Abstract
Background
Schizophrenia (SCZ) is a heterogeneous neuropsychiatric disorder for which current treatment has insufficient efficacy and severe adverse effects. The modifiable gut microbiome might be a potential target for intervention to improve neurobiological functions through the gut-microbiome-brain axis.
Methods
In this case-control study, gut microbiota of 132 patients with SCZ and increased waist circumference were compared with gut microbiota of two age- and sex-matched control groups, composed of 132 healthy individuals and 132 individuals with metabolic syndrome. Shotgun sequencing was used to characterize fecal samples at the taxonomic and functional levels. Cognition of the patients with SCZ was evaluated using the Brief Assessment of Cognition instrument.
Results
SCZ gut microbiota differed significantly from those of healthy control subjects and individuals with metabolic syndrome in terms of richness and global composition. SCZ gut microbiota were notably enriched in Flavonifractor plautii, Collinsella aerofaciens, Bilophila wadsworthia, and Sellimonas intestinalis, while depleted in Faecalibacterium prausnitzii, Ruminococcus lactaris, Ruminococcus bicirculans, and Veillonella rogosae. Functional potential of the gut microbiota accounted for 11% of cognition variability. In particular, the bacterial functional module for synthesizing tyrosine, a precursor for dopamine, was in SCZ cases positively associated with cognitive score (ρ = 0.34, q ≤ .1).
Conclusions
Overall, this study shows that the gut microbiome of patients with SCZ differs greatly from that of healthy control subjects or individuals with metabolic syndrome. Cognitive function of patients with SCZ is associated with the potential for gut bacterial biosynthesis of tyrosine, a precursor for dopamine, suggesting that gut microbiota might be an intervention target for alleviation of cognitive dysfunction in SCZ.
Keywords: Case-control study, Cognition, Gut microbiome, Metagenomics, Schizophrenia, Tyrosine biosynthesis
Schizophrenia (SCZ) is a heterogeneous neuropsychiatric disorder characterized by hallucinations, delusions, social withdrawal, anhedonia, and cognitive impairment, with the first disease manifestation usually occurring in adolescence (1). SCZ is a highly heritable, polygenic disorder, but at present the molecular genetics of the disorder are only partially explained (2). Recently, there has been increasing interest in how gut microbiota relate to mental health (3), moving focus of apprehension away from the brain. The link is plausible, because psychiatric and gut disorders are epidemiologically linked (4) and antipsychotic drugs have antibacterial properties (5). There are several suggested pathways by which the microbiota harbored by the gut interact with the brain, such as modulation of the immune system, increased permeability of the gut-brain barrier, nervus vagus activation, or metabolization of neurotransmitters (6,7). The current medical treatment for SCZ consists of antipsychotic medication, which has limited effect on symptom severity but has adverse effects, such as blunting of affect, lethargy, and metabolic dysregulation, leading to obesity and cardiovascular disease. Despite decades of biological optimism, the anticipated breakthrough in understanding SCZ and developing new targets for treatment has been disappointing and calls for radically new thinking.
So far, the field linking microbiota to SCZ has moved extensively in only a few years, from case-control studies of taxonomic and functional alterations to translational studies where candidate microbes are transferred to rodents (8,9). Despite increased evidence of microbiota alterations across all psychiatric disorders (10), replicability of findings is limited. Several features point to limitations in the basic studies, suggesting gaps in knowledge. Eleven cross-sectional studies (8,11, 12, 13, 14, 15, 16, 17, 18, 19, 20) of gut microbiota from patients with SCZ have been published, reporting somewhat contradictory findings, the number of patients ranging from 10 to 90 (21). The poor replicability may be explained by high risks of false positive findings (type 1 errors), false negatives (type 2 errors), and substantial heterogeneity across studies. The high risk of type 1 errors is due to small samples and lack of preregistrations. The heterogeneity concerns methods to collect samples, different clinical inclusion criteria, different techniques to sequence and analyze data, and different research questions, because some aim to identify biomarkers for diagnostic purposes while others search for causal links that can be targets for future treatment of SCZ. While seeming similar, the difference in these two questions concerns controlling for confounders. A specific risk in SCZ studies is the confounding effects of lifestyle factors and psychotropic medication, and proper methods of deconfounding are a prerequisite for casual inference (22).
Another limitation in the field of biological psychiatry is the lack of valid diagnostic categories. While genetic research, acknowledging overlapping genetics, moves toward transdiagnostic designs (23,24), the Research Domain Criteria initiative (25) suggests to focus on symptom domains rather than arbitrary categories.
In this large study, we aim at identifying taxonomic and functional alterations in stool microbiota (as a proxy of gut microbiota) of patients with SCZ using novel methods to adjust for medication and metabolic features and link these to levels of psychotic symptoms, negative symptoms, cognition, and global functioning.
Methods and Materials
Study Population
Individuals with SCZ (n = 132) and increased waist circumference (which is a common but not guaranteed comorbidity) were recruited from a lifestyle trial, CHANGE (26), conducted at university hospitals in Aarhus and Copenhagen, Denmark from 2012 to 2014. Diagnoses were confirmed by structured diagnostic interviews by trained clinicians. Psychometric measures at study entry included the Scale for the Assessment of Negative Symptoms (SANS) and Scale for the Assessment of Positive Symptoms instruments (27), a measure of cognition (Brief Assessment of Cognition in Schizophrenia [BACS]) (28), and the Global Assessment of Functioning (GAF) (29). The study of SCZ cases was approved by the Ethical Committees of the Capital Region of Denmark (H-4-2012-051).
Healthy control (HC) subjects who reported no acute or chronic medical disorders were selected among age- and sex-matched individuals from the population-based Danish Study of Functional Disorders (n = 74) (30). The Danish Study of Functional Disorders was approved by the Ethical Committees of the Capital Region of Denmark (H-3-2012-015). Another sample of healthy individuals by self-report (included in the overall HC group) was phenotyped at Novo Nordisk Foundation Center for Basic Metabolic Research, University of Copenhagen (unpublished; n = 58). The study was approved by the Ethical Committees of the Capital Region of Denmark (H-3-2012-145). A second, dysmetabolic control group matched with SCZ cases on body mass index (BMI) and sex was selected among nondiabetic samples from the MetaHIT project (n = 132) (31) (Table 1; Table S1 in Supplement 1). This control group is referred to as the metabolic syndrome (MS) group. The MetaHIT subcohort was approved by the Ethical Committees of the Capital Region of Denmark (H-C-2008-017). All study participants gave informed consent, and all examinations were done in accordance with the Helsinki Declaration. Details are provided in Supplement 1.
Table 1.
Characteristics of the Three Study Samples
| Characteristics | SCZ, n = 132 | HC, n = 132 | MS, n = 132 | pHealthy | pDysmetabolic |
|---|---|---|---|---|---|
| BMI, kg/m2, Mean (SD) | 35 (6.2) | 24 (3.8) | 34 (4.5) | <.001 | .21 |
| Age, Years, Mean (SD) | 41 (12) | 40 (12) | 57 (7.7) | .65 | <.001 |
| Sex, Female/Male, n | 73/59 | 75/57 | 71/57 | .9 | 1 |
| Smoking Status, Current/Never/Previous, n | 64/38/30 | 16/70/44 | 14/61/57 | <.001 | <.001 |
For quantitative variables (BMI and age), p values are obtained from Wilcoxon tests; for qualitative variables (sex, smoking status), they are obtained from χ2 tests. pHealthy compares SCZ and HC, pDysmetabolic compares SCZ and MS.
BMI, body mass index; HC, healthy control; MS, metabolic syndrome; SCZ, schizophrenia.
Metagenomics Profiling
Total DNA extracted from stool samples was sequenced and the sequencing reads used to assess the presence and abundance of 10.4 million genes from the integrated gene catalog 2 (32,33) and 8.4 million genes from the oral gene catalog (34), as detailed in Supplement 1. The genes from both catalogs were organized into metagenomics species (MGSs), the abundance of which was deduced from the cognate gene abundances (see Supplement 1).
Statistical Analysis
All statistical analyses were performed using R version 3.6.0 (35). Contrasts in quantitative variables were performed using the Wilcoxon test to compare two groups or Kruskal-Wallis followed by post hoc Dunn tests to compare more than two groups. Associations between categorical variables were performed using the χ2 test. Correlations between variables were performed using Spearman’s test. The Benjamini-Hochberg method was used for multiple testing correction. Unless stated otherwise, a corrected p value was considered significant when ≤ .1. Effect size was computed as Cliff’s Delta using the package effsize version 0.7.4 (36).
Bray-Curtis dissimilarity was computed on the log10-transformed MGS table with the package vegan version 2.5.7 (37). Principal coordinates analysis was performed on the Bray-Curtis dissimilarity with the package ade4 version 1.7.16 (38). Bray-Curtis dissimilarity variance between groups was then analyzed by permutational multivariate analysis of variance with the function adonis from the package vegan. Shannon index was computed with the function diversity from the package vegan.
Covariate Deconfounding
Covariate deconfounding was performed on each metagenomics feature with the R package metadeconfoundR version 0.1.5 (39) (see Supplement 1). The pipeline was run on four different partitions of the dataset: 1) SCZ cases versus HC subjects with Anatomical Therapeutic Chemical code drug dosages (i.e., quantitative, encoded as defined daily doses) in addition to age, BMI, sex, and smoking status (these four variables were accounted for in all four runs); 2) SCZ cases versus HC subjects with drug groups as binary trait (medication class taken/not taken) (Figure S1 in Supplement 1); 3) SCZ cases versus dysmetabolic control group (MS) with Anatomical Therapeutic Chemical code dosages as above; 4) SCZ cases versus MS control subjects with drug groups as binary variables, as above. A feature was considered confounded if it was found confounded in at least one of the datasets.
Contrasted Metagenomics Features
We performed the Kruskal-Wallis test followed by post hoc Dunn tests on each metagenomics feature to assess microbiome contrast between SCZ cases, HC subjects, and control subjects with MS. Contrasted features were selected as follows: 1) false discovery rate ≤ 0.1 for Kruskal-Wallis test; 2) p ≤ .05 for Dunn test in the SCZ/HC comparison and in the SCZ/MS comparison; 3) same directionality in SCZ/HC and SCZ/MS comparisons regarding status (SCZ or control); and 4) not confounded by covariates.
Status Prediction
We predicted the class of each individual (SCZ vs. HC or SCZ vs. MS) using an elastic net regression with the R package caret version 6.0.84 (40). Input was the log10-transformed, scaled, and centered MGS table. MGSs present in <10% of individuals were filtered out. The dataset was split into a training set (75%) and a test set (25%). Models were fit on the training set under cross-validation and then used to predict class of the testing set. The area under the receiver operating characteristic curve was computed on the test set. The entire procedure (splitting, fitting, predicting) was repeated 100 times.
Modeling Cognition Score
Random forests were used to predict cognition score (BACS) using the R packages VSURF version 1.1.0 (41) and randomForest version 4.6.14 (42). Explanatory variables consisted of either functional modules, bioclinical variables (including medication), or the combination of both. Functional modules were log-transformed, centered, and scaled. Those present in <10% of individuals were filtered out. Quantitative bioclinical variables were centered and scaled. The validation scheme consisted of a 5-fold cross-validation repeated 20 times. The variable selection procedure is based on a preliminary ranking of the explanatory variables using the random forests permutation-based score of importance and proceeds using a stepwise forward strategy for variable introduction. The increase of mean square error was the metric used to assess the importance of the variables.
Results
Taxonomic Analyses of Gut Microbiota
Fecal samples were analyzed from 132 SCZ cases and two age- and sex-matched control groups consisting of 132 HC individuals and 132 individuals with MS (Table 1).
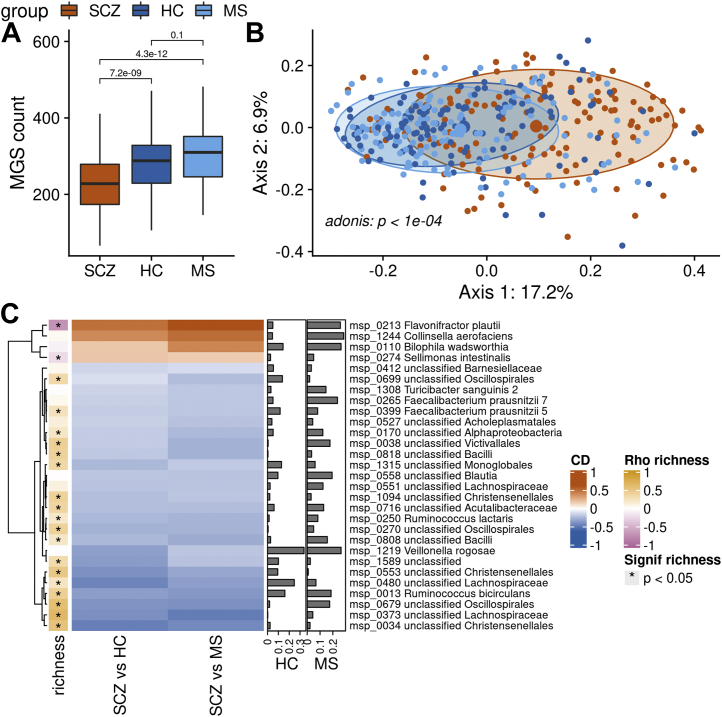
Patients with SCZ showed substantially lower MGS richness, which was decreased by 21% to 24% relative to the two control groups (Figure 1A). The decrease in richness was still significant after deconfounding for BMI, age, smoking status, sex, and medication. Results were similar using the Shannon index as alpha-diversity measure (Figure S2 in Supplement 1). Principal coordinates analysis revealed a clear difference in composition of gut microbiota from patients with SCZ as compared with the two control groups (p = 1 × 10−4, permutational multivariate analysis of variance) (Figure 1B). Consistently, we were able to predict the class of each individual based on the MGS abundance using elastic net regression with a mean area under the receiver operating characteristic curve of 0.86 ± 0.04 (SCZ vs. HC) and 0.92 ± 0.03 (SCZ vs. MS) (Figure S3A, B in Supplement 1). Univariate analyses showed that out of 600 MGSs present in at least 10% of individuals, 202 (34%) differed in abundance between SCZ cases and HC individuals or individuals with MS. Among these, 31 (15%) were enriched in SCZ and 171 (85%) were depleted. After deconfounding for medication, BMI, age, and smoking status, 17 MGSs remained enriched in SCZ and 104 MGSs remained depleted in SCZ (referred to as SCZ-MGS and control-MGS, respectively). Most (85%) correlated significantly with MGS richness in the SCZ cohort (p ≤ .05, Spearman’s correlation). SCZ-MGSs correlated inversely with richness, while control-MGSs correlated positively, in line with the decrease in richness observed in the SCZ gut microbiota as compared with HC and MS gut microbiota.
Figure 1.
Contrast in MGS richness and taxonomic composition. (A) Difference in richness of MGS between patients with SCZ, HC subjects, and individuals with MS. p values from Wilcoxon tests are displayed. Boxes represent the median IQRs between the first and third quartiles; whiskers represent the lowest or highest values within 1.5 times IQR from the first or third quartiles. (B) Principal coordinates analysis ordination of the Bray-Curtis dissimilarity matrix computed on the MGS abundance from patients with SCZ and control subjects (HC and MS). (C) Heatmap of the effect size (CD) of MGSs significantly contrasted between SCZ and control subjects after covariate deconfounding and selected with elastic net regression to predict status (red and blue denote MGSs enriched in SCZ and control subjects, respectively; scale is shown on the right side of the panel). The right bars show the variable importance of each MGS in the status prediction (for HC vs. SCZ or MS vs. SCZ). The left colored bar shows Spearman’s correlations of each MGS with MGS richness (computed in patients with SCZ only). Red = more abundant in patients with SCZ; blue = more abundant in control subjects. CD, Cliff’s Delta; HC, healthy control; IQR, interquartile range; MGS, metagenomic species; MS, metabolic syndrome; SCZ, schizophrenia.
SCZ-MGSs that were also selected by the elastic net regression to predict the individual status included Flavonifractor plautii, Collinsella aerofaciens, Bilophila wadsworthia, and Sellimonas intestinalis. In contrast, control-MGSs selected by the model included two subspecies of Faecalibacterium prausnitzii, Turicibacter sanguinis, Ruminococcus lactaris, Veillonella rogosae, and Ruminococcus bicirculans (Figure 1C). All contrasted MGSs are listed in Table S1 in Supplement 1.
Of note, only two oral bacteria were part of the differentially abundant MGSs before deconfounding: V. rogosae, more abundant in control subjects, and Streptococcus vestibularis, more abundant in SCZ cases but confounded by smoking and medication status. This shows that there was no increased invasion of oral species in the SCZ gut microbiota in our cohort.
Summing up all MGS abundance according to their taxonomic annotation, we found that, at the phylum level, Actinobacteriota and Desulfobacterota were more abundant in SCZ. At the family level, Tannerellaceae, Lachnospiraceae, Desulfovibrionaceae, Coriobacteriaceae, Actinomycetaceae, and Lactobacillaceae were more abundant in SCZ cases while Victivallaceae was depleted. At the genus level, Parabacteroides, Lachnoclostridium, Lawsonibacter, Enterocloster, Collinsella, Flavonifractor, Evtepia, Absiella-Longicatena, Lactobacillus, Pauljensenia, Sellimonas, and Tyzzerella were more abundant in SCZ cases while Ruminococcus, Eisenbergiella, Coprococcus, Acetatifactor, Angelakisella, and Marvinbryantia were depleted (Figure S4A, C in Supplement 1).
Functional Analyses of Gut Microbiota
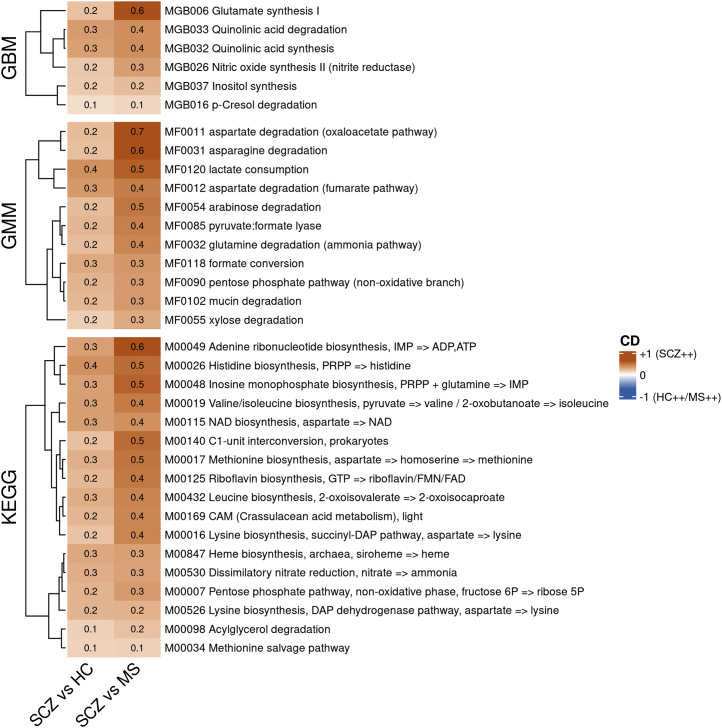
To assess potential for bacterial synthesis of various metabolites, we carried out module-level analysis. Of the 44 gut-brain modules present in at least 10% of individuals, 10 were differentially abundant between SCZ cases and control subjects (both HC and MS), and six remained significantly contrasted after drug and other covariates deconfounding (Figure 2, top). Among these, glutamate synthesis, quinolinic acid degradation and synthesis, inositol synthesis, nitric oxide synthesis (nitrite reductase), and p-cresol degradation modules were all more abundant in patients with SCZ.
Figure 2.
Contrast in functional potential. Heatmaps of the effect size (CD) of the gut metagenome-predicted functional modules (generated from GBMs, GMMs, and KEGG pathway modules) that are contrasted between patients with SCZ and control subjects (both HC and MS) after covariate deconfounding. Figures displayed in the cell give the exact CD. SCZ++ = more abundant in patients with SCZ; HC++/MS++ = more abundant in control subjects. CD, Cliff’s Delta; GBM, gut-brain module; GMM, gut-metabolic module; HC, healthy control; KEGG, Kyoto Encyclopedia of Genes and Genomes; MS, metabolic syndrome; SCZ, schizophrenia.
Considering other module types, we found 11 gut-metabolic modules and 17 Kyoto Encyclopedia of Genes and Genomes modules that were enriched in SCZ cases as compared with HC subjects and individuals with MS after deconfounding for covariates. Among Kyoto Encyclopedia of Genes and Genomes modules, 7 of 17 were related to essential amino acid synthesis: lysine (two distinct pathways), methionine (two distinct pathways), valine/isoleucine, histidine, and leucine (Figure 2, bottom).
Association Between Gut Microbiota, SCZ Diagnosis, and Psychopathology Scores
Most patients with SCZ were diagnosed as ICD-10 F.20 (n = 120, 90.9%) (Table S1 in Supplement 1). The second most numerous diagnosis group was ICD-10 F.25 (n = 10, 7.5%). There was no significant difference in beta-diversity regarding these two groups (Figure S5 in Supplement 1).
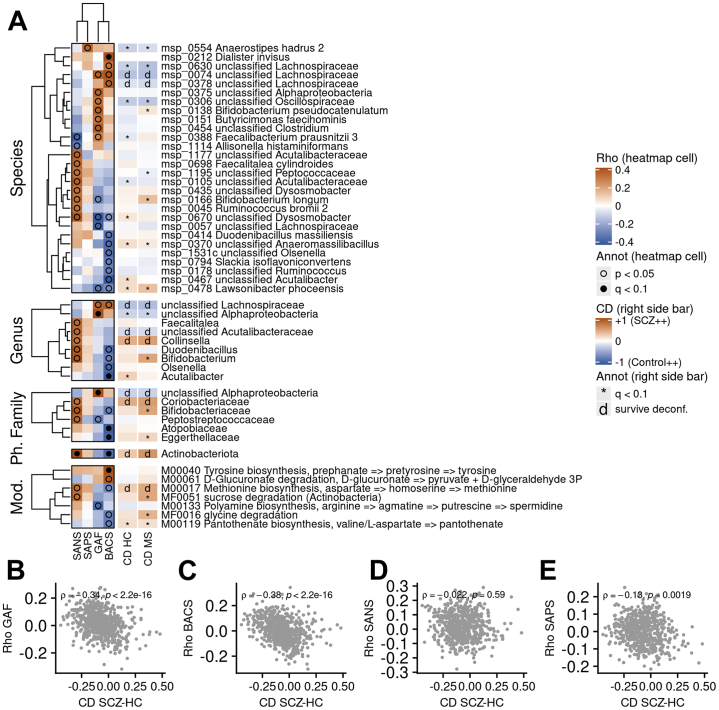
Metrics of GAF, BACS, and SANS correlated to different microbiome taxonomic features (MGS, genus, family, and phylum) (Figure 3A). Features positively associated with GAF and BACS in cases tended to be more abundant in HC subjects, and conversely (ρ = −0.34, and −0.38, respectively; p < 2.2 × 10−16) (Figure 3B, C), suggesting that the drift of the microbiome composition from that found in HC subjects toward that found in SCZ cases is associated with a decrease of cognition. In contrast, SANS and the Scale for the Assessment of Positive Symptoms showed no or only modest gut associations, respectively (Figure 3D, E).
Figure 3.
Association between psychopathological scores of SCZ cases and metagenomics features. (A) Heatmap of Spearman’s correlations between scores for psychopathology and cognitive functions (SAPS, SANS, GAF, BACS) and metagenomics features (from top to bottom: MGS, genus, family, phylum, functional modules) in cases with SCZ. Only features with at least one p value under .01 are displayed. Black dots denote correlations with false discovery rate ≤ 0.1, while empty circles indicate correlation with p ≤ .05. The right-side bars indicate CD (effect size) of the feature in the SCZ/HC contrast or in the SCZ/MS contrast (red: more abundant in SCZ; blue: more abundant in HC or MS). Asterisk indicates that the feature was significantly different in the given comparison (either SCZ/HC or SCZ/MS) but was confounded; d means that the feature was still contrasted after covariate deconfounding. (B–E) Spearman’s correlations of MGSs with (B) GAF, (C) BACS, (D) SANS, and (E) SAPS according to CD between SCZ cases and HC subjects. Correlations are computed using the SCZ cases only. The coefficient from Spearman’s correlation along with its p value is displayed. Annot, annotation; BACS, Brief Assessment of Cognition in Schizophrenia; CD, Cliff’s Delta; GAF, Global Assessment of Functioning; HC, healthy control; MGS, metagenomic species; MS, metabolic syndrome; Mod., modules; Ph., phylum; SANS, Scale for the Assessment of Negative Symptoms; SAPS, Scale for the Assessment of Positive Symptoms; SCZ, schizophrenia.
BACS was positively associated with abundance of the species Dialister invisus and inversely associated with the genus Acutalibacter, the families Atopobiaceae and Eggerthellaceae, and the phylum Actinobacteriota (q ≤ .1). GAF metrics were associated with a genus and to a family assigned to Alphaproteobacteria. Consistently, the phylum Actinobacteriota and the unclassified family assigned to Alphaproteobacteria remained more abundant in SCZ cases after covariate deconfounding. Having a closer look at the Bifidobacterium genus, reported to be more abundant in SCZ (11), we found that it correlated positively with SANS and negatively with BACS (p ≤ .05), although it was significantly more abundant in SCZ only when considering the MS cohort as control subjects.
At the functional module level, BACS was positively associated with the metagenome-predicted tyrosine biosynthesis potential (ρ = 0.34, q ≤ .1) (Figure 3A). Bacterial potentials for methionine biosynthesis (from aspartate), more abundant in SCZ after covariate deconfounding, correlated inversely to BACS (p ≤ .05).
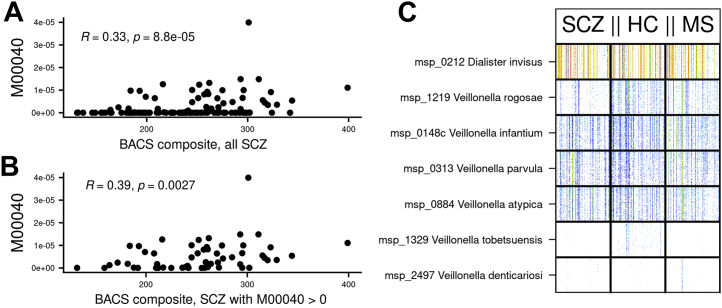
To further analyze correlations with cognition, we examined the proportion of the BACS variance explained by a combination of bioclinical variables (including medication) and functional microbial modules. The functional modules and the bioclinical variables could account on average for 11% and 11% of BACS variability, respectively. The combination of both inputs with bioclinical variables accounted for only a small but significant additional explanation (14% of BACS variability) (Figure S6A in Supplement 1). With functional modules or their combination with bioclinical variables as input, the module that contributed most to explanation of BACS variability was tyrosine biosynthesis, selected in 100% and 91% of tests, respectively (Figure S6B, C in Supplement 1). Only two other modules were also selected in both cases: aspartate degradation (fumarate pathway) and lysine biosynthesis.
The microbial tyrosine biosynthesis potential from prephanate was the only module significantly associated with BACS after correction for multiple testing and deconfounding for covariates (age, BMI, smoking status, and drugs). Furthermore, when considering only the individuals who carried the module (n = 54), the correlation was even stronger than when considering all individuals (ρ = 0.39, p = .0027 and ρ = 0.33, p = 8.8 × 10−5, respectively), suggesting that the relationship is not driven by the zero values (Figure 4A, B). The tyrosine biosynthesis module was not significantly differential between SCZ, HC, and MS groups, neither in presence (43%, 46%, 39%, respectively, p = .46) nor in abundance (p = .5). The module was encoded in seven species: D. invisus and six Veillonella (V. infantium, V. parvula, V. rogosae, V. atypica, V. tobetsuensis, and V. denticariosi) (Figure 4C); all but V. tobetsuensis and V. denticariosi were present in more than 10% of individuals. V. rogosae was more abundant in HC individuals and individuals with MS, even after deconfounding; V. infantium was more abundant in HC subjects as compared with patients with SCZ but not in individuals with MS, and the remaining MGSs were not contrasted. The most abundant of these MGSs was D. invisus, which contributed 98% of the module abundance (<1% for each of the other MGSs). Consistently, D. invisus was also positively correlated to BACS (ρ = 0.34, p = 5.7 × 10−5).
Figure 4.
Relationship between tyrosine biosynthesis potential of gut microbiota and BACS in SCZ cases. Spearman’s correlation displaying abundance of gut metagenome-predicted tyrosine biosynthesis module (M00040) in function of BACS composite in (A) all SCZ cases (n = 132) or (B) in SCZ cases with a strictly positive abundance only of the tyrosine biosynthesis module (n = 57). Coefficients of Spearman’s correlation along with the associated p value are shown. (C) Bar codes of MGSs involved in the gut metagenome-predicted tyrosine biosynthesis module (M00040). Individuals are in columns and MGSs in rows; each bar indicates the abundance of 50 marker genes of a species in an individual, and the colors indicate the abundance (white, absence; red, relative abundance of 1 × 10−4). Individuals are sorted first by status (patients with SCZ on the left, HC subjects in the middle, individuals with MS on the right) and then by increasing richness (MGS count). BACS, Brief Assessment of Cognition in Schizophrenia; HC, healthy control; MGS, metagenomic species; MS, metabolic syndrome; SCZ, schizophrenia.
Discussion
To our knowledge, this is the largest study on alteration of the gut microbiota, aiming at identifying potential new targets to treat cognitive deficits, in patients with SCZ.
Very few studies have reported associations between gut microbiota and clinical features in SCZ, which is surprising because these may be more promising than relying on diagnostic criteria outlining groups with high clinical heterogeneity. Cognition is a core feature and cognitive deficits are a currently untreatable symptom that substantially hinders recovery in SCZ. We show that abundances of the tyrosine biosynthesis module and of its main driver, D. invisus, are positively associated with measures of cognition in SCZ cases. Tyrosine has previously been associated with cognition in healthy individuals and in the pathophysiology of SCZ (43,44), in line with what we found. We did not identify any other studies reporting associations to cognition in SCZ.
We found only modest associations between psychotic symptoms and any taxa or functional modules. Li et al. (19) reported a positive association between Succinivibrio and the Positive and Negative Syndrome Scale, while Nguyen et al. (12) found that increased negative symptoms were associated with decreased abundance of family Ruminococcaceae. Schwarz et al. (15) investigated first-episode patients and found that the Lactobacillus group was associated with positive symptoms and global assessment of functioning, while Lachnospiracacae and Ruminococcaceae were negatively correlated with negative symptoms.
As expected, we found gut microbiome diversity and richness to be lower in SCZ cases. Both indices have been suggested to be beneficial to the host, and low diversity has been found across various disorders (45), even if the clinical significance is not well described. Nine of the SCZ studies included gut diversity, of which eight reported significant differences. However, a recent meta-analysis found that diversity across psychiatric disorders was well preserved, while richness was generally low (10).
The relative abundance of microbial taxa is reported at seven levels: kingdom, phylum, class, order, family, genus, and species. Our most important findings are highlighted here and discussed in perspective of earlier findings. At the genus level, Lachnoclostridium, Enterocloster, and Lawsonibacter were more abundant in SCZ cases while Coprococcus, Ruminococcus, and Eisenbergiella were depleted. Depletion of Coprococcus (14) and Ruminococcus (19) replicated earlier studies. At the family level, we found that Lachnospiraceae was more abundant in SCZ. This finding opposes three previous reports (8,14,20). The link between Lachnospiraceae and SCZ, a short-chain fatty acid–producing family, has been hypothesized to go through an alteration of the gut, triggering proinflammatory immune response (20). The conflicting findings may be explained by different strategies for deconfounding or by differences in geographical origin, because all three studies reporting decreased levels originate from China. At the phylum level, Actinobacteriota and Desulfobacteriota were enriched in SCZ cases. Higher abundance of Actinobacteriota was observed in other studies of severe mental disorders (46, 47, 48) and in two previous SCZ studies (11,19).
Identifying contrasting functional modules may be a more promising step forward, because these can be linked to metabolic pathways already hypothesized to be involved in the pathophysiology of SCZ. We identified six functional gut-brain modules more abundant in SCZ, namely glutamate synthesis, quinolinic acid degradation and synthesis, p-cresol degradation, inositol synthesis, and nitric oxide synthesis (nitrite reductase). Of these, pathways involved in quinolinic acid may be of special interest in relation to SCZ. There is growing evidence that the immune system plays a major role in SCZ pathology (49) and the pathway involving kynurenine is suggested as a possible link between peripheral immune activation and central neurotransmitters. Kynurenine is metabolized into several neuroactive metabolites, including quinolinic acid, which is neurotoxic (50). This is an indirect replication of another recent study, where fecal samples from SCZ cases were transplanted to pathogen-free mice, observing elevation of the kynurenine–kynurenic acid pathway of tryptophan metabolism with increased degradation in serum and the central nervous system and an increased dopamine level in the prefrontal cortex (8). Six studies reported findings at the functional level (11,14,16, 17, 18, 19). Ma et al. (17) found no differences, while Pan et al. (18) identified several functional modules that differed, including lipoic acid metabolism, steroid hormone biosynthesis, and alpha-linolenic acid metabolism. Similarly, Li et al. (19) reported several differential functional modules. Xu et al. (16) especially highlighted the finding of increased glutamate synthase. Shen et al. (14) found that pathways including vitamin B12 and fatty acids differed.
The overall inconsistency in reported results should lead to increased attention to cooperation, preregistration, and reporting guidelines as proposed by Knight et al. (51), which will likely decrease false discovery rates and increase the comparability of data in future studies.
A major strength of this study is the careful adjustment for two major confounders, metabolic disease and psychotropic medication, and the large sample size. Yet, our study has limitations, including the specificity of our cohort (overweight and obese patients with SCZ mainly with F.20 diagnosis) and the cross-sectional study design, which does not allow for causal inference. In addition, nonreported covariates such as diet, past use of antibiotics, drug or alcohol abuse, poor oral hygiene, and deviation in actual lifestyle from what is reported are likely to contribute to some residual confounding. Furthermore, our study focused only on bacterial genetic content and did not measure or analyze nonbacterial organisms (such as phages) or fecal metabolites, which could have provided new insights into the role of gut microbiota in SCZ. Finally, it should be stressed that our analyses are exploratory because the analyses were not preplanned, and our study may be statistically underpowered to detect important differences. Thus, our findings need confirmation in independent studies.
Conclusions
We show that the gut microbiome of SCZ exhibits reduced species richness compared with both normal weight healthy individuals and metabolically matched control subjects without SCZ. We find further indications that the kynurenine pathway is involved, because two gut-brain modules involving the neurotoxic metabolite quinolinic acid were more abundant in SCZ. With this, we suggest that parts of the gut-microbiome-brain axis involving quinolinic acid should be further explored in future studies, preferentially involving experimental manipulation and longitudinal observations to allow for causal inferences. Similarly, we show that tyrosine biosynthesis may be important for cognition in SCZ and suggest that it should be explored further.
Acknowledgments and Disclosures
The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation (to THH, TN, YF, HV, TH, OP). This work was supported in part by Metagenopolis (Grant No. ANR-11-DPBS-0001 [to FT, ELC, SF, MB, FPO, NP, NG, FL, SDE]). SKF was supported by the German Centre for Cardiovascular Research and by the BMBF German Ministry of Education and Research. TB was supported by DFG grant KFO339.
SDE and FT had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. OP and SDE conceived the research idea. SDE, TH, and OP designed the analytical plan. HS, THH, TN, TJ, and HV recruited and examined study participants. Microbial DNA extraction and sequencing, library preparation, and sequencing quality assurance were supervised by SDE and undertaken by FL and NG. FT conducted bioinformatics analyses supported by SF, ELC, FPO, MB, and NP. FT, TB, LJ, and SKF undertook drug deconfounding of microbiome data. HS and FT wrote the draft manuscript. OP was responsible for overall supervision, edited and revised the manuscript, and is the guarantor of this study. All authors contributed to critically revising the manuscript for important intellectual content and approved the final version of the manuscript.
We thank Benoit Quinquis for his help in generating the sequences and Franck Gauthier for his help in processing sequences and formatting figures and manuscript.
Data are available in a public open access repository. Metagenomics reads generated in this study are available (without conditions of reuse) under the accession numbers PRJEB41217, PRJEB41786, and PRJEB41787 at the European Nucleotide Archive in the European Bioinformatics Institute (https://www.ebi.ac.uk).
The authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2022.01.009.
Contributor Information
Oluf Pedersen, Email: oluf@sund.ku.dk.
Stanislav D. Ehrlich, Email: s.ehrlich@ucl.ac.uk.
Supplementary Material
References
- 1.van Os J., Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 2.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Ripke S., Walters J.T.R., O’Donovan M.C. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv. 2020 doi: 10.1101/2020.09.12.20192922. [DOI] [Google Scholar]
- 3.Bastiaanssen T.F.S., Cowan C.S.M., Claesson M.J., Dinan T.G., Cryan J.F. Making sense of . . . the microbiome in psychiatry. Int J Neuropsychopharmacol. 2019;22:37–52. doi: 10.1093/ijnp/pyy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fadgyas-Stanculete M., Buga A.M., Popa-Wagner A., Dumitrascu D.L. The relationship between irritable bowel syndrome and psychiatric disorders: From molecular changes to clinical manifestations. J Mol Psychiatry. 2014;2:4. doi: 10.1186/2049-9256-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan A.P., Crowley J.J., Nonneman R.J., Quackenbush C.R., Miller C.N., Ryan A.K., et al. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsythe P., Kunze W., Bienenstock J. Moody microbes or fecal phrenology: What do we know about the microbiota-gut-brain axis? BMC Med. 2016;14:58. doi: 10.1186/s12916-016-0604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Severance E.G., Prandovszky E., Castiglione J., Yolken R.H. Gastroenterology issues in schizophrenia: Why the gut matters. Curr Psychiatry Rep. 2015;17:27. doi: 10.1007/s11920-015-0574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng P., Zeng B., Liu M., Chen J., Pan J., Han Y., et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice [published correction appears in Sci Adv 2019; 5:eaay2759] Sci Adv. 2019;5 doi: 10.1126/sciadv.aau8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu F., Guo R., Wang W., Ju Y., Wang Q., Ma Q., et al. Transplantation of microbiota from drug-free patients with schizophrenia causes schizophrenia-like abnormal behaviors and dysregulated kynurenine metabolism in mice. Mol Psychiatry. 2020;25:2905–2918. doi: 10.1038/s41380-019-0475-4. [DOI] [PubMed] [Google Scholar]
- 10.Nikolova V.L., Hall M.R.B., Hall L.J., Cleare A.J., Stone J.M., Young A.H. Perturbations in gut microbiota composition in psychiatric disorders: A review and meta-analysis [published correction appears in JAMA Psychiatry 2022; 79:87] JAMA Psychiatry. 2021;78:1343–1354. doi: 10.1001/jamapsychiatry.2021.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu F., Ju Y., Wang W., Wang Q., Guo R., Ma Q., et al. Metagenome-wide association of gut microbiome features for schizophrenia. Nat Commun. 2020;11:1612. doi: 10.1038/s41467-020-15457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen T.T., Kosciolek T., Maldonado Y., Daly R.E., Martin A.S., McDonald D., et al. Differences in gut microbiome composition between persons with chronic schizophrenia and healthy comparison subjects. Schizophr Res. 2019;204:23–29. doi: 10.1016/j.schres.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan X., Zhang P., Wang Y., Liu Y., Li X., Kumar B.U., et al. Changes in metabolism and microbiota after 24-week risperidone treatment in drug naïve, normal weight patients with first episode schizophrenia. Schizophr Res. 2018;201:299–306. doi: 10.1016/j.schres.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Shen Y., Xu J., Li Z., Huang Y., Yuan Y., Wang J., et al. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: A cross-sectional study. Schizophr Res. 2018;197:470–477. doi: 10.1016/j.schres.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Schwarz E., Maukonen J., Hyytiäinen T., Kieseppä T., Orešič M., Sabunciyan S., et al. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr Res. 2018;192:398–403. doi: 10.1016/j.schres.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Xu R., Wu B., Liang J., He F., Gu W., Li K., et al. Altered gut microbiota and mucosal immunity in patients with schizophrenia. Brain Behav Immun. 2020;85:120–127. doi: 10.1016/j.bbi.2019.06.039. [DOI] [PubMed] [Google Scholar]
- 17.Ma X., Asif H., Dai L., He Y., Zheng W., Wang D., et al. Alteration of the gut microbiome in first-episode drug-naïve and chronic medicated schizophrenia correlate with regional brain volumes. J Psychiatr Res. 2020;123:136–144. doi: 10.1016/j.jpsychires.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Pan R., Zhang X., Gao J., Yi W., Wei Q., Su H. Analysis of the diversity of intestinal microbiome and its potential value as a biomarker in patients with schizophrenia: A cohort study. Psychiatry Res. 2020;291:113260. doi: 10.1016/j.psychres.2020.113260. [DOI] [PubMed] [Google Scholar]
- 19.Li S., Zhuo M., Huang X., Huang Y., Zhou J., Xiong D., et al. Altered gut microbiota associated with symptom severity in schizophrenia. PeerJ. 2020;8 doi: 10.7717/peerj.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Pan L.Y., Zhang Z., Zhou Y.Y., Jiang H.Y., Ruan B. Analysis of gut mycobiota in first-episode, drug-naïve Chinese patients with schizophrenia: A pilot study. Behav Brain Res. 2020;379:112374. doi: 10.1016/j.bbr.2019.112374. [DOI] [PubMed] [Google Scholar]
- 21.Vindegaard N., Speyer H., Nordentoft M., Rasmussen S., Benros M.E. Gut microbial changes of patients with psychotic and affective disorders: A systematic review. Schizophr Res. 2021;234:1–10. doi: 10.1016/j.schres.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Hernán M.A., Hsu J., Healy B. A second chance to get causal inference right: A classification of data science tasks. Chance. 2019;32:42–49. [Google Scholar]
- 23.Yuan N., Chen Y., Xia Y., Dai J., Liu C. Inflammation-related biomarkers in major psychiatric disorders: A cross-disorder assessment of reproducibility and specificity in 43 meta-analyses. Transl Psychiatry. 2019;9:233. doi: 10.1038/s41398-019-0570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee S.H., Ripke S., Neale B.M., Faraone S.V., Purcell S.M., et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carcone D., Ruocco A.C. Six years of research on the National Institute of Mental Health’s Research Domain Criteria (RDoC) initiative: A systematic review. Front Cell Neurosci. 2017;11:46. doi: 10.3389/fncel.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speyer H., Christian Brix Nørgaard H., Birk M., Karlsen M., Storch Jakobsen A., Pedersen K., et al. The CHANGE trial: No superiority of lifestyle coaching plus care coordination plus treatment as usual compared to treatment as usual alone in reducing risk of cardiovascular disease in adults with schizophrenia spectrum disorders and abdominal obesity. World Psychiatry. 2016;15:155–165. doi: 10.1002/wps.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreasen N.C. Methods for assessing positive and negative symptoms. Mod Probl Pharmacopsychiatry. 1990;24:73–88. doi: 10.1159/000418013. [DOI] [PubMed] [Google Scholar]
- 28.Keefe R.S.E., Goldberg T.E., Harvey P.D., Gold J.M., Poe M.P., Coughenour L. The Brief Assessment of Cognition in Schizophrenia: Reliability, sensitivity, and comparison with a standard neurocognitive battery. Schizophr Res. 2004;68:283–297. doi: 10.1016/j.schres.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen G., Hagtvet K.A., Karterud S. Generalizability studies of the Global Assessment of Functioning-Split version. Compr Psychiatry. 2007;48:88–94. doi: 10.1016/j.comppsych.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Dantoft T.M., Ebstrup J.F., Linneberg A., Skovbjerg S., Madsen A.L., Mehlsen J., et al. Cohort description: The Danish study of Functional Disorders. Clin Epidemiol. 2017;9:127–139. doi: 10.2147/CLEP.S129335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 32.Wen C., Zheng Z., Shao T., Liu L., Xie Z., Le Chatelier E., et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis [published correction appears in Genome Biol 2017; 18:214] Genome Biol. 2017;18:142. doi: 10.1186/s13059-017-1271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plaza Onate F., Pons N., Gauthier F., Almeida M., Ehrlich S.D., Le Chatelier E. Updated Metagenomic Species Pan-genomes (MSPs) of the human gastrointestinal microbiota. Portail Data INRAE. 2021. Available at: [DOI]
- 34.Le Chatelier E., Almeida M., Plaza Oñate F., Pons N., Gauthier F., Ghozlane A., et al. A catalog of genes and species of the human oral microbiota. Portail Data INRAE. 2021. Available at: [DOI]
- 35.R Core Team A language and environment for statistical computing. 2019. https://www.R-project.org/ Available at:
- 36.Torchiano M. Effsize - A package for efficient effect size computation. 2016. Available at: [DOI]
- 37.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. Vegan: Community ecology package. 2019. http://cran.r-project.org/package=vegan Available at:
- 38.Thioulouse J., Dray S., Dufour A.B., Siberchicot A., Jombart T., Pavoine S. Springer; New York: 2018. Multivariate Analysis of Ecological Data with ade4. [Google Scholar]
- 39.Birkner T. MetadeconfoundR Release for Documentation of the MetaDrugs Analysis as Part of the MetaCardis Consortium. https://github.com/TillBirkner/metadeconfoundR_V0.1.5_doc Available at:
- 40.Kuhn M., Wing J., Weston S., Williams A., Keefer C., Engelhardt A., et al. caret: Classification and regression training. 2021. https://cran.r-project.org/web/packages/caret/index.html Available at:
- 41.Genuer R., Poggi J.M., Tuleau-Malot C. VSURF: Variable selection using random forests. 2019. https://cran.r-project.org/package=VSURF Available at:
- 42.Liaw A., Wiener M. Classification and regression by randomForest. R News. 2002;2/3:18–22. [Google Scholar]
- 43.Jongkees B.J., Hommel B., Kühn S., Colzato L.S. Effect of tyrosine supplementation on clinical and healthy populations under stress or cognitive demands—A review. J Psychiatr Res. 2015;70:50–57. doi: 10.1016/j.jpsychires.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Bjerkenstedt L., Farde L., Terenius L., Edman G., Venizelos N., Wiesel F.A. Support for limited brain availability of tyrosine in patients with schizophrenia. Int J Neuropsychopharmacol. 2006;9:247–255. doi: 10.1017/S1461145705005638. [DOI] [PubMed] [Google Scholar]
- 45.Gerritsen J., Smidt H., Rijkers G.T., de Vos W.M. Intestinal microbiota in human health and disease: The impact of probiotics. Genes Nutr. 2011;6:209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Painold A., Mörkl S., Kashofer K., Halwachs B., Dalkner N., Bengesser S., et al. A step ahead: Exploring the gut microbiota in inpatients with bipolar disorder during a depressive episode. Bipolar Disord. 2019;21:40–49. doi: 10.1111/bdi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z., Li J., Gui S., Zhou C., Chen J., Yang C., et al. Comparative metaproteomics analysis shows altered fecal microbiota signatures in patients with major depressive disorder. Neuroreport. 2018;29:417–425. doi: 10.1097/WNR.0000000000000985. [DOI] [PubMed] [Google Scholar]
- 48.Zheng P., Zeng B., Zhou C., Liu M., Fang Z., Xu X., et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21:786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 49.Müller N., Weidinger E., Leitner B., Schwarz M.J. The role of inflammation in schizophrenia. Front Neurosci. 2015;9:372. doi: 10.3389/fnins.2015.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marx W., McGuinness A.J., Rocks T., Ruusunen A., Cleminson J., Walker A.J., et al. The kynurenine pathway in major depressive disorder, bipolar disorder, and schizophrenia: A meta-analysis of 101 studies. Mol Psychiatry. 2021;26:4158–4178. doi: 10.1038/s41380-020-00951-9. [DOI] [PubMed] [Google Scholar]
- 51.Knight R., Vrbanac A., Taylor B.C., Aksenov A., Callewaert C., Debelius J., et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.