Abstract
Background
Phylogenetic relationships of the genus Hapalemur remains controversial, particularly within the Hapalemur griseus species group. In order to obtain more information on the taxonomic status within this genus, and particularly in the cytogenetic distinct subspecies group of Hapalemur griseus, 357 bp sequence of cytochrome b and 438 bp of 12S mitochondrial DNAs were analyzed on a sample of animals captured in areas extending from the north to the south-east of Madagascar. This sample covers all cytogenetically defined types recognized of the genus Hapalemur.
Results
Phylogenetic trees and distances analyses demonstrate a first emergence of Hapalemur simus followed by H. aureus which is the sister clade of the H. griseus subspecies. Hapalemur griseus is composed of 4 subspecies separated into two clades. The first contains H. g. griseus, H. g. alaotrensis and H. g. occidentalis. The second consists of H. g. meridionalis. A new chromosomal polymorphic variant from the region of Ranomafana, H. griseus ssp, has been analysed and was found in both clades.
Conclusions
Our results support the raising of H. g. meridionalis to the specific rank H. meridionalis, while neither cytogenetic nor molecular evidences support the raising of H. g. alaotrensis to a species rank despite its morphological characteristics. The new cytotype H. g. ssp which has been previously characterized by cytogenetic studies contains animals clustering either with the group of Hapalemur griseus griseus or with that of Hapalemur meridionalis. This suggests the existence of an ancestral polymorphism or an introgression of mitochondrial DNA between subspecies.
Background
The genus Hapalemur of the Malagasy Prosimian family Lemuridae comprises three clearly different species: Hapalemur griseus[1], H. simus[2] and H. aureus[3]. Classification of different types within the H. griseus group has been controversial, particularly in the light of the variations existing in coat colour [4]. Systematic cytogenetic studies led to the description of four subspecies of H. griseus characterised by different karyotypes: H. g. griseus (HGG) (2N = 54) [5,6], initially considered as the most widespread and supposed to live in the Eastern forest of Madagascar from the north to the south [7]; H. g. occidentalis (HGO) (2N = 58) [6] restricted in the north-west and the eastern part of Madagascar as east as Maroantsetra [4,8]; H. g. meridionalis (HGM) (2N = 54) [9] found in the south, near Fort Dauphin, and differing from H. g. griseus in one metacentric chromosome ; H. g. alaotrensis (HGA) (2N = 54) [6], restricted to the bed of the Alaotra lake, with a karyotype similar to HGG containing more juxtacentromeric heterochromatin, and characterized by a larger body size [4]. In addition, a new variant of HG, H. griseus ssp (HGssp), has recently been described. This type is characterized by a chromosomal polymorphism (2N = 54, 55 and 56) and is living in the area of Ranomafana-Kianjavato [8].
The comparison of the karyotypes of the different Hapalemur led to a phylogenetic tree characterised by an earlier emergence of H. simus (HSI), followed by H. aureus (HAU) which represents the sister clade of the different groups of the H. griseus subspecies [8,10]. As molecular studies were fruitfully used for taxonomic investigations of several taxa, including lemurs [11-15], and as no large morphological differences could be found between the cytogenetically close subspecies of H. griseus, mitochondrial cytochrome b and 12S DNA analyses were performed in order to determinate more accurately their species and/or subspecies status.
Results
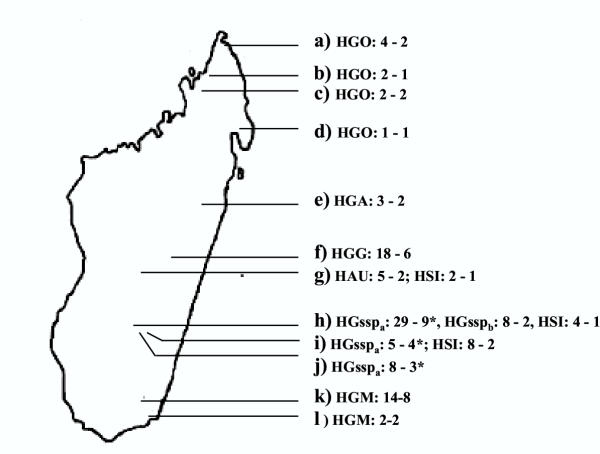
A total of 115 animals covering all the genus Hapalemur were captured in bamboo forests extending from the north to the south of Madagascar. The different capture areas of each species and subspecies as the number of animals are indicated in Fig. 1.
Figure 1.
Map of Madagascar showing the capture locations and/or the origin of the different Hapalemur species and subspecies, a) Analamera, b) Ambato, c) Ambakoany, d) Maroantsetra, e) Alaotra lake, f) Maromiza, g) Tsimbazaza zoo, h) Ranomafana, i) AmbolomavoJ) Kianjavato, k) Andohahela, l) Mandena. Abbreviations : HSI = Hapalemur simus, HAU = H. aureus, HGM = H. griseus meridionalis, HGsspb= H. griseus sspb, HGsspa= H. griseus sspa, HGG = H. g. griseus, HGA = H. g. alaotrensis, HGO = H. g. occidentalis. The first number behind each taxon represents the number of animals captured and the second, the number of haplotype found in this area. *The haplotype HGsspa02 is present in h, i and j; Hgsspa03 in h and j; Hgsspa07 in h and i; HGsspa10 in h and i.
The PCR amplification of the cytochrome b resulted in a 357 bp fragment. Alignments using blast search demonstrated that this fragment corresponds to the beginning of the mitochondrial cytochrome b gene. All the sequences were aligned and the substitutions were scored. Animals sharing the same sequences were regrouped into a same haplotype. These different haplotypes are summarised in Fig. 1 and table 2. Consensus sequences constructed with the most common sequence in each species and subspecies (not shown) demonstrated that transition substitutions occur more frequently than transversions (an average of eight transitions for 1 transversion) and gaps are absent. As expected [16], most of the substitutions occur on the third position of the codon (54 out of 64 substitutions), while the first position changed nine times and the second once. The portion amplified comprises the four invariant codons considered necessary for cytochrome b function [17]. Changes were sometimes noticed in the third base of the codon in these positions, but the amino acid remains the same so that the functionality of the cytochrome b is not affected.
Table 2.
Haplotypes and GenBank accession numbers of Hapalemur cytochrome b sequences.
| Species and subspecies/number of individuals | Haplotype/number of individuals | GenBank accession numbers |
| Hapalemur simus (HSI)/14 | 01/3 | AJ428977 |
| 02/1 | AJ428978 | |
| 03/4 | AJ428979 | |
| 04/6 | AJ428980 | |
| Hapalemur aureus (HAU) / 5 | 01/4 | AJ428957 |
| 02/1 | AJ428958 | |
| Hapalemur griseus meridionalis (HGM)/16 | 01/1 | AJ428959 |
| 02/1 | AJ428960 | |
| 03/1 | AJ428961 | |
| 04/1 | AJ428962 | |
| 05/1 | AJ428963 | |
| 06/1 | AJ428964 | |
| 07/1 | AJ428965 | |
| 08/7 | AJ428966 | |
| 09/1 | AJ428967 | |
| 10/1 | AJ428968 | |
| Hapalemur griseus ssp (HGssp) / 50 | 01/7 | AJ429054 |
| 02/12 | AJ429055 | |
| 03/3 | AJ429056 | |
| 04/2 | AJ429057 | |
| 05/6 | AJ429058 | |
| 06/1 | AJ429059 | |
| 07/6 | AJ429060 | |
| 08/5 | AJ429061 | |
| 09/1 | AJ429062 | |
| 10/2 | AJ429063 | |
| 11/2 | AJ429064 | |
| 12/1 | AJ429065 | |
| 13/1 | AJ429066 | |
| 14/1 | AJ429067 | |
| Hapalemur griseus alaotrensis (HGA) / 3 | 01/1 | AJ428969 |
| 02/2 | AJ428970 | |
| Hapalemur griseus griseus HGG)/18 | 01/1 | AJ428971 |
| 02/12 | AJ428972 | |
| 03/1 | AJ428973 | |
| 04/1 | AJ428974 | |
| 05/1 | AJ428975 | |
| 06/2 | AJ428976 | |
| Hapalemur griseus occidentalis (HGO) / 9 | 01/2 | AJ428982 |
| 02/2 | AJ428983 | |
| 03/1 | AJ428984 | |
| 04/1 | AJ428985 | |
| 05/2 | AJ428986 | |
| 06/1 | AJ428987 | |
| Eulemur macaco flavifrons (EMF)/11 | 01/11 | AJ428981 |
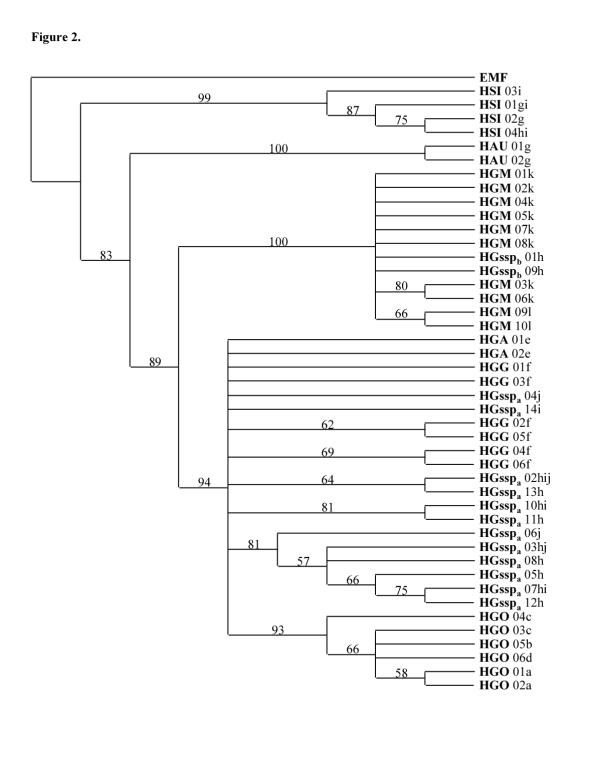
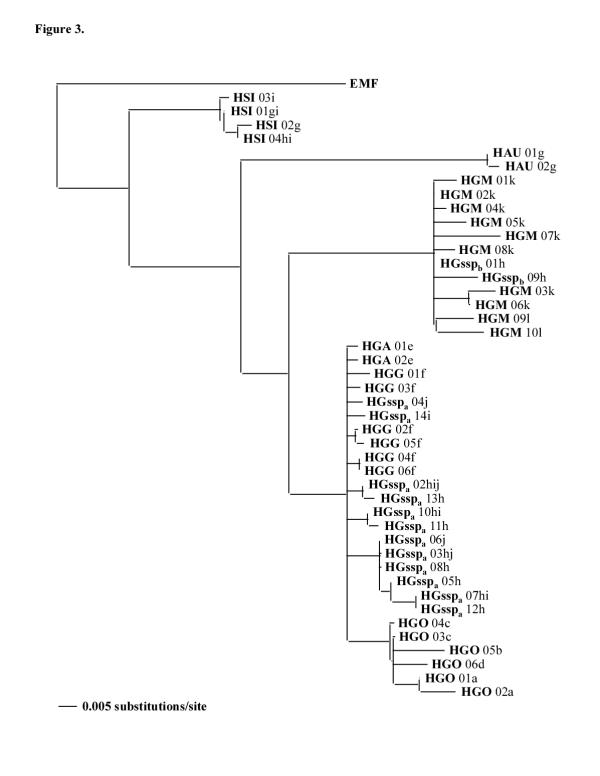
Kimura two parameter distances [18] and absolute distances calculated with the different haplotypes (Table 1) as phylogenetic trees constructed with the computer program Pylogenetic Analysis Using Parsimony (PAUP) 4.0b.4a [19] (Figs 2, 3, 4, 5) were used to compare the different Hapalemur species and subspecies. Neighbor-joining analyses of cytochrome b haplotypes demonstrated that H. simus emerged first followed by H. aureus which is the sister clade of the H. griseus subspecies. Inside H. griseus subspecies, two subclades are present and the H. griseus ssp (HGssp) are distributed in both the subclades (Figs 2, 3). For this reason, the H. griseus ssp were separated in HGsspa and Hgsspb. The first subclade (S1) contains HGA, HGG, HGsspa and HGO. The second subclade (S2) is composed of HGM and HGsspb. Maximum parsimony analyses (data not shown) demonstrated the same topology, unless the bootstrap support for S1 dropped from 94 to 66.
Table 1.
Lower and higher values of Kimura two parameters distances (under the diagonal) and absolute distances (above the diagonal) between haplotypes of Hapalemur species and subspecies. For abbreviations see Figure 1.
| HSI | HAU | HGM | HGsspb | HGA | HGG | HGsspa | HGO | |
| HSI | 0–7 0–0.01991 | 36 42 | 38 46 | 38 42 | 30 35 | 30 36 | 30 39 | 31 40 |
| HAU | 0.11024 0.13002 | 0–2 0–0.00563 | 41 48 | 42 46 | 31 34 | 30 36 | 30 36 | 34 39 |
| HGM | 0.11797 0.14524 | 0.12893 0.15338 | 0–9 0–0.02565 | 0 10 | 23 28 | 21 29 | 21 34 | 25 32 |
| HGsspb | 0.11797 0.13085 | 0.13290 0.14524 | 0.00000 0.02855 | 0–5 0–0.01414 | 22 26 | 21 27 | 21 32 | 25 30 |
| HGA | 0.09098 0.11075 | 0.09431 0.10393 | 0.06558 0.08459 | 0.06558 0.07747 | 0–1 0–0.00281 | 1 3 | 2 9 | 4 10 |
| HGG | 0.09098 0.11075 | 0.09098 0.11075 | 0.06240 0.08789 | 0.06240 0.08092 | 0.00281 0.00847 | 0–4 0–0.01133 | 3 10 | 5 11 |
| HGsspa | 0.09098 0.12087 | 0.09098 0.11049 | 0.06240 0.10473 | 0.06240 0.09719 | 0.00563 0.02580 | 0.00847 0.02875 | 0–10 0–0.02868 | 5 13 |
| HGO | 0.09431 0.12469 | 0.10445 0.12117 | 0.07522 0.09767 | 0.07522 0.09055 | 0.01133 0.02883 | 0.01421 0.03180 | 0.01421 0.03781 | 0–6 0–0.01705 |
Figure 2.
Phylogenetic tree based on 357 bp sequences of the cytochrome b gene. Bootstrap method with neighbor joining search and Kimura two parameter distance correction obtained with 10000 replications are used. Only nodes with a bootstrap value greater than 50% are indicated. The tree is rooted using an Eulemur macaco flavifrons (EMF) sequence and the comparisons are made between the haplotypes. The letter behind each taxon indicates the capture locality. For abbreviations see Figure 1.
Figure 3.
Phylogram based on 357 bp sequences of the cytochrome b gene. Bootstrap method with neighbor joining search and Kimura two parameter distance correction obtained with 10000 replications are used. The tree is rooted using an Eulemur macaco flavifrons (EMF) sequence and the comparisons are made between the haplotypes. The letter behind each taxon indicates the capture locality. For abbreviations see Figure 1.
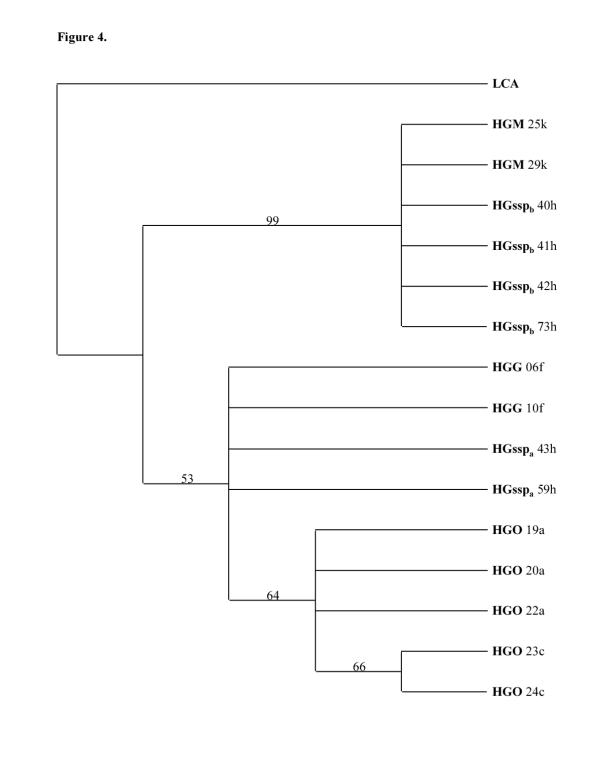
Figure 4.
Phylogenetic tree based on a 438 bp 12S mitochondrial DNA sequences. Bootstrap method with neighbor joining search and Kimura two parameter distance correction obtained with 10000 replications are used. Only nodes with a bootstrap value greater than 50% are indicated. The tree is rooted using a Lemur catta (LCA) sequence (EMBL, accession n°A Y012130) The letter behind each taxon indicates the capture locality. For abbreviations see Figure 1.
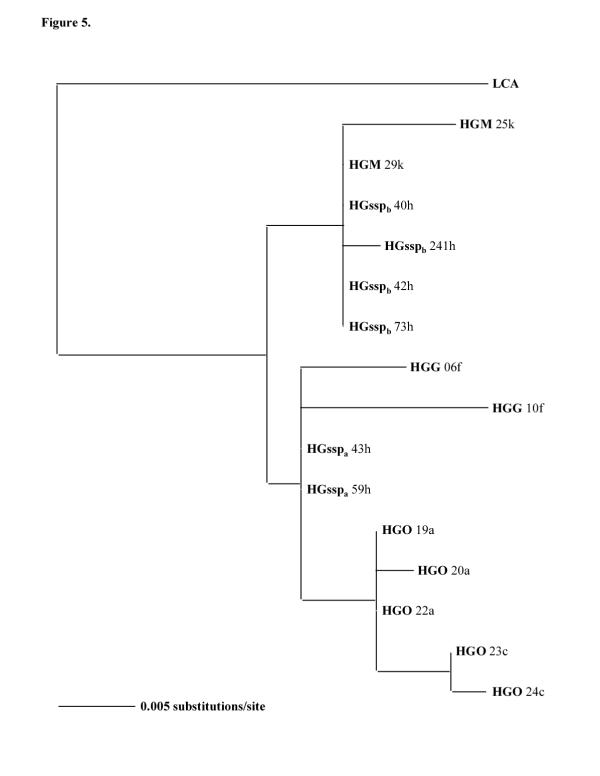
Figure 5.
Phylogram based on a 438 bp 12S mitochondrial DNA sequences. Bootstrap method with neighbor joining search and Kimura two parameter distance correction obtained with 10000 replications are used. The tree is rooted using a Lemur catta (LCA) sequence (EMBL, accession n°A Y012130) The letter behind each taxon indicates the capture locality. For abbreviations see Figure 1.
The use of the Kimura two parameter method (Table 1) shows inside S1 a very close distance between HGA and HGG (ranging from 0.00281 to 0.00847), while a larger distance occurs between HGG and HGsspa (0.00847 to 0.02875) as well as between HGG and HGO (0.01412 to 0.03180). In S2 (Table 1), short distances are observed between HGM and HGsspb (0–0.02855), (in this subclade, the sequence of HGM 02 and HGssp 01 are identical). Larger distances ranging from 0.10473 (HGsspa/HGM) to 0.06240 (HGM-HGsspb/HGG-HGsspa) separates S1 from S2 (Table 1).
In order to confirm the position of H. griseus ssp inside the H. griseus subclades, a portion of 12S mitochondrial DNA was amplified and sequenced. The resulted 438 bp fragments were aligned, and the sequences were analyzed with the program PAUP. The trees obtained with HGO, HGG, HGssp and HGM 12S sequences (Figs 4, 5) also demonstrated a separation of HGssp into two subclades. The first (HGsspa), which is composed of animals clustering with HGO and HGG when cytochrome b sequences are analyzed (Figs 2, 3), also clusters with HGO and HGG when 12S sequences are employed (Figs 4, 5). The second (HGsspb), constructed with animals clustering with HGM when cytochrome b sequences are analyzed (Figs 2, 3), also clusters with HGM when 12S sequences are employed (Figs 4, 5).
Discussion
During the last decade, comparisons of mitochondrial DNA sequences have been very useful for the analyses of the phylogenetic species relationships including lemurs [11-15,20]. In our study, cytochrome b and 12S mtDNA sequences were used, in order to clarify the position of each species and subspecies in the genus Hapalemur. Despite the fact that we analysed only short sequences, the genetic distances are considered to be relevant because of the large number of animals involved, since large sample size reduces errors in the estimation of evolutionary relatedness [11,21].
Comparisons of cytochrome b sequences demonstrated that pairwise genetic distances between H. simus and H. aureus and the other Hapalemur (distances ranging from 0.09098 to 0.15338) are the highest. The corresponding phylogram also shows that these taxa are well separated. Our molecular data thus strongly support the species status for H. simus, H. aureus and H. griseus.
Among the H. griseus, the genetic divergence observed between H. g. meridionalis and the HGG, HGA, HGsspa and HGO (ranging from 0.06240 to 0.10473) is markedly higher than all other values of the same level of intra H. griseus comparison (ranging from 0.00281 to 0.03781). The phylogram also clearly separates the H. g. meridionalis from other H. griseus. This high genetic divergence in the ranging of those observed between species of lemurs, would support the classification of H. g. meridionalis in a separate species "H. meridionalis" despite the relatively small chromosomal differences observed [9].
Lower distances (ranging from 0.01133 to 0.03781) are observed between H. griseus and H. g. occidentalis. Moreover, these two forms cluster in the same clade. So, despite the cytogenetic differences existing between these two forms, which differ by two chromosomal rearrangements [10], we propose the maintenance of the subspecies status for H. g. occidentalis.
On the basis of our analyses, H. g. alaotrensis is indistinguishable from H. g. griseus. The very short genetic distances found between H. g. griseus and H. g. alaotrensis (0.00281 to 0.00847) suggests a combination of these two groups into a single subspecies. However, the differences of the body sizes [4] and the differences in the content of heterochromatin found in both karyotypes [22] support the separation of H. g. griseus and H. g. alaotrensis in separate subspecies [23], but in no case in species apart as it has been previously proposed [24].
The systematic position of HGssp is more difficult to establish when cytogenetic and molecular data are compared. Cytogenetic data demonstrated the existence of an unique chromosomal polymorphic subspecies characterized by two karyotypes, 2N = 56 and 2N = 54, and their hybrids 2N = 55 [8]. In the view of molecular data, HGssp contains two groups, HGsspa and HGsspb, separated by an important genetic distance (0.06240–0.09719) in the range of those observed between species. Each of these two groups contains both karyotypes, 2N = 56 and 2N = 54, as well as their hybrids. The HGsspa are separated from HGG, HGA and HGO by a genetic distance in the range of those observed between subspecies (0.00563–0.03781), while HGsspb appears similar to HGM. Moreover, the HGsspb haplotypes are mixed with those of HGM. Taking into account these results, HGsspa should be considered as belonging to the group of HGG, HGA and HGO, and HGsspb as belonging to the group of HGM.
The existence of two well-separated clades among the Hapalemur griseus ssp originating from the Ranomafana region could possibly resulted from either recent mitochondrial DNA introgression or ancestral polymorphism. As the boundaries between HGM and HGssp are still unknown, we could hypothesize that the limits could be close to Ranomafana, allowing hybridization between these two forms. A transfer of mitochrondrial DNA from HGM into the HGssp population could have occurred through a matrilinear process resulting in the HGsspb haplotype. New investigations in areas located in the south of Ranomafana should thus allow the finding of HGM populations containing introgressed HGssp mitochondrial DNAs, unless this transfer occurs only from HGM to HGssp. A second hypothesis is that an ancestral polymorphic population containing both HGM and HGssp haplotypes and a chromosomal polymorphism have diverged in two separated populations. In the population of Ranomafana, the HGM and HGssp haplotypes as well as the chromosomal polymorphism were maintained. In the second population, only the HGM haplotype remained present, and a gain of a large block of heterochromatin gave rise to the karyotype characteristic of the HGM [9].
The comparisons of the phylogenetic trees based on mitochondrial DNA sequences with those previously obtained from cytogenetic data are only partially concordant. In both trees, H. simus emerges first, followed by H. aureus and then by the different H. griseus forms. The cytogenetic data allow to propose an evolutionary tree in which H. g. occidentalis emerges first, followed by H. g. griseus, and H. g. meridionalis[10], whereas on the tree based on cytochrome b and 12S sequences, H. g. meridionalis appears as a sister clade of the other H. griseus. This difference may be related to the short distances observed inside each clade which allowed no branching order.
Conclusions
Our molecular studies of the Hapalemur genus raise the question of the classification of H. g. meridionalis in the species status H. meridionalis. They also confirm the subspecies status of H. g. occidentalis and the absence of arguments in favour of the classification of H. g. alaotrensis as a separate species. The sequencing of the Hapalemur griseus ssp originating from Ranomafana reveals animals clustering either with the H. meridionalis or with the group of Hapalemur griseus griseus. No monophyletic clade could be determined in this new cytotype, so that the taxonomic status of the Hapalemur griseus ssp remains undefined. As our molecular data did not match the branching sequence within the H. griseus group based on cytogenetic data, further investigations including nuclear DNA will be necessary in order to resolve this issue.
Materials and methods
Animal studied
Six survey were organised from 1997 to 2001 allowing the capture of 115 animals in bamboo forests extending from the north to the south of Madagascar. Animals were captured using blowpipe projections and then sexed, weighted and measured. Skin samples were cut off under general anaesthesia with a 2 mg/kg injection of ketamine solution (Ketalar® Parke-Davis) and conserved deep frozen in liquid nitrogen. The different capture areas and the number of animals are indicated in Figure 1. As the H. griseus taxonomy is essentially based on cytogenetic criteria, karyotypes were made in order to confirm the species and subspecies rank. From the north to the south, the following subspecies were caught: a total of nine H. g. occidentalis (four at Analamera (a), two at Ambato (b), two at Ambakoany (c), and one at Maroantsetra (d)). Three H. g. alaotrensis at the Alaotra lake (e); 18 H. g. griseus at Maromiza (f); 50 H. griseus ssp. (37 at Ranomafana (h), five at Ambolomavo (i), eight at Kianjavato (j)); 16 H. g. meridionalis (14 in Andohahela (k) and two in Mandena (l)). In addition, 12 H. simus (four from Ranomafana (h), and eight from Ambolomavo (i)), as well as two H. simus and five H. aureus from the Zoological Park of Tsimbazaza (g) were studied. Animals were released at their capture location, immediately after recovery from anaesthesia, except two H. g. occidentalis captured in Ambato as well as three H. g. alaotrensis which are kept in the Zoological Park of Mulhouse and one H. g. occidentalis from Maroan setra which is kept in the private Zoological Park of Mandraka. As outgroup, we used a sample of Eulemur macaco flavifrons, from one of the individuals captured in the Sahamalaza forest for an earlier study [25].
DNA extraction
DNA samples were extracted from the skin biopsies using the standard proteinase K digestion followed by a phenol chloroform extraction as described by Sambrook et al.[26] with minor modifications. Small pieces of skin (~9mm2) were suspended in 200μl of extraction buffer (Tris 0.2M pH 8.4; KCl 0.5M; proteinase K: 5 mg/ml), and incubated at 37°C overnight. The samples were than heated at 95°C and mixed with an equal volume (200μl) of phenol/chloroform (1/1). After centrifugation (6 mn at 8500 g) the aqueous part was precipitated with 2 volumes of absolute ethanol at -20°C in presence of 1/10 (V/V) of ammonium acetate 5M. After centrifugation (5 mn at 8500 g), pellets were rinsed with 200μl of 70% ethanol and dried at 37°C. Pellets were resuspended in sterile double-distilled water and the concentration of the DNAs were measured by absorption at 260 nm. DNA samples were then stored at -30°C.
Amplification conditions
Cytochrome b
The polymerase chain reaction (PCR) was employed to generate a double-stranded fragment of 357 bp corresponding to a part of the mitochondrial cytochrome b gene. Each amplification was performed in the presence of 20 mM Tris-HCl pH 8.4, 50 mM KCl, 1.5mM MgCl2 (Gibco BRL), 200 μg BSA, 0.5 μM of each primer, 5U of Taq DNA polymerase (Perkin Elmer Cetus), 200 μM of each dNTP (Boehringer Mannheim) and 360ng of template DNA in a volume of 200 μl. The following primers derived from those described by Kocher et al.[27] were employed: Pr181 (5'-CCATCCAACATGTCAGCATGATGAAA-3') and Pr182 (5'-CCCTCAGAATGATATTTGTCCTCA-3'). Reactions were done in a Perkin Elmer Cetus DNA thermocycler 480 as follows: predenaturation (7 mn at 93 °C) and 35 cycles of denaturation (30 s at 94°C), annealing (45 s at 42°C), extension (1 mn 30 s at 72°C), followed by a final extension step (10 mn at 72°C).
12S mitochondrial DNA
The same PCR conditions than those described for the cytochrome b amplifications were applied, with a concentration of 2 mM MgCl2 and the following set of primers: Pr179 (5'-AAACTAGGATTAGATACCCTATTAT-3') and Pr180 (5'-AAGAGCGACGGGCGATGTGT-3'). Amplifications were performed in a Perkin Elmer Cetus DNA thermocycler 480 as follows: predenaturation (10 mn at 94°C) and 40 cycles of denaturation (30 s at 94°C), annealing (45 s at 45°C), extension (1 mn 30 s at 72°C), followed by a final extension step (10 mn at 72°C).
Sequences
Amplification products were electrophoresed on 1.3% agarose gels in TBE buffer (Tris base 87 mM, boric acid 89 mM, EDTA 2 mM, pH 8.0) in the presence of 0.5 μg/ml of ethidium bromide. Electrophoresis were performed at 200 mA for 2 h in TBE. Gels were than examined and photographed under UV light with a Polaroid system. The major bands on the gels were cut and centrifuged (8500 g 10 mn) in order to recover the DNA fragments [28]. The fragments were then precipitated with 0.1 volumes of ammonium acetate 5 M and 2 volumes of absolute ethanol at -20°C. After centrifugation (8500 g 10-mn), the pellets were air dried, resuspended in 50 μl of sterile bidistilled water and sequenced on an automatic ABI PRISM sequencer with the Taq dye deoxy terminator cycle sequencing kit. Each sample was sequenced from 5' to 3' and 3' to 5'.
Cytochrome b sequences were aligned and for each taxa, similar sequences were grouped under one haplotype. These haplotypes and the EMBL GenBank accession numbers are listed in table 2. EMBL GenBank accession numbers for the 12S sequences are listed in table 3.
Table 3.
GenBank accession numbers of Hapalemur 12S sequences.
| Species/subspecies | N° | GenBank accession numbers |
| Hapalemur griseus meridionalis (HGM) | 25 | AJ429205 |
| 29* | AJ429206 | |
| Hapalemur griseus ssp (HGssp) | 41 | AJ429207 |
| 43 | AJ429208 | |
| 59 | AJ429209 | |
| Hapalemur griseus griseus (HGG) | 06 | AJ429210 |
| 10 | AJ429211 | |
| Hapalemur griseus occidentalis (HGO) | 19 ** | AJ429212 |
| 20 | AJ429213 | |
| 23 | AJ429214 | |
| 24 | AJ429215 |
* 12S sequence of HGM29 is similar to HGssp40, HGssp42 and HGssp73. ** 12S sequence of HGO19 is similar to HGO22.
The aligned sequences were analysed using neighbor-joining and maximum parsimony methods with the computer program Pylogenetic Analysis Using Parsimony (PAUP) 4.0b.4a [19]. Genetic distances (d) measured in neighbor-joining analyses were calculated using the Kimura two parameter method [18] with the following formula: d=-1/21n [(1-2P-Q)Xsqr(1-2Q)], with P=transitions/positions scored and Q=transversions/positions scored. For neighbor-joining and maximum parsimony analyses, bootstraps of 10000 replicates were performed to examine the relative support of each relationship in the resultant topologies. Maximum parsimony trees were calculated via fast stepwise addition with random addition sequence.
Acknowledgments
Acknowledgements
This work was partially supported by the Association Européenne pour l'Etude et la Conservation des Lémuriens (A.E.E.C.L.), the DREIF-MENESR (EA3428), and the Institut de Génétique et de Biologie Moléculaire et Cellulaire (Pr. P. Chambon). The authors thank the Commission Tripartite of the Malagasy Government and the Ministère pour la Production Animale et des Eaux et Forêts for their permission to capture the animals, to take samples and for delivery CITES. Thanks are also due to Dr J. Ganzhorn for the correction of the manuscript, Mrs M. Lavaux for her secretarial assistance, Dr B. Ravaoarimanana and M. Hauwy for their helpful comments, and to S. Vicaire (IGBMC, Strasbourg) for sequencing the cytochrome b and 12S fragments. Special thanks for insight comments on the cladograms are due to Dr. D. Montagnon.
Contributor Information
Jean-Luc Fausser, Email: marguerite.lavaux@embryo-ulp.u-strasbg.fr.
Prosper Prosper, Email: marguerite.lavaux@embryo-ulp.u-strasbg.fr.
Giuseppe Donati, Email: gdonati@deee.unipi.it.
Jean-Baptiste Ramanamanjato, Email: jb-rama@dts.mg.
Yves Rumpler, Email: marguerite.lavaux@embryo-ulp.u-strasbg.fr.
References
- Link JA. Lemur griseus. Beytr Naturg. 1785. p. 65.
- Gray JE. Notes on Hapalemur (Prolemur) simus, a new species lately living in the gardens of the society. Proc Zool Soc Lond. 1870. pp. 828–831.
- Meier B, Albignac R, Peyrieras A, Rumpler Y, Wright P. A new species of Hapalemur (primates) from south east Madagascar. Folia Primatol. 1987;48:211–215. doi: 10.1159/000156299. [DOI] [PubMed] [Google Scholar]
- Petter JJ, Albignac R, Rumpler Y. Mammifères lémuriens (primates prosimiens). In: ORSTOM, CNRS, editor. Faune de Madagascar. Vol. 44 1977. [Google Scholar]
- Chu EHY, Swomley BA. Chromosomes of lemurine lemurs. Science. 1961;133:1925–1926. [PubMed] [Google Scholar]
- Rumpler Y, Albignac R. Cytogenetic study of the endemic Malagasy lemur: Hapalemur, I. Geoffroy, 1851. J Hum Evol. 1973;2:267–270. doi: 10.1002/ajpa.1330380220. [DOI] [PubMed] [Google Scholar]
- Tattersall I. The primates of Madagascar. New York, Columbia University Press. 1982.
- Rumpler Y, Prosper , Hauwy M, Rabarivola C, Rakotoarisoa G, Dutrillaux B. Chromosomal evolution of the Hapalemur griseus subspecies (Malagasy Prosimian), including a new chromosomal polymorphic variant. Chromosome Research (accepted) [DOI] [PubMed]
- Warter S, Randrianasolo G, Dutrillaux B, Rumpler Y. Cytogenetic study of a new subspecies of Hapalemur griseus. Folia Primatol. 1987;48:50–55. [Google Scholar]
- Rumpler Y, Warter S, Ishak B. Chromosomal evolution in prosimians. Hum Evol. 1989;4:157–170. [Google Scholar]
- Montagnon D, Ravaoarimanana B, Rumpler Y. Taxonomic relationships and sampling effects among Lepilemuridae and Lemuridae using partial cytochrome b gene. CR Acad Sci Paris/Life Sci Serie III. 2001;334:647–656. doi: 10.1016/S0764-4469(01)01331-2. [DOI] [PubMed] [Google Scholar]
- Montagnon D, Ravaoarimanana B, Rakotosamimana B, Rumpler Y. Ancient DNA from Megaladapis edwardsi (Malagasy Subfossil): Preliminary results using partial cytochrome b sequence. Folia Primatol. 2001;72:30–32. doi: 10.1159/000049916. [DOI] [PubMed] [Google Scholar]
- Pastorini J, Forstner MRJ, Martin RD. Relationships among Brown lemurs (Eulemur fulvus) based on mitochondrial DNA sequences. Mol Phylogenet Evol. 2000;16:418–429. doi: 10.1006/mpev.2000.0796. [DOI] [PubMed] [Google Scholar]
- Pastorini J, Martin RD, Ehresmann P, Zimmerman E, Forstner MRJ. Molecular phylogeny of the lemur family Cheirogaleidae (primates) based on mitochondrial DNA sequences. Mol Phylogenet Evol. 2001;19:45–56. doi: 10.1006/mpev.2000.0904. [DOI] [PubMed] [Google Scholar]
- Wyner Y, DeSalle R, Absher R. Phylogeny and character behaviour in the family lemuridae. Mol Phylogenet Evol. 2000;15:124–134. doi: 10.1006/mpev.1999.0723. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Vilgalys R, Ruvolo M. Molecular evolutionary dynamics of cytochrome b in strepsirrhine primates: the phylogenetic significance of third-position transvertion. Mol Biol Evol. 1996;13:1339–1350. doi: 10.1093/oxfordjournals.molbev.a025580. [DOI] [PubMed] [Google Scholar]
- Howell N, Gilbert K. Mutational analysis of the mouse mitochondrial cytochrome b gene. J Mol Biol. 1988;203:607–618. doi: 10.1016/0022-2836(88)90195-7. [DOI] [PubMed] [Google Scholar]
- Kimura MA. Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:11–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Swofford D. PAUP*4.0b.4a. Phylogenetic analysis using parsimony* (and other methods), Sunderland, MA, Sinauer Associates. 2000.
- Hayasaka K, Fujii K, Horai S. Molecular phylogeny of macaques: Implication of nucleotide sequences from a 896-base pair region of mitochondrial DNA. Mol Biol Evol. 1996;13:1044–1053. doi: 10.1093/oxfordjournals.molbev.a025655. [DOI] [PubMed] [Google Scholar]
- Martin AP, Bailey D, Kessing D, Palumbi SR. Accuracy of estimating genetic distances between species from short sequences of mitochondrial DNA. Mol Biol Evol. 1990;7:485–488. doi: 10.1093/oxfordjournals.molbev.a040621. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Dutrillaux B. Chromosomal evolution in Malagasy lemurs. III. Chromosome banding studies in the genus Hapalemur and the species Lemur catta. Cytogenet Cell Genet. 1978;21:201–211. doi: 10.1159/000130897. [DOI] [PubMed] [Google Scholar]
- Rumpler Y. The significance of chromosomal studies in the systematics of the Malagasy lemurs. In: Tattersall I, Sussman RW, editor. Lemur Biology . New York, Plenum Press; 1975. pp. 25–40. [Google Scholar]
- Groves CP. A theory of human and primate evolution. Oxford, Clarendon press. 1989.
- Fausser JL, Rabarivola C, Meir B, Hahn T, Rumpler Y. Genetic comparison between different populations of Eulemur macaco flavifrons in Northwest Madagascar using RAPD markers. Am J Primatol. 2000;51:249–255. doi: 10.1002/1098-2345(200008)51:4<249::AID-AJP4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor, Cold Spring Harbor press. 2 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989;86:6196–6200. [Google Scholar]
- Tautz D, Rentz M. An optimized free-squeeze method for the recovery of DNA fragments from agarose gels. Anal Bio chem. 1983;132:14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]