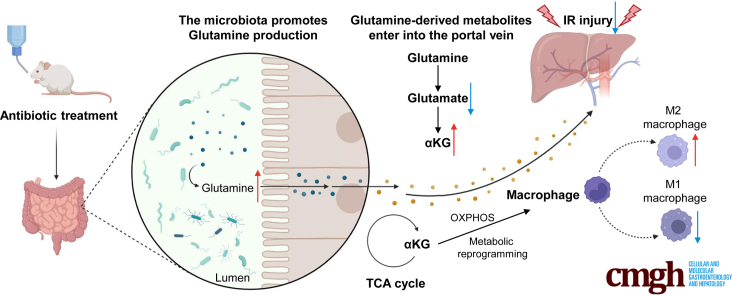
Abstract
Background & Aims
Many studies have revealed crucial roles of the gut microbiota and its metabolites in liver disease progression. However, the mechanism underlying their effects on liver ischemia/reperfusion (I/R) injury remain largely unknown. Here, we investigate the function of gut microbiota and its metabolites in liver I/R injury.
Methods
C57BL/6 mice was pretreated with an antibiotic cocktail. Then, we used multi-omics detection methods including 16s rRNA sequencing, ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC–MS/MS) to explore the changes of gut microbiota and metabolites in both feces and portal blood to reveal the mechanism of their protective effect in liver I/R injury.
Results
We found that antibiotic pretreatment (ABX) could significantly reduce the severity of I/R-induced hepatic injury, and this effect could be transferred to germ-free mice by fecal microbiota transplantation (FMT), suggesting a protective role of the gut microbiota depletion. During I/R, the rates of serum α-ketoglutarate (αKG) production and glutamate reduction, downstream products of gut microbiota-derived glutamine, were more significant in the ABX mice. Then, we showed that αKG could promote alternative (M2) macrophage activation through oxidative phosphorylation, and oligomycin A could inhibit M2 macrophage polarization and reversed this protective effect.
Conclusions
These findings show that the gut microbiota and its metabolites play critical roles in hepatic I/R injury by modulating macrophage metabolic reprogramming. Potential therapies that target macrophage metabolism, including antibiotic therapies and novel immunometabolism modulators, can be exploited for the treatment of liver I/R injury.
Keywords: Liver Ischemia/Reperfusion Injury, Microbiota, Glutamine, a-ketoglutarate, Immunometabolism, Macrophage Reprogramming
Graphical abstract
Summary.
In this article, we found antibiotic pretreatment could protect against liver I/R injury through altered gut microbiota, glutamine level, and glutamine downstream products αKG in circulation. αKG as a checkpoint could promote macrophage metabolic reprogramming and change the proportion of M1 and M2 macrophages.
Liver ischemia/reperfusion (I/R) injury is a common complication of liver resection, transplantation, trauma, and hemorrhagic shock.1,2 I/R injury involves 2 consecutive stages of hypoxia-induced damage and proinflammatory immune-mediated reperfusion injury; this is a complex process that includes innate–adaptive immune crosstalk and the activation of diverse cellular cascades. During I/R, Kupffer cells (KCs), which are hepatic macrophages located in the liver sinusoid, are activated. KCs act as sentinels that attract circulating macrophages, neutrophils, natural killer cells, and platelets. Then, recruited macrophages become one of the main cell types that attack liver parenchymal cells, such as hepatocytes and liver sinusoidal endothelial cells.3, 4, 5 Therefore, the role of macrophages in hepatic I/R injury cannot be ignored.
The human gut microbiome, which contains the second largest genome in the human body,6 plays an important role in the maintenance of host health, homeostasis, and responses to pathologic stimuli. In addition, metabolites associated with the gut microbiome, such as short-chain fatty acids, are either produced or transferred by intestinal microflora and contribute to many diseases.7 Indeed, the liver and gut are closely connected in terms of anatomy and physiological function through the enterohepatic circulation. Many studies have shown that the gut microbiota and its metabolites can affect different kinds of liver diseases, including oxidative liver injury, nonalcoholic fatty liver disease, nonalcoholic steatohepatitis and liver cirrhosis progression, and hepatocellular carcinoma.8, 9, 10, 11, 12, 13, 14 Moreover, Nakamura et al15 demonstrated that modification of the intestinal microbiota through antibiotic pretreatment could reduce liver transplantation–mediated damage by relieving endoplasmic reticulum stress and increasing liver autophagy in both a mouse model and a large clinical cohort. However, whether the gut microbiota can exert an effect on hepatic I/R injury through its metabolites is still worth studying.
Macrophages are characterized by their innate plasticity in polarization. They can acquire different activation states in response to endogenous danger stimuli and then promote either inflammation (M1) or tissue repair (M2).16 The known mechanisms underlying the regulation of macrophage polarization include cytokine cascades, metabolic reprogramming, and epigenetic modification.17, 18, 19, 20 Among these mechanisms, metabolic reprogramming is emerging as a key regulator of macrophage polarization. M1 macrophages mainly rely on aerobic glycolysis, which is also called the Warburg effect, whereas M2 macrophages that possess an intact tricarboxylic acid (TCA) cycle and electron transport chain (ETC) can produce adenosine triphosphate (ATP) through oxidative phosphorylation (OXPHOS). It is worth noting that fatty acid oxidation (FAO) is important for fueling the TCA cycle in M2 macrophages.17,21, 22, 23 Thus, the immunometabolism and polarization of macrophages play an important role in host homeostasis and the development of various diseases.
In this study, we used an antibiotic cocktail to modify the gut microbiota. Intriguingly, the antibiotic pretreatment (ABX) group showed good protective effects against hepatic I/R injury. Then, we found that this protection against liver I/R injury could be transferred to germ-free (GF) mice by fecal microbiota transplantation (FMT), indicating that the gut microbiota, instead of antibiotics, exerted this protective effect. Because altered gut microbiota cause changes in metabolites, we used ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC–MS/MS) to explore the changes in the metabolomics of feces, blood, and liver (data not shown). The results revealed that glutamine and α-ketoglutarate (αKG) were present in higher concentrations in feces and blood, respectively, in the ABX group after hepatic I/R injury. In addition, the levels of intermediate products of the TCA cycle and pyruvate produced by glycolysis were also increased. Thus, the number of M2 macrophages who favor oxidative metabolism in the livers of mice treated with antibiotics or supplemented with dimethyl αKG was increased in our study. Thus, we concluded that elevated glutamine levels in the intestine cause an increase in αKG levels in the blood, αKG can promote macrophage M2 polarization by fueling the TCA cycle, and finally, the increased number of M2 macrophages can repair hepatic I/R injury in vivo. Furthermore, we used oligomycin A, which can block OXPHOS and the ETC by inhibiting H+-ATP synthase. As expected, the protective effect of ABX on hepatic I/R injury and the increased numbers of M2 macrophages were simultaneously reversed. This study established a connection among the gut microbiota, its metabolites, and liver I/R injury. In addition, we aimed to explore potential therapies that target macrophage metabolism, including antibiotic therapies and novel immunometabolism modulators, for use in the treatment of hepatic I/R injury.
Results
Antibiotic Pretreatment Could Protect the Liver From Ischemia and Reperfusion Injury
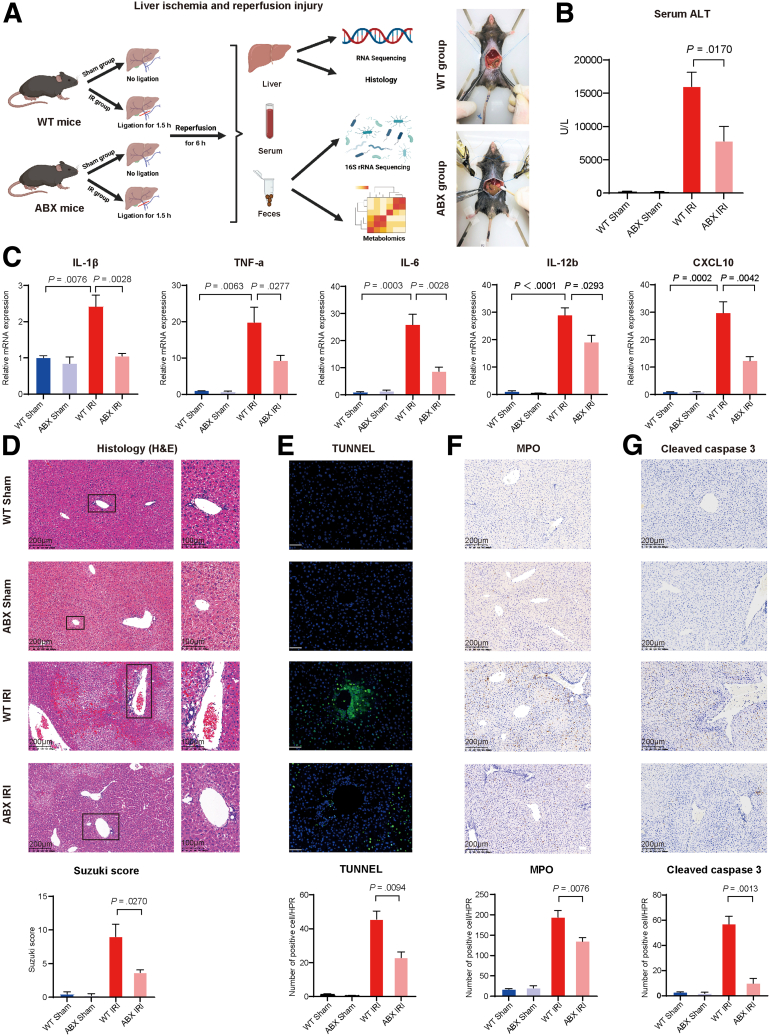
Wild-type (WT) mice and quadruple ABX mice were randomly divided into a sham group and a hepatic I/R surgery group. Then, we aimed to explore whether ABX could play a role in the mouse model of liver I/R that was established with 1.5 hours of warm ischemia, followed by 6-hour reperfusion. The liver, blood, and feces of each group of mice were collected for future study (Figure 1A). As Figure 1B shows, compared with that of the WT mice, the serum alanine aminotransferase (ALT) level of the ABX mice was decreased by 50% from approximately 15,000 U/L to 7500 U/L (P = .0170). Then, the expression of different inflammatory factors was detected by real-time polymerase chain reaction. The mRNA levels of interleukin (IL)-1β (P = .0028), tumor necrosis factor-α (P = .0277), IL-6 (P = .0028), IL-12b (P = .0293), and CXCL10 (P = .0042) were all decreased in the ABX group (Figure 1C). Moreover, we conducted histologic analysis of liver tissues. H&E staining and quantitative Suzuki scoring showed that the necrosis area, degree of congestion and edema, and vacuole-like lesions were alleviated in ABX mice (Figure 1D, P = .0270). Deoxyuridine-5´-triphosphate biotin nick end labeling (TUNEL) staining (green fluorescence represents necrosis/apoptosis in liver tissue) and immunohistochemical staining of myeloperoxidase (MPO) and cleaved caspase-3 (brown particles represent positive cells) demonstrated that inflammation and necrosis of the liver were decreased in the ABX group according to both qualitative and quantitative data (Figure 1E–G, P = .0094, P = .0076, and P = .0013, respectively.)
Figure 1.
Antibiotic pretreatment could protect the liver from I/R injury. (A) Pattern diagram and intraoperative photos of hepatic I/R injury. WT mice or ABX mice were randomly divided into 2 groups. One group was the sham group (WT Sham, n = 4; ABX Sham, n = 4), and the other group was subjected to liver I/R surgery (WT IRI, n = 10; ABX IRI, n= 10). (B) Serum ALT levels of the 4 groups. (C) Relative mRNA expression of inflammatory cytokines Il1β, Tnfα, Il6, Il12b, and Cxcl10. (D) H&E staining and Suzuki’s quantitative score. (E) TUNEL staining and semiquantitative analysis. Scale bars, 100 μm. (F) Immunochemical staining of MPO and semiquantitative analysis. (G) Immunochemical staining of cleaved caspase 3 and semiquantitative analysis. For all data, statistical comparisons were carried out by Student t test. P < .05 indicates significant differences.
Antibiotics Could Lead to Changes in the Fecal Microbiota
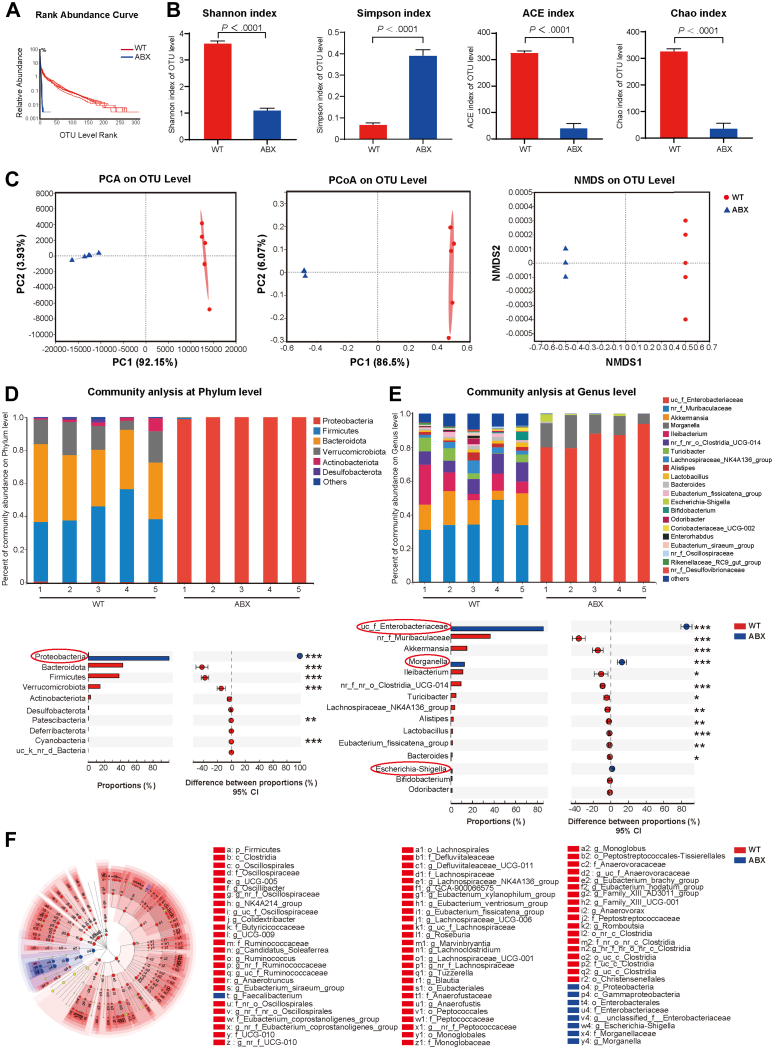
Because of the protective effect of antibiotic pretreatment on a mouse model of liver I/R and the close relationship between gut microbiota and liver diseases mediated by the enterohepatic axis, we used 16S rRNA sequencing technology to explore how the gut microbiota changed after 4 weeks of quadruple antibiotic pretreatment. Sequencing analysis of fecal microbes resulted in 565,702 raw reads. On the basis of the 97% similarity level, all the effective reads were clustered into operational taxonomic units. The rank abundance curve demonstrated that the relative abundance of fecal microbiota was greatly reduced in the ABX group compared with the WT group (Figure 2A). As shown in Figure 2B of the α diversity analysis, the Shannon index, ACE index, and Chao index, which are positively correlated with species diversity, all decreased significantly in the ABX group. However, the Simpson index, which shows opposite trends to the indicators listed above, was significantly increased after 4 weeks of antibiotic pretreatment. Qualitative analysis showed that the Shannon index, ACE index, and Chao index were decreased by 69.5%, 87.5%, and 88.8%, respectively (all P<.0001), whereas the Simpson index was increased by 470.7% (P<.0001). According to β diversity analysis, as shown in Figure 2C, principal component analysis (PCA), principal coordinate analysis (PCoA), and nonmetric multidimensional scaling (NMDS) analysis revealed a distinct clustering of fecal microbiota between the 2 groups (analysis of similarities, P = .003). Next, we further studied the composition of the fecal microbiota community at the phylum and genus levels. According to the analysis of species differences at the phylum level, only the abundance of Proteobacteria was increased in the ABX group (P<.001). Among the Proteobacteria phyla, the abundances of the unclassified_f_Enterobacteriaceae and Morganella genera were significantly increased (both P < .001), and the abundance of the Escherichia-Shigella genus was also increased, but the difference was not statistically significant (Figure 2D and E). These results suggested that antibiotic pretreatment could reduce the abundance, richness, and diversity of fecal microbiota, but it led to an increase in the amount of the Proteobacteria phylum and the 2 unclassified_f_Enterobacteriaceae and Morganella genera. Finally, linear discriminant analysis effect size also indicated that unclassified_f_Enterobacteriaceae, Morganella, Escherichia-Shigella, and Faecailbacterium dominated in the ABX group (Figure 2F).
Figure 2.
Antibiotic pretreatment could reduce the abundance, diversity, and richness of the gut microbiota and change its composition. (A) Rank abundance curve of WT and ABX mice. (B) α diversity comparison: Shannon index, Simpson index, ACE index, and Chao index of WT and ABX mice. (C) β diversity of clustering analysis: PCA, PCoA, and NMDS analysis of WT and ABX mice at the operational taxonomic unit (OTU) level. (D and E) Comparison of composition of gut microbiota at the phylum level (D) and at the genus level (E) between WT and ABX mice. (F) Linear discriminant analysis (LDA) effect size (LEfSe) analysis with | LDA | ≥2. Fecal samples from WT (n = 5) and ABX (n = 5) mice were analyzed by 16S rRNA sequencing. Statistical comparisons were carried out by Student t test and ANOSIM analysis except LEfSe analysis using the nonparametric Kruskal–Wallis sum-rank test and Wilcoxon rank-sum test. P < .05 indicates significant differences.
The Protective Effect of Antibiotics on Hepatic I/R Injury Could Be Transferred Through the Gut Microbiota
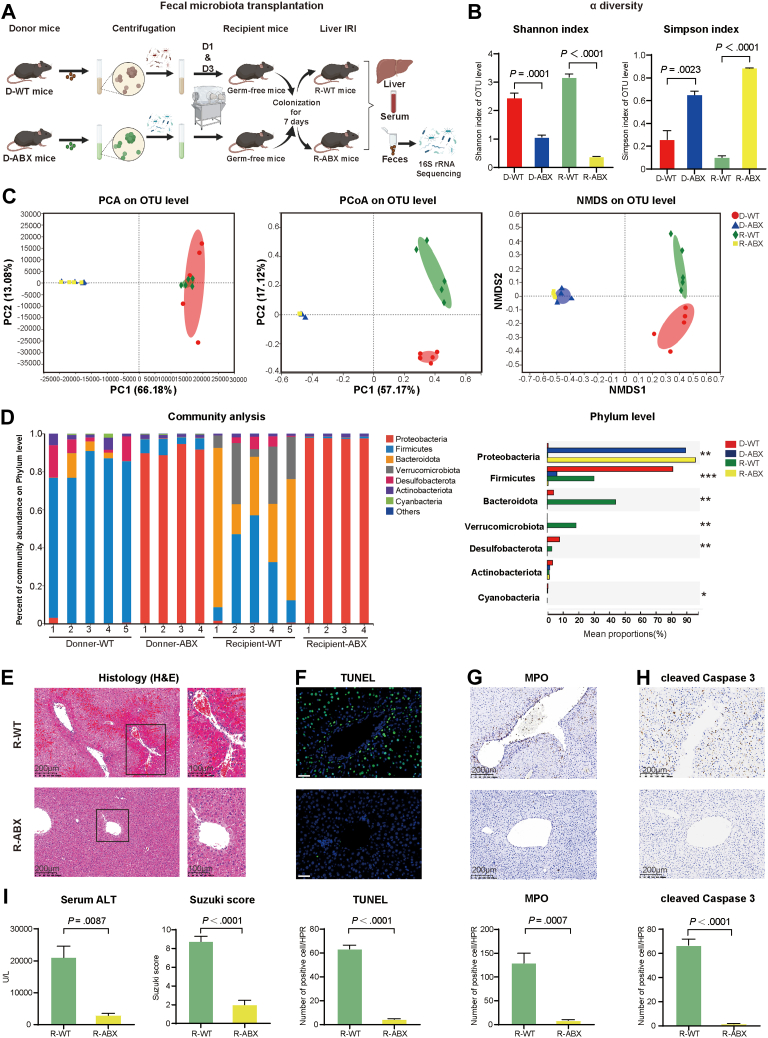
To determine the impact of the gut microbiota on the mouse model of liver I/R, we colonized GF mice with the fecal microbiota from WT mice and ABX mice to exclude the interference of antibiotics. After 4 weeks of antibiotic pretreatment, we collected the feces of WT mice (D-WT) and ABX mice (D-ABX) and then transferred the fecal microbiota to GF mice (named the R-WT group and R-ABX group, respectively) on the first and third days during the 7 days of colonization. After successful colonization with fecal microbiota, we performed liver I/R surgery on the recipient mice, and then liver, serum, and fecal samples were all collected for future study (Figure 3A). As Figure 3B shows, consistent with the donor mice, the α diversity, according to Shannon index and Simpson index, was decreased in the R-ABX mice compared with the R-WT mice (both P<.0001). In terms of β diversity, PCA showed that the D-WT (red) and R-WT (green) groups gathered into one cluster, whereas the D-ABX (blue) and R-ABX (yellow) groups clustered into another, indicating that fecal transplantation was successful. However, the D-WT and R-WT groups exhibited a certain deviation in PCoA and NMDS analysis, and the D-ABX and R-ABX groups still gathered well into one cluster (Figure 3C). Then, we analyzed the components of the fecal bacteria at the phylum level. Consistent with the previous results, the Proteobacteria phylum was increased in both the D-ABX group and R-ABX group compared with the D-WT group and R-WT group (Figure 3D, both P < .0001, Kruskal-Wallis H test, P < .01). Ultimately, microbiota transplantation was sufficient to simulate the phenotype observed in the ABX mice, as evidenced by serum measurements and multiple histologic analyses. We measured serum ALT levels, which are the key marker for detecting liver damage, and these levels were decreased from 20,000 U/L in the R-WT group to approximately 2000 U/L in the R-ABX group (Figure 3I, P = .0087). In addition, H&E and TUNEL staining (both P < .0001) and immunochemical staining of MPO (P = .0007) and cleaved caspase 3 (P < .0001) all indicated that the degree of damage and necrosis of the liver were reduced in the R-ABX group (Figure 3E–H). These data demonstrated that changes in the gut microbiota played a protective role in hepatic I/R injury, thus eliminating a role of antibiotics in the protective effect.
Figure 3.
Protective effect of antibiotics on hepatic I/R injury could be transferred to GF mice in FMT experiments. (A) Pattern diagram and protocol of FMT experiment. We transferred the fecal microbiota of WT (D-WT, n = 5) and ABX (D-ABX, n = 4) mice to GF mice (R-WT, n = 5; R-ABX, n = 4). One mouse in each of the D-ABX and R-ABX groups died unexpectedly. (B) α diversity comparison: Shannon index and Simpson index of the 4 groups. (C) β diversity of clustering analysis: PCA, PCoA, and NMDS analysis of the 4 groups at the OTU level. (D) Comparison of composition of gut microbiota at phylum level in the 4 groups. (E) H&E staining and Suzuki’s quantitative score of the R-WT and R-ABX groups. (F) TUNEL staining and semiquantitative analysis of R-WT and R-ABX groups. Scale bars, 100 μm. (G and H) Immunochemical staining of MPO (G) and cleaved caspase 3 (H) in the R-WT and R-ABX groups. (I) Serum ALT levels of R-WT and R-ABX groups. For all data, statistical comparisons between 2 groups were carried out by Student t test. For comparisons among 4 groups, the Kruskal–Wallis H test was used. P < .05 indicates significant differences.
Alterations in Gut Microbiota Led to an Increase in Glutamine Concentrations in Fecal Metabolites
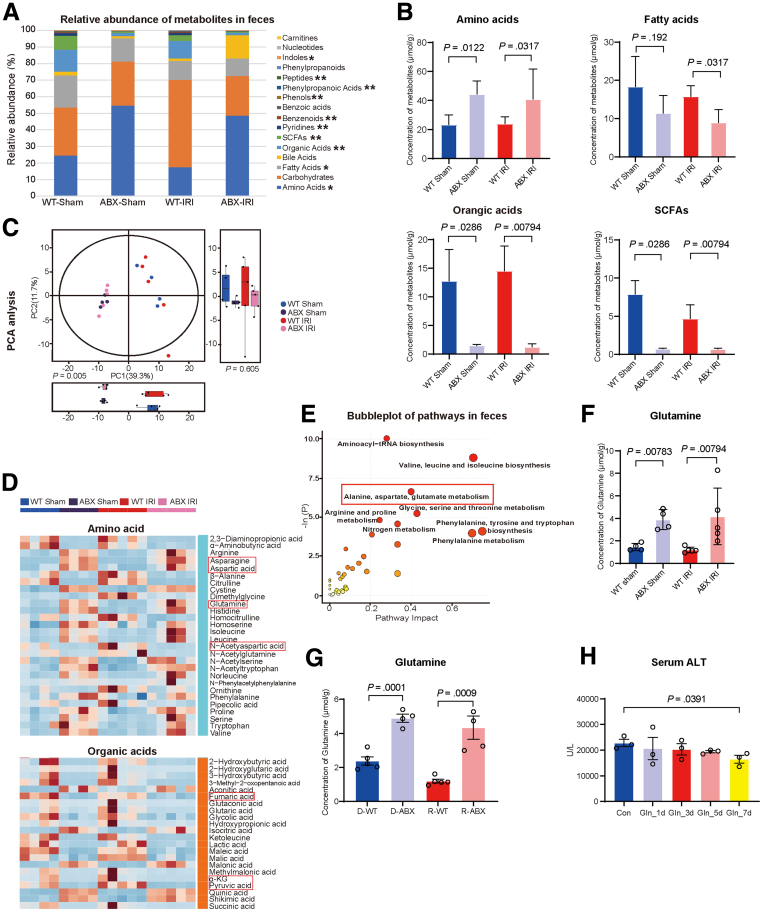
The profound influence of the gut microbiota on the homeostasis and pathogenesis of the host is strongly related to a series of complex interactions, especially the microbiome-metabolism-host axis.24,25 To assess metabolic changes caused by antibiotic pretreatment under both sham and liver I/R conditions, a targeted metabolome profile was generated on fecal samples by UPLC–MS/MS system. According to the major components of fecal metabolites, amino acids occupied the largest proportion, and these levels were increased in ABX mice (WT Sham: ABX Sham, 23.92 ± 6.71 μmol/g: 44.29 ± 9.23 μmol/g; P = .0122; WT IRI: ABX IRI, 23.87 ± 4.65 μmol/g: 44.40 ± 21.29 μmol/g; P = .0317). The other 3 metabolites that were different in the 4 groups and occupied a larger proportion were fatty acids (WT Sham: ABX Sham, 18.32 ± 7.89 μmol/g: 11.38 ± 4.62 μmol/g, P = .192; WT IRI: ABX IRI, 15.76 ± 2.83 μmol/g: 9.61 ± 3.28 μmol/g, P = .0317), organic acids (WT Sham: ABX Sham, 12.81 ± 5.58 μmol/g: 1.50 ±0.22 μmol/g, P = .0286; WT IRI: ABX IRI, 14.67 ± 4.56 μmol/g: 1.35 ± 0.57 μmol/g, P = .00794), and short-chain fatty acids (WT Sham: ABX Sham, 7.89 ± 1.78 μmol/g: 0.74 ± 0.073 μmol/g, P = .0286; WT IRI: ABX, IRI 4.69 ± 1.82 μmol/g: 70.78 μmol/g: 0.74 ± 0.073 μmol/g). The levels of all of these metabolites were all decreased in the ABX mice (Figure 4A and B). Distinctive clustering of metabolites was apparent between WT and ABX mice according to PCA, indicating that altered gut microbiota could lead to changes in fecal metabolites (Figure 4C). On the basis of the bubble chart of hierarchical clustering analysis (Figure 4E) and heatmap of differential metabolites (Figure 4D), the alanine, aspartate, and glutamate metabolism pathway has become one of the main pathways of interest. Studies have shown that glutamine plays a protective role in I/R injury in both isolated rat livers and in vivo rat models.26,27 In addition, glutamine and the gut microbiota maintain the local and systemic health of the host through a bidirectional interaction.28,29 As Figure 4F shows, the levels of glutamine, which is one of the biomarkers of the alanine, aspartate, and glutamate metabolism pathway, were significantly increased in the ABX group (WT Sham: ABX Sham, 1.46 ± 0.32 μmol/g: 3.89 ± 0.89 μmol/g, P = .00783; WT IRI: ABX IRI, 1.20 ± 0.25 μmol/g: 4.17 ± 2.51 μmol/g, P = .00794). To further prove the correlation between altered gut microbiota and fecal glutamine, the concentration of glutamine in feces of the 4 groups in FMT experiment (D-WT, D-ABX, R-WT, and R-ABX) was also detected by metabolome sequencing. As Figure 4G shows, the level of fecal glutamine in R-ABX group was significantly higher than that of R-WT group after FMT (R-WT: R-ABX, 1.2 ± 0.12 μmol/g: 4.04 ± 0.65 μmol/g, P = .0009), which is similar to their donor group (D-WT: D-ABX, 2.38 ± 0.26 μmol/g: 4.89 ± 0.27 μmol/g, P = .0001). That means the elevated level of fecal glutamine was indeed due to the altered gut microbiota after 4 weeks of antibiotics pretreatment. Finally, to demonstrate the critical protective role of fecal glutamine in liver I/R injury, mice were given glutamine gavage of different lengths of time (1, 3, 5, and 7 days) before liver I/R surgery. Seven days of glutamine gavage exhibited protective effect against hepatic I/R injury because the level of serum ALT was decreased in Gln_7d group compared with Con group (Figure 4H; Con: Gln_7d, 22,783.33 ± 1730.73 U/L: 16,516.67 ± 1858.20 U/L, P = .0391). Thus, these findings indicated that the increase in fecal glutamine levels was associated with the changes in the gut microbiota caused by antibiotic pretreatment.
Figure 4.
Altered gut microbiota could lead to changes in fecal metabolites, especially the concentration of glutamine. (A) Relative abundance of metabolites in the feces of the 4 groups. Samples from WT Sham (n = 4), ABX Sham (n = 4), WT IRI (n = 5), and ABX IRI (n = 5) groups were sequenced for fecal metabolomics. (B) Concentrations of amino acids, fatty acids, organic acids, and short-chain fatty acids (SCFAs) in the feces of the 4 groups. (C) PCA of the 4 groups. (D) Heatmap of comparison of concentrations of fecal amino acids and organic acids in the 4 groups. (E) Bubble chart of cluster analysis of metabolic pathways in feces. (F) Fecal concentration of glutamine in the 4 groups. (G) Fecal concentration of glutamine in 4 groups of FMT (D-WT, D-ABX, R-WT, and R-ABX). (H) Serum ALT of Con and glutamine gavage pretreatment groups for different time durations (n = 3). For all data, statistical comparisons between 2 groups were carried out by Student t test. P < .05 indicates significant differences.
Glutamine Derived From the Gut Microbiota Increased the Serum Concentration of Its Downstream Product αKG
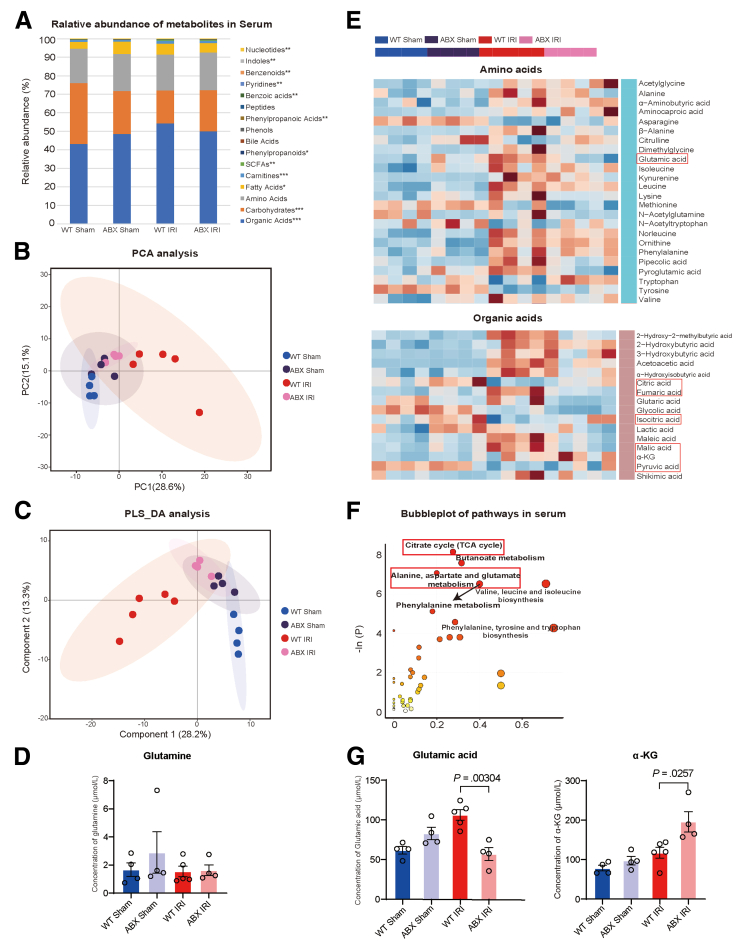
To further determine the changes in serum metabolites to determine which metabolite enters the liver through the portal vein and plays a direct protective role against I/R injury, a targeted metabolome profile was also generated with serum samples. Similarly, we obtained blood samples from sham and hepatic I/R mice in both the WT and ABX groups. According to the analysis of major serum metabolite components, organic acids are the largest and statistically significant (WT Sham: ABX Sham: WT IRI: ABX IRI, 9395.83 ± 1211.49 μmol/L: 10,281.44 ± 218.84 μmol/L: 13,444.75 ± 1149.02 μmol/L: 10,580.99 ± 901.18 μmol/L, P = .011) (Figure 5A). The metabolites of the 4 groups were divided into 4 different clusters by PCA and partial least squares-discriminant analysis, but it seems that the ABX Sham and ABX IRI groups have a closer relationship (Figure 5B and C). Because the concentration of glutamine was not different between the WT IRI and ABX IRI groups (Figure 5D; 571.89 ± 171.30 μmol/L: 545.00 ± 187.09 μmol/L, P = .56), its downstream products glutamate and αKG became the focus of our attention. Indeed, on the basis of the differential metabolite heatmap (Figure 5E) and pathway cluster analysis (Figure 5F), pathways related to glutamine metabolism, including the TCA cycle and alanine, aspartate, and glutamate metabolism pathways, have a large impact on serum metabolites. Compared with those in the WT IRI group, glutamate levels were significantly decreased (106.24 ± 15.24 μmol/L: 56.98 ± 16.25, P = .00304), and αKG levels were increased (117.45 ± 31.09 μmol/L: 195.51 ± 51.74, P = .0025) in the ABX IRI group (Figure 5G), indicating that glutamine is converted into αKG through glutamate after entering the blood. These results showed that increasing the concentration of glutamine in feces led to an increase in serum αKG levels via glutaminolysis.
Figure 5.
Fecal glutamine could increase the serum concentration of αKG. (A) Relative abundance of metabolites in the serum of the 4 groups. Samples from the WT Sham (n = 4), ABX Sham (n = 4), WT IRI (n = 5), and ABX IRI (n = 5) groups were sequenced to assess serum metabolites, but one sample from the ABX IRI group was removed from the analysis because of high heterogeneity. (B and C) PCA (B) and partial least squares-discriminant analysis (PLS_DA) (C) analysis of the 4 groups. (D) Concentrations of serum glutamine in 4 groups. (E) Heatmap of comparison of serum concentrations of amino acids and organic acids in the 4 groups. (F) Bubble chart of cluster analysis of metabolic pathways in blood. (G) Concentration of serum glutamate and αKG in the 4 groups. For all data, statistical comparisons between 2 groups were carried out by Student t test. P < .05 indicates significant differences.
Serum αKG Cooperated With Glycolysis-derived Pyruvate and Synergistically Fueled the TCA Cycle
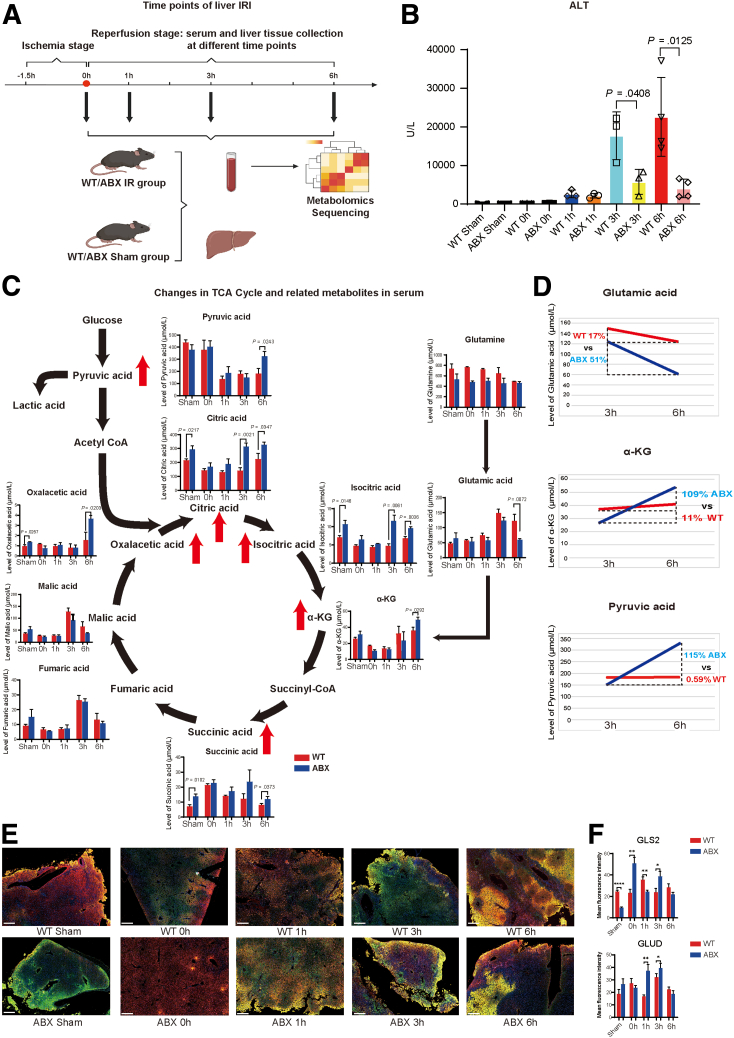
We next investigated the kinetics of serum levels of ALT and various metabolites during hepatic I/R. After 1.5 hours of ischemia, liver and blood samples were collected at 0, 1, 3, and 6 hours of reperfusion (Figure 6A). As Figure 6B shows, the serum ALT levels of the WT and ABX groups continued to rise and peaked at 6 hours. The protective effect of antibiotic pretreatment on liver IRI appeared at 3 hours after reperfusion (17,716.67 ± 6154.34 U/L: 5766.67 ± 3226.58 U/L, P = .0408) and became more pronounced at 6 hours in the reperfusion phase (24,700 ± 11,459.49: 4062.5 ± 2388.65, P = .0125). Furthermore, serum metabolites associated with the TCA cycle and glycolysis pathway were investigated by using UPLC–MS/MS system (Figure 6C). Similar to previous results, glutamate levels were significantly decreased (P = .0072) after 6 hours of reperfusion, and serum glutamine levels did not exhibit significant changes at any time point. The levels of αKG (P = .0292 at 6 hours after reperfusion) and other intermediate products in the TCA cycle, including citrate, isocitrate, succinate, and oxaloacetate, were increased at least one time point after hepatic I/R. Furthermore, the levels of the glycolysis product pyruvate were increased at 6 hours after reperfusion (P = .0243), indicating that αKG pulled the glycolysis pathway and then cooperated with pyruvate to promote progression of the TCA cycle. Moreover, we calculated the rate of change in the glutamate, αKG, and pyruvate levels between 3 and 6 hours of reperfusion (Figure 6D). The concentration of glutamate was decreased by 51% in the ABX group compared with 17% in the WT group. However, the concentrations of αKG and pyruvate were increased by 109% and 115%, respectively, with 11% and 0.59% in the WT group. Furthermore, to elucidate the causal relationship between the conversion of gut-derived glutamine to serum αKG by glutaminolysis, the liver tissue sections of different time points during IR process in both WT and ABX groups were used to detect the changes of expression of enzyme glutaminase 2 (GLS2) (which is mainly expressed in liver tissue) and glutamate dehydrogenase 1/2 (GLUD). These 2 enzymes mediate the conversion of glutamine to αKG via glutamate. As was shown in Figure 6E and F, GLS2 was highly expressed in ABX group in the beginning of reperfusion (0 hour, P = .0036) and 3 hours after (P = .0344). In the meantime, the expression of GLUD was increased gradually from 0 to 3 hours after reperfusion and peaked at 3 hours. It was also highly expressed in ABX group at both 1 hour (P = .0051) and 3 hours (P = .0181) time points compared with that of WT group. Thus, the results indicated that serum αKG could fuel the progression of the TCA cycle by pulling glycolysis. The increase in pyruvate may passively occur as a consequence of the increase in αKG.
Figure 6.
Kinetics of αKG and TCA cycle-related metabolites were altered by antibiotic pretreatment. (A) Pattern diagram and protocol of the kinetics experiments after liver I/R surgery. Liver and blood specimens from the sham and I/R groups were collected at different time points in the reperfusion phase (for each group, n = 3–4). (B) Serum ALT levels of each group. (C) Dynamic changes in the levels of αKG, TCA cycle-related metabolites, and glycolysis-related metabolites. (D) Rate of change in serum glutamate, αKG, and pyruvate levels from 3 to 6 hours in the reperfusion phase. (E) Immunofluorescence colocalization of GLS2 and GLUD in liver tissue sections in different time point after reperfusion (GLS2-Cy3-red fluorescence, GLUD-488 fluorescence). The legend is 200 μm. (F) Quantitative statistical analysis of GLS2 and GLUD expression by using mean fluorescence intensity. For all data, statistical comparisons between 2 groups were carried out by Student t test. P < .05 indicates significant differences.
The Increase in the αKG Levels Could Promote Macrophage Metabolic Reprogramming to Support the M2 Phenotype via OXPHOS
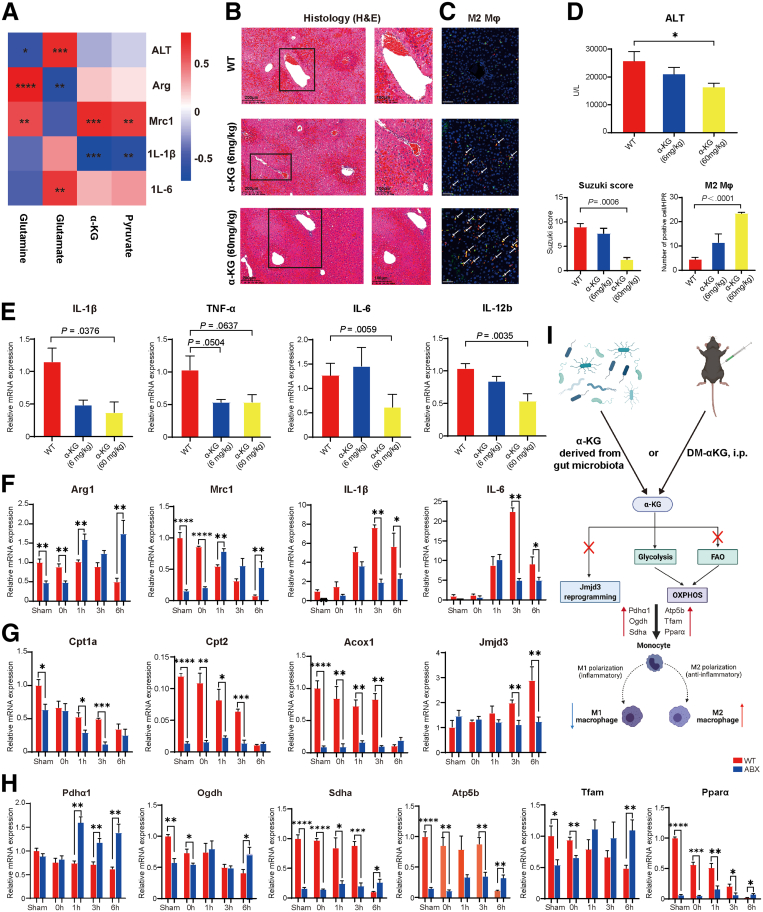
Previous studies have proven that αKG plays an important role in the activation of M2 macrophages by increasing FAO and Jmjd3-dependent epigenetic reprogramming.30 In addition, αKG could also protect against hepatic I/R injury in an orthotopic liver transplantation model in rats by shifting the M1/M2 balance of KCs.31 Thus, to assess the relationship between αKG and the M2 macrophage phenotype in the liver I/R model, we first performed a correlation analysis between the serum concentration of αKG, including the concentrations of other relevant metabolites such as glutamine in feces, glutamate, αKG, and pyruvate in serum, and the clinical indicator ALT, and the expression of M1/M2 macrophage marker genes (Figure 7A). The data showed that serum αKG levels were positively correlated with the expression of the M2 marker gene Mrc1 (r = 0.75, P = .000335) and negatively correlated with the expression of the M1 inflammatory gene Il1b (r = –0.75, P = .000377). In addition, fecal glutamine levels were negatively correlated with ALT levels (r = –0.56, P = .0165) and positively correlated with the expression of the two M2 macrophage genes Arg1 (r = 0.84, P = .0000149) and Mrc1 (r = 0.60, P = .00825). Then, mice were intraperitoneally injected with phosphate-buffered saline (PBS) as WT or DM-αKG (a cell-permeable analog of αKG) at different doses (6 mg/kg or 60 mg/kg) 1 hour before I/R surgery to further explore the relationship between αKG levels and the M1/M2 macrophage phenotype. Compared with those in the WT group, the serum ALT levels were decreased and statistically significant in the αKG (60 mg/kg) group (25,737.5 ± 3378.3: 16,400 ± 1326.2 U/L, P = .0477) (Figure 7D). According to H&E staining, as shown in Figure 7B, a greater level of protection against liver I/R injury was achieved with a higher dose of αKG (60 mg/kg) in both qualitative and quantitative analyses (P = .0006). Furthermore, immunofluorescence showed that the number of CD68+/CD163+ macrophages (M2 macrophages) was increased to a greater extent in the αKG (60 mg/kg) group than in the WT group (P < .0001) (Figure 7C). In addition, the mRNA expression of the M1 macrophage markers Il1β (P = .0376), Tnfα (P = .0637), Il6 (P = .0059), and Il12b (P = .0035) was decreased in the αKG (60 mg/kg) group (Figure 7E). These results demonstrated that αKG supplementation could protect against I/R injury by shifting the M1/M2 balance. Next, we explored M1/M2 macrophage polarization and performed in-depth mechanistic research by returning to kinetics models of WT and ABX mice. The results showed that the mRNA expression of Arg1 (P = .0068) and Mrc1 (P = .0012) was significantly increased in the ABX group at least at 6 hours. Moreover, the expression of Il1β (P = .0492) and Il6 (P = .0488) was decreased in the ABX group after 6 hours of reperfusion. However, it seems that the difference at 3 hours was more obvious (Figure 7F). Because epigenetic and metabolic signaling play an important role in macrophage reprogramming, we conducted an in-depth study on the mechanism by which αKG changes the polarization of macrophages. As shown in Figure 7G, the mRNA expression of FAO markers, including Cpt1a, Cpt2, and Acox1, was decreased from 0 to 3 hours during the reperfusion stage, and these differences became statistically nonsignificant at 6 hours. In addition, the expression of Jmjd3, which is an αKG-dependent specific demethylase of histone H3 lysine 27 (H3K27me3) that promotes macrophage M2 polarization, was also decreased at 3 hours (P = .0061) and 6 hours (P = .0059). Therefore, FAO- and Jmjd3-dependent epigenetic pathways were excluded. Finally, the expression of OXPHOS markers, including Pdhα1 (P = .0028), Ogdh (P = .0198), Sdhα (P = .0354), Atp5b (P = .0028), Tfam (P = .0061), and Pparα (P = .0236), was increased at least at 6 hours (Figure 7H). Consistent with the metabolome results, the above results proved that serum αKG and pyruvate synergistically promoted the OXPHOS metabolic pathway by pulling glycolysis and shifting the balance of M1/M2 macrophages.
Figure 7.
Serum αKG could lead to macrophage metabolic reprogramming to promote M2 phenotype differentiation via the OXPHOS metabolic pathway. (A) Heatmap of the correlation analysis between the fecal levels of glutamine, serum glutamate, αKG, and pyruvate and the disease indicator ALT as well as the expression of the M1 or M2 gene markers Il1β, Il6, Arg1, and Mrc1. (B) H&E staining and Suzuki’s quantitative score of the WT, αKG (6 mg/kg, n = 5), and αKG (60 mg/kg, n = 5) groups. (C) Immunofluorescence double labeling of M2 macrophages and semiquantitative analysis. Scale bars, 100 μm. (D) Serum ALT levels of the 3 groups. (E) Relative mRNA expression of the M1 marker genes Il1β, Tnfα, Il6, and Il12b in the 3 groups. (F) Relative mRNA expression of the M2 marker genes Arg1 and Mrc1 and the M1 marker genes Il1β and Il6 at different time points. (G) Relative mRNA expression of the FAO metabolic pathway-related genes Cpt1a and Cpt2 and the epigenetic gene Jmjd3. (H) Relative mRNA expression of the OXPHOS metabolic pathway-related genes Pdhα1, Ogdh, Sdhα, Atp5b, Tfam, and Pparα. The polymerase chain reaction data above show the kinetics of macrophage M1/M2 marker expression, n = 3–4 for each group. (I) Mechanistic diagram of how αKG can protect the liver from I/R injury. For all data, statistical comparisons between 2 groups were carried out by Student t test. Correlation comparison was performed by Spearman’s correlation analysis. P < .05 indicates significant differences.
Application of Oligomycin A to Inhibit the OXPHOS Pathway Reversed the Protective Effect of Antibiotics on Hepatic I/R Injury
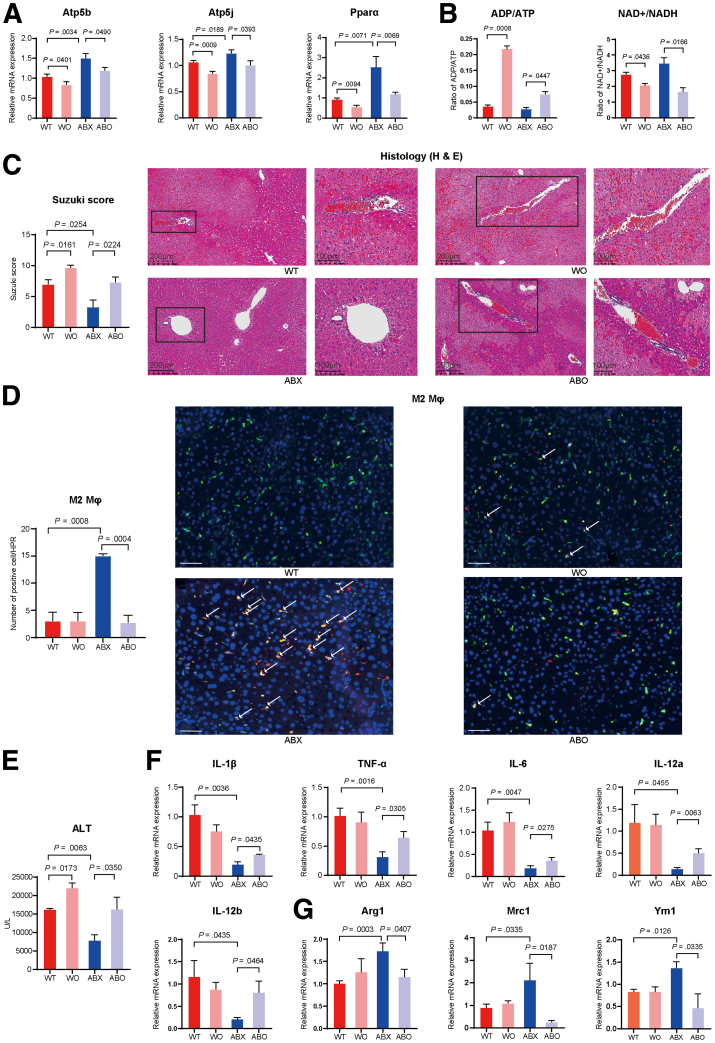
Oligomycin A, which is an inhibitor of H+-ATP synthase, can effectively block the metabolic pathway of OXPHOS.32 Thus, to determine the importance of the activation of the OXPHOS pathway by αKG in the polarization of M2 macrophages, WT or ABX mice were divided into 2 groups; one group was injected with PBS, and the other was injected intraperitoneally with 0.5 mg/kg oligomycin A 1 hour before hepatic I/R surgery. The expression of OXPHOS pathway markers, including Atp5b (WT: P = .0401; ABX: P = .0490), Atp5j (WT: P = .0009; ABX: P = .0393), and Pparα (WT: P = .0094; ABX: P = .0069), was effectively inhibited by oligomycin A in both the WT and ABX groups (Figure 8A). The adenosine diphosphate/ATP ratio was increased in the oligomycin A treatment group (WT: P = .0008; ABX: P = .0477). In addition, the NAD+/NADH ratio was decreased after oligomycin A injection (WT: P = .0436; ABX: P = .0166) (Figure 8B). These results all indicated that the OXPHOS pathway was effectively blocked by oligomycin A. Next, we studied whether the protective effect of antibiotics could be reversed by inhibiting the OXPHOS pathway. Compared with that of the ABX group, the serum ALT level of the ABX mice injected with oligomycin A (ABO) was increased (P = .0350), and the ALT level of the WT mice injected with oligomycin A was also higher than that of the WT group (P = .0173) (Figure 8E). H&E staining showed that the degree of liver damage in the ABO group was more serious than that in the ABX group (P = .0224), as was that in the WT group (P = .0161) (Figure 8C). Next, we further investigated the changes in the M1/M2 phenotype of macrophages. Immunofluorescence showed that the number of CD68+/CD163+ macrophages was significantly decreased in the ABO group compared with the ABX group (P = .0004) (Figure 8D). The relative mRNA expression of M1 macrophage marker genes, including Il1β (P = .0435), Tnfα (P = .0305), Il6 (P = .0275), Il12a (P = .0063), and Il12b (P = .0464), was increased in the ABO group (Figure 8F). However, the relative expression of M2 macrophage markers, including Arg1 (P = .0407), Mrc1 (P = .0187), and Ym1 (P = .0335), was decreased after inhibition of the OXPHOS metabolic pathway (Figure 8G). Thus, these results showed that the application of oligomycin A to inhibit the OXPHOS pathway could reverse the protective effect of antibiotics on hepatic I/R injury by shifting the balance of M1 and M2 macrophage phenotypes.
Figure 8.
Administration of oligomycin A reversed the protective effect of antibiotic pretreatment on hepatic I/R injury. (A) Relative mRNA expression of Atp5b and Atp5j in the WT, WO, ABX, and ABO groups (n = 5 per group). (B) Changes in the levels of serum nucleic acid metabolomics, including ADP/ATP and NAD+/NADH. (C) H&E staining and Suzuki’s quantitative score. (D) Immunofluorescence double labeling of M2 macrophages and semiquantitative analysis. Scale bars, 100 μm. (E) Serum ALT levels in the 4 groups. (F) Relative mRNA expression of the M1 marker genes Il1β, Tnfα, Il6, Il12a, and Il12b in the 4 groups. (G) Relative mRNA expression of the M2 marker genes Arg1, Mrc1, and Ym1 in the 4 groups. For all data, statistical comparisons between 2 groups were carried out by Student t test. P < .05 indicates significant differences.
αKG Could Increase BMDMs M2 Activation and Inhibit M1 Polarization in Vitro and This Effect Was Reversed by Oligomycin A
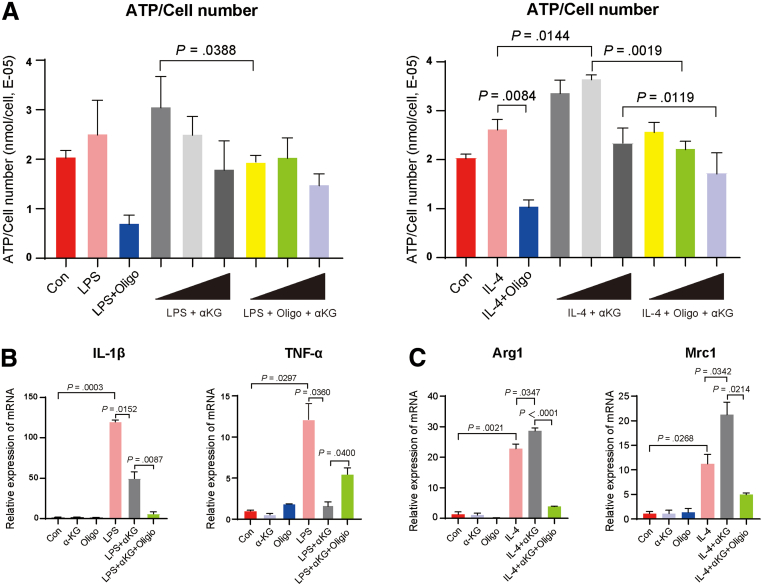
To further prove the effect of αKG, which is the critical metabolite we found in portal vein blood metabolomics above to shift the balance of macrophage M1/M2 polarization, we extracted mouse bone marrow derived macrophages (BMDMs) to see the direct effect of αKG in vitro and using oligomycin A to verify the mechanism that αKG supported macrophage M2 phenotype via OXPHOS pathway. Gradient doses of αKG (0.1, 1, and 10 μmol/L) were used to explore the optimal concentration to increase ATP production of BMDMs at the beginning. As Figure 9A shows, in IL-4 induced M2 macrophage related groups, 1 μmol/L of αKG could efficiently increase ATP level compared with only IL-4 stimulation group (P = .0144). Oligomycin A could decrease ATP level in both 1 and 10 μmol/L αKG stimulation group (P = .0019 and .0119, respectively). Although the effect of αKG and oligomycin A in lipopolysaccharide-induced M1 macrophage related groups was not as good as that of M2 macrophage, we could still see the trends of αKG to increase ATP level in the lower dose, and oligomycin A could reverse this effect. We speculated that this is because M1 macrophages mainly rely on glycolysis rather than OXPHOS pathway as their main energy source. Then, 1 μmol/L of αKG was chosen as the optimum concentration to see its effect on inducing metabolic reprogramming to promote the polarization of macrophage in M2 direction. According to Figure 9C, the expression of both M2 markers Arg1 and Mrc1 was increased in the IL-4 + αKG group compared with only IL-4 stimulation group (P = .0347 and .0342, respectively), and oligomycin A could sufficiently inhibit this effect. Both Arg1 and Mrc1 expression were decreased in IL-4 + Oligo + αKG group (P<.0001 and P = .0214, respectively). As for lipopolysaccharide-induced M1 macrophage, the expression of IL-1β and TNF-α were decreased as what we expected in lipopolysaccharide + αKG group (P = .0152 and 0.0360, respectively). However, oligomycin A did not reverse the effect of αKG on decreasing IL-1β expression. Therefore, all the results of in vitro BMDM experiment showed that αKG could shift the balance between M1 and M2 polarization via macrophage metabolic reprogramming, and oligomycin A reversed this effect by blocking OXPHOS pathway.
Figure 9.
Exogenous αKG supplement could promote BMDM M2 activation and inhibit M1 polarization. (A) On the left, the value of ATP production normalized by cell number of M1 macrophage-related group: Con, LPS stimulation group, LPS + oligomycin A group, LPS + dose gradient of αKG (0.1, 1, 10 μmol/L), and LPS + oligomycin A + dose gradient of αKG (0.1, 1, 10 μmol/L) (n = 3). On the right, the value of ATP normalized by cell number of M2 macrophage-related group: Con, IL-4 stimulation, IL-4 + oligomycin A, IL-4 + dose gradient of αKG (0.1, 1, 10 μmol/L), and IL-4 + oligomycin A + dose gradient of αKG (0.1, 1, 10 μmol/L) (n = 3). After 1 μmol/L of αKG was chosen as the optimal concentration to increase ATP production of BMDMs, M1 and M2 markers were used to perform macrophage phenotyping by quantitative polymerase chain reaction. (B) Relative mRNA expression of the M1 marker genes Il1β, Tnfα in LPS induced-M1 macrophage related group. (C) Relative mRNA expression of the M2 marker genes Arg1, Mrc1 in IL-4 induced-M2 macrophage related group. For all data, statistical comparisons between 2 groups were carried out by Student t test. P < .05 indicates significant differences. LPS, lipopolysaccharide.
Discussion
The gut microbiota and its metabolites play an important role in host physiology and body homeostasis as well as in various liver diseases.8, 9, 10, 11, 12, 13, 14 However, few studies have shown the relationship between the gut microbiota, its metabolites, and liver I/R injury, which is a real challenge of liver surgery and transplantation.2 In recent studies, Li et al33 found that oral ABX cocktail pretreatment could enhance liver IR injury in the morning and demonstrated that gut microbiota played a role in regulating the diurnal variation of liver IR injury of mice. But in a larger basic and clinical cohort study, Nakamura et al15 proved the benefits of ABX pretreatment on liver I/R injury and proposed the therapeutic strategy of gut microbiome. Thus, the relationship between antibiotics treatment, altered gut microbiota, and liver I/R injury are complex and worthy of further study.
Consistent with research of Nakamura et al,15 we found that quadruple antibiotic pretreatment could efficiently alleviate liver I/R injury. Because the composition and function of the gut microbiota were undoubtedly influenced by antibiotic intervention, we next used 16S rRNA sequencing to detect which changes occur in the gut microenvironment.34 The results indicated that the relative abundance, diversity, and richness of the fecal microbiota were decreased in ABX mice. Specific to the bacterial composition, the microbiota of ABX mice include increased abundance of the phylum Proteobacteria and the genera unclassified_f_Enterobacteriaceae and Morganella. As early as the end of the 20th century, studies showed that Escherichia coli expresses enzymes for glutamine synthesis.35,36 Although 16S rRNA sequencing cannot provide accurate results at the species level, the increase in uc_f_Enterobacteriaceae could partly explain the increase in the level of the metabolite glutamine in fecal samples. Furthermore, to exclude the effect of antibiotics on liver I/R, FMT experiments were conducted and provided strong evidence to prove our results. We transferred fecal microbiota from both WT and ABX mice to GF mice during 7 days of colonization and then investigated whether the bacteria were successfully colonized. According to the results, the R-ABX mice exhibited better protection against hepatic I/R injury than their donors because serum ALT levels were decreased to a greater extent after FMT. Thus, we concluded that FMT could efficiently transfer the protective effect of antibiotics on hepatic I/R injury, and the changes in the gut microbiota were the key players in reducing hepatic I/R-associated damage. At the same time, to verify whether the altered microbiota composition and/or function are responsible in the increased concentration of fecal glutamine, glutamine of the 4 groups (D-WT, D-ABX, R-WT, and R-ABX) was detected again by targeted metabolome sequencing. Similar to what we found in donor WT and ABX groups, the fecal glutamine level of R-ABX group was also significantly increased compared with R-WT group. That means the increase of glutamine in feces was indeed due to the changes of gut microbiota after antibiotics pretreatment. Furthermore, to support our conclusion that glutamine was at least one of the critical metabolites that play their role in protecting liver I/R injury, mice were gavaged with glutamine for different duration (1, 3, 5, and 7 days) before liver surgery. The result of serum ALT level showed that the long-term (7 days) exogenous glutamine supplementation could reduce the degree of liver damage. Thus, glutamine alone was sufficient to produce protective effect as we observed in antibiotics intervention.
Abundant evidence has revealed that there is a close bidirectional relationship between the gut microbiota and its metabolites.7 Thus, we used UPLC–MS/MS targeted metabolomic system to investigate the changes in fecal, serum, and hepatic metabolites in response to altered gut bacteria. First, for fecal metabolomics, glutamine became the focus of our research based on hierarchical clustering analysis. It is worth noting that valine, leucine, and isoleucine biosynthesis was also a key metabolic pathway, as shown in the bubble chart. Indeed, studies have proven that branched-chain amino acids can protect against hepatic I/R injury in rats by attenuating KC activation.37 Thus, we suggest that the increased level of branched-chain amino acids is also a reason why antibiotics have a protective effect on hepatic I/R injury. For serum metabolomics, the downstream decomposition products of glutamine, glutamate, and αKG naturally attracted our attention. The results showed that compared with those in the WT IRI group, the concentration of αKG was increased, but glutamate level was decreased, in ABX IRI group, suggesting that glutamine was broken down after entering the blood circulation. Furthermore, we measured the levels of serum αKG and TCA cycle-related metabolites at different time points. The results showed that αKG fueled the progression of the TCA cycle by promoting the glycolysis pathway, as indicated by the increased level of pyruvate at 6 hours. Although there may be other metabolites that play a role in protection against liver I/R injury, we finally chose the glutamine-αKG metabolic pathway by comprehensively considering the metabolomics results in both the feces and serum. At the same time, we established the causal relation between the conversion of gut-derived glutamine to serum αKG via glutamate, which is also called glutaminolysis. First, we traced back to the source of this pathway, glutamine, and we demonstrated the increase of glutamine in feces of ABX mice. Next, glutamine from the intestine would be collected in the portal vein blood, resulting in the high expression of downstream glutaminase GLS2 and glutamate dehydrogenase GLUD in the whole liver tissue, which would eventually lead to the increase of αKG in the blood. The more expression of GLS2 and GLUD, the more this pathway was activated. Thus, glutamate and αKG showed an inverse change in serum metabolomics. But in fact, the level of the upstream source substrate glutamine in blood did not decrease despite the activation of the glutamine decomposition pathway. We speculated that this was due to continuous supplementation of gut-derived glutamine in ABX group.
Immunometabolism, which examines the contribution of diverse metabolic pathways to the development, fate, and behavior of immune cells, has become an increasingly burgeoning research field in recent years.23,38,39 Liu et al30 showed that αKG can promote the M2 phenotype of macrophages via the FAO metabolic pathway and Jmjd3-dependent epigenetic reprogramming. However, in our liver I/R injury mouse model, we found that the number of M2 macrophages, which are responsible for anti-inflammatory processes and tissue repair, was increased by OXPHOS but not by the FAO or Jmjd3-dependent epigenetic pathways. This was due to the increase in serum αKG levels that passively caused an increase in pyruvate levels in the glycolysis pathway and then coordinately promoted progression of the TCA cycle to induce M2 macrophage polarization via the OXPHOS pathway. Moreover, Cheng et al31 proved that DM-αKG could attenuate hepatic I/R injury in a rat OLT model by regulating KC polarization. However, in our study, we focused on the finding that αKG, which is produced by the breakdown of glutamine in the intestine, could protect against liver I/R injury in mice by promoting macrophage metabolite reprogramming. We used both the different models and intervention methods to increase αKG production and described the different mechanisms by which αKG affects macrophage polarization. Finally, using oligomycin A to inhibit H+-ATP synthase efficiently reversed the protective effect of antibiotics on hepatic I/R injury. In the meantime, we also performed mouse BMDMs in vitro to see the direct effect of αKG on increasing ATP production and causing macrophage metabolic reprogramming to promote M2 macrophage activation. Consistent with in vivo experimental results, αKG could efficiently increase ATP level and change the balance between M1 and M2 polarization.
In summary, we provide a link among the gut microbiota, its metabolites, and macrophage polarization and present evidence for potential therapies that target macrophage metabolism, including antibiotic therapies and novel immunometabolism modulators, to overcome the challenge of liver I/R injury in clinical practice.
Methods
Mice
WT C57BL/6JGpt male mice (8–10 weeks old) and GF C57BL/6JGpt (8–10 weeks old) mice were purchased from GemPharmatech (Shanghai, China). WT animals were housed under specific pathogen-free conditions in a temperature-controlled room (22°C–24°C) with a normal light–dark cycle, and the animals were given free access to food and water. GF mice were maintained in sterile isolators under the care of the Emory Gnotobiotic core. For the ABX experiment, WT mice were administered a combination of vancomycin (1 g/L, Sigma Aldrich, cat. #1404-93-9), ampicillin (1 g/L, Sigma Aldrich, cat. #69-53-4), kanamycin (1 g/L, Sigma Aldrich, cat. # 25389-94-0), and metronidazole (1 g/L, Sigma Aldrich, cat. #443-48-1) in their drinking water for 4 months as previously described.40 Fecal samples were collected in fresh sterile tubes, instantly frozen in liquid nitrogen, and stored at –80°C. These stools were later subjected to 16S rRNA sequencing, metabonomic analysis, and FMT experiments. In the FMT experiment, the feces of 10 mice, including 5 WT mice and 5 ABX mice, were collected. For fecal transplantation experiments, 100 mg of stool was resuspended in 1 mL PBS, homogenized, and centrifuged at 500g for 3 minutes, and the bacterial supernatant was collected. Within 10 minutes of preparing the bacterial solution, recipient mice were gavaged with 200 μL of the supernatant on the first and third days during the 7 days of colonization.40,41 Then, all the feces of the donor and recipient mice were collected for bacteriological analysis and metabolome sequencing. For glutamine gavage experiment (Sigma Aldrich, cat. #1294808), 15 mice were randomly divided into 5 groups as control and glutamine gavage for different durations (1, 3, 5, and 7 days). The intragastric dose of each group was 25 mg/kg. All the experimental protocols were in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD) and were approved by the School of Medicine, Shanghai Jiao Tong University.
Liver Ischemia and Reperfusion Surgery
Male C57BL/6JGpt mice, including WT mice, ABX mice, and GF mice colonized by FMT, were randomly divided into a sham group and a liver IR group. We performed a partial liver IR injury operation as described previously.42,43 In brief, the arterial/portal vessels to the cephalad lobes were clamped for 90 minutes. Vascular occlusion was not performed in the sham group. For the drug intervention experiment, mice were injected intraperitoneally with different gradient doses of DM-αKG (6 mg/kg or 60 mg/kg, Sigma Aldrich, cat. #349631) or oligomycin A (0.5 mg/kg, MedChemExpress, cat. #1404-19-9) 1 hour before liver IR surgery. The mice were killed at various time points after reperfusion; liver and serum samples were collected for analysis.
Cell
BMDMs were collected and cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 20% L929 cell (one kind of mouse fibroblast cell line, ATCC, cat. #CCL1) culture supernatant for differentiation for 7 days at 37°C, 5% of CO2 culture. On day 7, differentiated mature BMDMs were replaced with Dulbecco modified Eagle medium (without L929 cell culture supernatant) overnight and then stimulated with 10 ng/mL lipopolysaccharide (Sigma Aldrich, cat. #L2630) to induce M1 polarization and 20 ng/mL IL-4 for M2 (Bioconcept, cat. #214-14) activation for 6 hours. In ATP measurement experiment, BMDMs were given a gradient dose of αKG at 0.1, 1, and 10 μmol/L (Sigma Aldrich, cat. #75890) with or without oligomycin A (5 μmol/L) while stimulating M1/M2 polarization. Then 1 μmol/L concentration of αKG was chosen as the optimal dose to perform polarization phenotype identification by using quantitative polymerase chain reaction.
ALT Measurement
The ALT levels in serum were measured by an ALT kit (Thermo Fisher, Waltham, MA, cat. #TR71121) according to the manufacturer’s instructions.
ATP Measurement
The 5 × 104 BMDMs were seeded in 100 μL of 10% fetal bovine serum Dulbecco modified Eagle medium in 96-well plates. After polarization with/without αKG and oligomycin A treatment, 100 μL of Cell titer-Glo luminiscence ATP reconstituted buffer and substrate (Promega, cat. #G7570) was added to each well, and luminescence was measured after 10 minutes following manufacturer’s instructions. A standard curve with ATP was performed using the same kit and following manufacturer’s instructions.
Real-Time Polymerase Chain Reaction
Total RNA was isolated with TRIzol (Sigma Aldrich, cat. #93289). One microgram of total RNA was reverse transcribed into cDNA with a PrimeScript Reverse Transcription Reagent Kit (Takara, Otsu, Shiga, Japan, cat. #RR037Q). Quantitative polymerase chain reaction was performed with a Step One Plus Real-Time PCR System (Thermo Fisher Scientific) with a Sybr Premix Ex Taq Kit (Takara, cat. #DRR420A). GAPDH was used as a reference. The relative mRNA expression of IL-1β, TNF-α, IL-6, IL-12a, IL-12b, and CXCL10, markers of M1/M2 macrophage phenotypes, and markers of metabolic pathways, including OXPHOS, FAO, and Jmjd3, was determined by 2-ΔΔCT calculation. The primers used in this experiment are as follows.
| Species | Gene | Forward primer (5´-3´) | Reverse primer (3´-5´) |
|---|---|---|---|
| Mouse | GAPDH | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| IL-1β | TCGCAGCAGCACATCAACAAGAG | TGCTCATGTCCTCATCCTGGAAGG | |
| TNF-α | ATGTCTCAGCCTCTTCTCATTC | GCTTGTCACTCGAATTTTGAGA | |
| IL-6 | CTCCCAACAGACCTGTCTATAC | CCATTGCACAACTCTTTTCTCA | |
| IL-12a | GACCTGTTTACCACTGGAACTA | GATCTGCTGATGGTTGTGATTC | |
| IL-12b | TGAGAAGTATTCAGTGTCCTGC | CTGTGAGTTCTTCAAAGGCTTC | |
| CXCL10 | CAACTGCATCCATATCGATGAC | GATTCCGGATTCAGACATCTCT | |
| Arg1 | CATATCTGCCAAAGACATCGTG | GACATCAAAGCTCAGGTGAATC | |
| Mrc1 | CCTATGAAAATTGGGCTTACGG | CTGACAAATCCAGTTGTTGAGG | |
| Ym1 | CAGTGTTCTGGTGAAGGAAATG | ACCCAGACTTGATTACGTCAAT | |
| Cpt1a | CTACATCACCCCAACCCATATT | GATCCCAGAAGACGAATAGGTT | |
| Cpt2 | TGTCTTTGATGTCCTCGATCAA | TCGGTTCTCACTGGTCAAATAA | |
| Acox1 | CCAATGCTGGTATCGAAGAATG | CGACTGAACCTGGTCATAGATT | |
| Jmjd3 | CCCCCATTTCAGCTGACTAA | CTGGACCAAGGGGTGTGTT |
| Species | Gene | Forward primer (5´-3´) | Reverse primer (3´-5´) |
|---|---|---|---|
| Mouse | Pdhα1 | GGTCTGTTTGACATTATACGGC | CGCACAAGATATCCATTCCATC |
| Ogdh | CAATCAGCTCTATGACTGCAAC | GGATTTGGGAGTGAAGACGATT | |
| Sdha | ACATCAGAACTACGCCTAAACA | TTTCACAGCCTTCTTGCAATAC | |
| Atp5b | GGGATTACCACCCATCCTAAAT | TACTTTCTGGCCTCTAACCAAG | |
| Atp5j | GCATTTGAAGAGGAACATTGGT | ATCTTGTCCACGAAGAGTTTCT | |
| Tfam | TCGCATCCCCTCGTCTATCAGTC | TGGGTAGCTGTTCTGTGGAAAATCG | |
| Pparα | GAGCTGCAAGATTCAGAAGAAG | GAATCTTTCAGGTCGTGTTCAC |
H&E Staining and TUNEL Assay
Liver tissues were embedded in paraffin, and serial 5-μm-thick sections were placed onto slides. After dewaxing and rehydration, the sections were stained with H&E according to routine protocols. For the TdT-mediated TUNEL assay, the sections were deparaffinized, and apoptotic cells were detected using the in situ BrdU-Red DNA fragmentation (TUNEL) assay kit (Abcam,cat. #ab66108) and counterstained with DAPI (Tocris, cat. #5748).
Immunohistochemistry
Paraffin liver sections were used to detect the activity of MPO and cleaved caspase 3. Sections were incubated overnight at 4°C with a monoclonal rabbit anti-MPO primary antibody (MPO, 1:500; Abcam, cat. #ab109116; RRID: AB_10865696) and a monoclonal rabbit anti-cleaved caspase-3 primary antibody (cleaved caspase-3, 1:400; Cell Signaling Technology, cat. #9661, RRID: AB_2341188). Then, the sections were rinsed with 0.01 mol/L PBS (pH 7.4) and incubated with biotinylated goat anti-rabbit immunoglobulin G (1:1000; Cell Signaling Technology, cat. #7074, RRID: AB_2099233) for 2 hours at room temperature. After washing, the tissues were incubated with streptavidin-horseradish peroxidase (1:4000; Sigma Aldrich, cat. #ab205718) for 2 hours at room temperature. Immunoreactivity was visualized by incubating the tissue sections in 0.01 mol/L PBS containing 0.05% 3′,3-diaminobenzidine tetrahydrochloride (Sigma Aldrich, cat. #D3939) and 0.003% hydrogen peroxide for 10 minutes in the dark. The sections were then stained with hematoxylin (Servicebio, China, cat. #G1004) and mounted. Control slides that were not incubated with primary antibodies were examined in all cases. Immunoreactive cells were stained yellow-brown. The numbers of positive cells were counted in 5 cross sections in each sample.
Immunofluorescence
Liver tissue sections were used to detect the number of M2 macrophages and changes in the expression of GLS2 and GLUD by double-labeling immunofluorescence. Liver sections were incubated with monoclonal rat anti-CD68 (CD68, 1:100; Abcam, cat. #ab53444, RRID: AB_869007) and monoclonal rabbit anti-CD163 (CD163, 1:500; Abcam, cat. #ab182422, RRID: AB_2753196) antibodies or monoclonal rat anti-CD68 (CD68, 1:100; Abcam) and monoclonal anti-CD86 antibodies (CD86, 1:100; Cell Signaling Technology, cat. #19589, RRID: AB_2892094) overnight at 4°C. The sections were then incubated with the corresponding secondary antibodies conjugated to Alexa Fluor 488 (1: 300, Cell Signaling Technology, cat. #4416, RRID: AB_10693769) or Alexa Fluor 546 (1: 300, Thermo Fisher Scientific, cat. #A-21430, RRID: AB_2535851) for 1 hour at room temperature. Similar to the experiment above, we applied polyclonal rabbit anti-GLS2 antibody (GLS2, 1:100; Abcam, cat. #ab113509, RRID: AB_10866157) and monoclonal rabbit anti-glutamate dehydrogenase 1/2 (GLUD, 1:1000; Cell Signaling Technology, cat. #12793, RRID: AB_2750880) incubation overnight at 4°C, followed by incubation with the fluorescent secondary antibodies Cy3 (1:100, Abcam, cat. #ab97075, RRID: AB_10679955) and Alexa Fluor 488 (1:500, Abcam, cat. #ab150077, RRID: AB_2630356) for 1 hour at room temperature. Control slides that were not incubated with primary antibodies were examined in all cases. For quantification, images were captured with a confocal microscope (LSM-710; Zeiss, Jena, Germany) by an investigator blinded to the treatment conditions and quantified using ImageJ software (National Institutes of Health).
16S rRNA Sequencing and Data Analysis
The fecal microbiota of the WT mice vs ABX mice as well as the donor and recipient WT and ABX mice in the FMT experiment were analyzed by 16S rRNA sequencing. DNA was extracted from the samples using a QIAamp Fast DNA Stool Mini Kit (Qiagen, CA, cat. #51604) according to the manufacturer’s instructions. The bacterial DNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific). Then, the 16S rRNA genes were amplified with the bacterial primer pair 338F-806R, which flanks the V3–V4 region, using FastPfu Polymerase. Amplicons were then purified by gel extraction (AxyPrep DNA Gel Extraction Kit, Axygen Biosciences, CA, cat. #AP-GX-250) and quantified using QuantiFluor-ST (Promega, Madison, WI). Paired-end sequencing was performed using an Illumina MiSeq system (Illumina, CA). High-throughput pyrosequencing of the polymerase chain reaction products was then performed on the free online platform of the Majorbio Cloud Platform (www.majorbio.com).
Sequencing reads were demultiplexed and filtered. Operational taxonomic units were selected at 97% similarity cutoff, and the identified taxonomy was then aligned using the Greengenes database (version 13.8). The relative species abundance in each group was evaluated on the basis of rank-abundance curves, and alpha-diversity indices (Shannon, Simpson, Chao, and ACE) were analyzed. For beta-diversity analysis, PCA, PCoA, and NMDS were performed using Quantitative Insights into Microbial Ecology (QIIME) 1.9.1. For the analysis of differences in bacterial composition, we used Student t test with P < .05 as a threshold. From phylum to genus, the biomarkers in the 2 groups were quantitatively analyzed by linear discriminant analysis effect size analysis. Linear discriminant analysis effect size analysis, with linear discriminant analysis threshold of >2, was performed using the nonparametric Kruskal–Wallis test to identify the most differentially abundant taxa.
Metabolomics
Fecal sample preparation
Fecal samples were thawed in an ice bath to reduce degradation. Approximately 5 mg of each sample was weighed and transferred to a new 1.5 mL tube. Then, 25 μL of water was added, the samples were homogenized with zirconium oxide beads for 3 minutes, and 120 μL of methanol containing internal standard was added to extract the metabolites. The samples were homogenized for another 3 minutes and then centrifuged at 18,000g for 20 minutes. Twenty microliters of the supernatants was transferred to a 96-well plate. The following procedures were performed on a Biomek 4000 workstation (Biomek 4000, Beckman Coulter, Inc, Brea, CA). Twenty microliters of freshly prepared derivative reagents was added to each well. The plate was sealed, and derivatization was carried out at 30°C for 60 minutes. After derivatization, 330 μL of ice-cold 50% methanol solution was added to dilute the samples. Then, the plate was stored at –20°C for 20 minutes, followed by centrifugation at 4000g at 4°C for 30 minutes. A total of 135 μL of each supernatant was transferred to a new 96-well plate with 10 μL internal standards in each well. Serial dilutions of the derivatized stock standards were added to the left wells. Finally, the plate was sealed for LC–MS analysis.
Serum sample preparation
Samples were thawed in an ice bath to reduce degradation. Then, 25 μL of plasma was added to a 96-well plate, and the plate was transferred to the Eppendorf epMotion Workstation (Eppendorf Inc, Hamburg, Germany). Then, 120 μL ice-cold methanol with partial internal standards was automatically added to each sample, and the samples were vortexed vigorously for 5 minutes. The plate was centrifuged at 4000g for 30 minutes (Allegra X-15R, Beckman Coulter, Inc, Indianapolis, IN).
Then, the plate was returned to the workstation. Thirty microliters of each supernatant was transferred to a clean 96-well plate, and 20 μL of freshly prepared derivative reagents was added to each well. The plate was sealed, and derivatization was carried out at 30°C for 60 minutes. After derivatization, 330 μL of ice-cold 50% methanol solution was added to dilute the samples. Then, the plate was stored at –20°C for 20 minutes, followed by centrifugation at 4000g at 4°C for 30 minutes. A total of 135 μL of each supernatant was transferred to a new 96-well plate with 10 μL internal standards in each well. Serial dilutions of the derivatized stock standards were added to the left wells. Finally, the plate was sealed for LC–MS analysis.
UPLC–MS/MS
A UPLC–MS/MS system (ACQUITY UPLC-Xevo TQ-S, Waters Corp, MA) was used to analyze 11 targeted metabolites of interest. We used UPLC columns, including an ACQUITY HPLC BEH C18 1.7 μm VanGuard precolumn (2.1 × 5 mm) and an ACQUITY HPLC BEH C18 1.7 μm analytical column (2.1 × 100 mm), to perform the chromatographic separation of fecal samples at a constant temperature of 40°C. The injection volume of sample was 5 μL. The mobile phases were as follows; eluent A was 0.1% formic acid in water, and eluent B was acetonitrile/IPA (70:30). The gradient elution conditions were as follows: 0–1 minute (5% B), 1–11 minutes (5%–78% B), 11–13.5 minutes (78%–95% B), 13.5–14 minutes (95%–100% B), 14–16 minutes (100% B), 16–16.1 minutes (100-5% B), and 16.1–18 min (5% B). The flow rate was set to 0.40 mL/min. For mass spectrometry, the conditions were as follows: capillary: 1.5 (ESI+), 2.0 (ESI-) Kv; source temperature: 150°C; desolvation temperature: 550°C; desolvation gas flow: 1000 L/h. The quality control samples were prepared along with the test samples and run after every 14 samples to ensure reproducibility.
For data analysis, the raw data files generated by UPLC–MS/MS were processed with MassLynx software (version 4.1, Waters, MA) to perform peak integration, calibration, and quantitation for each metabolite. The powerful R studio package was used for statistical analysis. PCA and partial least square discriminant analysis were used for the classification and identification of differentially altered metabolites. Potential biomarkers of differentially expressed metabolites were characterized by variables with a variable influence on projection >1 and P < .05 in Student t test or Wilcoxon test. Z-transform was conducted to observe the distribution of different metabolites between groups. Pathway enrichment analysis was performed using pathway impact and hypergeometric tests.
Quantification and Statistical Analysis
All the data are presented as the mean ± standard deviation. Differences between 2 groups were analyzed by Student t test or Wilcoxon test. For comparison of data between multiple groups, the Kruskal–Wallis H test was used. Statistical analyses were performed using GraphPad Prism v 8.0 (USA) software. All the results were considered statistically significant at P < .05. The methods for the data analysis of fecal microbiota and metabolomics have been described above. Statistical parameters and the number of samples are stated in the figure legends.
Acknowledgments
CRediT Authorship Contributions
Tianfei Lu (Conceptualization: Equal; Funding acquisition: Equal; Supervision: Lead; Writing – review & editing: Lead)
Qing Li (Data curation: Lead; Formal analysis: Lead; Writing – original draft: Lead)
Weiwei Lin (Methodology: Equal; Resources: Equal)
Xianzhe Zhao (Methodology: Equal; Resources: Supporting)
Fu Li (Funding acquisition: Equal; Methodology: Equal; Resources: Supporting)
Jianmei Ji (Methodology: Equal; Resources: Equal)
Yu Zhang (Funding acquisition: Equal; Methodology: Equal; Resources: Equal; Writing – review & editing: Supporting)
Ning Xu, PhD (Funding acquisition: Lead; Writing – review & editing: Supporting)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by China National Science and Technology Major Project: grant no. 2018ZX10302 206-006-006 (NX); National Natural Science Foundation of China: grant no. 82170665 (TL), no. 82170673 (YZ), no. 81904017 (FL); and Natural Science Foundation exploration project of Zhejiang Province: grant no. LY21H030010 (YZ).
References
- 1.Bi J., Zhang J., Ren Y., et al. Irisin alleviates liver ischemia-reperfusion injury by inhibiting excessive mitochondrial fission, promoting mitochondrial biogenesis and decreasing oxidative stress. Redox Biol. 2019;20:296–306. doi: 10.1016/j.redox.2018.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhai Y., Petrowsky H., Hong J.C., et al. Ischaemia-reperfusion injury in liver transplantation: from bench to bedside. Nat Rev Gastroenterol Hepatol. 2013;10:79–89. doi: 10.1038/nrgastro.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner C., Galluzzi L., Kepp O., et al. Decoding cell death signals in liver inflammation. J Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 4.Peralta C., Jimenez-Castro M.B., Gracia-Sancho J. Hepatic ischemia and reperfusion injury: effects on the liver sinusoidal milieu. J Hepatol. 2013;59:1094–1106. doi: 10.1016/j.jhep.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Ye L., He S., Mao X., et al. Effect of hepatic macrophage polarization and apoptosis on liver ischemia and reperfusion injury during liver transplantation. Front Immunol. 2020;11:1193. doi: 10.3389/fimmu.2020.01193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaudel C., Sokol H. The gut microbiota at the service of immunometabolism. Cell Metab. 2020;32:514–523. doi: 10.1016/j.cmet.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Gong S., Lan T., Zeng L., et al. Gut microbiota mediates diurnal variation of acetaminophen induced acute liver injury in mice. J Hepatol. 2018;69:51–59. doi: 10.1016/j.jhep.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolodziejczyk A.A., Zheng D., Shibolet O., et al. The role of the microbiome in NAFLD and NASH. EMBO Mol Med. 2019;11 doi: 10.15252/emmm.201809302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouries J., Brescia P., Silvestri A., et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J Hepatol. 2019;71:1216–1228. doi: 10.1016/j.jhep.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saeedi B.J., Liu K.H., Owens J.A., et al. Gut-resident Lactobacilli activate hepatic Nrf2 and protect against oxidative liver injury. Cell Metab. 2020;31:956–968 e5. doi: 10.1016/j.cmet.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwabe R.F., Greten T.F. Gut microbiome in HCC: mechanisms, diagnosis and therapy. J Hepatol. 2020;72:230–238. doi: 10.1016/j.jhep.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Yu L.X., Schwabe R.F. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527–539. doi: 10.1038/nrgastro.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Coker O.O., Chu E.S., et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut. 2021;70:761–774. doi: 10.1136/gutjnl-2019-319664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura K., Kageyama S., Ito T., et al. Antibiotic pretreatment alleviates liver transplant damage in mice and humans. J Clin Invest. 2019;129:3420–3434. doi: 10.1172/JCI127550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Kasmi K.C., Stenmark K.R. Contribution of metabolic reprogramming to macrophage plasticity and function. Semin Immunol. 2015;27:267–275. doi: 10.1016/j.smim.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon S., Martinez F.O. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Locati M., Curtale G., Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan A.T., Goldrath A.W., Glass C.K. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity. 2017;46:714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouton A.J., Li X., Hall M.E., et al. Obesity, hypertension, and cardiac dysfunction: novel roles of immunometabolism in macrophage activation and inflammation. Circ Res. 2020;126:789–806. doi: 10.1161/CIRCRESAHA.119.312321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neill L.A., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Bossche J., O’Neill L.A., Menon D. Macrophage immunometabolism: where are we (going)? Trends Immunol. 2017;38:395–406. doi: 10.1016/j.it.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Wang Y., Ni Y., et al. Gut microbiome fermentation determines the efficacy of exercise for diabetes prevention. Cell Metab. 2020;31:77–91 e5. doi: 10.1016/j.cmet.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Zheng X., Xie G., Zhao A., et al. The footprints of gut microbial-mammalian co-metabolism. J Proteome Res. 2011;10:5512–5522. doi: 10.1021/pr2007945. [DOI] [PubMed] [Google Scholar]
- 26.Schuster H., Blanc M.C., Bonnefont-Rousselot D., et al. Protective effects of glutamine dipeptide and alpha-tocopherol against ischemia-reperfusion injury in the isolated rat liver. Clin Nutr. 2009;28:331–337. doi: 10.1016/j.clnu.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Szijarto A., Hahn O., Batmunkh E., et al. Short-term alanyl-glutamine dipeptide pretreatment in liver ischemia-reperfusion model: effects on microcirculation and antioxidant status in rats. Clin Nutr. 2007;26:640–648. doi: 10.1016/j.clnu.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Ma N., Ma X. Dietary amino acids and the gut-microbiome-immune axis: physiological metabolism and therapeutic prospects. Compr Rev Food Sci Food Saf. 2019;18:221–242. doi: 10.1111/1541-4337.12401. [DOI] [PubMed] [Google Scholar]
- 29.Perna S., Alalwan T.A., Alaali Z., et al. The role of glutamine in the complex interaction between gut microbiota and health: a narrative review. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20205232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P.S., Wang H., Li X., et al. Alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 31.Cheng M.X., Cao D., Chen Y., et al. alpha-ketoglutarate attenuates ischemia-reperfusion injury of liver graft in rats. Biomed Pharmacother. 2019;111:1141–1146. doi: 10.1016/j.biopha.2018.12.149. [DOI] [PubMed] [Google Scholar]
- 32.Shchepina L.A., Pletjushkina O.Y., Avetisyan A.V., et al. Oligomycin A, inhibitor of the F0 part of H+-ATP-synthase, suppresses the TNF-induced apoptosis. Oncogene. 2002;21:8149–8157. doi: 10.1038/sj.onc.1206053. [DOI] [PubMed] [Google Scholar]
- 33.Li R., Xie L., Li L., et al. The gut microbial metabolite, 3,4-dihydroxyphenylpropionic acid, alleviates hepatic ischemia/reperfusion injury via mitigation of macrophage pro-inflammatory activity in mice. Acta Pharm Sin B. 2022;12:182–196. doi: 10.1016/j.apsb.2021.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ianiro G., Tilg H., Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 35.Jiang P., Peliska J.A., Ninfa A.J. The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry. 1998;37:12802–12810. doi: 10.1021/bi980666u. [DOI] [PubMed] [Google Scholar]
- 36.Jiang P., Mayo A.E., Ninfa A.J. Escherichia coli glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49): kinetic characterization of regulation by PII, PII-UMP, glutamine, and alpha-ketoglutarate. Biochemistry. 2007;46:4133–4146. doi: 10.1021/bi0620510. [DOI] [PubMed] [Google Scholar]
- 37.Kitagawa T., Yokoyama Y., Kokuryo T., et al. Protective effects of branched-chain amino acids on hepatic ischemia-reperfusion-induced liver injury in rats: a direct attenuation of Kupffer cell activation. Am J Physiol Gastrointest Liver Physiol. 2013;304:G346–G355. doi: 10.1152/ajpgi.00391.2012. [DOI] [PubMed] [Google Scholar]
- 38.Murray P.J., Rathmell J., Pearce E. SnapShot: immunometabolism. Cell Metab. 2015;22:190–190 e1. doi: 10.1016/j.cmet.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Makowski L., Chaib M., Rathmell J.C. Immunometabolism: from basic mechanisms to translation. Immunol Rev. 2020;295:5–14. doi: 10.1111/imr.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thaiss C.A., Zeevi D., Levy M., et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 41.Zeng S.L., Li S.Z., Xiao P.T., et al. Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci Adv. 2020;6 doi: 10.1126/sciadv.aax6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji H., Liu Y., Zhang Y., et al. T-cell immunoglobulin and mucin domain 4 (TIM-4) signaling in innate immune-mediated liver ischemia-reperfusion injury. Hepatology. 2014;60:2052–2064. doi: 10.1002/hep.27334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y., Lu T., Zhang C., et al. Activation of YAP attenuates hepatic damage and fibrosis in liver ischemia-reperfusion injury. J Hepatol. 2019;71:719–730. doi: 10.1016/j.jhep.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]