This diagnostic study aims to determine if use of the HER2DX genomic assay (Reveal Genomics) in pretreatment baseline tissue samples of patients with ERBB2-positive breast cancer is associated with response to neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab.
Key Points
Question
Can the HER2DX genomic assay (Reveal Genomics) predict response to neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab in early-stage ERBB2-positive breast cancer?
Findings
In this diagnostic study of 155 patients with ERBB2 (formerly HER2)-positive breast cancer, the assay-reported pathologic complete response (pCR) score showed statistically significant association with pCR following trastuzumab-based chemotherapy independently of pertuzumab use. More importantly, a statistically significant increase in pCR rates with the addition of pertuzumab was only observed in assay-reported pCR-high disease, which represented 1 of 3 patients with ERBB2-positive breast cancer.
Meaning
This assay might provide meaningful clinical information to guide therapeutic decisions regarding the use of trastuzumab-based chemotherapy with or without pertuzumab in the neoadjuvant setting.
Abstract
Importance
Biomarkers to guide the use of pertuzumab in the treatment of early-stage ERBB2 (formerly HER2)-positive breast cancer beyond simple ERBB2 status are needed.
Objective
To determine if use of the HER2DX genomic assay (Reveal Genomics) in pretreatment baseline tissue samples of patients with ERBB2-positive breast cancer is associated with response to neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab.
Design, Setting, and Participants
This is a retrospective diagnostic/prognostic analysis of a multicenter academic observational study in Spain performed during 2018 to 2022 (GOM-HGUGM-2018-05). In addition, a combined analysis with 2 previously reported trials of neoadjuvant cohorts with results from the assay (DAPHNe and I-SPY2) was performed. All patients had stage I to III ERBB2-positive breast cancer, signed informed consent, and had available formalin-fixed paraffin-embedded tumor specimens obtained prior to starting therapy.
Exposures
Patients received intravenous trastuzumab, 8 mg/kg, loading dose, followed by 6 mg/kg every 3 weeks in combination with intravenous docetaxel, 75 mg/m2, every 3 weeks and intravenous carboplatin area under the curve of 6 every 3 weeks for 6 cycles, or this regimen plus intravenous pertuzumab, 840 mg, loading dose, followed by an intravenous 420-mg dose every 3 weeks for 6 cycles.
Main Outcome and Measures
Association of baseline assay-reported pathologic complete response (pCR) score with pCR in the breast and axilla, as well as association of baseline assay-reported pCR score with response to pertuzumab.
Results
The assay was evaluated in 155 patients with ERBB2-positive breast cancer (mean [range] age, 50.3 [26-78] years). Clinical T1 to T2 and node-positive disease was present in 113 (72.9%) and 99 (63.9%) patients, respectively, and 105 (67.7%) tumors were hormone receptor positive. The overall pCR rate was 57.4% (95% CI, 49.2%-65.2%). The proportion of patients in the assay-reported pCR-low, pCR-medium, and pCR-high groups was 53 (34.2%), 54 (34.8%), and 48 (31.0%), respectively. In the multivariable analysis, the assay-reported pCR score (as a continuous variable from 0-100) showed a statistically significant association with pCR (odds ratio [OR] per 10-unit increase, 1.43; 95% CI, 1.22-1.70; P < .001). The pCR rates in the assay-reported pCR-high and pCR-low groups were 75.0% and 28.3%, respectively (OR, 7.85; 95% CI, 2.67-24.91; P < .001). In the combined analysis (n = 282), an increase in pCR rate due to pertuzumab was found in the assay-reported pCR-high tumors (OR, 5.36; 95% CI, 1.89-15.20; P < .001) but not in the assay-reported pCR-low tumors (OR, 0.86; 95% CI, 0.30-2.46; P = .77). A statistically significant interaction between the assay-reported pCR score and the effect of pertuzumab in pCR was observed.
Conclusions and Relevance
This diagnostic/prognostic study demonstrated that the genomic assay predicted pCR following neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab. This assay could guide therapeutic decisions regarding the use of neoadjuvant pertuzumab.
Introduction
Pertuzumab is approved for treatment of early and advanced ERBB2 (formerly HER2)-positive breast cancer.1,2 In early disease, the addition of pertuzumab to trastuzumab-based chemotherapy increases pathologic complete response (pCR) rates.3 In the NeoSphere phase 2 trial,3 pertuzumab and trastuzumab plus docetaxel showed an improvement in the pCR rate compared with trastuzumab plus docetaxel (Δ, 16.8%). Moreover, the addition of 1 year of pertuzumab to trastuzumab-based chemotherapy improved invasive disease–free survival (6-year survival, 91% vs 88%).4,5 Of note, the benefit was restricted to node-positive disease, and no overall survival benefit was observed.5 Overall, the benefits of pertuzumab in early-stage ERBB2-positive disease are modest.
HER2DX (Reveal Genomics) is a clinically available genomic test that provides 2 scores to predict long-term prognosis (ie, risk score) and likelihood of pCR (ie, pCR score) in early ERBB2-positive breast cancer.6 The 27-gene assay integrates clinical and biological information tracking immune response, luminal differentiation, tumor proliferation, and expression of the ERBB2 amplicon.6 This diagnostic/prognostic study aims to determine if use of this genomic assay in pretreatment baseline tissue samples is associated with response to neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab.
Methods
GOM-HGUGM-2018-05 Cohort
GOM-HGUGM-2018-05 (hereafter, GOM) is a prospective observational study of consecutive patients with stage I to III ERBB2-positive breast cancer treated with neoadjuvant therapy across 7 hospitals in Spain. Patients received 6 cycles of intravenous docetaxel, 75 mg/m2, every 3 weeks in combination with intravenous carboplatin area under the curve of 6 every 3 weeks and intravenous trastuzumab, 8 mg/kg, loading dose followed by 6 mg/kg every 3 weeks (TCH). Once neoadjuvant pertuzumab was reimbursed in Spain, most patients received TCH in combination with intravenous pertuzumab, 840 mg, loading dose, followed by an intravenous 420-mg dose every 3 weeks (TCHP).
This study followed the Standards for Reporting of Diagnostic Accuracy (STARD) reporting guideline7 and was approved by an ethics committee at Hospital General Universitario Gregorio Marañón. Patients signed written informed consent.
DAPHNe and I-SPY2 Cohorts
Ninety-eight patients in DAPHNe, a prospective single-arm phase 2 study, were treated with preoperative paclitaxel, 80 mg/m2, weekly for 12 weeks in combination with trastuzumab and pertuzumab.8 The HER2DX results in DAPHNe are reported elsewhere.9
The I-SPY2 study10 adaptively randomized 128 patients with stage II to III ERBB2-positive breast cancer to 4 cycles of intravenous T-DM1, 3.6 mg/kg, every 3 weeks in combination with pertuzumab (n = 52); paclitaxel, trastuzumab, and pertuzumab (n = 45); or a control arm of paclitaxel and trastuzumab (n = 31). Patients received 4 cycles of doxorubicin, 60 mg/m2, and cyclophosphamide, 600 mg/m2, intravenously, every 2 to 3 weeks, before surgery. The primary results of HER2DX in I-SPY2 have been reported elsewhere.6
HER2DX
In the GOM and DAPHNe cohorts, the HER2DX standardized assay was performed from pretreatment baseline samples, as previously described.6 Preestablished cutoffs were used for each score. In I-SPY2, HER2DX was applied onto publicly available microarray data (GSE181574) from 127 patients,10 as previously described.6
Statistical Analysis
The primary objective was to evaluate the association between the HER2DX-reported pCR score and pCR in the breast and axilla. Univariable and multivariable logistic regression models were used. To build the multivariable model, the least absolute shrinkage and selection operator regression was used for variable selection. The C statistic was calculated to determine the discrimination capacity of HER2DX.
The secondary objective was to evaluate the ability of HER2DX to predict response to neoadjuvant pertuzumab. To accomplish this goal, a combined patient-level analysis of 3 cohorts (GOM, DAPHNe, I-SPY2) was undertaken. Pooled odds ratios (ORs) and 95% CIs were calculated with random effect models using the DerSimonian-Laird method, and the I2 was reported to estimate the percentage of total variability due to between-cohort heterogeneity. Interaction tests, used to evaluate the different pertuzumab effect according to HER2DX-reported pCR groups, were adjusted by cohort. Across the 3 cohorts, pCR was defined as ypT0/isN0. The significance level was set to a 2-sided α = .05. Statistical computations were carried out in R, version 4.0.3 (R Foundation for Statistical Computing).
Results
GOM Cohort Characteristics
As of June 2022, 155 patients with available pretreatment baseline RNA had enrolled in the study (Table 1 and eTable 1 and eFigure 1 in Supplement 1). Briefly, the mean (range) age of patients was 50.3 (26-78) years, and 85 patients (55.2%) were premenopausal. Clinical T1 to T2 disease was present in 113 (72.9%) patients, clinical node-positive disease (cN1-cN3) was present in 99 (63.9%) patients, and 105 (67.7%) tumors were hormone receptor positive. Sixty-seven (43.2%) and 88 (56.8%) patients received TCH and TCHP, respectively. The overall pCR rate was 57.4% (95% CI, 49.2%-65.2%): 52.2% (95% CI, 39.8%-64.4%) among those receiving TCH and 61.4% (95% CI, 50.3%-71.4%) among those receiving TCHP.
Table 1. Patient Characteristics Among the GOM Neoadjuvant Cohort (N = 155).
| Characteristic | No. (%) |
|---|---|
| Pathological response in breast and axilla | |
| Complete response | 89 (57.4) |
| Residual disease | 66 (42.6) |
| Hormone receptor status | |
| Positive | 105 (67.7) |
| Negative | 50 (32.3) |
| Intrinsic subtype | |
| Luminal A | 38 (24.5) |
| Luminal B | 26 (16.8) |
| ERBB2 enriched | 80 (51.6) |
| Basallike | 8 (5.2) |
| Normallike | 3 (1.9) |
Abbreviation: GOM, GOM-HGUGM-2018-05 trial.
Assay-Reported pCR Score in the GOM Cohort
The assay-reported pCR score (range, 0-100) showed a statistically significant association with pCR (OR per 10-unit increase, 1.43; 95% CI, 1.22-1.70; P < .001) after adjusting for treatment and clinicopathological factors (Table 2). The pCR rates in the assay-reported pCR-high and pCR-low groups were 75.0% and 28.3%, respectively (OR, 7.85; 95% CI, 2.67-24.91; P < .001). In patients treated with TCHP, the pCR rates in the assay-reported pCR-high and pCR-low groups were 85.7% and 27.3%, respectively (OR, 16.0; 95% CI, 4.72-67.09; P < .001). The C statistics for the assay-reported pCR score (as a continuous variable) were 0.746 (all population) and 0.812 (TCHP).
Table 2. Association of Pretreatment Baseline Variables With Response in 155 Patients With ERBB2-Positive Early-Stage Breast Cancer Treated With Neoadjuvant TCH or TCHP in the GOM-HGUGM-2018-05 Cohort.
| Characteristic | Patients, No. | pCR rate | Univariate model | Multivariable model | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |||
| HER2DX pCR score (10-unit increase) | 155 | NA | 1.39 (1.23-1.60) | <.001 | 1.43 (1.22-1.70) | <.001 |
| HER2DX pCR score groups | ||||||
| Low | 53 | 28.3% | 1 [Reference] | NA | 1 [Reference] | NAa |
| Medium | 54 | 70.4% | 6.02 (2.67-14.27) | <.001 | 6.58 (2.50-18.75) | <.001a |
| High | 48 | 75.0% | 7.60 (3.22-19.09) | <.001 | 7.85 (2.67-24.91) | <.001a |
| Clinical tumor stage | ||||||
| cT1-cT2 | 113 | 60.2% | 1 [Reference] | NA | NA | NA |
| cT3-cT4 | 42 | 50.0% | 0.66 (0.32-1.35) | .26 | NA | NA |
| Clinical nodal stage | ||||||
| cN0 | 56 | 69.6% | 1 [Reference] | NA | 1 [Reference] | NA |
| cN1-cN3 | 99 | 50.5% | 0.44 (0.22-0.88) | .02 | 0.36 (0.15-0.81) | .02 |
| PAM50 | ||||||
| ERBB2 enriched | 80 | 68.8% | 1 [Reference] | NA | 1 [Reference] | NA |
| Non-ERBB2 enriched | 75 | 45.3% | 0.38 (0.19-0.72) | .004 | 0.65 (0.26-1.62) | .35 |
| Treatment | ||||||
| TCH | 67 | 52.2% | 1 [Reference] | NA | 1 [Reference] | NA |
| TCHP | 88 | 61.4% | 1.45 (0.76-2.77) | .26 | 1.97 (0.90-4.44) | .09 |
| Hormone receptor status | ||||||
| Positive | 105 | 51.4% | 1 [Reference] | NA | NA | NA |
| Negative | 50 | 70.0% | 2.20 (1.09-4.60) | .03 | NA | NA |
| Age (10-unit increase) | 155 | NA | 0.71 (0.51-0.98) | .04 | 0.70 (0.47-1.01) | .07 |
| Ki-67 IHC (10-unit increase) | 155 | NA | 1.04 (0.89-1.23) | .60 | 0.82 (0.66-1.01) | .07 |
Abbreviations: IHC, immunohistochemistry; NA, not applicable; OR, odds ratio; pCR, pathologic complete response; TCH, docetaxel, carboplatin, trastuzumab; TCHP, docetaxel, carboplatin, trastuzumab, pertuzumab.
A separate multivariable model has been performed using HER2DX-reported pCR score groups instead of HER2DX-reported pCR score. To avoid multicollinearity, HER2DX-reported pCR score groups and HER2DX-reported pCR score cannot be included in the same model.
Assay-Reported pCR Score and Pertuzumab Response
A total of 264 (72.9%) and 98 (27.1%) patients received and did not receive neoadjuvant pertuzumab, respectively (eTables 1-3 in Supplement 1). No statistically significant difference in pCR rates was found across the 3 studies. The overall pCR rates in patients treated with and without pertuzumab were 59.8% and 43.9%, respectively (Δ, 15.9%; OR, 2.09; 95% CI, 1.26-3.52; P = .005; eFigure 2 in Supplement 1). The pCR rates with and without pertuzumab differed according to assay-reported pCR score (eFigure 2 in Supplement 1). In patients with assay-reported pCR-high, pCR-medium, and pCR-low disease, the difference in pCR rates (with pertuzumab vs without pertuzumab) were 34.4%, 12.7%, and 0.2% in favor of pertuzumab, respectively.
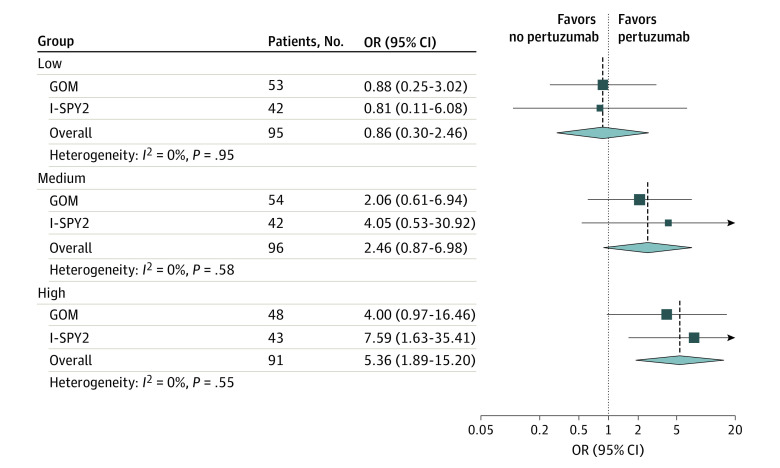
In the combined patient-level analysis of GOM and I-SPY2 cohorts (Figure and eFigures 2 and 3 in Supplement 1), an increase in pCR rate associated with pertuzumab was found in assay-reported pCR-high tumors (OR, 5.36; 95% CI, 1.89-15.20; P < .001) but not in assay-reported pCR-low tumors (OR, 0.86; 95% CI, 0.30-2.46; P = .77). A statistically significant interaction was observed between the assay-reported pCR-high group vs pCR-medium and pCR-low groups, and the pCR-high group vs the pCR-low group (eFigure 2 in Supplement 1).
Figure. Association of HER2DX Pathologic Complete Response (pCR) Groups With Response to Pertuzumab in a Combined Patient-Level Analysis (N = 282).
Data are from the neoadjuvant cohorts in the GOM-HGUGM-2018-05 (GOM) and I-SPY2 trials. OR indicates odds ratio.
Discussion
To our knowledge, this is the first study to demonstrate that the HER2DX-reported pCR score predicts response to neoadjuvant pertuzumab. A potential biological explanation is that the assay-reported pCR-high disease is composed of ERBB2-positive tumors that have the highest expression and activity of ERBB2 and/or the highest infiltration of B and T immune cells,6 all of which are biological features previously associated with pertuzumab response.11,12 In contrast, the pCR rate in the assay-reported pCR-low disease is low (ie, <25%) and does not increase with pertuzumab. Less clear is the value of pertuzumab in the assay-reported pCR-medium group.
Pertuzumab is approved for treatment of clinically high-risk ERBB2-positive breast cancer. However, the absolute increase in pCR rates in unselected patients with stage II to III disease in the NeoSphere trial is less than 20%.3 In addition, the absolute increase in invasive disease–free survival when 1 year of pertuzumab is added to trastuzumab-based chemotherapy is small, except in node-positive disease (Δ, 4.9% at 8 years).4,5 These modest results from the NeoSphere and APHINITY trials have led many countries to decline reimbursement of pertuzumab in early-stage disease or to limit its use in the adjuvant setting if the cancer is node positive. Thus, a biomarker such as the present genomic assay, which can help identify patients who will benefit the most from neoadjuvant pertuzumab, might be of clinical value.
Limitations
The retrospective nature of this study and the lack of randomization and long-term survival outcomes represent the main limitations. Another limitation is that the assay was evaluated in silico in the I-SPY2 cohort and that we did not address if the type of chemotherapy backbone mattered.
Conclusions
This diagnostic/prognostic study showed that the HER2DX genomic assay can predict pCR following neoadjuvant trastuzumab-based chemotherapy with or without pertuzumab. This assay could guide therapeutic decisions regarding the use of neoadjuvant pertuzumab.
eTable 1. Patient characteristics of the GOM neoadjuvant cohort according to treatment (TCH versus TCHP)
eTable 2. Main patient population of the DAPHNe trial cohort and HER2DX sub-population
eTable 3. Main patient population of the ISPY-2 trial cohort with HER2DX data
eFigure 1. CONSORT diagram of the GOM cohort
eFigure 2. Association of HER2DX pCR score with response to pertuzumab in a combined patient-level analysis from GOM, DAPHNe and ISPY-2 neoadjuvant cohorts (n=362)
eFigure 3. Relationship between HER2DX pCR score, treatment and pCR
Data Sharing Statement
References
- 1.Martínez-Sáez O, Prat A. Current and future management of HER2-positive metastatic breast cancer. JCO Oncol Pract. 2021;17(10):594-604. doi: 10.1200/OP.21.00172 [DOI] [PubMed] [Google Scholar]
- 2.Tarantino P, Trapani D, Curigliano G. Mastering the use of novel anti-HER2 treatment options. JCO Oncol Pract. 2021;17(10):605-606. doi: 10.1200/OP.21.00216 [DOI] [PubMed] [Google Scholar]
- 3.Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25-32. doi: 10.1016/S1470-2045(11)70336-9 [DOI] [PubMed] [Google Scholar]
- 4.Piccart M, Procter M, Fumagalli D, et al. ; APHINITY Steering Committee and Investigators . Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer in the APHINITY trial: 6 years’ follow-up. J Clin Oncol. 2021;39(13):1448-1457. doi: 10.1200/JCO.20.01204 [DOI] [PubMed] [Google Scholar]
- 5.Loibl S, Jassem J, Sonnenblick A, et al. VP6-2022: adjuvant pertuzumab and trastuzumab in patients with early HER-2 positive breast cancer in APHINITY: 8.4 years' follow-up. Ann Oncol. 2022;33(9):986-987. doi: 10.1016/j.annonc.2022.06.009 [DOI] [Google Scholar]
- 6.Prat A, Guarneri V, Pascual T, et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer. EBioMedicine. 2022;75:103801. doi: 10.1016/j.ebiom.2021.103801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossuyt PM, Reitsma JB, Bruns DE, et al. ; STARD Group . STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waks AG, Desai NV, Li T, et al. A prospective trial of treatment de-escalation following neoadjuvant paclitaxel/trastuzumab/pertuzumab in HER2-positive breast cancer. NPJ Breast Cancer. 2022;8(1):63. doi: 10.1038/s41523-022-00429-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waks AG, Ogayo ER, Parè L, et al. Assessment of the HER2DX assay in patients with ERBB2-positive breast cancer treated with neoadjuvant paclitaxel, trastuzumab, and pertuzumab. JAMA Oncol. Published online April 13, 2023. doi: 10.1001/jamaoncol.2023.0181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark AS, Yau C, Wolf DM, et al. Neoadjuvant T-DM1/pertuzumab and paclitaxel/trastuzumab/pertuzumab for HER2+ breast cancer in the adaptively randomized I-SPY2 trial. Nat Commun. 2021;12(1):6428. doi: 10.1038/s41467-021-26019-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bianchini G, Kiermaier A, Bianchi GV, et al. Biomarker analysis of the NeoSphere study: pertuzumab, trastuzumab, and docetaxel versus trastuzumab plus docetaxel, pertuzumab plus trastuzumab, or pertuzumab plus docetaxel for the neoadjuvant treatment of HER2-positive breast cancer. Breast Cancer Res. 2017;19(1):16. doi: 10.1186/s13058-017-0806-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianchini G, Pusztai L, Pienkowski T, et al. Immune modulation of pathologic complete response after neoadjuvant HER2-directed therapies in the NeoSphere trial. Ann Oncol. 2015;26(12):2429-2436. doi: 10.1093/annonc/mdv395 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Patient characteristics of the GOM neoadjuvant cohort according to treatment (TCH versus TCHP)
eTable 2. Main patient population of the DAPHNe trial cohort and HER2DX sub-population
eTable 3. Main patient population of the ISPY-2 trial cohort with HER2DX data
eFigure 1. CONSORT diagram of the GOM cohort
eFigure 2. Association of HER2DX pCR score with response to pertuzumab in a combined patient-level analysis from GOM, DAPHNe and ISPY-2 neoadjuvant cohorts (n=362)
eFigure 3. Relationship between HER2DX pCR score, treatment and pCR
Data Sharing Statement