Abstract
We compared the phylogenetic compositions of marine planktonic archaeal populations in different marine provinces. Samples from eight different environments were collected at two depths (surface and aphotic zone), and 16 genetic libraries of PCR-amplified archaeal 16S rRNA genes were constructed. The libraries were analyzed by using a three-step hierarchical approach. Membrane hybridization experiments revealed that most of the archaeal clones were affiliated with one of the two groups of marine archaea described previously, crenarchaeotal group I and euryarchaeotal group II. One of the 2,328 ribosomal DNA clones analyzed was related to a different euryarchaeal lineage, which was recently recovered from deep-water marine plankton. In temperate regions (Pacific Ocean, Atlantic Ocean, and Mediterranean Sea) both major groups were found at the two depths investigated; group II predominated at the surface, and group I predominated at depth. In Antarctic and subantarctic waters group II was practically absent. The clonal compositions of archaeal libraries were investigated by performing a restriction fragment length polymorphism (RFLP) analysis with two tetrameric restriction enzymes, which defined discrete operational taxonomic units (OTUs). The OTUs defined in this way were phylogenetically consistent; clones belonging to the same OTU were closely related. The clonal diversity as determined by the RFLP analysis was low, and most libraries were dominated by only one or two OTUs. Some OTUs were found in samples obtained from very distant places, indicating that some phylotypes were ubiquitous. A tree containing one example of each OTU detected was constructed, and this tree revealed that there were several clusters within archaeal group I and group II. The members of some of these clusters had different depth distributions.
The past few decades of research in marine microbial ecology have revealed that prokaryotes are important components of the marine plankton. In addition to accounting for bulk biomass and activity, prokaryotes have central roles in mediating a variety of different biogeochemical cycles (2, 13). Determining the specific prokaryote composition of marine water, however, has been hindered by a lack of techniques for studying microbial community structure in situ. Therefore, little is known about which microbial species are responsible for the biomass and activities measured in the field and about the spatial distribution and temporal dynamics of these species. In the last few years, molecular techniques based on the use of 16S rRNA gene sequences as phylogenetic markers have begun to provide information about the identities of microorganisms in natural and complex systems (1, 49). The marine picoplankton assemblage was one of the first assemblages to be investigated, and the results obtained revealed that most marine prokaryotes were undescribed species that had not been cultivated (5, 8, 16, 32). Uncultured and undescribed microorganisms seem to be present and even dominant in many different environments (1, 36).
Of the different uncultivated organisms detected in marine plankton by molecular techniques, new types of archaea were perhaps the most unexpected. The Archaea, the Bacteria, and the Eucarya are the three lineages of life, and the Archaea is composed of the kingdom Crenarchaeota and the kingdom Euryarchaeota (50). Recently, a third kingdom, the Korarchaeota, has been proposed based on sequences retrieved from hot spring environments (3, 46). Cultured crenarchaeotes are extreme thermophiles and are generally obligate anaerobes with sulfur-dependent metabolism. However, many rRNA genes of apparently nonthermophilic crenarchaeotes have been retrieved from different ecosystems (6). Marine crenarchaeotes (referred to as group I archaea) have been found in different geographic areas and at different depths in the plankton community (8, 9, 15, 17, 26, 47) and also in marine and freshwater sediments (23, 24, 31, 46) and marine animals (28, 41). The sequences of group I archaea form a separate cluster related to clusters of sequences recovered from freshwater sediments (43), forest soils (22), and other terrestrial environments (4, 6, 21). The euryarchaeotes that have been cultured are metabolically more diverse than crenarchaeotes and include extreme halophiles, methanogens, and some sulfur-metabolizing thermophiles. Euryarchaeotal sequences have been retrieved from different marine plankton samples (8, 9, 16, 17, 26, 47) and form a cluster referred to as the group II archaea, which also includes sequences obtained from marine sediments (31) and the digestive tracts of fishes (47). Recently, a new euryarchaeotal group (referred to as group III archaea) has been detected in deep-water marine plankton (17). The sequences that are most similar to the group III archaea are sequences recovered in coastal marine sediments (33, 48).
A few studies dealing with the diversity, abundance, and ecology of marine planktonic archaea have been performed. These studies revealed that marine archaea could be important components of the prokaryotic assemblage and could account for up to 20 to 30% of the total picoplankton rRNA in Antarctic coastal waters (9, 27, 34, 35) and in Pacific coastal waters (26) and up to 40 to 60% of the prokaryote counts in California and Mediterranean waters (18). In some studies marine archaea exhibited temporal dynamics, and the level of these organisms decreased during the austral spring in Antarctic coastal waters (27, 34). They also exhibited spatial differences and generally were more abundant below the photic zone (17, 18, 26, 27). In the Santa Barbara Channel, each group dominated the archaeal assemblage in a different region of the water column (26), whereas the group II archaea were generally scarce in Antarctic waters (10, 27, 35). Archaeal composition has been studied by sequencing some clones in genetic libraries (8, 17, 26). However, in these studies the authors examined only a few marine areas, and the genetic libraries have not been systematically analyzed.
The aim of this study was to determine whether the patterns of archaeal distribution that have been observed are a general feature of the world's oceans or a peculiarity of the few samples analyzed to date. In order to do this, we collected samples from eight regions, including the Atlantic Ocean, the Pacific Ocean, the Southern Ocean, and the Mediterranean Sea, and at two depths (the surface and the aphotic zone). Libraries of archaeal 16S rRNA genes were constructed from these samples, thoroughly analyzed, and compared. Overall, we determined the marine archaeal group affiliations for approximately 100 clones per library by performing membrane hybridization experiments and the putative phylogenetic affiliations of approximately 40 clones per library by performing a restriction fragment length polymorphism (RFLP) analysis, as well as 48 new archaeal sequences. In this study we systematically examined the distribution of group I and group II archaea in different marine regions and at different depths (and the presence of other archaeal groups) and determined whether similar types of marine archaea were found in all systems and whether particular types had depth-specific distributions.
MATERIALS AND METHODS
Sampling and nucleic acid extraction.
Samples were collected from different marine locations (Table 1), including the Atlantic Ocean (North Atlantic and Cantabrian Sea), the Mediterranean Sea (Alboran Sea), the Pacific Ocean (Santa Barbara Channel), and the Southern Ocean (three stations across Drake Passage and a coastal station in the Antarctic Peninsula area). Seawater samples from different depths were collected with Niskin bottles attached to a rosette and a conductivity-temperature-depth sensor. The sample from Arthur Harbor, Antarctica, was collected and processed as described by DeLong et al. (10). Chlorophyll a concentrations were determined by fluorometry (37), and prokaryote abundance was determined by epifluorescence microscopy (40) or by flow cytometry (19). Microbial biomass was collected with 0.2-μm-pore-size Sterivex filter units (Durapore; Millipore) by filtering approximately 20 liters of seawater through a prefilter and the Sterivex filter unit in succession with a peristaltic pump. The prefilters used were 0.8-μm-pore-size polycarbonate filters and 1.0- and 1.6-μm-nominal-pore-size glass fiber filters (Table 1). The Sterivex units were filled with 1.8 ml of lysis buffer (40 mM EDTA, 50 mM Tris-HCl, 0.75 M sucrose) and stored at −20°C. Nucleic acids were extracted by digesting preparations with lysozyme, proteinase K, and sodium dodecyl sulfate, extracting the nucleic acids with phenol-chloroform-isoamyl alcohol, and then desalting and concentrating the nucleic acids with a Centricon-100 concentrator (26). The integrity of the DNA extracted was checked by agarose gel electrophoresis. DNA yields were quantified by a Hoescht dye fluorescence assay (38). Nucleic acid extracts were stored at −70°C until they were analyzed.
TABLE 1.
Marine regions sampled during the present study
| Marine region | Type of site | Maximal depth | Coordinates | Date(s) (mo/day/yr) | Size fraction analyzed | Library code |
|---|---|---|---|---|---|---|
| North Atlantic Ocean | Offshore | 2,850 | 59°30′N, 21°08′W | 6/14/98 | 0.2–1.6 μm | AT |
| Cantabrian Sea, Atlantic Ocean | Slope | 132 | 43°42′N, 6°09′W | 1/17/98 | 0.2–1.6 μm | CA |
| Alboran Sea, Mediterranean Sea | Offshore | 941 | 36°15′N, 4°15′W | 11/9/97 | 0.2–1.6 μm | ME |
| Santa Barbara Channel, Pacific Ocean | Coastal | 522 | 34°15′N, 119°54′W | 12/3/94 | 0.2–1.0 μm | SB |
| Drake Passage, north of subantarctic front | Offshore | 4,520 | 55°39′S, 58°02′W | 2/18/98 | 0.2–0.8 μm | DN |
| Drake Passage, subantarctic frontal region | Offshore | 3,461 | 56°18′S, 57°38′W | 2/17/98 | 0.2–0.8 μm | DF |
| Drake Passage, south of polar front | Offshore | 3,074 | 60°40′S, 54°53′W | 2/13/98 | 0.2–0.8 μm | DS |
| Arthur Harbor (Anvers Island), Antarctica | Coastal | 20 | 64°46′S, 64°04′W | 8/4/96, 8/16/96 | 30 kDa–1.6 μm | AM |
Ribosomal DNA (rDNA) clone libraries.
Archaeal 16S rRNA genes were amplified by PCR by using different combinations of archaeon-specific primers 20f, 21f, and 958r and universal primer 1392r (8, 28). Each PCR mixture (100 μl) contained 10 ng of natural DNA as a template, 10 to 15 pmol of each primer, 20 nmol of each deoxynucleoside triphosphate, 2.5 U of Taq DNA polymerase (GIBCO BRL), and the PCR buffer supplied with the enzyme. PCR were performed with a Genius (Techne) thermocycler by using the following conditions: an initial denaturation step consisting of 94°C for 3 min, 30 cycles consisting of 94°C for 45 s, 55°C for 45 s, and 72°C for 60 s, and a final elongation step consisting of 72°C for 5 min. The products of two to four independent PCR were combined before cloning in order to reduce the potential bias in separate reactions (39). The PCR fragments were cloned by using a TA cloning kit (Invitrogen) as recommended by the manufacturer. Between 100 and 300 putative positive colonies were transferred to multiwell plates (12 by 8 wells) containing Luria-Bertani medium with 7% glycerol and stored at −70°C.
Membrane hybridization experiments.
rDNA clones were transferred to three nylon membranes (Amersham) on Luria-Bertani agar and incubated overnight at 37°C, until visible colonies appeared. The colonies were lysed as previously described (26), and the nucleic acids were immobilized on the membranes by UV cross-linking. Each membrane was then hybridized overnight at 45°C with one of the following three probes (26): a universal probe for archaea (probe Arch-915), a probe specific for marine crenarchaeotal group I (probe GI-554), and a probe specific for marine euryarchaeotal group II (probe GII-554). The high-stringency wash temperatures were 56°C for Arch-915 and 40°C for GI-554 and GII-554. For some libraries (the SB and AM libraries [Table 2]), the probes were labeled with 32P, the hybridization and washing conditions were the conditions described previously (26), and the signal was detected by exposure to autoradiographic film. For the other libraries, the probes were labeled with fluorescein isothiocyanate, and for hybridization and washing we used the reagents and procedures recommended by Boehringer Mannheim. A colorimetric method was used to amplify and detect the hybridized probes. Membranes were incubated with an anti-fluorescein isothiocyanate antibody conjugated with the enzyme horseradish peroxidase (anti-fluorescein-POD Fab fragments; Boehringer Mannheim) and then with a chromogenic substrate (BM blue POD substrate, precipitating; Boehringer Mannheim), which in the presence of the enzyme precipitated and formed a permanent, dark blue spot.
TABLE 2.
Physical and biological parameters for the samples from which the libraries were generated
| Marine region | Depth (m) | Temp (°C) | Salinity (‰) | Chlorophyll concn (μg liter−1) | Prokaryote concn (105 cells ml−1) | DNA yield (μg liter−1) | Library(ies)a |
|---|---|---|---|---|---|---|---|
| North Atlantic Ocean | 5 | 11.8 | 35.33 | 0.85 | 4.18 | 1.30 | AT-5 |
| 200 | 8.8 | 35.29 | 0.04 | 1.16 | 0.33 | AT-200 | |
| Cantabrian Sea | 15 | 14.3 | 35.67 | 0.50 | 5.60 | 0.69 | CA-15 |
| Alboran Sea | 5 | 18.1 | 36.56 | 0.98 | 6.35 | 0.73 | ME-5 |
| 450 | 13.2 | 38.50 | 1.46 | 0.11 | ME-450 | ||
| Santa Barbara Channel | 0 | 13.9 | 33.41 | SB-0 | |||
| 200 | 9.0 | 34.05 | SB-200, SB-200B | ||||
| Drake Passage, north | 5 | 6.9 | 34.12 | 0.35 | 3.34 | 0.31 | DN-5 |
| 200 | 4.9 | 34.21 | 0.04 | 1.41 | 0.19 | DN-200 | |
| Drake Passage, frontal zone | 5 | 7.2 | 34.10 | 0.45 | 5.46 | 0.33 | DF-5 |
| 200 | 4.9 | 34.20 | 0.06 | 1.33 | 0.12 | DF-200 | |
| Drake Passage, south | 5 | 0.7 | 34.14 | 0.43 | 2.01 | 0.27 | DS-5 |
| 200 | 0.2 | 34.37 | 0.05 | 3.13 | 0.09 | DS-200 | |
| Arthur Harbor (4 August 1996)b | 20 | −1.8 | 33.70 | 0.10 | 1.50 | AM-20A | |
| Arthur Harbor (16 August 1996) | 20 | AM-20B |
Most libraries were derived from amplicons produced with primers 21f and 958r. The exceptions were the SB-200B library (derived from amplicons produced with primers 20f and 1392r) and the AM-20A and AM-20B libraries (derived from amplicons produced with primers 20f and 958r).
Average data for September 1996.
RFLP analysis and sequencing.
Archaeal inserts from selected clones were PCR amplified with primers 21f and 958r. The PCR products (length, approximately 915 bp) were subjected to separate enzymatic digestions overnight at 37°C with the tetrameric restriction enzymes HaeIII and RsaI (GIBCO BRL) and subsequently electrophoresed in a 2.5% low-melting-point agarose gel for 2 to 4 h at 60 to 80 V. A 50-bp DNA ladder (GIBCO BRL) was included in the gel, and the sizes of DNA fragments larger than 25 bp were determined; these fragments were used for analysis. RFLP analyses were performed separately for group I and group II clones (affiliations had been determined previously by membrane hybridization analysis). The RFLP patterns obtained with each enzyme were identified by using three-part designations; the first part indicated the enzyme (H, HaeIII; R, RsaI), the second part indicated the archaeal group (I, group I; II, group II), and the third part indicated the actual pattern (patterns A to T). Clones that produced identical patterns with both enzymes were grouped into discrete operational taxonomic units (OTUs), which were also identified by three-part designations; the first part indicated the affiliation of the clone with one of the two groups (I, group I; II, group II), the second part (a letter) indicated the RFLP pattern obtained with HaeIII, and the third part (a letter) indicated the RFLP pattern obtained with RsaI.
At least one clone that was representative of each OTU defined was partially sequenced with a Thermo Sequenase dye terminator cycle sequencing kit (Amersham). Sequences that were at least 631 bp long (Escherichia coli positions 45 to 737) were obtained for 26 group I clones, 21 group II clones, and 1 group III clone. Clones were designated by using the library code (Table 2) and a number in parentheses (which was preceded by P when the clone had mismatches with group I or group II probes). For the clones from the Santa Barbara libraries we used the designations which were submitted to GenBank (26), but we obtained longer sequences (SB95-1 to SB95-50 corresponded to the SB-0 library, and SB95-51 to SB95-90 corresponded to the SB-200B library). All sequences were subjected to the Ribosomal Database Project command CHECK_CHIMERA (25), and two of the sequences were found to be chimeric artifacts and were excluded from all analyses. The whole insert of group III clone ME-450 (P9) was sequenced.
Sequences that exactly matched the group I or group II probe sequences were retrieved from the GenBank database (searched on 7 May 1999). Nonthermophilic crenarchaeotes other than members of the marine cluster (6) always had some mismatches in the target regions of the probes and, therefore, were not selected. Two marine group III clones (17) were also retrieved. A GDE format file (all marine archaea) with the sequences in our libraries aligned with sequences retrieved from GenBank can be obtained through anonymous ftp at cucafera.icm.csic.es in the directory pub/massana.
Phylogenetic analysis.
Phylogenetic analyses were performed with the software GDE 2.2 and Treetool 2.0.1 obtained from the Ribosomal Database Project (25) and the software package Phylip, version 3.5 (11). Sequences were manually aligned by using a GDE file. Distance matrices were calculated with DNAdist (Phylip) by using the Kimura two-parameter model and by assuming that the transition/transversion ratio was 2.0. Trees were inferred by performing a neighbor-joining analysis (Phylip) and were edited with Treetool. A maximum-likelihood analysis was performed with DNAml (Phylip). A bootstrap neighbor-joining analysis was performed with 100 replicates with random taxon addition. A bootstrap maximum-likelihood analysis was performed with 50 replicates by using only 27 significant sequences.
A dendrogram based on the information obtained from the RFLP analysis was constructed. First, we designed a binary matrix with the values 1 and 0, which represented the presence and the absence of restriction sites in each OTU, respectively. The binary matrix was used to construct a similarity matrix with Jaccard's dichotomy coefficient with the software SYSTAT 5.2.1. The similarity matrix was converted to a distance matrix by subtracting each coefficient in the matrix from one. The distance matrix was then used to generate a dendrogram with the unweighted pair group method with arithmetic averages (UPGMA) implemented in the neighbor subprogram of Phylip and edited in Treetool.
Nucleotide sequence accession numbers.
Sequences were deposited in the GenBank database under the following accession numbers: AF223111 for clone AT-5 (1); AF223112 for AT-200 (1); AF223113 for AT-200 (7); AF223114 for CA-15 (P18); AF223115 for ME-450 (5); AF223116 for ME-450 (9); AF223117 for ME-450 (20); AF223118 for ME-450 (P3); AF223119 for ME-450 (P5); AF223120 for SB95-1; AF223121 for SB95-20; AF223122 for DN-5 (1); AF223123 for DN-200 (1); AF223124 for DS-5 (1); AF223125 for DS-5 (P21); AF223126 for AM-20A (101); AF223127 for AM-20A (102); AF223128 for AM-20A (103); AF223129 for AM-20A (104); AF223130 for AM-20A (117); AF223131 for AT-5 (21); AF223132 for AT-5 (P24); AF223133 for AT-200 (29); AF223134 for AT-200 (P25); AF223135 for CA-15 (22); AF223136 for CA-15 (23); AF223137 for CA-15 (27); AF223138 for CA-15 (32); AF223139 for CA-15 (P4); AF223140 for ME-450 (21); AF223141 for ME-450 (30); AF223142 for ME-450 (38); AF223143 for ME-450 (P14); AF223144 for SB95-35; AF223145 for SB95-48; AF223146 for SB95-87; AF223147 for DF-5 (21); AF223148 for AM-20A (122); AF223149 for AM-20A (123); and AF223150 for ME-450 (P9).
RESULTS
Sampling sites and genetic libraries generated.
We collected samples from eight different marine areas (Table 1), including the Atlantic Ocean, the Mediterranean Sea, the Pacific Ocean, and the Southern Ocean. Stations were located on the continental shelf and the continental slope and offshore. In general, we analyzed two depths, the surface and the aphotic zone. Some physical and biological parameters of the samples are shown in Table 2. The samples collected had wide ranges of temperatures (−1.8 to 18°C), salinities (33.37 to 38.50‰), surface chlorophyll a concentrations (0.10 to 0.98 μg liter−1), and prokaryote concentrations (1.16 × 105 to 6.35 × 105 cells ml−1). The planktonic microbial biomass in the samples was collected on filters, and the total nucleic acids were extracted. We determined (data not shown) that most free-living prokaryotes were present in the size fraction analyzed. The DNA yields ranged from 0.09 to 1.30 μg of DNA liter of seawater−1 (Table 2) and were several times greater in the surface water than in the deeper water at all stations. Nucleic acid extracts were used for PCR amplification of partial 16S rRNA genes in which archaeon-specific primers were used. Amplification was obtained for all of the samples tested, which confirmed that marine archaea were ubiquitous in the plankton. A genetic library of archaeal genes was generated for each sample (Table 2). Of the 16 libraries which we analyzed, 11 were new in this study, whereas the 5 other libraries have been described previously (SB libraries were described by Massana et al. [26]; AM libraries were described by DeLong et al. [10]). These libraries were analyzed and compared by using a hierarchical approach that included membrane hybridization, RFLP analysis, and sequence analysis.
Analysis of archaeal libraries.
In the membrane hybridization experiments, most of the archaeal clones hybridized with either group I or group II probes (Table 3), indicating that these groups accounted for the bulk of the marine archaea in our samples. The nine archaeal clones that did not hybridize with these probes were sequenced. Four of these clones were affiliated with group I, and four were affiliated with group II (Table 3) but exhibited between one and three mismatches in the target regions of the probes. The sequence of the remaining clone, ME-450 (P9), was very similar to two sequences belonging to group III marine archaea (17). The fact that only 8 of 2,327 group I or group II clones exhibited mismatches with their respective probes indicates that these probes are well suited for studying marine archaea and detecting new groups in plankton.
TABLE 3.
Analysis of the genetic libraries by membrane hybridization with archaeal, group I, and group II probesa
| Library | No. of clones belonging to:
|
OTU(s) of other archaea | |||
|---|---|---|---|---|---|
| Archaea | Group I | Group II | Other archaea | ||
| AT-5 | 95 | 22 | 72 | 1 | II-CC |
| AT-200 | 91 | 69 | 21 | 1 | II-JK |
| CA-15 | 198 | 93 | 103 | 2 | I-AA, II-EC |
| ME-5 | 74 | 7 | 67 | 0 | |
| ME-450 | 196 | 173 | 19 | 4 | I-BD, I-ND, II-JK, III-AA |
| SB-0 | 110 | 29 | 81 | 0 | |
| SB-200 | 276 | 237 | 39 | 0 | |
| SB-200B | 189 | 175 | 14 | 0 | |
| DN-5 | 89 | 84 | 5 | 0 | |
| DN-200 | 92 | 92 | 0 | 0 | |
| DF-5 | 85 | 47 | 38 | 0 | |
| DF-200 | 90 | 90 | 0 | 0 | |
| DS-5 | 92 | 90 | 1 | 1 | I-AA |
| DS-200 | 92 | 92 | 0 | 0 | |
| AM-20A | 276 | 273 | 3 | 0 | |
| AM-20B | 283 | 277 | 6 | 0 | |
| Total | 2,328 | 1,850 | 469 | 9 | |
We analyzed the relative abundance of group I and group II clones in different libraries (Table 3). Previously, we described the dominance of group II clones at the surface and the dominance of group I clones at a depth of 200 m in the Santa Barbara Channel (26) (Table 3). A similar trend was observed in the other two temperate marine areas examined (Table 3), the North Atlantic Ocean (77 and 24% group II clones at depths of 5 and 200 m, respectively) and the Mediterranean Sea (90 and 10% group II clones at depths of 5 and 450 m, respectively), whereas the sample from the Cantabrian Sea, which was obtained at a depth of 15 m, contained similar amounts of the two groups. The Southern Ocean samples, on the other hand, exhibited a different pattern. Previously, we observed a scarcity of group II clones in coastal Antarctic libraries (10) (Table 3). The data obtained for three stations in the Drake Passage were consistent with this finding; group I clones were the dominant clones at depths of 5 and 200 m in most cases, and group II clones were virtually absent (Table 3). The only exception was the DF-5 library, in which 45% of the clones belonged to group II. The conclusion that emerged from these data is that the group I archaea are the dominant archaea throughout the water column in the Southern Ocean and below the surface in temperate regions, whereas the group II archaea are the dominant archaea at the surface in temperate regions.
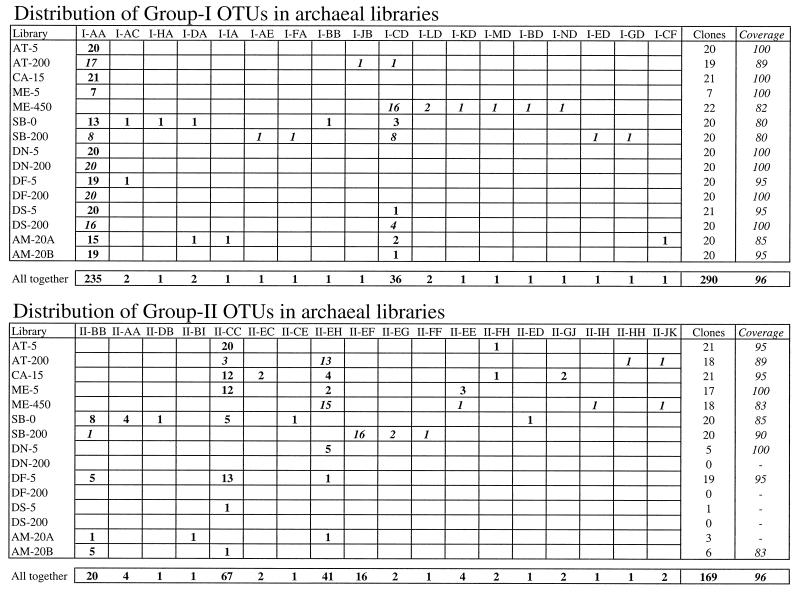
We randomly chose approximately 20 group I clones and 20 group II clones from each library and compared them by performing an RFLP analysis. Each clone was assigned to a single OTU based on the patterns obtained with two tetrameric restriction enzymes. We detected 18 different OTUs in the 290 group I clones analyzed and 18 different OTUs in the 169 group II clones analyzed (Fig. 1). The eight group I and II clones with mismatches in the target region (Table 3) are also included in Fig. 1. The distribution of OTUs in the different libraries revealed that a few OTUs were abundant and widespread, whereas most OTUs appeared only once. Coverage values (20), which were estimates of the proportion of the assemblage sampled by our approach, were always very high, even though only 20 clones were analyzed in most cases (Fig. 1). OTU I-AA dominated the group I clones in most libraries and represented 81% of all of the group I clones analyzed (Fig. 1). This OTU was widely distributed at the different sampling sites and at different depths (0 and 200 m). The second most abundant group I OTU was OTU I-CD (12% of group I clones), which dominated the ME-450 library, codominated the SB-200 library, and appeared in six other libraries (Fig. 1). The group II clones were grouped into four dominant OTUs. OTU II-CC was the most abundant OTU (40% of group II clones); this OTU appeared in most libraries and was the dominant OTU in four of the five surface libraries. OTU II-BB (12% of group II clones) was the dominant OTU in one surface library (SB-0). Deep-water libraries were dominated by OTU II-EH (AT-200 and ME-450) or OTU II-EF (SB-200), which represented 24 and 9% of all group II clones, respectively.
FIG. 1.
Distribution of group I and group II OTUs among the libraries analyzed. The figure shows the number of clones belonging to each OTU (in italics for deep libraries), the number of clones analyzed, and the coverage values for each library. The last row calculates the same parameters but considers all the clones in each group as belonging to the same assemblage.
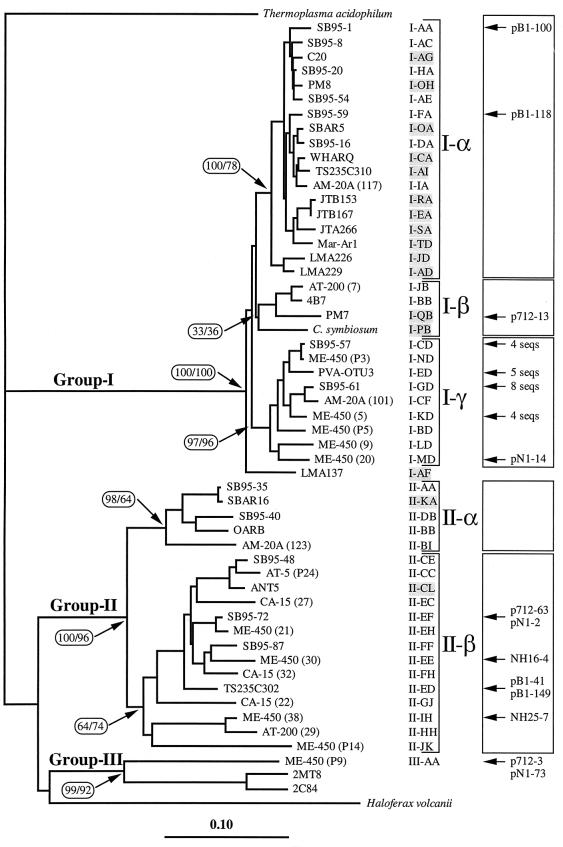
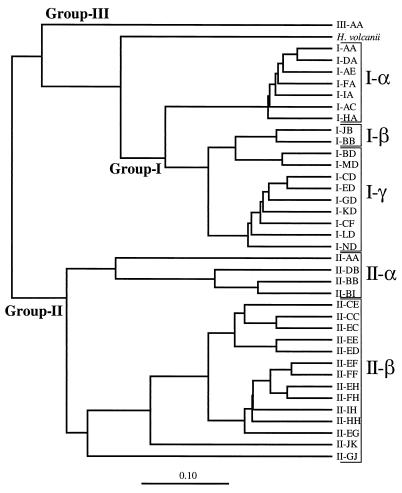
Finally, we partially sequenced one clone from each OTU and constructed a phylogenetic tree by performing a neighbor-joining analysis (Fig. 2). In this tree we also included sequences retrieved from GenBank which represented new OTUs after a computer-simulated RFLP analysis was performed (Table 4). The only OTU not represented in this tree was OTU II-EG; the corresponding sequence was too short and almost identical to the sequence of clone SB95-72. Since we chose one representative from each OTU described at the moment of the analysis (considering both clones from our libraries and clones from the GenBank database), we believe that this tree is fairly representative of the genetic diversity of marine archaea. In general, the differences among sequences belonging to group II were greater than the differences among sequences belonging to group I. For the group I sequences LMA137 was the most distant from the other clones, and there were three distinct clusters. Most sequences in cluster I-α produced either RFLP pattern H-I-A or RFLP pattern R-I-A, all of the sequences in cluster I-β produced RFLP pattern R-I-B, and most of the sequences in cluster I-γ produced RFLP pattern R-I-D. Group II sequences formed two clusters (Fig. 2). The robustness of the topology of the tree was confirmed by the results of a maximum-likelihood analysis (data not shown), and the only difference observed was poorer definition of cluster I-β. The bootstrap values obtained in the neighbor-joining and maximum-likelihood analyses revealed that all of the clusters except cluster I-β were very consistent (Fig. 2).
FIG. 2.
Phylogenetic tree for marine archaea inferred from DNAdist and a neighbor-joining analysis by using 631 bp (E. coli positions 45 to 737). One clone of each OTU is shown on the tree. OTUs that appear only in GenBank clones are shaded. The affiliations of the clones of Fuhrman et al. are indicated on the right (the four sequences most similar to OTU I-CD are pB1-47, pB1-80, pB1-124, and pB1-151; the five sequences most similar to OTU I-ED are pB1-22, NH49-8, nH49-14, p712-12, and p712-24; the eight sequences most similar to OTU I-GD are NH25-1, NH25-13, NH49-4, NH49-9, pB1-53, pB1-123, pN1-56, and p712-37; and the four sequences most similar to OTU I-KD are pB1-92, pN1-10, pN1-27, and pN1-43). Bootstrap values (percentages) for the neighbor-joining and maximum-likelihood analyses are indicated for the most significant nodes. Scale bar = 0.10 change per sequence position.
TABLE 4.
GenBank clones that include the sequence region from position 21 to position 958 and match group I or group II probes
| Clone or organism | OTU | Origin | Reference |
|---|---|---|---|
| SBAR5 | I-OAa | Marine plankton, surface | 8 |
| SBAR12 | I-AA | Marine plankton, surface | 8 |
| SBAR1A | II-EE | Marine plankton, surface | 8 |
| SBAR16 | II-KA | Marine plankton, surface | 8 |
| WHARQ | I-CA | Marine plankton, surface | 8 |
| WHARN | II-CL | Marine plankton, surface | 8 |
| OARB | II-BB | Marine plankton, surface | 9 |
| ANT12 | I-AA | Marine plankton, surface | 9 |
| ANT5 | II-CL | Marine plankton, surface | 9 |
| FB7 | I-BB | Marine plankton, 200 m | 4 |
| Cenarchaeum symbiosum | I-PB | Sponge endosymbiont | 41 |
| PVA-OTU2 | I-CD | Hydrothermal vent, 1,000 m | 31 |
| PVA-OTU3 | I-ED | Hydrothermal vent, 1,000 m | 31 |
| PVA-OTU4 | I-CD | Hydrothermal vent, 1,000 m | 31 |
| PVA-OTU1 | II-FH | Hydrothermal vent, 1,000 m | 31 |
| LMA137 | I-AF | Freshwater sediment, 100 m | 24 |
| LMA226 | I-JD | Freshwater sediment, 100 m | 24 |
| LMA229 | I-AD | Freshwater sediment, 100 m | 24 |
| LMA238 | I-AF | Freshwater sediment, 100 m | 24 |
| FF619 | II-EH | Marine fish digestive tract | 47 |
| FF620 | II-EH | Marine fish digestive tract | 47 |
| FIN625 | II-FH | Marine fish digestive tract | 47 |
| TS10C286 | I-CD | Marine particles | 47 |
| TS10C299 | I-CD | Marine particles | 47 |
| TS235C306 | I-CD | Marine particles | 47 |
| TS235C310 | I-AI | Marine particles | 47 |
| TS10C294 | II-FH | Marine particles | 47 |
| TS10C298 | II-EH | Marine particles | 47 |
| TS235C302 | II-ED | Marine particles | 47 |
| JTA47 | I-AA | Marine sediments, 6,400 m | —b |
| JTA266 | I-SA | Marine sediments, 6,400 m | — |
| JTB153 | I-RA | Marine sediments, 6,400 m | — |
| JTB167 | I-EA | Marine sediments, 6,400 m | — |
| JTB168 | I-CA | Marine sediments, 6,400 m | — |
| Mar-Ar-1 | I-TD | Marine sediments, 11,000 m | 23 |
| Mar-Ar-11 | I-TD | Marine sediments, 11,000 m | 23 |
| Mar-Ar-15 | I-RA | Marine sediments, 11,000 m | 23 |
| C6 | I-AA | Marine plankton, 500 m | — |
| C20 | I-AG | Marine plankton, 500 m | — |
| C33 | I-CA | Marine plankton, 500 m | — |
| C46 | I-AA | Marine plankton, 500 m | — |
| C48 | I-AA | Marine plankton, 500 m | — |
| PM7 | I-QB | Marine plankton, 500 m | — |
| PM8 | I-QH | Marine plankton, 500 m | — |
Boldface type indicates an OTU that was not represented in our libraries.
—, data obtained from GenBank (unpublished data).
The ecological significance of clusters was evaluated by determining the affiliation of surface and deep-water clones. Most surface group I clones in our libraries (160 of 169 clones) belonged to cluster I-α. A majority of the deep-water group I clones (83 of 121 clones) also belonged to cluster I-α, but a significant number of clones (37 clones) belonged to cluster I-γ and 1 clone belonged to cluster I-β. The majority of the clones in cluster I-γ (82% of the clones) belonged to deep-water libraries, suggesting that the members of this cluster are adapted to live in the aphotic zone. Most of the group II clones (88 of 113 surface clones and 55 of 56 deep-water clones) belonged to cluster II-β. Cluster II-α contained 25 surface clones and only one deep-water clone, and therefore the members of this cluster seem to be adapted to surface water. Cluster II-β could be subdivided into a subcluster that was formed by OTUs II-CC, II-CE, and II-EC and contained most of the surface clones (67 surface clones and 3 deep-water clones) and a subcluster which contained the 21 surface clones and 51 deep-water clones which belonged to the remaining OTUs in the cluster.
Some marine archaeal clones retrieved from GenBank were not assigned to any OTU since they did not include the 21/958 sequence region. These clones were found by Fuhrman et al. in 16S rDNA genetic libraries obtained with universal primers from deep-sea samples (15–17). Nevertheless, a 200-bp fragment of these clones (sometimes the whole sequence that was available) was aligned with our sequences, and a phylogenetic tree was constructed (data not shown): this allowed us to determine the positions of these clones in the tree shown in Fig. 2. Based on the short fragments compared, these clones were always very similar to some of the clones shown in the tree in Fig. 2, and their affiliation in clusters was consistent with the deep-sea origin of the libraries. Thus, a majority of the group I clones (22 of 25 clones) belonged to cluster I-γ, and all of the group II clones belonged to cluster I-β. Our group III sequence was very similar (98.2% similarity) to two sequences described previously (17), which clearly defined archaeal group III. The closest relatives of this group are environmental sequences retrieved from marine sediments (clones 2MT8 and 2C84 in Fig. 2 [33, 48]), although the distances were significantly greater (average level of similarity, 75.6%).
Validation of the RFLP analysis of archaeal libraries.
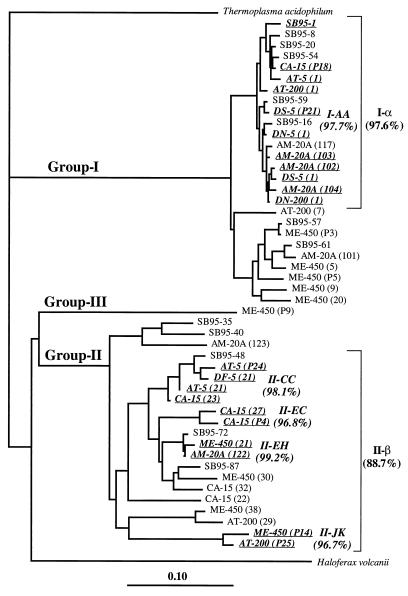
In this study we used a RFLP approach to define archaeal OTUs and to compare genetic libraries. In order to determine the phylogenetic consistency and breadth of the OTUs defined, different clones belonging to the same OTU were sequenced and added to a tree containing all of the clones in our libraries (Fig. 3). For the most abundant OTU, OTU I-AA, we sequenced 11 clones obtained from different places and depths (Fig. 3). All of the sequences obtained were very similar (97.7% similarity), and they were distributed throughout cluster I-α, which also included six other OTUs. Four OTU II-CC clones obtained from different places were sequenced, and they grouped together closely (98.1% similarity) (Fig. 3), like the other three examples examined. We estimated that clones belonging to the same OTU had an average level of similarity of 97.7%. This is a very high value, considering that a similarity value greater than 97% for the whole 16S rRNA gene is used to indicate that organisms belong to the same species. Our findings indicate that the RFLP analysis placed clones in the phylogenetic tree very consistently.
FIG. 3.
Phylogenetic analysis of all of the marine archaeal sequences in our libraries. The tree was constructed as described in the legend to Fig. 2. Clones belonging to the same OTU are underlined, and the average similarity values are indicated.
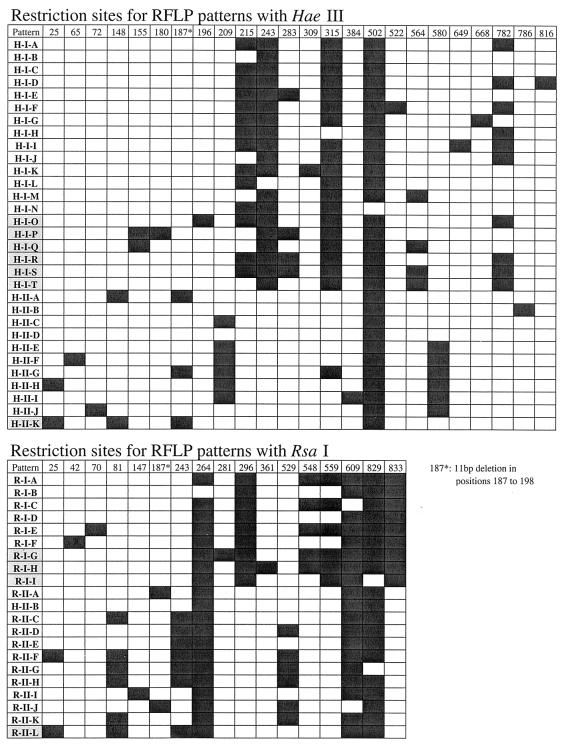
The RFLP analysis was also validated by constructing a dendrogram for the OTUs by using the information derived from RFLP analysis instead of sequences. Figure 4 shows a map of the restriction sites for each RFLP pattern obtained with both enzymes. We identified 20 group I patterns and 11 group II patterns with HaeIII and 9 group I patterns and 12 group II patterns with RsaI (Fig. 4). Then we constructed a dendrogram by considering the presence or absence of restriction sites in the OTUs detected in our libraries (Fig. 5). In this tree, the OTUs grouped together and formed a structure identical to that shown in Fig. 2; the three marine archaeal groups were clearly separated, and the same group I and group II clusters were identified. Only two OTUs (OTUs I-BD and I-MD) belonging to cluster I-γ (Fig. 2) appeared in cluster I-β instead (Fig. 5). This could be explained by the poorer definition of cluster I-β (lower bootstrap values in Fig. 2). Therefore, the results of this analysis indicated that the information obtained by RFLP analysis was powerful enough to group almost all clones in their corresponding clusters.
FIG. 4.
Map of restriction sites for the enzymes HaeIII and RsaI for the RFLP patterns detected in clones from our libraries and retrieved from the GenBank database (patterns exclusive of GenBank clones are shaded). For convenience, the positions of restriction sites are referred to the sequence of clone SB95-57. The DNA fragments that formed each RFLP pattern can be determined by subtracting two consecutive restriction sites (for example, pattern H-I-A was formed by the 215-, 28-, 72-, 187-, 280-, and 132-bp fragments).
FIG. 5.
Dendrogram generated by using the restriction sites of each OTU found in our libraries. A distance matrix was calculated from a binary matrix based on the presence of restriction sites, and the dendrogram was inferred by using the unweighted pair group method with arithmetic averages.
DISCUSSION
Significance and limitations of the methodological approach used.
Marine archaea obtained from different regions of the world's oceans were compared by analyzing genetic libraries of 16S rRNA genes. This approach has been widely used to describe the microbial diversity in different habitats (4, 5, 31) and allows workers to study uncultivated prokaryotes, such as the marine archaea (8, 15). However, it is known that clonal representation in genetic libraries can be biased, particularly due to the PCR step (39, 45), and may not reflect the relative levels of organisms in a sample (but see reference 12). The biases can affect the results at two levels; some sequences may be present but not be amplified, and particular types may be over- or underamplified. Using different primer sets (universal primers [17] and different combinations of archaeal primers [Table 3]) resulted in retrieval of similar archaeal phylotypes. This suggests that we amplified all of the archaeal phylotypes present in the samples, since it is unlikely that the different primer sets were biased against the same phylotypes. We studied the quantitative aspect of the libraries by monitoring the relative levels of group I and group II clones. In one case, parallel libraries (SB-200 and SB-200B) were examined with two primer sets, and similar results were obtained (Table 3). The genetic libraries obtained with universal primers revealed that group II clones (6 of 33 archaeal clones) were less prevalent in libraries obtained from Atlantic and Pacific deep-water samples (17). Moreover, the relative levels of group I and group II phylotypes in some samples were determined independently by performing quantitative rRNA hybridization experiments. In one case, both approaches were used for the same samples (26), and there was very good agreement in the relative levels of both groups. Other experiments revealed that the group I signal was dominant throughout the water column in Antarctic waters (27, 35). Therefore, our results based on comparisons of the compositions and levels of particular clone types in the libraries could be somewhat biased, but several pieces of evidence indicate that our main conclusions remain valid.
Genetic libraries were analyzed by using three techniques in succession. The first technique, membrane hybridization, facilitated quick analysis of many clones (2,328 clones), but the information obtained was limited and could be used only to assign each archaeal clone to one of the broad groups. The second technique, RFLP analysis, allowed us to affiliate a moderately high number of clones (460 clones) with defined OTUs. The third technique was the most labor-intensive technique (48 clones were partially sequenced) and was used to determine primary sequences, which were the most useful data for determining the relationships of new clones to each other and to clones in large and well-established databases. To compare the clonal compositions of different libraries, we used the RFLP approach, which was faster, easier, and cheaper than sequencing (in fact, there was an order of magnitude difference in the number of clones analyzed). However, in the RFLP analysis a small fraction of the sequence information was used, and the consistency of the OTUs defined had to be evaluated. We found that clones belonging to the same OTU were closely related (Fig. 3), even though they originated from very widely separated areas or different depths. Moreover, a dendrogram constructed by using the restriction sites (Fig. 5) had the same topology as the phylogenetic tree constructed by using sequences (Fig. 2), indicating that the RFLP analysis was informative enough to determine the relationships of the different OTUs. This suggests that the RFLP analysis performed here was adequate to compare the different libraries. This is in agreement with the results of a computer-simulated study in which the researchers obtained a fairly good representation of bacterial diversity by using three tetrameric restriction enzymes (30).
Distribution of archaea in the water column.
The pattern found in California coastal waters (group II phylotypes are the dominant phylotypes at the surface and group I phylotypes are the dominant phylotypes at depth [26]) was also found in the Mediterranean Sea and in the North Atlantic Ocean (Table 3). In contrast, in Southern Ocean samples group I phylotypes were the dominant phylotypes at both depths, and group II phylotypes were almost absent (Table 3). We concluded that group I phylotypes are ubiquitous and are abundant and often the dominant phylotypes in most marine waters, whereas group II phylotypes are the dominant phylotypes only at the surface in temperate regions. The two groups are very distantly related phylogenetically and seem to occupy different ecological niches. At the surface, where group II clones are the predominant clones based on archaeal sequences, the relative level of archaea is lower (18, 26, 27, 35), but the specific activity of the prokaryotic assemblage is higher and there is an active food web based on algal photosynthesis. Recently, it has been suggested that instead of being planktonic, group II archaea could originate from the digestive tracts of very common fishes (47). In deeper, dark waters where group I organisms predominate, the relative archaeal level is higher (18, 26, 35), but the activity of the prokaryotic assemblage is lower (27) and largely dependent on sedimenting material. We still do not know what the role of archaea in the marine plankton is, but defining the distribution of the different groups should help elucidate some aspects of the ecology of these organisms.
Recently, Fuhrman and Davis (17) described euryarchaeotal group III archaea which accounted for 2 of 33 archaeal clones in deep-water libraries in the Pacific and Atlantic oceans. Membrane hybridization experiments were also aimed at detecting the possible presence of this type or other new types of archaea in the plankton. Most of the clones in our libraries belonged to either the group I archaea or the group II archaea, and only one clone belonged to group III (Table 3). The latter clone originated from our deepest sampling site in the Mediterranean Sea (depth, 450 m). Therefore, group III archaea seem to be rare, but they are widespread and are found among the deepest marine plankton (at depths below 400 m).
The phylogenetic tree that reflected the genetic diversity of marine archaea revealed the existence of distinct clusters (Fig. 2), a common occurrence when marine bacterial assemblages are investigated (5, 16, 32). Similar clusters in marine archaeal group I have been identified previously (6, 17). Closely related phylotypes belonging to different but neighboring clusters can occupy distinct ecological niches and replace each other vertically in the marine water column (14, 29) or temporally during seasonal succession in a meromictic lake (7). In fact, the clusters of marine archaea detected reflect a depth-specific distribution. Cluster I-α appears to contain shallow-water phylotypes (a majority of the surface clones and many 200-m clones in our libraries), whereas cluster I-γ contains most clones obtained from deep-water samples, including clones in our library collected at 450 m and clones in other libraries collected at 500 to 3,000 m (17). Similarly, cluster II-α is composed of clones obtained from surface samples, and cluster II-β contains surface clones (in a smaller subcluster [see above]) and clones obtained from 200-m samples or samples collected at greater depths.
Ubiquity and limited diversity of marine planktonic archaea.
At the level of resolution of the RFLP analysis of rRNA genes, planktonic archaeal diversity appeared to be remarkably limited. In general, only one or two OTUs dominated each library, and some other OTUs were represented by very few clones (Fig. 1). When the data were investigated more carefully, the less abundant clones were sometimes found to belong to the same phylogenetic unit. This was the case for all OTUs belonging to cluster I-α, which were not significantly different from the most abundant OTU, OTU I-AA (Fig. 3). PCR biases probably do not explain the limited diversity, since according to the kinetic model (45) PCR biases tend to favor overrepresentation of rare types, thus increasing the diversity sampled. The relatively low archaeal diversity in worldwide marine plankton assemblages contrasts with the higher archaeal diversity found in other environments, such as marine hydrothermal vent sediments (31), salt marsh sediments (33), soils (4), and hot springs (3). Also, genetic libraries generated with the other microbial components of the marine plankton revealed much greater diversity of both bacteria (5, 16, 32) and eukarya (42; B. Díez, unpublished results).
The comparison of OTUs from different libraries revealed that the dominant types were present in most of the samples examined (Fig. 1), suggesting that a limited number of archaeal taxa dominate the archaeal plankton of the oceans and are very widespread. This is particularly true for OTU I-AA, which was the dominant OTU in most libraries and was retrieved from all systems studied, both at the surface and at a depth of 200 m. Furthermore, many clones in the database also belong to this OTU (Table 4). This OTU was scarce or absent from the libraries obtained from the greatest depths (ME-450 and the libraries of Fuhrman et al.), in which OTU I-CD or its relatives dominated. The most abundant group II OTU, OTU II-CC, was also found at the surface in most areas, whereas deep-water libraries from widely separated places were dominated by two closely related OTUs, OTUs II-EH and II-EF (which differed by only one restriction site) (Fig. 4). Although similar archaeal phylotypes have been recovered from marine sediments (23, 31, 46), these phylotypes may in fact have originated from the plankton. On the other hand, with a few exceptions, these specific archaeal types are rare in soils, freshwater sediments, or hot springs or are absent in these habitats (see reference 6 for a review). Considering all this, it is now apparent that these archaea are typical components of marine planktonic assemblages.
Our results show that the patterns of archaeal diversity detected in a few samples in previous studies are applicable to large areas of the oceans of the world. Despite the differences in temperature, chlorophyll a concentration, and prokaryote abundance of the samples analyzed and the enormous distances that separate our sampling sites, very similar prokaryote sequences were amplified from all of the samples, indicating that a few cosmopolitan phylotypes are the dominant phylotypes in marine archaeal assemblages.
ACKNOWLEDGMENTS
We thank Ricardo Anadón and Mario Quevedo (University of Oviedo), Antoni Calafat (University of Barcelona), and Josep M. Gasol (ICM) for making sampling possible. The laboratory assistance of Eduardo Balbuena, Núria Molist, and Beatriz Díez is appreciated. We thank Emilio Ortega-Casamayor for helpful discussions.
This work was financed by EU project MIDAS (MAS3-CT97-0154) and DGICYT project PB95-0222-C02-01 to C.P.A. and by National Science Foundation grants OCE95-29804 and OPP94-18442 to E.F.D. Sampling in the North Atlantic was financed by NERC project ACSOE-NAE and CICYT project MAR97-1875-E, sampling in the Alboran Sea was financed by EU project MATER (MAS3-CT96-0051), sampling along the Cantabrian coast was financed by the project “Control a largo plazo de las condiciones Químico-Biológicas en la plataforma Continental Asturiana” (U. Oviedo-IEO), and sampling in the Drake Passage was financed by CICYT project E-DOVETAIL (ANT96-0866).
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 3.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britschgi T B, Giovannoni S J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;57:1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley D H, Graber J R, Schmidt T M. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl Environ Microbiol. 1998;64:4333–4339. doi: 10.1128/aem.64.11.4333-4339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casamayor E O, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong E F, Wu K Y, Prézelin B B, Jovine R V M. High abundance of archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 10.DeLong E F, King L L, Massana R, Cittone H, Murray A E, Schleper C, Wakeham S G. Dibiphytanyl ether lipids in nonthermophilic crenarchaeotes. Appl Environ Microbiol. 1998;64:1133–1138. doi: 10.1128/aem.64.3.1133-1138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. Phylip—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 12.Felske A, Akkermans A D L, De Vos W M. Quantification of 16S rRNA in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl Environ Microbiol. 1998;64:4581–4587. doi: 10.1128/aem.64.11.4581-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenchel T. Marine plankton food chains. Annu Rev Ecol Syst. 1988;19:19–38. [Google Scholar]
- 14.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuhrman J A, Davis A A. Widespread archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- 18.Fuhrman J A, Ouverney C C. Marine microbial diversity studied via 16S rRNA sequences: cloning results from coastal waters and counting of native archaea with fluorescent single cell probes. Aquat Ecol. 1998;32:3–15. [Google Scholar]
- 19.Gasol J M, del Giorgio P A. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. 2000. Sci. Mar., in press. [Google Scholar]
- 20.Good I J. The population frequencies of species and the estimation of the population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 21.Hershberger K L, Barns S M, Reysenbach A-L, Dawson S C, Pace N R. Wide diversity of crenarchaeota. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 22.Jurgens G, Lindström K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soils. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato C, Li L, Tamaoka J, Horikoshi K. Molecular analyses of the sediment of the 11000-m deep Mariana Trench. Extremophiles. 1997;1:117–123. doi: 10.1007/s007920050024. [DOI] [PubMed] [Google Scholar]
- 24.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massana R, Murray A E, Preston C M, Delong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massana R, Taylor L T, Murray A E, Wu K Y, Jeffrey W H, DeLong E F. Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antarctica, during early spring. Limnol Oceanogr. 1988;43:607–617. [Google Scholar]
- 28.McInerney J O, Wilkinson M, Patching J W, Embley T M, Powell R. Recovery and phylogenetic analysis of novel archaeal rRNA sequences from a deep-sea deposit feeder. Appl Environ Microbiol. 1995;61:1646–1648. doi: 10.1128/aem.61.4.1646-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1988;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 30.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. Diversity of deep-sea hydrothermal vent archaea from Loihi Seamount, Hawaii. Deep-Sea Res. 1998;45:303–317. [Google Scholar]
- 32.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 33.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of Archaea in sediment samples from coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray A E, Wu K Y, Moyer C L, Karl D M, DeLong E F. Evidence for circumpolar distribution of planktonic archaea in the Southern Ocean. Aquat Microb Ecol. 1999;18:263–273. [Google Scholar]
- 36.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 37.Parsons T R, Maita Y, Lalli C M. A manual of chemical and biological methods for seawater analysis. 1st ed. New York, N.Y: Pergamon Press; 1984. [Google Scholar]
- 38.Paul J H, Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl Environ Microbiol. 1982;43:1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 41.Preston C M, Wu K Y, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappé M S, Suzuki M T, Vergin K L, Giovannoni S J. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl Environ Microbiol. 1998;64:294–303. doi: 10.1128/aem.64.1.294-303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schleper C, Holben W, Klenk H-P. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein J, Marsh T L, Wu K Y, Shizuya H, DeLong E F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takai K, Sako Y. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol Ecol. 1999;28:177–188. [Google Scholar]
- 47.van der Maarel M J E C, Artz R R E, Haanstra R, Forney L J. Association of marine archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vetriani C, Reysenbach A-L, Doré J. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol Lett. 1998;161:83–88. doi: 10.1111/j.1574-6968.1998.tb12932.x. [DOI] [PubMed] [Google Scholar]
- 49.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:219–286. [Google Scholar]
- 50.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]