Abstract
Background
In December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged with a high transmissibility rate and resulted in numerous negative impacts on global life. Preventive measures such as face masks, social distancing, and vaccination helped control the pandemic. Nonetheless, the emergence of SARS-CoV-2 variants, such as Omega and Delta, as well as coronavirus disease 2019 (COVID-19) reinfection, raise additional concerns. Therefore, this study aimed to determine the overall prevalence of reinfection on global and regional scales.
Methods
A systematic search was conducted across three databases, PubMed, Scopus, and ProQuest Central, including all articles pertaining to COVID-19 reinfection without language restriction. After critical appraisal and qualitative synthesis of the identified relevant articles, a meta-analysis considering random effects was used to pool the studies.
Results
We included 52 studies conducted between 2019 and 2022, with a total sample size of 3,623,655 patients. The overall prevalence of COVID-19 reinfection was 4.2% (95% confidence interval [CI]: 3.7–4.8%; n = 52), with high heterogeneity between studies. Africa had the highest prevalence of 4.7% (95% CI: 1.9–7.5%; n = 3), whereas Oceania and America had lower estimates of 0.3% (95% CI: 0.2–0.4%; n = 1) and 1% (95% CI: 0.8–1.3%; n = 7), respectively. The prevalence of reinfection in Europe and Asia was 1.2% (95% CI: 0.8–1.5%; n = 8) and 3.8% (95% CI: 3.4–4.3%; n = 43), respectively. Studies that used a combined type of specimen had the highest prevalence of 7.6% (95% CI: 5.8–9.5%; n = 15) compared with those that used oropharyngeal or nasopharyngeal swabs only that had lower estimates of 6.7% (95% CI: 4.8–8.5%; n = 8), and 3.4% (95% CI: 2.8–4.0%; n = 12) respectively.
Conclusion
COVID-19 reinfection occurs with varying prevalence worldwide, with the highest occurring in Africa. Therefore, preventive measures, including vaccination, should be emphasized to ensure control of the pandemic.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12889-023-15626-7.
Keywords: COVID-19, Reinfection, Prevalence
Background
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as the 2019 novel coronavirus (2019-nCoV) [1, 2]. It emerged in December 2019 with a very high transmissibility rate. The first case was reported in Wuhan City, Hubei Province, China [1]. The World Health Organization (WHO) declared it a pandemic on March 11, 2020 [1, 3].
Before the 2019 pandemic, other epidemics of coronaviruses have been reported, including SARS-CoV-1 and Middle East Respiratory Syndrome coronavirus (MERS-CoV), which have a high fatality rate compared with that of SARS-CoV-2 [4–8]. These infections were rapidly controlled before they became a global emergency [9–13]. Approximately 80% of SARS-CoV-2 infections are asymptomatic [14]. Human-to-human transmission occurs by respiratory droplets, close contact, and possibly aerosol and fecal–oral contact [14–16].
Globally, more than 650 million COVID-19 cases and 6.6 million deaths have been reported [17]. Europe has reported over 41.3% of all global cases, followed by the Americas (28.4%), Western Pacific (15.9%), Southeast Asia (9.3%), East Mediterranean (3.6%), and Africa (1.4%) [17].
The basic reproduction number of COVID-19 is estimated to be between 1.4 and 2.4, with an average incubation period of 4–5 days, while the recovery rate is 98.8% [17–19]. However, severity and recovery depend on various factors in which an increased risk is found in older individuals or those with underlying conditions such as cancer, diabetes mellitus, cardiovascular diseases, and chronic respiratory diseases [20–25].
SARS-CoV-2 continues to change over time, and some variants have raised global health concerns because they are associated with increased risks of transmission, clinical worsening of the disease, or resistance to containment measures [26, 27]. Therefore, they are collectively named variants of concern (VOC) [26]. Among these variants, we cite Alpha, Beta, Gamma, Delta, and Omicron, the recently circulating VOC [26, 28]. The Omicron variant has the capacity to skip acquired immunity from prior infection, hence increasing the risk of reinfection [29–32]. Other factors that increase COVID-19 reinfection include female sex, older age, underlying comorbidity, unvaccinated status, and being a healthcare provider [33–36]. Compared with that of the initial COVID-19 infection, the risk of case fatality in reinfected cases decreased by 68% [33].
Various efforts have been made to control this pandemic, such as vaccines, social distancing, wearing of face mask, hand hygiene, and isolation of infected patients [37–42]. Despite these efforts, some cases of reinfection have been reported since the first wave of COVID-19 [43–45].
To date, there is no conventional definition of COVID-19 reinfection. European countries consider a range of 45–90 days from a previously confirmed infection to a newly confirmed infection, whereas the European Center for Disease Control (ECDC) proposes a period of more than 60 days after the first infection [46]. The American Center for Disease Control (CDC) recommends that reinfection be considered after a sufficient period has elapsed for immunity to mount up [47].
Different other authors consider reinfection when SARS-CoV-2 is detected 90 days after the initial or prior infection, whereas before that period, it is usually considered as relapse, reactivation, or re-positivity of the initial SARS-CoV-2 infection [48–51]. Dafna et al. defined re-positivity, reinfection, and relapse based on both clinical and epidemiological aspects [51]. They clinically defined reinfection as a recurrence of clinical symptoms and a positive polymerase chain reaction (PCR) after 90 days of the previous infection or within 90 days if there was a symptom—free period and two recorded negative PCR [51].
In addition to the lack of a conventional definition of reinfection, evidence of the occurrence of SARS-CoV-2 reinfection is still limited. This systematic review and meta-analysis determined the current prevalence of COVID-19 reinfection at global and regional levels, considering a longer follow-up period.
Methods
Study design
Systematic review and meta-analysis were reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA).
Search strategy
A digital search was conducted on June 14, 2022, using three databases: PUBMED, SCOPUS, and ProQuest Central. The search strategy used for each database is shown in the (Additional file 1: Table S1). The search was restricted to studies published from 2019 (when the first case of COVID-19 occurred). We considered all observational studies that reported sufficient data to compute the prevalence of COVID-19 reinfection without language, age, or sex restriction. Case reports and series were excluded because they could not provide the denominator for the calculation of prevalence. For this study, COVID-19 reinfection was defined as the development of a new COVID-19 infection after a previous infection that was declared cured. The references of the included articles were scrutinized as potential sources for additional studies.
Data management and study selection
The titles and abstracts of the relevant articles retrieved from the databases were transferred to the Rayyan online software, which was used to organize the literature search results. After the removal of duplicates, the selection of articles based on title and abstract was performed independently by three reviewers (UJ, YN, and DE), with disagreements being addressed through discussions to reach a consensus. Subsequently, the final inclusion of articles was decided based on the full texts independently assessed by the three authors.
Data extraction and quality assessment
From relevant eligible articles, the following data were extracted: author name, year of publication, country (where the study was conducted), study design, type of used data, definition of reinfection, total number of participants with primary infection, number of reinfected patients, population of the study, type of specimen, vaccination status considerations, and mean/median age of the study participants. The outcome was that people previously infected with SARS-CoV-2 subsequently developed a new infection after being declared as cured.
The National Institutes of Health (NIH) quality assessment tool for cohort and cross-sectional studies was used to evaluate the methodological quality and risk of bias of the included articles. The 14 items of the NIH tool were used independently by three authors, UJ, NY, and ED, for the evaluation of every study, and a consensus was reached through discussions. Each item was scored as 0 if the condition was not met or 1 if the condition was met. Articles with a final overall score less than or equal to 4 were categorized as being of poor quality (high risk of bias), 5–10 as being of fair quality (medium risk of bias), and 11 and above as being of good quality (low risk of bias). More details on each question can be found in the Additional file 1 and online [52].
Data analysis
We used STATA/SE software version 17.0 for data analysis. Random-effects meta-analysis using the command “metaprop” was used to calculate the pooled prevalence of COVID-19 reinfection and 95% confidence interval (CI). To assess the sources and contribution of numerous factors to heterogeneity, we conducted univariable meta-regression and subgroup analysis by region (country), type of study, and type of specimen. Throat and oropharyngeal swabs were considered equivalent in the analysis.
I2 was used to check for heterogeneity between studies, and r2 was reported to refer to the proportion of variance explained by the covariates. We performed a sensitivity analysis to explore the effect of individual studies on the pooled estimate by eliminating the studies one by one and checking if there was substantial variation in the pooled estimates. Funnel plots and Egger’s test were used to check for publication bias. To determine temporal variations in the magnitude and direction of the pooled association estimate, we also performed a random-effects cumulative meta-analysis. The studies were first organized according to the year of publication and then sequentially included in the analysis in chronological order. The pooled estimates were updated as each study was added.
Results
Study selection
The search strategy and secondary bibliographic search yielded 1,419 relevant articles. After screening and removing duplicates, 52 studies with a total sample of 3,623,655 patients were finally included in the systematic review and meta-analysis (Fig. 1).
Fig. 1.
Flowchart
Characteristics and quality of the studies included in the meta-analysis
Fifty-two studies included in this meta-analysis were published between 2019 and 2022, 24 (46.1%) were conducted in China. Thirty-three studies (63.5%) were cohort studies, and 19 (36.5%) were cross-sectional studies. We observed that cohort studies used data from active surveillance and testing while cross-sectional studies used data from routine laboratory testing.
The definition of COVID-19 reinfection differed from author to author; some authors considered reinfection as a new positive PCR following two consecutive negative PCR taken after primary infection [53–66]; some other studies considered reinfection as a new infection after 90 days of primary infection or after initial testing [29, 34, 36, 67–73]. Other studies considered reinfection as a new positive PCR 30–60 days after the first cured infection [74–78], and for others, the definition was retesting positive following prior complete recovery and/or after discharge [79–101].
Sixteen studies (30.8%) did not specify the type of samples used for PCR retesting. Among the studies that reported information on the type of sample, the most frequently used samples were nasopharyngeal swabs, which were solely used in 12 (23.1%) studies, and oropharyngeal samples, which were solely used in 8 studies (15.4%). Fifteen studies (28.8%) combined two or more samples (nasopharyngeal, oropharyngeal, sputum, and fecal samples), and nasal samples were used in only one study (1.9%) (Table 1).
Table 1.
Characteristics of the studies included in the meta-analysis
| Author name | Year of publication | Type of Study | Type of used data | Country | Definition of reinfection | Re-infection cases | Primary infection cases | Specimen |
|---|---|---|---|---|---|---|---|---|
| Ai Tang Xiao [53] | 2020 | Cross-sectional | Routine testing | China | Positive RT-PCR after two consecutive negative results | 15 | 70 | Throat swab |
| Anna Jeffery-Smith [102] | 2021 | Cohort | Active Surveillance / testing | England | Positive RT-PCR after at least 90 days after previous SARS-CoV-2 infection | 10 | 656 | Nose and throat swab |
| Anne Rivelli [34] | 2022 | Cohort | Active Surveillance / testing | USA | Positive PCR after 90 days of primary infection | 159 | 2625 | Nasopharyngeal, oropharyngeal, nasal |
| Bo Yuan [79] | 2020 | Cohort | Active Surveillance / testing | China | Retest positive after complete recovery | 20 | 182 | Nasopharyngeal and anal swab |
| Cheryl Cohen [68] | 2022 | Cohort | Active Surveillance / testing | South-Africa | Positive RT-PCR after at least 90 days after previous SARS-CoV-2 infection | 87 | 749 | Nasal swab |
| Christian Hansen [54] | 2021 | Cross-sectional | Routine testing | Denmark | Positive PCR test in two COVID-19 surge | 72 | 11,068 | Throat swab |
| Efren Murillo-Zamora [103] | 2021 | Cohort | Active Surveillance / testing | Mexique | Reappearance of symptoms at least 28 days after the initial confirmed infection | 210 | 99,993 | Not clearly specified |
| Emilie Finch [104] | 2022 | Cohort | Active Surveillance / testing | USA | Positive PCR test more than 30 days after initial seropositive result | 14 | 309 | Not clearly specified |
| Fabiánová K [105] | 2021 | Cross-sectional | Routine testing | Czech Republic | Positive RT-PCR after at least 90 days after negative PCR test | 28 | 16,582 | Not clearly specified |
| Fariba Zare [106] | 2021 | Cohort | Active Surveillance / testing | Iran | Positive PCR test more than 30 days after the first positive test | 10 | 4039 | Nasopharyngeal and pharyngeal swab |
| Flacco Maria Elena [36] | 2021 | Cohort | Active Surveillance / testing | Italy | Positive RT-PCR with at least 90 days, and with ≥ 2 consecutive negative tests between periods | 24 | 7173 | Nasopharyngeal swab |
| Flacco Maria Elena [107] | 2022 | Cohort | Active Surveillance / testing | Italy | 2 Positive RT-PCR samples detected 45 days or more apart, with at least 1 intermediate negative PCR test | 729 | 119,266 | Nasopharyngeal swab |
| Guangming Ye [81] | 2020 | Cohort study | Active Surveillance / testing | China | Positive swab sample after recovery/discharge | 5 | 55 | Not clearly specified |
| Godwin E. Akpan [108] | 2022 | Cross-sectional | Routine testing | Liberia | Two positive RT-PCR samples detected 90 days or more apart, with at least 1 intermediate negative PCR test | 13 | 5459 | Not clearly specified |
| Hou-wei Du [109] | 2020 | Cohort | Active Surveillance / testing | China | Positive 2019 nCov nucleic acid test during follow-up after discharge | 3 | 126 | Nasopharyngeal and oropharyngeal swab |
| Hui Zhu [110] | 2020 | Cross-sectional | Routine testing | China | Positive RT-PCR after two consecutive negative results separated by at least 24 h | 17 | 98 | Nasopharyngeal swab or sputum |
| Ji Zhou [111] | 2020 | Cohort | Active Surveillance / testing | China | PCR during follow-up after prior SARS-Cov-2 infection | 23 | 345 | oropharyngeal swab |
| Jia Huang [112] | 2020 | Cohort | Active Surveillance / testing | China | Positive test after discharge | 69 | 414 | Nasopharyngeal swab |
| Jianghong An [56] | 2020 | Cohort | Active Surveillance / testing | China | Positive RT-PCR during follow-up after two consecutive negative results separated by at least 24 h | 38 | 262 | Nasopharyngeal and anal swab |
| Jie Chen [57] | 2020 | Cohort | Active Surveillance / testing | China | Positive RT-PCR during follow-up after two consecutive negative results separated by at least 24 h | 81 | 1067 | Throat swab |
| Jing Lu [113] | 2020 | Cross-sectional | Routine testing | China | Positive RT-PCR after two consecutive negative results separated by at least 24 h | 87 | 619 | Nasopharyngeal, throat and anal swab |
| Jing Yuan [62] | 2020 | Cohort | Active Surveillance / testing | China | Positive RT-PCR during follow-up after two consecutive negative results separated by at least 24 h | 25 | 172 | Nasopharyngeal and anal swab |
| Jinru Wu [114] | 2020 | Cross-sectional | Routine testing | China | Positive RT-PCR after discharge | 10 | 60 | Nasopharyngeal and anal swab |
| Ju Zhang [115] | 2021 | Cohort | Active Surveillance / testing | China | Positive SARS-Cov-2 test after negative PCR test between the two infection periods | 6 | 273 | Not clearly specified |
| Juliet R.C. Pulliam [116] | 2022 | Cross-sectional | Routine testing | South-Africa | Positive RT-PCR with at least 90 days after negative PCR test | 105,353 | 2,942,248 | Not clearly specified |
| Justin Wong [117] | 2020 | Cross-sectional | Routine testing | Brunei | Positive PCR during the study follow-up after 2 consecutive negative PCR at the discharge | 21 | 106 | Nasopharyngeal swab |
| Laith J. Abu Raddad [118] | 2021 | Cross-sectional | Routine testing | Qatar | After 45 days of the initial positive swab test | 243 | 133,266 | Nasopharyngeal and oropharyngeal swab |
| Lawandi A [71] | 2022 | Cohort | Active Surveillance / testing | USA | Positive RT-PCR with at least 90 days after negative qPCR test | 253 | 131,773 | Not clearly specified |
| Lei Pan [119] | 2021 | Cross-sectional | Routine testing | China | PCR redetected after discharge | 20 | 1350 | Throat or sputum swab |
| Leidi A [87] | 2022 | Cohort | Active Surveillance / testing | Switzerland | PCR during follow-up after prior SARS-Cov-2 infection | 5 | 498 | Nasopharyngeal and oropharyngeal swab |
| Maolu Tian [60] | 2020 | Cohort | Active Surveillance / testing | China | PCR during follow-up after two consecutive negative results | 20 | 147 | Oropharyngeal swab |
| Muhammad Syafiq Abdullah [88] | 2020 | Cohort | Active Surveillance / testing | Brunei | Retest positive after discharge | 27 | 138 | Nasopharyngeal and throat swab |
| Naila A Shaheen [89] | 2022 | Cohort | Active Surveillance / testing | Saudi Arabia | Positive RT-PCR tests during follow-up | 132 | 35,288 | Not clearly specified |
| Philippe Brouqui [72] | 2021 | Cohort | Active Surveillance / testing | France | Positive RT-PCR with at least 90 days after negative qPCR test | 46 | 6771 | Nasopharyngeal swab |
| Pilz Stefan [120] | 2021 | Cross-sectional | Routine testing | Austria | Positive SARS-Cov-2 tests after discharge | 40 | 14,840 | Not clearly specified |
| Rujun Hu [121] | 2020 | Cross-sectional | Routine testing | China | Recurrent positive RT-PCR after discharge | 11 | 69 | Nasopharyngeal swab |
| Salehi-Vaziri M [122] | 2021 | Cohort | Active Surveillance / testing | Iran | Positive rRT-PCR during the follow-up after the initial infection | 5 | 1492 | Not clearly specified |
| Sezanur Rahman [123] | 2022 | Cohort | Active Surveillance / testing | Bangladesh | Positive rRT-PCR during the follow-up after the initial infection | 38 | 750 | Nasopharyngeal swab |
| Sheehan M Meghan [73] | 2021 | Cohort | Active Surveillance / testing | USA | Positive RT-PCR with at least 90 days after previous SARS-CoV-2 infection | 62 | 1278 | not clearly specified |
| Shiua Luo [124] | 2020 | Cross-sectional | Routine testing | China | Symptoms and positive PCR tests after discharge | 13 | 1673 | Throat swab |
| Sivan Gazit [95] | 2022 | Cohort | Active Surveillance / testing | Israel | Positive PCR test during the follow-up period | 1374 | 86,275 | Not clearly specified |
| Tao Liu [58] | 2020 | Cross-sectional | Routine testing | China | Positive RT-PCR after two consecutive negative results separated by at least 24 h | 11 | 150 | Throat swab |
| Valeria Cento [125] | 2020 | Cohort | Active Surveillance / testing | Italy | Positive RT-PCR during follow-up after the discharge | 264 | 2521 | Nasopharyngeal swab |
| Wang Deng [65] | 2020 | Cross-sectional | Routine testing | China | Positive RT-PCR after the discharge | 61 | 576 | Nasopharyngeal and anal swab |
| Wang Xingyu [66] | 2020 | Cohort | Active Surveillance / testing | China | Positive SARS-Cov-2 test during the follow-up after the discharge | 8 | 131 | Not clearly specified |
| William R. Hartman [97] | 2020 | Cohort | Active Surveillance / testing | USA | Positive RT-PCR after 14 days of symptoms free | 11 | 86 | Nasopharyngeal swab |
| Yan Dong [126] | 2021 | Cross-sectional | Routine testing | China | Positive nucleic acid after the discharge | 60 | 742 | Nasopharyngeal swab |
| You Zou [59] | 2020 | Cohort | Active Surveillance / testing | China | Recurrent positive PCR after discharge with 2 consecutive negative tests | 53 | 257 | Throat swab |
| Youjiang Li [127] | 2020 | Cohort | Active Surveillance / testing | China | Positive PCR during follow-up after the discharge | 4 | 13 | Sputum, oral, nasal, and fecal swabs |
| Yun-Jung Kang [100] | 2020 | Cross-sectional | Routine testing | South Korea | Positive PCR after with discharge undetectable COVID-19 virus | 292 | 8922 | Not clearly specified |
| Zheng Jiazhen [101] | 2020 | Cohort | Active Surveillance / testing | China | Positive nucleic acid test during follow-up after discharge | 27 | 285 | Nasopharyngeal swab |
| Xiao Dong [78] | 2021 | Cross-sectional | Routine testing | USA | Positive RT-PCR with at least 60 days after 2 consecutive negative SARS-CoV-2 tests | 23 | 690 | Not clearly specified |
All studies were conducted either in adults or in both adults and children, with the participants’ ages ranging from 0 to 100 years. No studies have considered vaccinated and unvaccinated participants separately.
The quality assessment results are presented in Table 2. Overall, 42 (80.8%) studies were of good quality, 10 (19.2%) were of fair quality, and no study was of poor quality.
Table 2.
Quality assessment (NIH Quality assessment tool)
| Author name | Q1 | Q2 | Q3 | Q4 | Q5 | Q6 | Q7 | Q8 | Q9 | Q10 | Q11 | Q12 | Q13 | Q14 | SCORE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ai Tang Xiao | 1 | 1 | 1 | 1 | na | 1 | 1 | Na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Anna Jeffery-Smith | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Bo Yuan | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Cheryl Cohen | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Christian Holm | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Efren Murillo-Zamora | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Emilie Finch | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 0 | 1 | na | 1 | 1 | 11 Good |
| Fabiánová K | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Fariba Zare | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Flacco Maria Elena | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Flacco Maria Elena | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Guangming Ye | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Godwin E. Akpan | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Hou-wei Du | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Hui Zhu | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Ji Zhou | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Jia Huang | 1 | 1 | 1 | 1 | na | 1 | 1 | Na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Jianghong An | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Jie Chen | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Jing Lu | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Jing Yuan | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Jinru Wu | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Ju Zhang | 0 | 0 | 1 | 0 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 8 Fair |
| Juliet R.C. Pulliam | 0 | 0 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 9 Fair |
| Justin Wong | 1 | 1 | 1 | 1 | na | 1 | 0 | na | 1 | 1 | 1 | na | 1 | 1 | 10 Fair |
| Laith J. Abu Raddad | 1 | 1 | 1 | 1 | na | 1 | 1 | 1 | 1 | 0 | 1 | na | 1 | 1 | 11 Good |
| Lawandi A | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Lei Pan | 1 | 1 | 1 | 1 | na | 1 | 1 | 1 | 1 | 0 | 1 | na | 1 | 1 | 11 Good |
| Leidi A | 1 | 1 | 1 | 0 | na | 1 | 1 | na | 1 | 0 | 1 | na | 1 | 1 | 9 Fair |
| Maolu Tian | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Muhammad Syafiq Abdullah | 0 | 1 | 1 | 1 | na | 1 | 0 | na | 1 | 1 | 1 | na | 1 | 1 | 9 Fair |
| Naila A Shaheen | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good | |
| Philippe Brouqui | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Pilz Stefan | 1 | 1 | 1 | 1 | na | 1 | 1 | Na | 1 | 1 | 1 | na | 1 | 1 | 10 Fair |
| Rujun Hu | 1 | 1 | 1 | 1 | na | 1 | 0 | na | 1 | 1 | 1 | na | 1 | 1 | 10 Fair |
| Salehi-Vaziri M | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Sezanur Rahman | 1 | 1 | 1 | 1 | na | 1 | 1 | Na | 1 | 1 | 1 | na | 1 | 0 | 10 Fair |
| Sheehan M Meghan | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Shiua Luo | 0 | 1 | 1 | 1 | na | 1 | 0 | na | 1 | 1 | 1 | na | 1 | 1 | 9 Fair |
| Sivan Gazit | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Tao Liu | 1 | 1 | 1 | 1 | na | 1 | 0 | na | 1 | 1 | 1 | na | 1 | 1 | 10 Fair |
| Valeria Cento | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Wang Deng | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Wang Xingyu | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| William R. Hartman | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Yan Dong | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| You Zou | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Youjiang Li | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Yun-Jung Kang | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Zheng Jiazhen | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Xiao Dong | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
| Anne Rivelli | 1 | 1 | 1 | 1 | na | 1 | 1 | na | 1 | 1 | 1 | na | 1 | 1 | 11 Good |
1: Yes
0: No
na not applicable
11–14: good quality
5–10: fair quality
0–4: poor quality
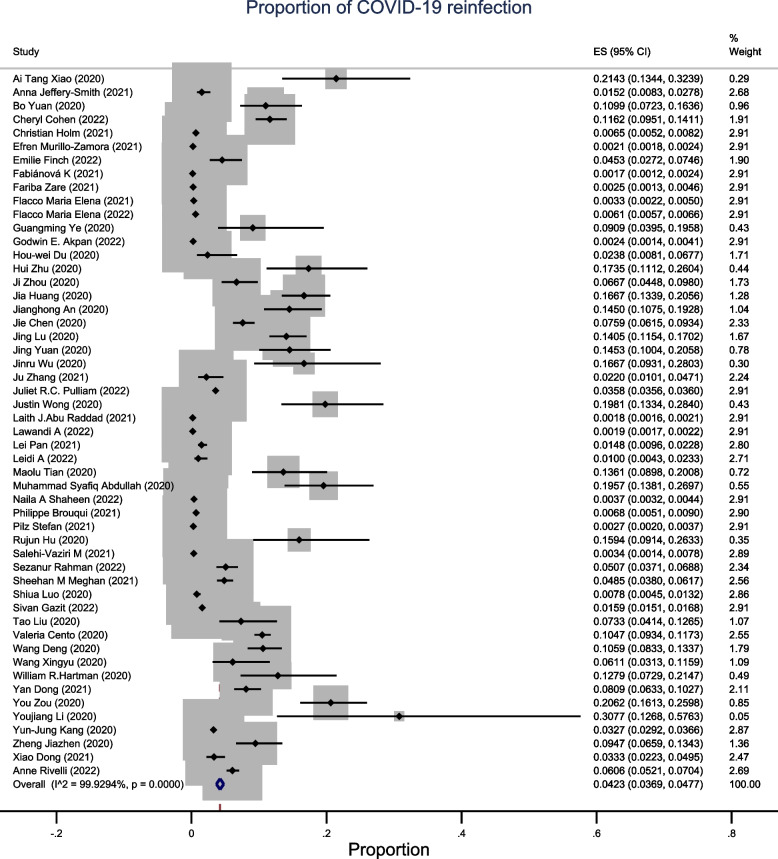
Prevalence of COVID-19 reinfection
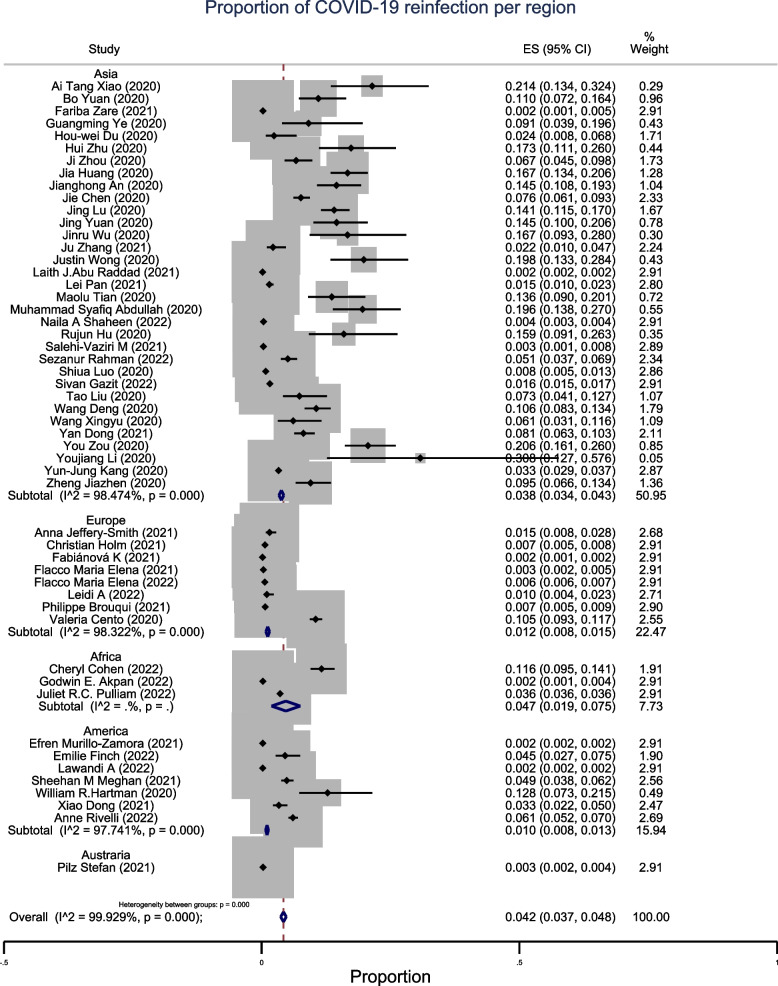
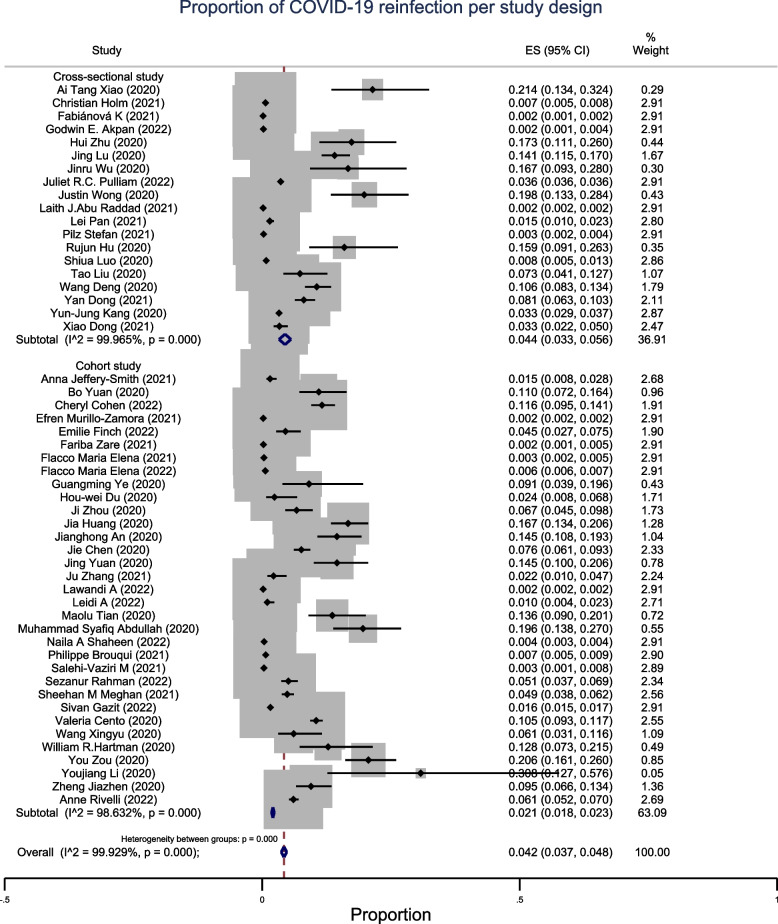
The pooled proportion of SARS-CoV reinfection was 4.2% (95% CI: 3.7–4.8%, n = 52), with high heterogeneity across continents (Fig. 2). The observed proportion of reinfection in studies conducted in Asia was 3.8% (95% CI: 3.4–4.3%; n = 33). In Europe, the proportion was 1.2% (95% CI: 0.8–1.5%; n = 8). The prevalence of reinfection was high in studies conducted in Africa, where the recorded proportion of reinfection was 4.7% (95% CI: 1.9–7.5%; n = 3). The proportion of reinfection in studies conducted in America was low at 1% (95% CI: 0.8–1.3%; n = 7) (Fig. 3). In the subgroup analysis by study design, cohort studies showed a proportion of SARS-CoV reinfection of 2.1% (95% CI: 1.8–2.3%; n = 33), below the overall pooled proportion of all studies. The proportion of reinfection recorded in the cross-sectional studies was 4.4% (95% CI: 3.3–5.6%; n = 19) (Fig. 4).
Fig. 2.
A meta-analysis of pooled estimates of recurrent COVID-19
Fig. 3.
Sub-group meta-analysis by region of pooled estimates of recurrent COVID-19
Fig. 4.
Sub-group meta-analysis by study design of pooled estimates of recurrent COVID-19
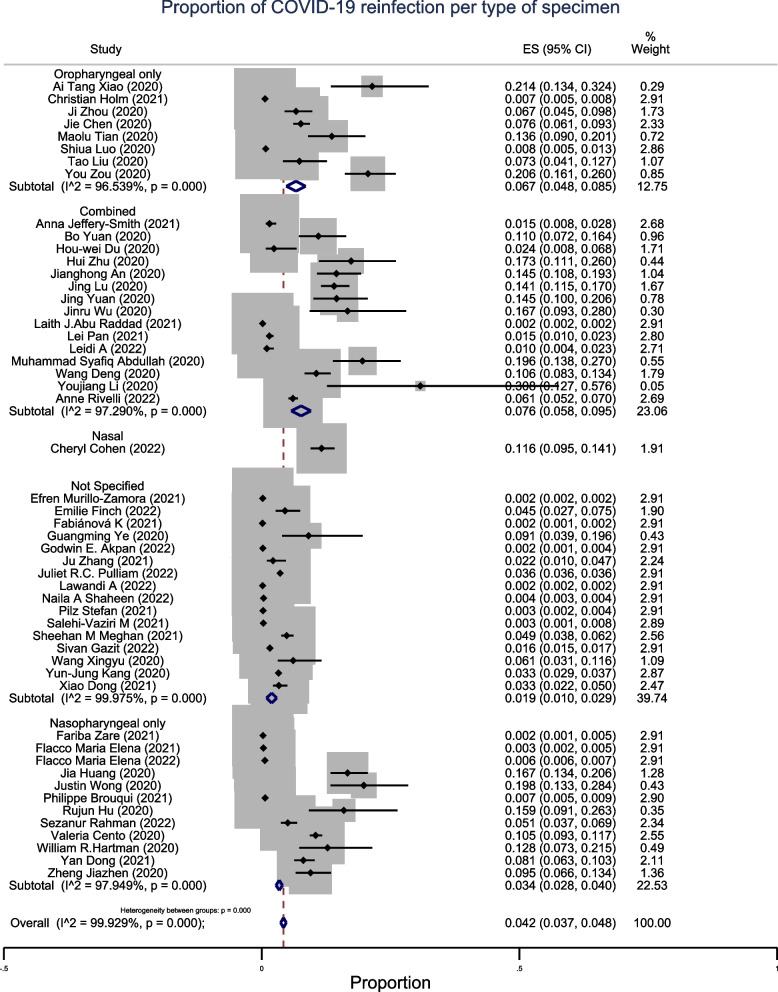
In the subgroup analysis by the type of specimen used for PCR retesting, we observed that the prevalence of reinfection of COVID-19 was 6.7% (95% CI: 4.8–8.5%; n = 8) in the studies that used oropharyngeal samples only. The prevalence was 3.4% (95% CI: 2.8–4.0%; n = 12) in studies that used nasopharyngeal samples only and 7.6% (95% CI: 5.8–9.5%; n = 16) in studies with combined samples. For studies that did not specify the type of sample used and those that used nasal swabs, the prevalence of COVID-19 reinfection was 1.9% (95% CI: 1.0–2.9%; n = 16) and 11.6% (95% CI: 9.5–14.1%; n = 1), respectively. (Fig. 5).
Fig. 5.
Sub-group meta-analysis by type of sample of pooled estimates of recurrent COVID-19
Sensitivity analysis, cumulative effect assessment, and publication bias
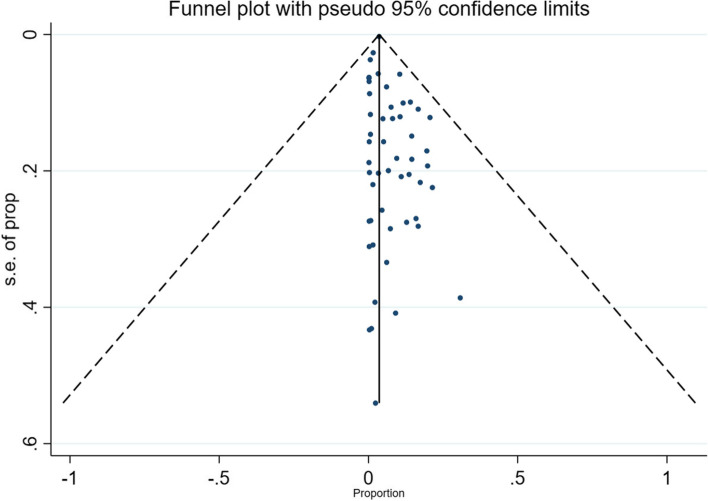
+ + Sensitivity analysis was performed to evaluate whether an individual study had a predominant effect on the overall pooled prevalence of COVID-19 reinfection. This was performed by consecutively removing one study at a time while repeating the analysis. A study by Youjiang Li showed a substantial influence on the pooled prevalence, and its removal increased the prevalence to 13.1%. There was no discernible difference in the direction and magnitude of the pooled estimates across the years according to the cumulative random-effects meta-analysis. The funnel plot showed almost no asymmetry, and the Egger test for publication bias was not statistically significant (P = 0.27), suggesting no publication bias (Fig. 6).
Fig. 6.
A funnel plot for publication bias
Discussion
This study determined the current prevalence of COVID-19 reinfection at global and regional levels. We found an overall proportion of COVID-19 reinfection of 4.2% (95% CI: 3.7–4.8%, n = 52).
Initially, it was debated whether it is possible to have COVID-19 reinfection [128]. The expert believed that infection with COVID-19 confers immunity, and that reinfection is less likely in previously infected people [129]. However, reinfection with COVID-19 is quite common, and naturally acquired immunity wanes within a few weeks of infection [129]. The reinfection was mostly observed with Omicron and its subvariants, which can evade natural immunity [129]. In a surveillance study conducted in South Africa, the Omicron variant was associated with an increased reinfection rate [29]. In contrast to other human coronaviruses, namely SARS-CoV, initially reported in Guangdong/China in 2002, and MERS-CoV, initially reported in Saudi Arabia in 2012, reinfection has not been reported as these infections were directly contained, leaving no time to document reinfection or recurrence [128]. However, reinfection was documented with other common coronaviruses within 4–48 weeks of primary infection (mean period of 37 weeks) [128, 130].
The prevalence of COVID-19 reinfection observed in this meta-analysis was low compared with that found in previous meta-analyses [131, 132]. This discrepancy in the prevalence of COVID-19 reinfection can be explained by these meta-analyses conducted early during the pandemic when few studies were available, in addition to the difference in the time of follow-up. A meta-analysis by Camilla et al. included 17 studies, and the follow-up period ranged from 1 to 60 days [131]. Another meta-analysis included 14 studies, and the median period from infection to recurrence ranged from 21 to 50 days [132]. This systematic review and meta-analysis included 52 relevant studies to determine the current prevalence of COVID-19 reinfection at the global and regional levels and considered a longer follow-up period.
There is a lack of a conventional definition of reinfection, and different authors of the studies included in existing meta-analyses have defined COVID-19 reinfection differently. Some authors have described cases of reinfection as continual viral shedding, whereas to ascertain reinfection, genomic sequencing should be performed to determine whether the second infecting virus is genetically different from the previous [51]. Zumrut et al. described a case of re-positivity of COVID-19 PCR after 27 days of a negative test, and it was inconclusive whether it was a reinfection or prolonged viral shedding, as no previous genotyping was performed for the first infection [133]. The CDC stated that viral shedding could be prolonged for up to 90 days, but even for severely infected persons, there was no replication-competent virus recovered after 10–20 days [134]; The ECDC proposed tests such as whole-genome sequencing and phylogenetic analysis to conclude on reinfection [48], and cases of genetically confirmed reinfection in a period of approximately 2 months from the previous infection have been reported in the literature [135, 136]. However, these genetic tests are not extensively available, and if considered widely, many cases of reinfection in many settings would not be recognized.
A higher proportion of recurrence of COVID-19 infection was observed in Africa (4.7%; 95% CI: 1.9–7.5%; n = 3) than that in other regions. This finding is unanticipated, as the incidence of COVID-19 in Africa is lower than that in other regions [17]. A review by Dufailu states that the incidence, hospitalization, and mortality rate of COVID-19 were lower in Africa than that in other continents [137].
This high rate of reinfection in Africa, where the pandemic was less severe, can be explained by the high population density in this region and the failure to implement pandemic control measures [137]. In addition, the low mortality rate observed in Africa, as well as the low incidence of severe cases of infection, should have led people to disregard restrictive measures and barrier actions against COVID-19, even in cases of previous infection, making reinfection an imminent occurrence [137, 138].
Other factors that should have influenced reinfection secondary to non-compliance with COVID-19 control measures by most of the population in Africa are the low vaccination rate, unsanitary conditions, lack of access to clean water, and lack of awareness [139]. A study by Wirsiy et al. published in 2020 mentioned that measures taken in Asia, Europe, and North America, such as physical (social) distance and consistent handwashing, were most difficult to implement in African countries where Internet connectivity was limited, population density, access to water was uneven, and social safety nets were limited [140].
On the other hand, this high prevalence observed in Africa is controversial, the study by Cohen et al., which has a high prevalence of COVID-19 reinfection used data from active surveillance and testing research compared to the other two African studies, which used data from routine testing. Active surveillance aims to detect every case. Cases are actively searched, tested, and followed—up, and their reporting is promoted; it provides the most complete, accurate, and timely information [141, 142] compared to passive surveillance, which relies on the analysis of data from reported cases and often lacks completeness [141, 142]. The difference between prevalence estimates obtained from routine laboratory testing vs active research surveillance data is less problematic in high-income countries, but particularly so in low- and middle-income countries, where testing capacity was correspondingly limited [143–146]. As gaps in testing were observed in Africa and more cases were missed [143–146], we would expect a difference between routine testing and active surveillance data. This, along with the relatively small number of studies from the continent, could have primarily raised the prevalence of COVID-19 reinfection, which is not necessarily real.
Oceania recorded the lowest prevalence of COVID-19 reinfection at 0.3% (95% CI: 0.2–0.4%; n = 1), but the sample size in this region was too small to predict the true estimates of reinfection.
Studies that used a combined type of specimens for PCR retesting had the highest prevalence of 7.6% (95% CI: 5.8–9.5%; n = 15). The studies that used nasopharyngeal specimens only had a lower prevalence of COVID-19 reinfection (3.4%; 95% CI: 3.7–4.8%; n = 12) compared with those that used only oropharyngeal specimens (6.7%; 95% CI: 4.8–8.5%; n = 8). These findings are the opposite of those observed in a meta-analysis by Azam et al., where the highest prevalence was found in studies that used a nasopharyngeal specimen only and the lowest in oropharyngeal specimens only [132].
These findings also disagree with those of a meta-analysis that assessed the positivity rate of COVID-19 using different types of specimens [147]. In the latter study, nasopharyngeal swabs showed a positivity rate of 45%, whereas that of oropharyngeal swabs was 7.6%, and the highest detection rate was found in bronchoalveolar fluids [147]. However, in agreement with the findings of this meta-analysis, other studies have shown that nasopharyngeal specimens yield a higher detection rate of COVID-19 than that of nasal or oropharyngeal swabs [148–151].
The discrepancy in these findings can be related to many studies considered in this meta-analysis that did not specify the type of specimen used for PCR retesting; however, this analysis considered throat and oropharyngeal swabs equivalent. Therefore, we hypothesize that combining two or more types of specimens should increase the detection rate of COVID-19, as substantiated by the findings of this study. However, more studies must validate this finding, and further meta-analyses will reduce this disagreement.
The difference between vaccinated and unvaccinated individuals in relation to the COVID-19 reinfection rate was not assessed because there was insufficient necessary information for this analysis. A prospective cohort study published in January 2022 by Sezanur Rahman revealed that new COVID-19 variants that emerged in 2021 could reinfect both naturally infected and vaccinated individuals and that being naturally infected confers better protection against COVID-19 for at least 6 months after primary infection [93]. Another systematic review indicated a risk of reinfection with COVID-19 in previously infected patients, including those vaccinated against the disease [152].
The severity of the reinfection was not addressed in this study. A systematic review conducted by Rubaid et al. in 2021 showed that the first infection and reinfection with COVID-19 showed a broadly similar pattern of clinical and management regimen but with a slightly higher severity among reinfected cases, evaluated by the need for mechanical ventilation and intensive care unit admission [153].
This meta-analysis has important implications for public health policies. The evidence of COVID-19 reinfection presented in this review should guide countries that demonstrate high reinfection prevalence to a future better preparedness for epidemic and pandemic diseases. Strategies should be set a priori on how preventive measures should be respected to prevent the spread of epidemics as well as reinfection.
The main strength of our study was the long period covered, which included studies from the first that reported reinfection in individuals pre-infected with COVID-19 until June 2022. This made it the first comprehensive meta-analysis to include many studies to date, which also considered the prevalence of COVID-19 reinfection by geographical area. In addition, most of the studies included in our analysis were of good quality.
The limitations of this study are that there is a difference in the definitions of reinfection used by the authors of the studies included in this meta-analysis, and it was difficult to compare findings across all studies. Additionally, no subgroup analysis was done by vaccination status or by definition of reinfection, and all sources of heterogeneity, such as the age group, were not explored because of a lack of appropriate related information among studies. Furthermore, the variability in sample size across the studies should have affected the prevalence and heterogeneity observed between studies.
Conclusion
The current evidence shows that COVID-19 reinfection occurs and has a prevalence that varies worldwide, with the highest prevalence occurring in Africa. Therefore, preventive measures, including vaccination, should be emphasized to ensure control of the pandemic. More studies are needed to understand the rate of COVID-19 reinfection in consideration of different variants as well as comparing reinfection among vaccinated and unvaccinated persons. Factors that increase this risk of reinfection have not been well identified; hence, further studies can help in further clarification and future planning of preventive interventions to control this pandemic.
Supplementary Information
Additional file 1: Table S1. Search strategy. Table S2. NIH Quality assessment tool questions.
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing
Abbreviations
- CDC
Center for Disease Control
- COVID-19
Coronavirus disease 2019
- ECDC
European Center for Disease Control
- MERS-CoV
Middle East Respiratory Syndrome coronavirus
- NIH
National Institute for Health
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- WHO
World Health Organization
Authors’ contributions
UJ, NY, ED carried out the literature review, titles and abstracts screening, data extraction, quality assessment, data analysis and interpretation of results and contributed to manuscript preparation. DC contributed to data analysis, interpretation of results, and critical revision of the article. KSF contributed to study conception and design, interpretation of results, critical revision of the article and supervision of the work. All authors reviewed the results and approved the final version of the manuscript.
Funding
None.
Availability of data and materials
The authors declare that the data supporting the findings of this study are presented within the article and its supplementary materials.
Declarations
Ethics approval and consent to participate
Not required.
Consent for publication
Not applicable.
Competing interests
Authors declared no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. Coronavirus disease (COVID-19) pandemic. 2022. Available from: https://www.who.int/europe/emergencies/situations/covid-19 [cited 2022 Sep 22].
- 2.WHO. Coronavirus disease (COVID-19). 2021. Available from: https://www.who.int/health-topics/coronavirus#tab=tab_1 [cited 2022 Sep 22].
- 3.WHO. WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020. 2020. Available from: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 [cited 2022 Sep 22].
- 4.Vijayanand P, Wilkins E, Woodhead M. Severe acute respiratory syndrome (SARS): a review. Clin Med. 2004;4(2):152–60. doi: 10.7861/clinmedicine.4-2-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Severe Acute Respiratory Syndrome (SARS). 2022. Available from: https://www.who.int/health-topics/severe-acute-respiratory-syndrome#tab=tab_1 [cited 2022 Sep 25].
- 6.Ksiaze TG, Erdman D, Goldsmith CS, Pere T, Emery S, Tong S, et al. Respiratory and enteric virus branch. N Engl J Med. 2003;348(20):1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 7.Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 8.Rat P, Olivier E, Dutot M. SARS-CoV-2 vs. SARS-CoV-1 management: antibiotics and inflammasome modulators potential. Eur Rev Med Pharmacol Sci. 2020;24:7880–5. doi: 10.26355/eurrev_202007_22293. [DOI] [PubMed] [Google Scholar]
- 9.Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J Biol Chem. 2020;295:12910–34. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asrani P, Hasan GM, Sohal SS, Hassan MI. Molecular basis of pathogenesis of coronaviruses: a comparative genomics approach to planetary health to prevent zoonotic outbreaks in the 21st century. OMICS. 2020;24(11):634–644. doi: 10.1089/omi.2020.0131. [DOI] [PubMed] [Google Scholar]
- 11.Ramadan N, Shaib H. Review middle east respiratory syndrome coronavirus (MERS-CoV): a review. Germs. 2019;9(1):35–42. doi: 10.18683/germs.2019.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azhar EI, Hui DSC, Memish ZA, Drosten C, Zumla A. The Middle East Respiratory Syndrome (MERS). Infectious Disease Clinics of North America. W.B. Saunders. 2019;33:891–905. [DOI] [PMC free article] [PubMed]
- 13.WHO. Middle East respiratory syndrome coronavirus (MERS-CoV). 2022. Available from: https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov) [cited 2022 Sep 25].
- 14.dos Santos WG. Natural history of COVID-19 and current knowledge on treatment therapeutic options. Biomed Pharmacother. 2020;129:110493. doi: 10.1016/j.biopha.2020.110493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sofi MS, Hamid A, Bhat SU. SARS-CoV-2: a critical review of its history, pathogenesis, transmission, diagnosis and treatment. Biosaf Health. 2020;2:217–25. doi: 10.1016/j.bsheal.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cevik M, Kuppalli K, Kindrachuk J, Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;23:371. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- 17.WHO. WHO Coronavirus (COVID-19) Dashboard. 2022. Available from: https://covid19.who.int/ [cited 2022 Sep 22].
- 18.Singh A, Chattopadhyay A. COVID-19 recovery rate and its association with development. Indian J Med Sci. 2020;29:1–8. [Google Scholar]
- 19.Achaiah NC, Subbarajasetty SB, Shetty RM. R0 and re of covid-19: Can we predict when the pandemic outbreak will be contained? Indian J Crit Care Med. 2020;24:1125–7. doi: 10.5005/jp-journals-10071-23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoaib N, Noureen N, Munir R, Shah FA, Ishtiaq N, Jamil N, et al. COVID-19 severity: studying the clinical and demographic risk factors for adverse outcomes. PLoS One. 2021;16:e0255999. doi: 10.1371/journal.pone.0255999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallo Marin B, Aghagoli G, Lavine K, Yang L, Siff EJ, Chiang SS, et al. Predictors of COVID-19 severity: a literature review. Rev Med Virol. 2021;31:1–10. doi: 10.1002/rmv.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y, Liu M, Chen Y, Shi S, Geng J, Tian J. Association between tuberculosis and COVID-19 severity and mortality: a rapid systematic review and meta-analysis. J Med Virol. 2021;93:194–6. doi: 10.1002/jmv.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Chen Y, Liu M, Shi S, Tian J. Impacts of immunosuppression and immunodeficiency on COVID-19: a systematic review and meta-analysis. J Infect. 2020;81:e93–5. doi: 10.1016/j.jinf.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81:e16–25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO. Tracking SARS-CoV-2 variants. 2022. Available from: https://www.who.int/activities/tracking-SARS-CoV-2-variants [cited 2022 Sep 25].
- 27.ECDC. SARS-CoV-2 variants of concern as of 22 September 2022. 2022. Available from: https://www.ecdc.europa.eu/en/covid-19/variants-concern [cited 2022 Sep 25].
- 28.Marco Cascella A, Rajnik M, Cuomo A, Dulebohn SC, di Napoli R. Features, Evaluation and Treatment Coronavirus (COVID-19). In: -StatPeals*NCBI Bookshelf. 2020. p. 1–17. Available from: https://www.ncbi.nlm.nih.gov/books/NBK554776/?report=printable. [PubMed]
- 29.Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science (1979) 2022;376(6593):eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altarawneh HN, Chemaitelly H, Hasan MR, Ayoub HH, Qassim S, AlMukdad S, et al. Protection against the Omicron Variant from Previous SARS-CoV-2 Infection. N Engl J Med. 2022;386(13):1288–1290. doi: 10.1056/NEJMc2200133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 2021; Available from: http://www.ncbi.nlm.nih.gov/pubmed/34909788.
- 32.Torjesen I. Covid-19: omicron variant is linked to steep rise in hospital admissions of very young children. BMJ. 2022;14(376):o110. doi: 10.1136/bmj.o110. [DOI] [PubMed] [Google Scholar]
- 33.Mensah AA, Lacy J, Stowe J, Seghezzo G, Sachdeva R, Simmons R, et al. Disease severity during SARS-COV-2 reinfection: a nationwide study. J Infect. 2022;84(4):542–550. doi: 10.1016/j.jinf.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards J, Rivelli A, Fitzpatrick V, Blair C, Copeland K. Incidence of COVID-19 reinfection among Midwestern healthcare employees. PLoS One. 2022;17:e0262164. doi: 10.1371/journal.pone.0262164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordström P, Ballin M, Nordström A. Risk of SARS-CoV-2 reinfection and COVID-19 hospitalisation in individuals with natural and hybrid immunity: a retrospective, total population cohort study in Sweden. Lancet Infect Dis. 2022;22(6):781–790. doi: 10.1016/S1473-3099(22)00143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flacco ME, Acuti Martellucci C, Soldato G, Carota R, Fazii P, Caponetti A, et al. Rate of reinfections after SARS-CoV-2 primary infection in the population of an Italian province: a cohort study. J Public Health. 2021;44(4):1–4. [DOI] [PMC free article] [PubMed]
- 37.WHO. Coronavirus disease (COVID-19). 2022. Available from: https://www.who.int/health-topics/coronavirus#tab=tab_3 [cited 2022 Sep 25].
- 38.Lotfi M, Hamblin MR, Rezaei N. COVID-19: transmission, prevention, and potential therapeutic opportunities. Clin Chim Acta. 2020;508:254–66. doi: 10.1016/j.cca.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascarella G, Strumia A, Piliego C, Bruno F, del Buono R, Costa F, et al. COVID-19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alsharif W, Qurashi A. Effectiveness of COVID-19 diagnosis and management tools: a review. Radiography. 2021;27:682–7. doi: 10.1016/j.radi.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochani RK, Kumar Ochani R, Asad A, Yasmin F, Shaikh S, Khalid H, et al. COVID-19 pandemic: from origins to outcomes. A comprehensive review of viral pathogenesis, clinical manifestations, diagnostic evaluation, and management. Infez Med. 2021;29:20–36. [PubMed] [Google Scholar]
- 42.Mouffak S, Shubbar Q, Saleh E, El-Awady R. Recent advances in management of COVID-19: a review. Biomed Pharmacother. 2021;143:107. doi: 10.1016/j.biopha.2021.112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren X, Zhou J, Guo J, Hao C, Zheng M, Zhang R, et al. Reinfection in patients with COVID-19: a systematic review. Glob Health Res Policy. 2022;7:12. doi: 10.1186/s41256-022-00245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Kaperak C, Sato T, Sakuraba A. COVID-19 reinfection: a rapid systematic review of case reports and case series. J Investig Med. 2021;69:1253–5. doi: 10.1136/jim-2021-001853. [DOI] [PubMed] [Google Scholar]
- 45.Tang X, Musa SS, Zhao S, He D. Reinfection or reactivation of severe acute respiratory syndrome Coronavirus 2: a systematic review. Front Public Health. 2021;9:663045. doi: 10.3389/fpubh.2021.663045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ECDC . Reinfection with SARS-CoV-2: implementation of a surveillance case definition within the EU/EEA Key messages. 2021. [Google Scholar]
- 47.CDC. Reinfections and COVID-19. 2022. Available from: https://www.cdc.gov/coronavirus/2019-ncov/your-health/reinfection.html [cited 2022 Sep 25].
- 48.ECDC . Reinfection with SARS-CoV-2: considerations for public health response. 2020. [Google Scholar]
- 49.UK Health Security Agency. COVID-19 daily dashboard amended to include reinfections. 2022. Available from: https://www.gov.uk/government/news/covid-19-daily-dashboard-amended-to-include-reinfections [cited 2022 Sep 25].
- 50.sciensano. Reinfection update October 2020. 2020. Available from: https://covid-19.sciensano.be/sites/default/files/Covid19/20201021_Advice_RAG%20reinfection.pdf [cited 2022 Sep 25].
- 51.Yahav D, Yelin D, Eckerle I, Eberhardt CS, Wang J, Cao B, et al. Definitions for coronavirus disease 2019 reinfection, relapse and PCR re-positivity. Clin Microbiol Infect. 2021;27:315–8. doi: 10.1016/j.cmi.2020.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.NHI. Study Quality Assessment Tools. Available from: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools [cited 2022 Dec 11].
- 53.Xiao AT, Tong YX, Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020;92:1755–6. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen CH, Michlmayr D, Gubbels SM, Mølbak K, Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397(10280):1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu H, Fu L, Jin Y, Shao J, Zhang S, Zheng N, et al. Clinical features of COVID-19 convalescent patients with re-positive nucleic acid detection. J Clin Lab Anal. 2020;34(7):e23392. doi: 10.1002/jcla.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An J, Liao X, Xiao T, Qian S, Yuan J, Ye H, et al. Clinical characteristics of recovered COVID-19 patients with re-detectable positive RNA test. Ann Transl Med. 2020;8(17):1084–1084. doi: 10.21037/atm-20-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Xiaoping Xu, Jing Hu, Chen Q, Fengfeng Xu, Liang H, et al. Clinical course and risk factors for recurrence of positive SARS-CoV-2 RNA: a retrospective cohort study from Wuhan, China. Aging. 2020;12(17):16675–16689. doi: 10.18632/aging.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu T, Wu S, Zeng G, Zhou F, Li Y, Guo F, et al. Recurrent positive SARS-CoV-2: Immune certificate may not be valid. J Med Virol. 2020;92:2384–6. doi: 10.1002/jmv.26074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou Y, Wang BR, Sun L, Xu S, Kong YG, Shen LJ, et al. The issue of recurrently positive patients who recovered from COVID-19 according to the current discharge criteria: investigation of patients from multiple medical institutions in Wuhan China. J Infect Dis. 2020;222(11):1784–1788. doi: 10.1093/infdis/jiaa301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian M, Long Y, Hong Y, Zhang X, Zha Y. The treatment and follow-up of ‘recurrence’ with discharged COVID-19 patients: data from Guizhou China. Environ Microbiol. 2020;22(8):3588–3592. doi: 10.1111/1462-2920.15156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Ding N, Ren L, Song R, Chen D, Zhao X, et al. COVID-19 reinfection in the presence of neutralizing antibodies. Natl Sci Rev. 2021;8(4):nwab006. doi: 10.1093/nsr/nwab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan J, Kou S, Liang Y, Zeng J, Pan Y, Liu L. Polymerase chain reaction assays reverted to positive in 25 discharged patients with COVID-19. Clin Infect Dis. 2020;71(16):2230–2232. doi: 10.1093/cid/ciaa398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu J, Peng J, Xiong Q, Liu Z, Lin H, Tan X, et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine. 2020;1:59. doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong J, Koh WC, Momin RN, Alikhan MF, Fadillah N, Naing L. Probable causes and risk factors for positive SARS-CoV-2 test in recovered patients: evidence from Brunei Darussalam. J Med Virol. 2020;92(11):2847–2851. doi: 10.1002/jmv.26199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng W, Guang TW, Yang M, Li JR, Jiang DP, Li CY, et al. Positive results for patients with COVID-19 discharged form hospital in Chongqing China. BMC Infect Dis. 2020;20(1):1–6. doi: 10.1186/s12879-020-05151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, Xu H, Jiang H, Wang L, Lu C, Wei X, et al. The clinical features and outcomes of discharged Coronavirus disease 2019 patients: a prospective cohort study. Oxford University Press. 2020;657–65. Available from: https://mc.manuscriptcentral.com/qjm. [DOI] [PMC free article] [PubMed]
- 67.Jeffery-Smith A, Rowland TAJ, Patel M, Whitaker H, Iyanger N, Williams SV, et al. Reinfection with new variants of SARS-CoV-2 after natural infection: a prospective observational cohort in 13 care homes in England. Lancet Healthy Longev. 2021;2(12):e811–9. doi: 10.1016/S2666-7568(21)00253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen C, Kleynhans J, von Gottberg A, McMorrow ML, Wolter N, Bhiman JN, et al. SARS-CoV-2 incidence, transmission, and reinfection in a rural and an urban setting: results of the PHIRST-C cohort study, South Africa, 2020–21. Lancet Infect Dis. 2022;22(6):821–834. doi: 10.1016/S1473-3099(22)00069-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fabianova K, Kyncl JVH, Kostalova J, Liptakova M, Orlikova H, Sebestova H, et al. COVID-19 reinfections. Epidemiol Mikrobiol Imunol. 2021;70(1):62–67. [PubMed] [Google Scholar]
- 70.Akpan GE, Bawo L, Amo-Addae M, Kennedy J, Wesseh CS, Whesseh F, et al. COVID-19 reinfection in Liberia: implication for improving disease surveillance. PLoS One. 2022;17:e0265768. doi: 10.1371/journal.pone.0265768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lawandi A, Warner S, Sun J, Demirkale CY, Danner RL, Klompas M, et al. Suspected severe acute respiratory syndrome Coronavirus 2 (SARS-COV-2) reinfections: incidence, predictors, and healthcare use among patients at 238 US healthcare facilities, 1 June 2020 to 28 February 2021. Clin Infect Dis. 2022;74(8):1489–1492. doi: 10.1093/cid/ciab671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brouqui P, Colson P, Melenotte C, Houhamdi L, Bedotto M, Devaux C, et al. COVID-19 re-infection. Eur J Clin Investig. 2021;51:631–2. doi: 10.1111/eci.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheehan Bs MM, Reddy AJ, Rothberg MB. Reinfection rates among patients who previously tested positive for COVID-19: a retrospective cohort study. Clin Infect Dis. 2021;73(10):1882–1886. doi: 10.1093/cid/ciab234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Finch E, Lowe R, Fischinger S, de St Aubin M, Siddiqui SM, Dayal D, et al. SARS-CoV-2 antibodies protect against reinfection for at least 6 months in a multicentre seroepidemiological workplace cohort. PLoS Biol. 2022;20(2):e3001531. doi: 10.1371/journal.pbio.3001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zare F, Teimouri M, Khosravi A, Rohani-Rasaf M, Chaman R, Hosseinzadeh A, et al. COVID-19 Reinfection in Shahroud, Iran A follow up Study. Epidemiol Infect. 2021;49:e159. doi: 10.1017/S095026882100087X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flacco ME, Soldato G, Acuti Martellucci C, di Martino G, Carota R, Caponetti A, et al. Risk of SARS-CoV-2 Reinfection 18 Months After Primary Infection: Population-Level Observational Study. Front Public Health. 2022;2:10. doi: 10.3389/fpubh.2022.884121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raddad LJA, Chemaitelly H, Malek JA, Ahmed Bsc AA, Mohamoud YA, Younuskunju S, et al. Assessment of the risk of SARS-CoV-2 reinfection in an intense re-exposure setting. 10.1101/2020.08.24.20179457.
- 78.Dong X, Zhou Y, Shu XOU, Bernstam EV, Stern R, Aronoff DM, et al. Comprehensive Characterization of COVID-19 Patients with Repeatedly Positive SARS-CoV-2 Tests Using a large U.S. Electronic Health Record Database. Microbiol Spectr. 2021;9(1):e0032721. doi: 10.1128/Spectrum.00327-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan B, Liu HQ, Yang ZR, Chen YX, Liu ZY, Zhang K, et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci Rep. 2020;10(1):11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murillo-Zamora E, Trujillo X, Huerta M, Ríos-Silva M, Aguilar-Sollano F, Mendoza-Cano O. Symptomatic SARS-COV-2 reinfection: healthcare workers and immunosuppressed individuals at high risk. BMC Infect Dis. 2021;21(1):923. doi: 10.1186/s12879-021-06643-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye G, Pan Z, Pan Y, Deng Q, Chen L, Li J, et al. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80(5):e14–e17. doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Du HW, Chen JN, Pan XB, Chen XL, Yixian-Zhang, Fang SF, et al. Prevalence and outcomes of re-positive nucleic acid tests in discharged COVID-19 patients. Eur J Clin Microbiol Infect Dis. 2021;40(2):413–7. doi: 10.1007/s10096-020-04024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou J, Zhang J, Zhou J, Yi H, Lin Z, Liu Y, et al. Clinical characteristics of re-positive COVID-19 patients in Huangshi, China: a retrospective cohort study. PLoS One. 2020;15:e0241896. doi: 10.1371/journal.pone.0241896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang J, Zheng L, Li Z, Hao S, Ye F, Chen J, et al. Recurrence of SARS-CoV-2 PCR positivity in COVID-19 patients: a single center experience and potential implications. 2020; 10.1101/2020.05.06.20089573.
- 85.Wu J, Liu X, Liu J, Liao H, Long S, Zhou N. Coronavirus Disease 2019 Test Results After Clinical Recovery and Hospital Discharge Among Patients in China | Enhanced Reader. JAMA Netw Open. 2020;3(5):e209759. doi: 10.1001/jamanetworkopen.2020.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan L, Wang R, Yu N, Hu C, Yan J, Zhang X, et al. Clinical characteristics of re-hospitalized COVID-19 patients with recurrent positive SARS-CoV-2 RNA: a retrospective study. Eur J Clin Microbiol Infect Dis. 2021;40:1245–52. doi: 10.1007/s10096-020-04151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leidi A, Koegler F, Dumont R, Dubos R, Zaballa ME, Piumatti G, et al. Risk of reinfection after seroconversion to SARS-CoV-2: A population-based propensity-score matched cohort study. Clin Infect Dis. 2022;74(4):622–629. doi: 10.1093/cid/ciab495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdullah MS, Chong PL, Asli R, Momin RN, Mani BI, Metussin D, et al. Post discharge positive re-tests in COVID-19: common but clinically non-significant. Infect Dis. 2020;52:743–5. doi: 10.1080/23744235.2020.1780309. [DOI] [PubMed] [Google Scholar]
- 89.Shaheen N, Sambas R, Alenezi M, Alharbi N, Aldibasi O, Bosaeed M. COVID-19 reinfection: a multicenter retrospective study in Saudi Arabia. Ann Thorac Med. 2022;17(2):81–86. doi: 10.4103/atm.atm_74_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pilz S, Chakeri A, Ioannidis JPA, Richter L, Theiler-Schwetz V, Trummer C, et al. SARS-CoV-2 re-infection risk in Austria. Eur J Clin Invest. 2021;51(4):520. doi: 10.1111/eci.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, et al. Positive RT-PCR Test Results in Patients Recovered from COVID-19. JAMA. 2020;323:1502–3. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salehi-Vaziri M, Pouriayevali MH, Fotouhi F, Jalali T, Banifazl M, Farahmand B, et al. SARS-CoV-2 re-infection rate in Iranian COVID-19 cases within one-year follow-up. Microb Pathog. 2021;1:161. doi: 10.1016/j.micpath.2021.105296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rahman S, Rahman MM, Miah M, Begum MN, Sarmin M, Mahfuz M, et al. COVID-19 reinfections among naturally infected and vaccinated individuals. Sci Rep. 2022;12(1):1438. doi: 10.1038/s41598-022-05325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo S, Guo Y, Zhang X, Xu H. A follow-up study of recovered patients with COVID-19 in Wuhan China. Int J Infect Dis. 2020;99:408–9. doi: 10.1016/j.ijid.2020.05.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gazit S, Shlezinger R, Perez G, Lotan R, Peretz A, Ben-Tov A, et al. The Incidence of SARS-CoV-2 Reinfection in Persons with Naturally Acquired Immunity With and Without Subsequent Receipt of a Single Dose of BNT162b2 Vaccine A Retrospective Cohort Study. Ann Intern Med. 2022;175(5):674–681. doi: 10.7326/M21-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cento V, Colagrossi L, Nava A, Lamberti A, Senatore S, Travi G, et al. Persistent positivity and fluctuations of SARS-CoV-2 RNA in clinically recovered COVID-19 patients. J Clean Prod. 2020;81:90–2. doi: 10.1016/j.jinf.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hartman WR, Hess AS, Connor JP. Persistent viral RNA shedding after COVID-19 symptom resolution in older convalescent plasma donors. Transfusion (Paris) 2020;60(10):2189–2191. doi: 10.1111/trf.15927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong Y, Huang H, Yang J, Yang L. Retrospective analysis on the clinical characteristics of patients who were reinfected with the Corona Virus in 2019. Am J Tranl Res. 2021;13(5):5505–5511. [PMC free article] [PubMed] [Google Scholar]
- 99.Li Y, Hu Y, Yu Y, Zhang X, Li B, Wu J, et al. Positive result of Sars-Cov-2 in faeces and sputum from discharged patients with COVID-19 in Yiwu. China J Med Virol. 2020;92(10):1938–1947. doi: 10.1002/jmv.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kang YJ. South Korea’s COVID-19 Infection Status: From the Perspective of Re-positive Test Results after Viral Clearance Evidenced by Negative Test Results. Disaster Med Public Health Prep. 2020;14(6):762–764. doi: 10.1017/dmp.2020.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng J, Zhou R, Chen F, Tang G, Wu K, Li F, et al. Incidence, clinical course and risk factor for recurrent pcr positivity in discharged covid-19 patients in guangzhou, china: a prospective cohort study. PLoS Negl Trop Dis. 2020;14(8):1–14. doi: 10.1371/journal.pntd.0008648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jeffery-Smith A, Rowland TAJ, Patel M, Whitaker H, Iyanger N, Williams SV, et al. Reinfection with new variants of SARS-CoV-2 after natural infection: a prospective observational cohort in 13 care homes in England. Lancet Healthy Longev. 2021;2(12):e811–e819. doi: 10.1016/S2666-7568(21)00253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murillo-Zamora E, Trujillo X, Huerta M, Ríos-Silva M, Aguilar-Sollano F, Mendoza-Cano O. Symptomatic SARS-COV-2 reinfection: healthcare workers and immunosuppressed individuals at high risk. BMC Infect Dis. 2021;21(1):1–5. doi: 10.1186/s12879-021-06643-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Finch E, Lowe R, Fischinger S, de St AM, Siddiqui SM, Dayal D, et al. SARS-CoV-2 antibodies protect against reinfection for at least 6 months in a multicentre seroepidemiological workplace cohort. PLoS Biol. 2022;20(2):1–12. doi: 10.1371/journal.pbio.3001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fabiánová K. COVID-19 reinfections. 2022. p. 6597. [Google Scholar]
- 106.Zare F, Teimouri M, Khosravi A, Rohani-Rasaf M, Chaman R, Hosseinzadeh A, et al. COVID-19 reinfection in Shahroud, Iran. Epidemiol Infect. 2021;149:1–12. doi: 10.1017/S095026882100087X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Flacco ME, Soldato G, Acuti Martellucci C, Di Martino G, Carota R, Caponetti A, et al. Risk of SARS-CoV-2 Reinfection 18 Months After Primary Infection: population-Level Observational Study. Front Public Health. 2022;2:1074. doi: 10.3389/fpubh.2022.884121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Akpan GE, Bawo L, Amo-Addae M, Kennedy J, Wesseh CS, Whesseh F, et al. COVID-19 reinfection in Liberia: implication for improving disease surveillance. PLoS One. 2022;17(3):e0265768. doi: 10.1371/journal.pone.0265768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Du HW, Chen JN, Pan XB, Chen XL, Yixian-Zhang, Shuang-Fang Fang, et al. Prevalence and outcomes of re-positive nucleic acid tests in discharged COVID-19 patients. Eur J Clin Microbiol Infect Dis. 2021;40(2):413–7. [DOI] [PMC free article] [PubMed]
- 110.Zhu H, Fu L, Jin Y, Shao J, Zhang S, Zheng N, et al. Clinical features of COVID-19 convalescent patients with re-positive nucleic acid detection. J Clin Lab Anal. 2020;34(7):1–10. doi: 10.1002/jcla.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou J, Zhang J, Zhou J, Yi H, Lin Z, Liu Y, et al. Clinical characteristics of re-positive COVID-19 patients in Huangshi, China: a retrospective cohort study. PLoS One. 2020;15:1–16. doi: 10.1371/journal.pone.0241896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang J. Recurrence of SARS-CoV-2 PCR positivity in COVID-19 patients: a single center experience and potential implications. Konstruksi Pemberitaan Stigma Anti-China pada Kasus Covid-19 di Kompas.com. 2020;68(1):1–12.
- 113.Lu J, Peng J, Xiong Q, Liu Z, Lin H, Tan X, et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine. 2020;59:102960. doi: 10.1016/j.ebiom.2020.102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu J, Liu X, Liu J, Liao H, Long S, Zhou N, et al. Coronavirus Disease 2019 Test Results after Clinical Recovery and Hospital Discharge among Patients in China. JAMA Netw Open. 2020;3:e209759. doi: 10.1001/jamanetworkopen.2020.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang J, Ding N, Ren L, Song R, Chen D, Zhao X, et al. COVID-19 reinfection in the presence of neutralizing antibodies. Natl Sci Rev. 2021;8(4):4–9. doi: 10.1093/nsr/nwab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wong J, Koh WC, Momin RN, Alikhan MF, Fadillah N, Naing L. Probable causes and risk factors for positive SARS-CoV-2 test in recovered patients: evidence from Brunei Darussalam. J Med Virol. 2020;92:2847–51. doi: 10.1002/jmv.26199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Abu-Raddad LJ, Chemaitelly H, Malek JA, Ahmed AA, Mohamoud YA, Younuskunju S, et al. Assessment of the Risk of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Reinfection in an Intense Reexposure Setting. Clin Infect Dis. 2021;73(7):e1830–e1840. doi: 10.1093/cid/ciaa1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pan L, Wang R, Yu N, Hu C, Yan J, Zhang X, et al. Clinical characteristics of re-hospitalized COVID-19 patients with recurrent positive SARS-CoV-2 RNA: a retrospective study. Eur J Clin Microbiol Infect Dis. 2021;40(6):1245–1252. doi: 10.1007/s10096-020-04151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pilz S, Chakeri A, Ioannidis JPA, Richter L, Theiler-Schwetz V, Trummer C, et al. SARS-CoV-2 re-infection risk in Austria. Eur J Clin Invest. 2021;51(4):1–7. doi: 10.1111/eci.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu R, Jiang Z, Gao H. Positive RT-PCR Test Results in Patients Recovered from COVID-19. JAMA. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Salehi-Vaziri M, Pouriayevali MH, Fotouhi F, Jalali T, Banifazl M, Farahmand B, et al. SARS-CoV-2 re-infection rate in Iranian COVID-19 cases within one-year follow-up. Microb Pathog. 2021;1(161):105296. doi: 10.1016/j.micpath.2021.105296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rahman S, Rahman MM, Miah M, Begum MN, Sarmin M, Mahfuz M, et al. COVID-19 reinfections among naturally infected and vaccinated individuals. Sci Rep. 2022;12(1):1–10. doi: 10.1038/s41598-022-05325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Luo S, Guo Y, Zhang X, Xu H. A follow-up study of recovered patients with COVID-19 in Wuhan, China. Int J Infect Dis. 2020;99:408–409. doi: 10.1016/j.ijid.2020.05.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cento V, Colagrossi L, Nava A, Anna Lamberti SS. Persistent positivity and fluctuations of SARS-CoV-2 RNA in clinically recovered COVID-19 patients. Elsevier. 2020;314:513–517. doi: 10.1016/j.jinf.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dong Y, Huang H, Yang J, Yang L. Retrospective analysis on the clinical characteristics of patients who were reinfected with the Corona Virus in 2019. Am J Transl Res. 2021;13(5):5505. [PMC free article] [PubMed] [Google Scholar]
- 127.Li Y, Hu Y, Yu Y, Zhang X, Li B, Wu J, et al. Positive result of Sars-Cov-2 in faeces and sputum from discharged patients with COVID-19 in Yiwu, China. J Med Virol. 2020;92:1938–47. doi: 10.1002/jmv.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Al-Tawfiq JA, Rabaan AA, Al-Omari A, al Mutair A, Al-Qahtani M, Tirupathi R. Learning from SARS and MERS: COVID-19 reinfection where we stand. Travel Med Infect Dis. 2021;41:1020. doi: 10.1016/j.tmaid.2021.102024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pilz S, Theiler-Schwetz V, Trummer C, Krause R, Ioannidis JPA. SARS-CoV-2 reinfections: overview of efficacy and duration of natural and hybrid immunity. Environ Res. 2022;209:112. doi: 10.1016/j.envres.2022.112911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Galanti M, Shaman J. Direct observation of repeated infections with endemic coronaviruses. J Infect Dis. 2021;223(3):409–415. doi: 10.1093/infdis/jiaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mattiuzzi C, Henry BM, Sanchis-Gomar F, Lippi G. Sars-cov-2 recurrent rna positivity after recovering from coronavirus disease 2019 (COVID-19): a meta-analysis. Acta Biomed. 2020;91(3):1–7. doi: 10.23750/abm.v91i3.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Azam M, Sulistiana R, Ratnawati M, Fibriana AI, Bahrudin U, Widyaningrum D, et al. Recurrent SARS-CoV-2 RNA positivity after COVID-19: a systematic review and meta-analysis. Sci Rep. 2020;10(1):20692. doi: 10.1038/s41598-020-77739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bal ZS, Ozkul A, Bilen M, Kurugol Z, Ozkinay F. The longest infectious virus shedding in a child infected with the G614 strain of SARS-COV-2. Pediatr Infect Dis J. 2021;40(7):E263–E265. doi: 10.1097/INF.0000000000003158. [DOI] [PubMed] [Google Scholar]
- 134.CDC. Ending Isolation and Precautions for People with COVID-19: Interim Guidance. 2022. Available from: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html [cited 2022 Sep 25].
- 135.Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21(1):52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Larson D, Brodniak SL, Voegtly LJ, Cer RZ, Glang LA, Malagon FJ, et al. A case of early reinfection with severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2021;73:2827–8. doi: 10.1093/cid/ciaa1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dufailu OA, Afriyie-Asante A, Gyan B, Kwabena DA, Yeboah H, Ntiakoh F, et al. COVID-19 in Africa: an ovarian victory? J Ovarian Res. 2021;14:1–3. doi: 10.1186/s13048-021-00820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Haider N, Osman AY, Gadzekpo A, Akipede GO, Asogun D, Ansumana R, et al. Lockdown measures in response to COVID-19 in nine sub-Saharan African countries. BMJ Global Health. 2020;5:e003319. doi: 10.1136/bmjgh-2020-003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.WHO. African countries scale back on COVID-19 measures. 2022. Available from: https://www.afro.who.int/news/african-countries-scale-back-covid-19-measures [cited 2022 Jul 1].
- 140.Wirsiy FS, Nkfusai CN, Ako-Arrey DE, Yeika EV, Manjong FT, Esther DK, et al. COVID–19 pandemic: implementing control measures in Africa using the “SHEF2” model. Pan Afr Med J. 2020;37:3. doi: 10.11604/pamj.supp.2020.37.1.24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nsubuga P, White ME, Thacker SB, Anderson MA, Blount SB, Broome C V, et al. Public Health Surveillance: A Tool for Targeting and Monitoring Interventions “What gets measured gets done.”-Anonymous. In: Disease Control Priorities in Developing Countries. 2006. Available from: www.who.int/csr/ihr/howtheywork/faq/en/#draft.
- 142.Murray J, Cohen AL. Infectious Disease Surveillance. In: International Encyclopedia of Public Health. 2nd ed. Geneva: Elsevier Inc.; 2016. p. 222–9.
- 143.Adepoju P. Closing Africa’s wide COVID-19 testing and vaccination gaps. Lancet Microbe. 2021;2(11):e573. doi: 10.1016/S2666-5247(21)00284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Seidu AA, Hagan JE, Ameyaw EK, Ahinkorah BO, Schack T. The role of testing in the fight against COVID-19: Current happenings in Africa and the way forward. Int J Infect Dis. 2020;98:237–40. doi: 10.1016/j.ijid.2020.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chitungo I, Dzobo M, Hlongwa M, Dzinamarira T. COVID-19: Unpacking the low number of cases in Africa. Public Health in Practice. 2020;1:100038. doi: 10.1016/j.puhip.2020.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kobia F, Gitaka J. COVID-19: Are Africa’s diagnostic challenges blunting response effectiveness? AAS Open Res. 2020;17(3):4. doi: 10.12688/aasopenres.13061.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bwire GM, Majigo MV, Njiro BJ, Mawazo A. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: a systematic review and meta-analysis. J Med Virol. 2021;93:719–25. doi: 10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nasiri K, Dimitrova A. Comparing saliva and nasopharyngeal swab specimens in the detection of COVID-19: a systematic review and meta-analysis. J Dental Sci. 2021;16:799–805. doi: 10.1016/j.jds.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang H, Liu Q, Hu J, Zhou M, Yu MQ, Li KY, et al. Nasopharyngeal Swabs Are More Sensitive Than Oropharyngeal Swabs for COVID-19 Diagnosis and Monitoring the SARS-CoV-2 Load. Front Med (Lausanne) 2020;18:7. doi: 10.3389/fmed.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang X, Tan L, Wang X, Liu W, Lu Y, Cheng L, et al. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int J Infect Dis. 2020;1(94):107–109. doi: 10.1016/j.ijid.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hitzenbichler F, Bauernfeind S, Salzberger B, Schmidt B, Wenzel JJ. Comparison of throat washings, nasopharyngeal swabs and oropharyngeal swabs for detection of sars-cov-2. Viruses. 2021;13(4):653. doi: 10.3390/v13040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Piri SM, Edalatfar M, Shool S, Jalalian MN, Tavakolpour S. A systematic review on the recurrence of SARS-CoV-2 virus: frequency, risk factors, and possible explanations. Infect Dis. 2021;53:315–24. doi: 10.1080/23744235.2020.1871066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dhillon RA, Qamar MA, Gilani JA, Irfan O, Waqar U, Sajid MI, et al. The mystery of COVID-19 reinfections: a global systematic review and meta-analysis. Ann Med Surg. 2021;72:103130. doi: 10.1016/j.amsu.2021.103130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Search strategy. Table S2. NIH Quality assessment tool questions.
Data Availability Statement
The authors declare that the data supporting the findings of this study are presented within the article and its supplementary materials.