Key Points
Question
Can the HER2DX assay predict pathologic complete response (pCR) in patients with early-stage ERBB2 (formerly HER2)–positive breast cancer who were treated with neoadjuvant paclitaxel, trastuzumab, and pertuzumab?
Findings
In this diagnostic study of biopsy specimens from 80 patients with early-stage ERBB2-positive breast cancer, the HER2DX assay was administered on baseline tumor biopsy specimens from patients treated during the phase 2 DAPHNe clinical trial. In a multivariable model that incorporated established predictive gene expression–based and clinicopathologic variables, including hormone receptor status, the HER2DX pCR likelihood score was significantly associated with pCR.
Meaning
The study results suggest that the HER2DX assay may help to optimize escalation or deescalation of neoadjuvant therapy in patients with early-stage ERBB2-positive breast cancer.
Abstract
Importance
Patients with early-stage ERBB2 (formerly HER2)–positive breast cancer (ERBB2+ BC) who experience a pathologic complete response (pCR) after receiving neoadjuvant therapy have favorable survival outcomes. Predicting the likelihood of pCR may help optimize neoadjuvant therapy.
Objective
To test the ability of the HER2DX assay to predict the likelihood of pCR in patients with early-stage ERBB2+ BC who are receiving deescalated neoadjuvant therapy.
Design, Setting, and Participants
In this diagnostic/prognostic study, the HER2DX assay was administered on pretreatment tumor biopsy samples from patients enrolled in the single-arm, multicenter, prospective phase 2 DAPHNe clinical trial who had newly diagnosed stage II to III ERBB2+ BC that was treated with neoadjuvant paclitaxel weekly for 12 weeks plus trastuzumab and pertuzumab every 3 weeks for 4 cycles.
Interventions and Exposures
The HER2DX assay is a classifier derived from gene expression and limited clinical features that provides 2 independent scores to predict prognosis and likelihood of pCR in patients with early-stage ERBB2+ BC. The assay was administered on baseline tumor samples from 80 of 97 patients (82.5%) in the DAPHNe trial.
Main Outcomes and Measures
The primary aim was to test the ability of the HER2DX pCR likelihood score (as a continuous variable from 0-100) to predict pCR (ypT0/isN0).
Results
Of 80 participants, 79 (98.8%) were women and there were 4 African American (5.0%), 6 Asian (7.5%), 4 Hispanic (5.0%), and 66 White individuals (82.5%); the mean (range) age was 50.3 (26.0-78.0) years. The HER2DX pCR score was significantly associated with pCR (odds ratio, 1.05; 95% CI, 1.03-1.08; P < .001). The pCR rates in the HER2DX high, medium, and low pCR score groups were 92.6%, 63.6%, and 29.0%, respectively (high vs low odds ratio, 30.6; P < .001). The HER2DX pCR score was significantly associated with pCR independently of hormone receptor status, ERBB2 immunohistochemistry score, HER2DX ERBB2 expression score, and prediction analysis of microarray 50 ERBB2-enriched subtype. The correlation between the HER2DX pCR score and prognostic risk score was weak (Pearson coefficient, −0.12). Performance of the risk score could not be assessed due to lack of recurrence events.
Conclusions and Relevance
The results of this diagnostic/prognostic study suggest that the HER2DX pCR score assay could predict pCR following treatment with deescalated neoadjuvant paclitaxel with trastuzumab and pertuzumab in patients with early-stage ERBB2+ BC. The HER2DX pCR score might guide therapeutic decisions by identifying patients who are candidates for deescalated or escalated approaches.
This clinical trial examines the ability of the HER2DX assay to predict the likelihood of pathologic complete response in patients with early-stage ERBB2–positive breast cancer who are receiving deescalated neoadjuvant therapy.
Introduction
Patients with stage II and III ERBB2-positive (ERBB2+; formerly HER2) breast cancer typically receive neoadjuvant chemotherapy with ERBB2-directed therapy. Those experiencing a pathologic complete response (pCR) have significantly better survival outcomes than those with residual invasive disease1,2; therefore, they may be candidates for deescalation of therapy. The HER2DX assay is a supervised learning algorithm that incorporates clinical information (tumor size, nodal staging) and 4 gene expression signatures (immune infiltration, tumor cell proliferation, luminal differentiation, and ERBB2 amplicon expression) to provide 2 independent scores with the potential to predict the likelihood of pCR (pCR score) and long-term prognosis (risk score).3,4
DAPHNe (NCT03716180) was a single-arm phase 2 trial in which patients with stage II to III ERBB2+ breast cancer received a deescalated neoadjuvant regimen comprising paclitaxel, trastuzumab, and pertuzumab (THP). The overall pCR rate was 56.7%.5 Patients received adjuvant ERBB2-directed therapy with or without further chemotherapy based on their response to the neoadjuvant regimen; adjuvant trastuzumab and pertuzumab only was recommended for patients who experienced pCR.5 In the present study, the HER2DX assay was administered on pretreatment tumor biopsy specimens to investigate the predictive value of the HER2DX pCR score, evaluate the HER2DX pCR score assay according to hormone receptor (HR) status, and explore the association between the predictive HER2DX pCR score and the prognostic HER2DX risk score. The prognostic value of the risk score could not be assessed given lack of recurrence events in the trial.
Methods
DAPHNe Patient Population and Trial Therapy
DAPHNe was a single-arm prospective phase 2 trial that enrolled patients with stage II to III ERBB2+ breast cancer. All patients received neoadjuvant THP for 4 cycles before surgery.5 Five patients (5.1%) experienced incomplete clinical response to THP and received additional neoadjuvant chemotherapy; they were excluded from this analysis. The degree of response to neoadjuvant therapy was quantified by the residual cancer burden (RCB) score6; pCR was defined as an RCB score of 0 (ypT0/isN0). All trial procedures were approved by the Dana-Farber/Harvard Cancer Center institutional review board, all patients provided written informed consent, and the study conformed to the Standards for Reporting of Diagnostic Accuracy (STARD) 2015 reporting guideline.7
HER2DX Assay
Development of the HER2DX assay was described previously.3 The HER2DX assay incorporates limited clinical features and expression of 27 genes that encompass immune infiltration, tumor cell proliferation, luminal differentiation, and ERBB2 amplicon expression signatures. The assay provides 2 independent scores to predict prognosis (risk score) and the likelihood of pCR (pCR score). The HER2DX assay also produces an ERBB2 score based on the level of ERBB2 gene expression. Scores are continuous, as well as subdivided into ordinal groups based on previously reported cut points.3
Application of HER2DX to DAPHNe Tumor Samples and Statistical Methods
Ribonucleic acid was extracted from baseline tumor biopsy specimens. The HER2DX assay was retrospectively evaluated centrally. Univariate and multivariable logistic regression analyses were used to investigate the association of each variable with pCR. The least absolute shrinkage and selection operator regression was used for variable selection in the multivariate model. Receiver operating characteristic (ROC) curves were used as a performance measure. Statistical analyses were performed in R, version 4.0.5 (R Foundation), and data imputation for the multivariable analysis was performed using multivariate imputation (mice R package). No adjustments were made for multiple comparisons, and the significance level was set to 2-sided α = .05.
Results
Patient and Tumor Characteristics
The HER2DX assay was administered for 80 of 98 patients (81.6%) enrolled and treated during the trial (eFigure in Supplement 1). Clinical T2 to T3 disease represented 65 cases (81.3%); 52 patients (65.0%) had clinical node-negative disease, and 56 (70.0%) had HR-positive disease. eTable 1 in Supplement 1 compares the HER2DX assay population with the overall trial population. The pCR rate among the HER2DX cohort was 60.0% (95% CI, 49.3%-70.7%). The pCR rate was 87.0% (95% CI, 79.6%-94.4%) in patients with HR-negative disease and 48.2% (95% CI, 37.2%-59.1%) in patients with HR-positive disease. The proportion of patients in the HER2DX pCR score high, medium, and low categories was 33.7%, 27.5%, and 38.8%, respectively.
Performance of the HER2DX pCR Score for pCR Prediction
The HER2DX pCR score as a continuous variable was significantly associated with pCR (odds ratio, 1.05; 95% CI, 1.03-1.08; P < .001), with a receiver operating characteristic curve area under the curve of 0.84 (95% CI, 0.75-0.92) for performance of HER2DX pCR score (Figure, A). The Figure depicts the expression of the 4 gene expression signatures across the HER2DX pCR score high, medium, and low groups. The pCR rates in the HER2DX pCR score high, medium, and low groups were 92.6%, 63.6%, and 29.0%, respectively (odds ratio of 30.6 for comparison of pCR score high vs pCR score low; 95% CI, 6.0-156.9; P < .001; Table). eTable 2 in Supplement 1 shows RCB categories according to HER2DX pCR score. In a univariable analysis evaluating standard clinicopathologic variables and various expression-based classifiers, there were multiple significant predictors of pCR status, including HER2DX pCR score, HER2DX ERBB2 score, prediction analysis of microarray 50 ERBB2-enriched status, ERBB2 immunohistochemistry status, and HR status. In a multivariable analysis, HER2DX pCR score and ERBB2 score were the only significant predictors of pCR (Table). The HER2DX pCR score performed well in HR-negative and HR-positive subpopulations (receiver operating characteristic curve areas under the curve of 0.857 and 0.762, respectively) (Figure, B and C). The pCR rates by HR status and HER2DX pCR score group are shown in eTable 3 in Supplement 1.
Figure. Performance and Genomic Features of the HER2DX Pathologic Complete Response (pCR) Score for Predicting pCR Following Neoadjuvant Paclitaxel, Trastuzumab, and Pertuzumab (THP) Therapy.
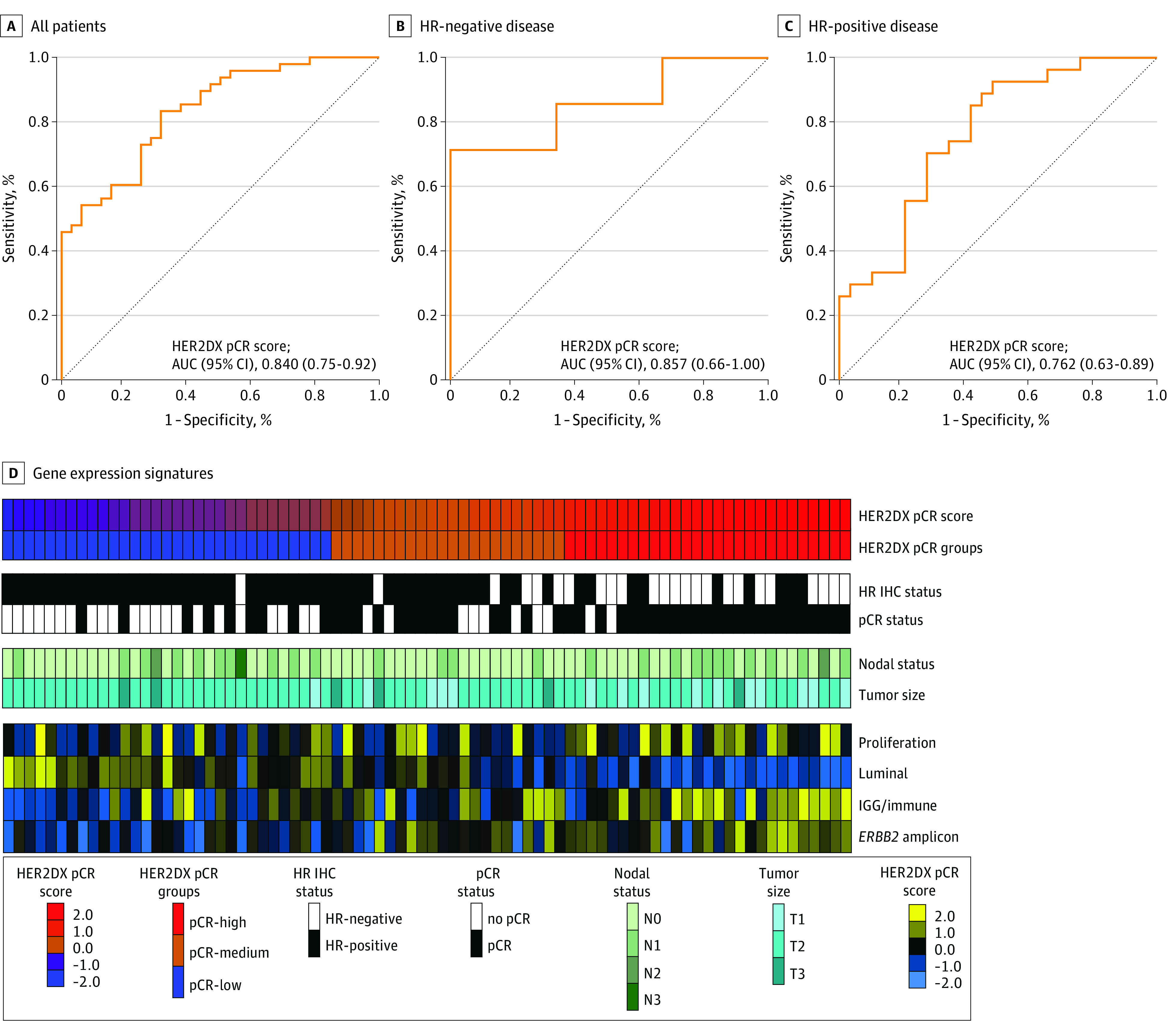
Receiver operating characteristic curve analysis of the HER2DX pCR score among all patients (A), hormone receptor (HR)–negative patients (B), and HR-positive patients (C). D, Expression of the 4 HER2DX gene expression signatures (immune, proliferation, luminal, and ERBB2 [formerly HER2] amplicon) across the HER2DX pCR score high, medium, and low groups. AUC indicates area under the curve; IGG immunoglobulin G; IHC, immunohistochemistry.
Table. Association of Pretreatment Baseline Variables With pCR in 80 Patients With ERBB2-Positive Early-Stage Breast Cancer Treated With Neoadjuvant THP in the DAPHNe Clinical Trial.
| Characteristic | No. | pCR rate, % | Univariable | Multivariable | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |||
| Overall cohort | 80 | 60.0 | ||||
| HER2DX pCR score (continuous variable) | 80 | NA | 1.05 (1.03-1.08) | <.001 | 1.03 (1.01-1.07) | .03 |
| HER2DX pCR score groups | ||||||
| Low | 31 | 29.0 | 1 [Reference] | NA | NA | NA |
| Medium | 22 | 63.6 | 4.30 (1.34-14.36) | .01 | NA | NA |
| High | 27 | 92.6 | 30.60 (1.30-156.90) | <.001 | NA | NA |
| HER2DX ERBB2 score (continuous variable) | 80 | NA | 1.05 (1.02-1.08) | <.001 | 1.03 (1.00-1.07) | .04 |
| HER2DX ERBB2 mRNA score | ||||||
| Low | 9 | 44.4 | 1 [Reference] | NA | NA | NA |
| Medium | 12 | 16.7 | 0.25 (0.03-1.86) | .18 | ||
| High | 59 | 71.2 | 3.09 (0.74-12-91) | .12 | ||
| Clinical tumor stage | ||||||
| cT1 | 15 | 80.0 | 1 [Reference] | NA | NA | NA |
| cT2-3 | 65 | 55.4 | 0.31 (0.08-1.20) | .09 | ||
| Clinical nodal stage | ||||||
| cN-negative | 52 | 59.6 | 1 [Reference] | NA | NA | NA |
| cN-positive | 28 | 60.7 | 1.05 (0.41-2.68) | .92 | ||
| PAM50 | ||||||
| Non–ERBB2-enriched | 34 | 35.3 | 1 [Reference] | NA | 1 [Reference] | NA |
| ERBB2-enriched | 46 | 78.3 | 6.6 (2.45-17.81) | <.001 | 2.05 (0.57-7.36) | .27 |
| ERBB2 IHC status | ||||||
| 2+ | 10 | 30.0 | 1 [Reference] | NA | NA | NA |
| 3+ | 68 | 66.2 | 4.57 (1.08-19.32) | .039 | NA | NA |
| Hormone receptor status | ||||||
| Positivea | 56 | 48.2 | 1 [Reference] | NA | 1 [Reference] | NA |
| Negative | 24 | 87.5 | 7.52 (2.01-28.10) | .003 | 1.79 (0.28-12.39) | .54 |
Abbreviations: IHC, immunohistochemistry; mRNA, messenger RNA; NA, not applicable; OR, odds ratio; PAM50, prediction analysis of microarray 50; pCR, pathologic complete response; THP, paclitaxel/trastuzumab/pertuzumab.
Hormone receptor–positive status was defined as estrogen receptor or progesterone receptor staining of 1% or greater in accordance with current guidelines from American Society of Clinical Oncology/College of American Pathologists.
HER2DX Risk Score Categories
The proportion of patients grouped into the HER2DX high vs low risk score groups was 48.7% and 51.3%, respectively. The correlation between the HER2DX pCR score and risk score was weak (Pearson coefficient, −0.12). With 19.1 (IQR, 15.2-22.5) months of median follow-up, there were no breast cancer recurrences,5 so HER2DX risk score performance could not be assessed.
Discussion
To our knowledge, the results of this study represent the first validation of HER2DX pCR score as a predictive assay in patients with ERBB2+ breast cancer who were treated with neoadjuvant THP. They also highlight the ability of HER2DX pCR score to outperform established predictive biomarkers, such as ERBB2-enriched subtype, and demonstrate the ability of HER2DX to predict pCR in HR-positive and HR-negative patient populations despite the well documented differences in responsiveness to neoadjuvant chemotherapy plus ERBB2-directed therapy between those subgroups.
Treatment with THP as a deescalated neoadjuvant regimen is the focus of 2 ongoing prospective clinical trials: CompassHER2-pCR (NCT04266249) and DECRESCENDO (NCT04675827). While the pCR-based deescalation approach is currently experimental, if the primary objectives of these 2 trials are met, neoadjuvant THP will likely become a standard regimen for patients with early-stage ERBB2+ breast cancer. The DAPHNe trial cohort, in combination with 2 other cohorts that included patients treated with neoadjuvant taxane/trastuzumab with or without pertuzumab, confirmed that a high HER2DX pCR score could predict a high likelihood of pCR following neoadjuvant THP and benefit from pertuzumab specifically.8 As adjuvant escalation options for ERBB2+ breast cancer may become further intensified, the customization of neoadjuvant therapy to optimize pCR is increasingly important. With further study as a predictive biomarker across neoadjuvant regimens, combined with further validation of the companion prognostic assay, HER2DX pCR score may identify candidates for deescalation beyond THP and escalation from THP in the neoadjuvant setting.
Limitations
This study had several limitations. Biomarker analyses were performed in retrospective fashion, and only a subset of the overall trial population was evaluable for the HER2DX biomarker (although this subpopulation appeared similar to the overall trial population). The HER2DX prognostic risk score could not be evaluated due to the lack of recurrence events in this cohort.
Conclusions
To date, genomic risk scores have only played a routine role in managing HR-positive/ERBB2-negative breast cancer. However, individualization of therapy is increasingly important in the management of early-stage ERBB2+ breast cancer as therapeutic options expand with newer agents being evaluated in ongoing clinical trials. Introduction of a pCR-predictive and long-term prognostic risk score is needed. The findings of this prognostic/diagnostic study indicate that prospective incorporation of HER2DX into escalation and deescalation trial designs can potentially further define the role for this assay in managing early-stage ERBB2+ breast cancer.
eTable 1. Main patient population of the DAPHNe trial cohort and HER2DX sub-population
eTable 2. HER2DX pCR likelihood score and residual cancer burden (RCB) category
eTable 3. HER2DX pCR likelihood score and hormone receptor (HR) IHC status
eFigure. Consort diagram
Data sharing statement
References
- 1.Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164-172. doi: 10.1016/S0140-6736(13)62422-8 [DOI] [PubMed] [Google Scholar]
- 2.Yau C, Osdoit M, van der Noordaa M, et al. ; I-SPY 2 Trial Consortium . Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet Oncol. 2022;23(1):149-160. doi: 10.1016/S1470-2045(21)00589-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A, Guarneri V, Pascual T, et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer. EBioMedicine. 2022;75:103801. doi: 10.1016/j.ebiom.2021.103801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Guarneri V, Paré L, et al. A multivariable prognostic score to guide systemic therapy in early-stage HER2-positive breast cancer: a retrospective study with an external evaluation. Lancet Oncol. 2020;21(11):1455-1464. doi: 10.1016/S1470-2045(20)30450-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waks AG, Desai NV, Li T, et al. A prospective trial of treatment de-escalation following neoadjuvant paclitaxel/trastuzumab/pertuzumab in HER2-positive breast cancer. NPJ Breast Cancer. 2022;8(1):63. doi: 10.1038/s41523-022-00429-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25(28):4414-4422. doi: 10.1200/JCO.2007.10.6823 [DOI] [PubMed] [Google Scholar]
- 7.Bossuyt PM, Reitsma JB, Bruns DE, et al. ; STARD Group . STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bueno-Muiño C, Echavarría I, López-Tarruella S, et al. Assessment of a genomic assay in patients with ERBB2-positive breast cancer following neoadjuvant trastuzumab-based chemotherapy with our without pertuzumab. JAMA Oncol. Published online April 27, 2023. doi: 10.1001/jamaoncol.2023.0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Main patient population of the DAPHNe trial cohort and HER2DX sub-population
eTable 2. HER2DX pCR likelihood score and residual cancer burden (RCB) category
eTable 3. HER2DX pCR likelihood score and hormone receptor (HR) IHC status
eFigure. Consort diagram
Data sharing statement


