMA | MM | PiggyBac | Lonza, G-Rex |
| CARAMBA | SLAMF7 | MM | Sleeping Beauty | Lonza, G-Rex |
| NCT03389035 | CD19 | B-ALL | Sleeping Beauty | Lonza, NA |
| NCT04213469 | CD19 | B-NHL | CRISPR/Cas9 knock-in (non-viral) | Lonza/Maxcyte, NA |
B-ALL, B-cell acute lymphoblastic leukemia; BCMA, B-cell maturation antigen; B-NHL, B-cell non-Hodgkin’s lymphoma; CAR, chimeric antigen receptor; CRISPR, clustered regularly interspaced short palindromic repeats; EGFR, epidermal growth factor receptor; MM, multiple myeloma; NSCLC, non-small-cell lung cancer; SLAMF7, SLAM family member 7.
Increasing the purity of non-viral CAR-T cells
To improve the purity and CAR-T cell yield in non-viral gene therapy, two strategies can be applied: surface marker-based enrichment and drug-based selection. Surface marker-based enrichment involves identifying cells that express a specific surface marker, such as truncated human EGFR, and then selecting them using an FDA-approved antibody, such as cetuximab.66 This method has been shown to be effective in enriching for anti-human epidermal growth factor receptor 2 (HER2) CAR-T cells in a clinical trial that used viral transduction at the Children’s Hospital in Seattle.24 Querques et al.15 utilized truncated EGFR (tEGFR) to enrich preclinical non-viral CD19 CAR-T cells manufactured using their Sleeping Beauty 100× protein. Drug-based selection, however, involves selecting cells that express a mutant form of the human dihydrofolate reductase enzyme (DHFR-FS), which confers resistance to the FDA-approved drug methotrexate.25 Poseida Therapeutics’ non-viral manufacturing platform uses the PiggyBac Transposon and methotrexate-based selection to enrich for CAR-DHFR-FS-positive cells.67
The potential for genotoxicity in non-viral CAR-T cell therapy
As non-viral CAR-T cell delivery approaches progress, methods for genotoxicity evaluation are lacking. Efforts are underway to develop techniques for detecting small insertions and deletions (indels), potential off-target DNA breaks, and endonuclease-mediated translocations.68
Sequencing techniques to detect on- and off-target insertion sites
Assessment of the location where transgenes are inserted in non-viral CAR-T cells is important for preventing disruption of genes that regulate tumor suppression or cell growth. Various in silico, in cellula, or in vitro assays are used to identify potential double-stranded break sites and off-target insertions. The primary methods for quantifying insertions are electrophoresis, tracking of indels by decomposition (TIDE), inference of CRISPR edits (ICE), and deep sequencing.
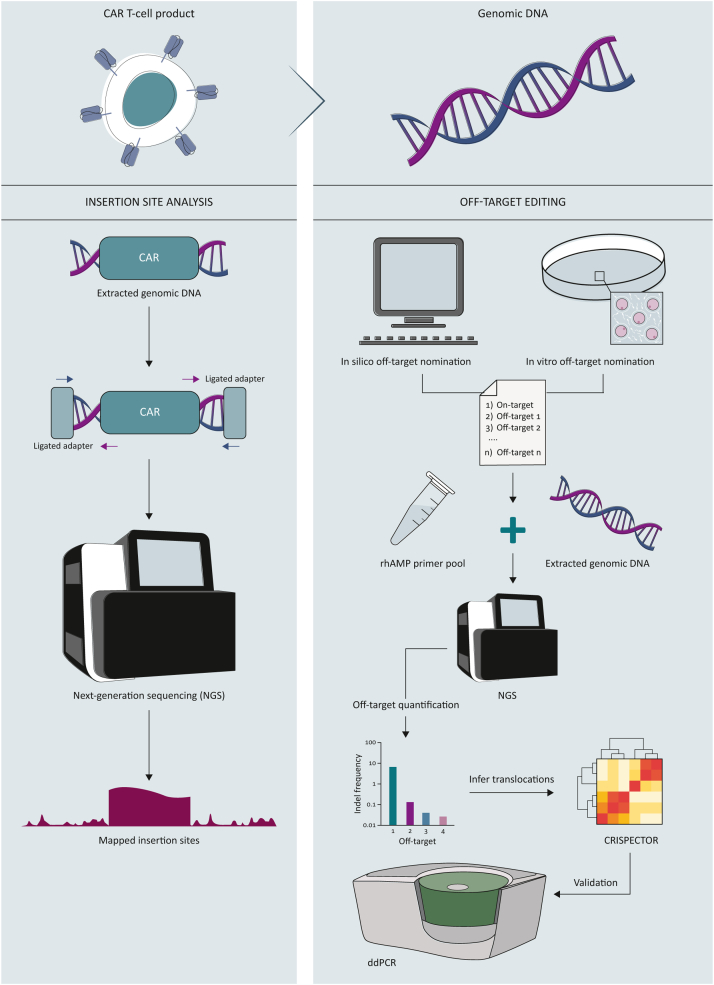
To determine insertion sites, two strategies are employed: sequence linearity and sequence proximity. The insertion sites identified through sequence linearity offer an in-depth understanding of the breakpoints, but the process is complex and involves multiple steps. It requires the use of linker or ligation-mediated PCR with primers that bind to both the transgene and the ligated adapter sequences, followed by secondary nested PCR and NGS. The sequencing reads are then mapped to the genome to identify the insertion sites.35,69,70 The targeted locus amplification approach is an alternative method that involves cross-linking genomic DNA, digesting it to obtain DNA circles, ligating the pieces, and carrying out inverse PCR starting from the known transgene. This creates a library that is subjected to NGS, which allows mapping of the sequencing reads to the corresponding insertion sites71 (Figure 3A). This approach has been widely established in the field and is less complex than the sequence linearity approach,47,48 but it offers less sequence resolution at the breakpoints.
Figure 3.
Assessment of potential genotoxic events in non-viral CAR-T cells via NGS. Created with BioRender.com.
CAR, chimeric antigen receptor; ddPCR, digital droplet polymerase chain reaction; NGS, next-generation sequencing; rhAMP,
Assessing off-target cutting and translocations
Identifying unintended cut sites after CRISPR editing is essential for evaluating its safety. This can be done using in vitro, in silico, or ex vivo prediction methods. Some of the commonly used sequencing techniques are GUIDE-seq, E-CRISP, Digenome-seq, SITE-seq, CIRCLE-seq, and DISCOVER-seq.72, 73, 74, 75 In silico prediction tools can determine off-target sites based on sequence similarity between the gRNA spacer and the human genome,76,77 but they do not quantify the editing frequency. To quantify the off-target cut sites, NGS is used, and rhAmpSeq is a popular assay that streamlines this process.78 The rhAmpSeq method uses primers that contain a single RNA base and a 3′ blocking moiety, which is cleaved when the RNA base of the primer hybridizes with the target DNA. This enables the primers to be activated and extended by the DNA polymerase to generate target amplicons, improving target specificity and eliminating primer dimers, and allowing for multiplex reactions.29,78
DSBs caused by CRISPR-based gene editing can result in translocations. When multiple editing events occur, ddPCR can be utilized to capture translocations between the on-target cut sites and is considered the gold standard for identifying even low-frequency translocations.79 Identifying translocations between on-target and off-target sites in an unbiased manner, however, remains challenging. A recent study by Amit et al.30 presented a computational method (CRISPECTOR) to extract translocation information from rhAmpSeq data. The candidate hits identified by CRISPECTOR can be further verified and monitored for clonal outgrowth during cultivation using ddPCR30,79 (Figure 3B).
To enhance the discrimination between on-target and off-target cutting, introducing mismatches in the gRNA sequence is one approach. Mismatches are typically limited to nucleotides at the end of the gRNA sequence. A study by Kath et al.19 reduced off-target cut sites in the TRAC (T-cell receptor alpha chain) locus by limiting the number of cut sites from four to one as determined by GUIDE-seq.19,80 Combining high-fidelity Cas9 mutants and mismatched gRNAs, however, can be difficult due to a reduction in on-target knock-out efficiency.19 An alternative solution is base editing, a technology that avoids the creation of DSBs altogether. Cas9 is fused with a cytidine or adenine deaminase, leading to point mutations and predictable base exchanges. These point mutations can create knock-out of the targeted gene by inducing stop mutations, and are targeted to specific sites through the use of gRNAs.81
Conclusions and perspectives
The field of non-viral transgene delivery into T cells has made significant advancements in recent years, with several platforms already being applied in clinical trials or being promising candidates for future clinical translation. These platforms offer a significant advantage over viral vectors by being more cost-efficient, thereby accelerating the development of innovative gene therapies. The integration of CRISPR/Cas-based editing techniques into these approaches provides unparalleled precision in genetic engineering, offering the ability to precisely insert or disrupt genes. Additionally, we have highlighted various processing techniques to facilitate the automated manufacturing of non-viral CAR-T cells and emphasized the importance of implementing measures to guarantee the safety and genetic stability of these therapies. These measures include the use of sequencing technologies for identifying off-target cut sites, as well as the application of computational methods to deconvolute translocations. With the increasing importance of non-viral transgene delivery and CRISPR/Cas-based editing in the field of gene therapy, it is vital that these technologies continue to evolve and be rigorously tested to ensure their safe and effective application in the clinic.
Acknowledgements
HBW is supported by the German Research Foundation (Deutsche Forschungsgemeinschaft). MA is supported in part by the award No. P30CA014089 from the U.S. National Cancer Institute (NCI).
Disclosure
HBW has received honoraria from Novartis and Miltenyi Biotec and travel support from Gilead and Maxcyte. SAF is on the scientific advisory board for FreshWind Bio and Alaunos Therapeutics and holds patents related to cellular immunotherapy. The remaining authors have no disclosures related to this work to report.
References
- 1.Neelapu S.S., Dickinson M., Munoz J., et al. Axicabtagene ciloleucel as first-line therapy in high-risk large B-cell lymphoma: the phase 2 ZUMA-12 trial. Nat Med. 2022;28(4):735–742. doi: 10.1038/s41591-022-01731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locke F.L., Miklos D.B., Jacobson C.A., et al. Axicabtagene ciloleucel as second-line therapy for large B-cell lymphoma. N Engl J Med. 2021;386(7):640–654. doi: 10.1056/NEJMoa2116133. [DOI] [PubMed] [Google Scholar]
- 3.Maude S.L., Laetsch T.W., Buechner J., et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378(5):439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munshi N.C., Anderson L.D., Shah N., et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N Engl J Med. 2021;384(8):705–716. doi: 10.1056/NEJMoa2024850. [DOI] [PubMed] [Google Scholar]
- 5.Majzner R.G., Rietberg S.P., Sotillo E., et al. Tuning the antigen density requirement for CAR T-cell activity. Cancer Discov. 2020;10(5):702–723. doi: 10.1158/2159-8290.CD-19-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baird J.H., Frank M.J., Craig J., et al. CD22-directed CAR T-cell therapy induces complete remissions in CD19-directed CAR-refractory large B-cell lymphoma. Blood. 2021;137(17):2321–2325. doi: 10.1182/blood.2020009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah N.N., Johnson B.D., Schneider D., et al. Bispecific anti-CD20, anti-CD19 CAR T cells for relapsed B cell malignancies: a phase 1 dose escalation and expansion trial. Nat Med. 2020;26(10):1569–1575. doi: 10.1038/s41591-020-1081-3. [DOI] [PubMed] [Google Scholar]
- 8.Lynn R.C., Weber E.W., Sotillo E., et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature. 2019;576(7786):293–300. doi: 10.1038/s41586-019-1805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avanzi M.P., Yeku O., Li X., et al. Engineered tumor-targeted T cells mediate enhanced anti-tumor efficacy both directly and through activation of the endogenous immune system. Cell Rep. 2018;23(7):2130–2141. doi: 10.1016/j.celrep.2018.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol Ther Oncolytics. 2016;3 doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukjanov V., Koutná I., Šimara P. CAR T-cell production using nonviral approaches. J Immunol Res. 2021;2021 doi: 10.1155/2021/6644685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozza M., De Roia A., Correia M.P., et al. A nonviral, nonintegrating DNA nanovector platform for the safe, rapid, and persistent manufacture of recombinant T cells. Sci Adv. 2021;7(16) doi: 10.1126/sciadv.abf1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parayath N.N., Stephan S.B., Koehne A.L., Nelson P.S., Stephan M.T. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat Commun. 2020;11(1):6080. doi: 10.1038/s41467-020-19486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnani C.F., Gaipa G., Lussana F., et al. Sleeping beauty-engineered CAR T cells achieve antileukemic activity without severe toxicities. J Clin Invest. 2020;130(11):6021–6033. doi: 10.1172/JCI138473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Querques I., Mades A., Zuliani C., et al. A highly soluble sleeping beauty transposase improves control of gene insertion. Nat Biotechnol. 2019;37(12):1502–1512. doi: 10.1038/s41587-019-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shy B.R., Vykunta V.S., Ha A., et al. High-yield genome engineering in primary cells using a hybrid ssDNA repair template and small-molecule cocktails. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monjezi R., Miskey C., Gogishvili T., et al. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia. 2017;31(1):186–194. doi: 10.1038/leu.2016.180. [DOI] [PubMed] [Google Scholar]
- 18.Mueller K.P., Piscopo N.J., Forsberg M.H., et al. Production and characterization of virus-free, CRISPR-CAR T cells capable of inducing solid tumor regression. J Immunother Cancer. 2022;10(9) doi: 10.1136/jitc-2021-004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kath J., Du W., Pruene A., et al. Pharmacological interventions enhance virus-free generation of TRAC-replaced CAR T cells. Mol Ther Methods Clin Dev. 2022;25:311–330. doi: 10.1016/j.omtm.2022.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Hu Y., Yang J., et al. Non-viral, specifically targeted CAR-T cells achieve high safety and efficacy in B-NHL. Nature. 2022;609:369–374. doi: 10.1038/s41586-022-05140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jing R., Jiao P., Chen J., et al. Cas9-cleavage sequences in size-reduced plasmids enhance nonviral genome targeting of CARs in primary human T cells. Small Methods. 2021;5(7) doi: 10.1002/smtd.202100071. [DOI] [PubMed] [Google Scholar]
- 22.Oh S.A., Senger K., Madireddi S., et al. High-efficiency nonviral CRISPR/Cas9-mediated gene editing of human T cells using plasmid donor DNA. J Exp Med. 2022;219(5) doi: 10.1084/jem.20211530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abou-el-Enein M., Elsallab M., Feldman S.A., et al. Scalable manufacturing of CAR T cells for cancer immunotherapy. Blood Cancer Discov. 2021;2(5):408–422. doi: 10.1158/2643-3230.BCD-21-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vitanza N.A., Johnson A.J., Wilson A.L., et al. Locoregional infusion of HER2-specific CAR T cells in children and young adults with recurrent or refractory CNS tumors: an interim analysis. Nat Med. 2021;27(9):1544–1552. doi: 10.1038/s41591-021-01404-8. [DOI] [PubMed] [Google Scholar]
- 25.Jonnalagadda M., Brown C.E., Chang W.C., Ostberg J.R., Forman S.J., Jensen M.C. Efficient selection of genetically modified human T cells using methotrexate-resistant human dihydrofolate reductase. Gene Ther. 2013;20(8):853–860. doi: 10.1038/gt.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunz A., Gern U., Schmitt A., et al. Optimized assessment of qPCR-based vector copy numbers as a safety parameter for GMP-grade CAR T cells and monitoring of frequency in patients. Mol Ther Methods Clin Dev. 2020;17:448–454. doi: 10.1016/j.omtm.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miskey C., Amberger M., Reiser M., et al. Genomic analyses of SLAMF7 CAR-T cells manufactured by sleeping beauty transposon gene transfer for immunotherapy of multiple myeloma. bioRxiv. 2019:675009. [Google Scholar]
- 28.Micklethwaite K.P., Gowrishankar K., Gloss B.S., et al. Investigation of product-derived lymphoma following infusion of piggyBac-modified CD19 chimeric antigen receptor T cells. Blood. 2021;138(16):1391–1405. doi: 10.1182/blood.2021010858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro J., Iancu O., Jacobi A.M., et al. Increasing CRISPR efficiency and measuring Its specificity in HSPCs using a clinically relevant system. Mol Ther Methods Clin Dev. 2020;17:1097–1107. doi: 10.1016/j.omtm.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amit I., Iancu O., Levy-Jurgenson A., et al. CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data. Nat Commun. 2021;12(1):3042. doi: 10.1038/s41467-021-22417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irving M., Lanitis E., Migliorini D., Ivics Z., Guedan S. Choosing the right tool for genetic engineering: clinical lessons from chimeric antigen receptor-T cells. Hum Gene Ther. 2021;32(19-20):1044–1058. doi: 10.1089/hum.2021.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moretti A., Ponzo M., Nicolette C.A., Tcherepanova I.Y., Biondi A., Magnani C.F. The past, present, and future of non-viral CAR T cells. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.867013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeNicola G.M., Karreth F.A., Adams D.J., Wong C.C. The utility of transposon mutagenesis for cancer studies in the era of genome editing. Genome Biol. 2015;16(1):229. doi: 10.1186/s13059-015-0794-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prommersberger S., Reiser M., Beckmann J., et al. CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free sleeping beauty gene transfer to treat multiple myeloma. Gene Ther. 2021;28(9):560–571. doi: 10.1038/s41434-021-00254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kebriaei P., Singh H., Huls M.H., et al. Phase I trials using sleeping beauty to generate CD19-specific CAR T cells. J Clin Invest. 2016;126:3363–3376. doi: 10.1172/JCI86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraser M.J., Smith G.E., Summers M.D. Acquisition of host cell DNA sequences by baculoviruses: relationship between host DNA insertions and FP mutants of autographa californica and galleria mellonella nuclear polyhedrosis viruses. J Virol. 1983;47(2):287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Zhang Z., Ding Y., et al. Phase I clinical trial of EGFR-specific CAR-T cells generated by the piggyBac transposon system in advanced relapsed/refractory non-small cell lung cancer patients. J Cancer Res Clin Oncol. 2021;147(12):3725–3734. doi: 10.1007/s00432-021-03613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slovin S.F., Dorff T.B., Falchook G.S., et al. Phase 1 study of P-PSMA-101 CAR-T cells in patients with metastatic castration-resistant prostate cancer (mCRPC) J Clin Oncol. 2022;40(suppl 6):98. [Google Scholar]
- 39.de Silva S., Mastrangelo M.A., Lotta L.T., Jr., Burris C.A., Federoff H.J., Bowers W.J. 794. Extending the transposable payload limit of the sleeping beauty (SB) transposon system using the Bipartite herpes simplex virus (HSV)/SB amplicon vector platform. Mol Ther. 2009;17(suppl 1):S304–S305. doi: 10.1038/gt.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bishop D.C., Clancy L.E., Simms R., et al. Development of CAR T-cell lymphoma in 2 of 10 patients effectively treated with piggyBac-modified CD19 CAR T cells. Blood. 2021;138(16):1504–1509. doi: 10.1182/blood.2021010813. [DOI] [PubMed] [Google Scholar]
- 41.Klompe S.E., Vo P.L.H., Halpin-Healy T.S., Sternberg S.H. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571(7764):219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann F.T., Kim M., Beh L.Y., et al. Selective TnsC recruitment enhances the fidelity of RNA-guided transposition. Nature. 2022;609(7926):384–393. doi: 10.1038/s41586-022-05059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mailankody S., Liedtke M., Sidana S., et al. Universal updated phase 1 data validates the feasibility of allogeneic anti-BCMA ALLO-715 therapy for relapsed/refractory multiple myeloma. Blood. 2021;138(suppl 1):651. [Google Scholar]
- 44.Qasim W., Amrolia P.J., Samarasinghe S., et al. First clinical application of Talen engineered universal CAR19 T cells in B-ALL. Blood. 2015;126(23):2046. [Google Scholar]
- 45.Ellis G.I., Sheppard N.C., Riley J.L. Genetic engineering of T cells for immunotherapy. Nat Rev Genet. 2021;22(7):427–447. doi: 10.1038/s41576-021-00329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eyquem J., Mansilla-Soto J., Giavridis T., et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth T.L., Puig-Saus C., Yu R., et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559(7714):405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGuirk J., Bachier C.R., Bishop M.R., et al. A phase 1 dose escalation and cohort expansion study of the safety and efficacy of allogeneic CRISPR-Cas9–engineered T cells (CTX110) in patients (Pts) with relapsed or refractory (R/R) B-cell malignancies (CARBON) J Clin Oncol. 2021;39(suppl 15):TPS7570. [Google Scholar]
- 50.Dar H., Henderson D., Padalia Z., P, et al. Preclinical development of CTX120, an allogeneic CAR-T cell targeting Bcma. Blood. 2018;132(suppl 1):1921. [Google Scholar]
- 51.Srivastava A., Mallela K.M.G., Deorkar N., Brophy G. Manufacturing challenges and rational formulation development for AAV viral vectors. J Pharm Sci. 2021;110(7):2609–2624. doi: 10.1016/j.xphs.2021.03.024. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen D.N., Roth T.L., Li P.J., et al. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat Biotechnol. 2020;38(1):44–49. doi: 10.1038/s41587-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wienert B., Nguyen D.N., Guenther A., et al. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nat Commun. 2020;11(1):2109. doi: 10.1038/s41467-020-15845-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu V.T., Weber T., Wefers B., et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 55.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18(8):495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li A., Kusuma G.D., Driscoll D., et al. Advances in automated cell washing and concentration. Cytotherapy. 2021;23(9):774–786. doi: 10.1016/j.jcyt.2021.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Li A., Barabadi M., McDonald H., et al. Improving cell viability using counterflow centrifugal elutriation. Cytotherapy. 2022;24(6):650–658. doi: 10.1016/j.jcyt.2022.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Vormittag P., Gunn R., Ghorashian S., Veraitch F.S. A guide to manufacturing CAR T cell therapies. Curr Opin Biotechnol. 2018;53:164–181. doi: 10.1016/j.copbio.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 59.Ghassemi S., Durgin J.S., Nunez-Cruz S., et al. Rapid manufacturing of non-activated potent CAR T cells. Nat Biomed Eng. 2022;6(2):118–128. doi: 10.1038/s41551-021-00842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pi C.H., Hornberger K., Dosa P., Hubel A. Understanding the freezing responses of T cells and other subsets of human peripheral blood mononuclear cells using DSMO-free cryoprotectants. Cytotherapy. 2020;22(5):291–300. doi: 10.1016/j.jcyt.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das R.K., O’Connor R.S., Grupp S.A., Barrett D.M. Lingering effects of chemotherapy on mature T cells impair proliferation. Blood Adv. 2020;4(19):4653–4664. doi: 10.1182/bloodadvances.2020001797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lock D., Monjezi R., Brandes C., et al. Automated, scaled, transposon-based production of CAR T cells. J Immunother Cancer. 2022;10(9) doi: 10.1136/jitc-2022-005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alzubi J., Lock D., Rhiel M., et al. Automated generation of gene-edited CAR T cells at clinical scale. Mol Ther Methods Clin Dev. 2021;20:379–388. doi: 10.1016/j.omtm.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ludwig J., Hirschel M. In: Chimeric Antigen Receptor T Cells: Development and Production. Swiech K., Malmegrim K.C.R., Picanço-Castro V., editors. Springer; New York, NY, USA: 2020. Methods and process optimization for large-scale CAR T expansion using the G-Rex cell culture platform; pp. 165–177. [DOI] [PubMed] [Google Scholar]
- 65.Smith T.A. In: Chimeric Antigen Receptor T Cells: Development and Production. Swiech K., Malmegrim K.C.R., Picanço-Castro V., editors. Springer; New York, NY, USA: 2020. CAR-T cell expansion in a Xuri cell expansion system W25; pp. 151–163. [Google Scholar]
- 66.Wang X., Chang W.C., Wong C.W., et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood. 2011;118(5):1255–1263. doi: 10.1182/blood-2011-02-337360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cranert S.A., Richter M., Tong M., et al. Manufacture of an allogeneic CAR-T stem cell memory product candidate for multiple myeloma, P-Bcma-ALLO1, is robust, reproducible and highly scalable. Blood. 2019;134(suppl 1):4445. [Google Scholar]
- 68.Blattner G., Cavazza A., Thrasher A.J., Turchiano G. Gene editing and genotoxicity: targeting the off-targets. Front Genome Ed. 2020;2 doi: 10.3389/fgeed.2020.613252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keng V.W., Villanueva A., Chiang D.Y., et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27(3):264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng Z., Liebers M., Zhelyazkova B., et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 71.de Vree PJP, de Wit E., Yilmaz M., et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol. 2014;32(10):1019–1025. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 72.Tsai S.Q., Zheng Z., Nguyen N.T., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14(6):607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beeke W., Wyman S.K., Richardson C.D., et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364(6437):286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Donnadieu E., Luu M., Alb M., et al. Time to evolve: predicting engineered T cell-associated toxicity with next-generation models. J Immunother Cancer. 2022;10(5) doi: 10.1136/jitc-2021-003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 2014;3(12):e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stemmer M., Thumberger T., del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobosy J.R., Rose S.D., Beltz K.R., et al. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol. 2011;11(1):80. doi: 10.1186/1472-6750-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stadtmauer E.A., Fraietta J.A., Davis M.M., et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481) doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiebking V., Lee C.M., Mostrel N., et al. Genome editing of donor-derived T-cells to generate allogenic chimeric antigen receptor-modified T cells: optimizing aβ T cell-depleted haploidentical hematopoietic stem cell transplantation. Haematologica. 2021;106(3):847–858. doi: 10.3324/haematol.2019.233882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuscu C., Parlak M., Tufan T., et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14(7):710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]