Abstract
Cr(VI) (chromate) is a widespread environmental contaminant. Bacterial chromate reductases can convert soluble and toxic chromate to the insoluble and less toxic Cr(III). Bioremediation can therefore be effective in removing chromate from the environment, especially if the bacterial propensity for such removal is enhanced by genetic and biochemical engineering. To clone the chromate reductase-encoding gene, we purified to homogeneity (>600-fold purification) and characterized a novel soluble chromate reductase from Pseudomonas putida, using ammonium sulfate precipitation (55 to 70%), anion-exchange chromatography (DEAE Sepharose CL-6B), chromatofocusing (Polybuffer exchanger 94), and gel filtration (Superose 12 HR 10/30). The enzyme activity was dependent on NADH or NADPH; the temperature and pH optima for chromate reduction were 80°C and 5, respectively; and the Km was 374 μM, with a Vmax of 1.72 μmol/min/mg of protein. Sulfate inhibited the enzyme activity noncompetitively. The reductase activity remained virtually unaltered after 30 min of exposure to 50°C; even exposure to higher temperatures did not immediately inactivate the enzyme. X-ray absorption near-edge-structure spectra showed quantitative conversion of chromate to Cr(III) during the enzyme reaction.
Cr(VI) (chromate) is a widespread industrial and nuclear waste. At the Department of Energy (DOE) sites, for example, it is the second most common heavy metal contaminant, ranging in concentration between 0.008 to 173 μM in groundwater and 98 nM to 76 mM in soil and sediments (15). Since soil water is stored in small capillary spaces, the latter may represent very high concentrations. Chromate is toxic, mutagenic, and probably carcinogenic (20, 22). It is also soluble and therefore does not remain confined to the site of initial contamination. Several bacteria possess chromate reductase activity that can convert chromate to Cr(III), which is much less toxic and less soluble, and thus reduction by these enzymes affords a means of chromate bioremediation.
Field studies have established that biostimulation of indigenous (wild-type) bacteria is an effective means of removing environmental pollutants (10, 11). In this method, nutrients are added to the environments, such as aquifers, to stimulate the growth of indigenous bacteria. Although this enhances the transforming ability of these bacteria, making them more effective agents of bioremediation, it also results in the generation of a large amount of biomass, which confines effective bioremediation to a narrow zone (10). Moreover, most contaminated sites, especially the DOE sites, contain several pollutants (15). The classical biostimulation approach (10) is likely to be of limited use at such sites, since individual enzymes as well as bacteria capable of remediating a given contaminant are inhibited by the presence of the other pollutants (19).
There is increasing recognition, therefore, that bioremediation can benefit from the application of molecular and bioengineering approaches, and an element of the DOE Natural and Accelerated Bioremediation Research Program is aimed at this objective. Thus, engineering of bacteria by using appropriate promoters can minimize the nutrient need and biomass production (8, 9). Furthermore, protein bioengineering can increase the ability, for example of chromate reductases, to function in the presence of other contaminants (19).
To improve bacterial chromate remediation by these approaches, it is necessary to clone the genes that encode chromate reductase activities. Once the promoters and other elements that regulate these genes are known, rational approaches can be devised to improve gene expression under in situ conditions. Cloned genes will also permit facile production of large quantities of pure enzymes using modern molecular methods. Easy availability of pure enzymes will permit detailed investigations of their kinetic and inhibition properties, leading to the identification of targets for improvement; it will also permit determination of high-resolution structure of these enzymes so that the desired improvements can be effectively attempted.
A chromate reductase activity was purified from Pseudomonas ambigua by Suzuki et al. in 1992 (18), and our initial aim was to study this enzyme. However, neither the enzyme nor the bacterium (which is not included in the American Type Culture Collection) was made available to us. Our resulting studies with the common environmental bacterium Pseudomonas putida led to the identification of a new chromate reductase activity in this bacterium that has not been reported previously (4, 18, 21). We describe here the purification to homogeneity and kinetic properties of this novel enzyme; a future report will deal with the molecular regulation of the encoding gene and the physiological role of the enzyme.
While this work was in progress, Suzuki et al. deposited the sequence of the gene encoding their enzyme in GenBank (accession number D83142). Our studies using PCR showed that this gene is absent from P. putida; this is supported by the fact that no homologue to the gene reported by Suzuki et al. is found in the genomic sequence recently put on the World Wide Web by The Institute of Genomic Research.
MATERIALS AND METHODS
Chromate reductase assays.
Chromate reductase activity was determined by measuring the decrease in chromate concentration during enzyme assays. Chromate was quantified, as described previously (2), by adding to the reaction mixture H2SO4 and 1,5-diphenylcarbazide to final concentrations of 0.1 M and 0.01%, respectively, and measuring the absorbance at 540 nm (A540), using a calibration curve relating chromate concentration (0 to 20 μM) to A540.
During purification, the enzyme was assayed at 30°C in 0.5-ml reaction mixtures containing (to a final concentration) 50 mM Tris-HCl buffer (pH 7.0) (T50 buffer), 0.05 mM K2CrO4, 0.1 mM NADH, and 0.1 to 0.3 ml of enzyme preparation. For the data presented in Table 1 and all the other assays, the K2CrO4 and NADH concentrations used were 0.5 and 1.0 mM, respectively; these concentrations were saturating and noninhibitory under these conditions. The optimal pH of the reductase activity was determined using three different buffers, and the possibility that the chemical composition of a given buffer rather than the pH influenced activity was checked by using buffers of overlapping ranges. The following buffers were used: citric acid-NaOH (pH 3 to 6), sodium phosphate (pH 6 to 7), and Tris-HCl (pH 7 to 9). At pH 6, the activity measured with citric acid-NaOH and sodium phosphate buffers was 390 and 330 nmol/min/mg at 50°C, respectively; at pH 7, with sodium phosphate and Tris-HCl buffers, it was 110 and 120 nmol/min/mg at 50°C, respectively. Little disappearance of Cr(VI) occurred in the absence of the enzyme at any pH. The enzyme heat stability was determined by exposing it, before testing, to specified temperatures. The enzyme was then cooled on ice and assayed for residual activity. Experiments relating to enzyme properties (end product, optimal activity, kinetics, inhibitions) were performed at 50°C and pH 5.0, using 50 mM citric acid-NaOH buffer (see Results). The incubation time for determining the optimal pH and temperature was 10 min.
TABLE 1.
Activity parameters at various steps of the chromate reductase purification
| Purification step | Vol (ml) | Total amt of protein (mg) | Total activity (U)a | Sp act (U/mg)ab | Purification factor (fold) |
|---|---|---|---|---|---|
| Crude extract | 302 | 8,760 | 7,446 | 0.85 | |
| Ultracentrifugationc | 407 | 6,198 | 13,016 | 2.1 | 2.5 |
| Ammonium sulfate (55–70%) | 40 | 880 | 3,168 | 3.6 | 4.2 |
| DEAE-Sepharose CL-6B | 3.3 | 6.6 | 1,041 | 158 | 186 |
| Polybuffer exchanger 94d | 1.2 | 0.24 | 78 | 325 | 382 |
| Superose 12 HR 10/30 | 3.0 | 0.03 | 16 | 533 | 627 |
One unit is defined as the amount of enzyme that converts 1 nmol of Cr(VI)/min at 30°C.
Measured in T50 buffer at 30°C.
150,000 × g for 90 min.
This represents peak 1 only.
Chromate reductase purification.
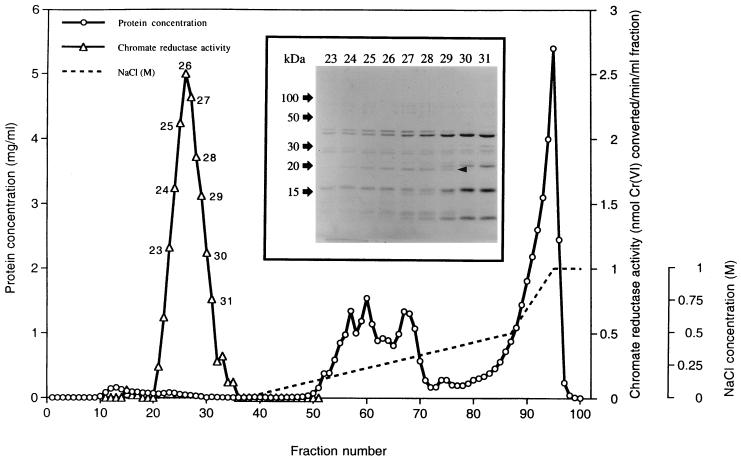
P. putida MK1 (6) (identical to strain ATCC 12633 except for resistance to rifampin) was grown in Luria-Bertani medium (500 ml of medium in 2-liter flasks at 30°C, shaken at 250 rpm) without chromate to an A660 of 2.5 to 3.0. The entire procedure was carried out twice in independent runs, using 20-liter cultures each time. Purification was carried out at 4°C. Following harvesting by centrifugation (10,000 × g for 30 min) and washing in T50 buffer, the cells were suspended in 320 ml of T50 buffer (A660, 80 to 100) and disrupted by passage through a French pressure cell (16,000 lb/in2). The lysate was centrifuged (13,500 × g for 30 min) to remove unbroken cells and debris, resulting in crude extract. This extract was further centrifuged (150,000 × g for 90 min) to remove membrane-associated material, generating the soluble extract. Measurements showed that chromate reductase activity was present exclusively in the soluble extract.
Purification to homogeneity entailed four further steps: ammonium sulfate fractionation, anion-exchange chromatography, chromatofocusing, and gel filtration. For the first-mentioned step, solid ammonium sulfate was added to the extract at various concentrations and the mixture was stirred for 2 h and centrifuged at 10,000 × g for 30 min. The fraction with the highest activity was dialyzed against T50 buffer and applied to an anion-exchange chromatography column (2.5 by 18 cm) containing DEAE Sepharose CL-6B, equilibrated with buffer of the same composition. Proteins were eluted at a flow rate of 30 ml/h: unbound ones with T50 buffer, moderately bound ones with a linear gradient of up to 0.5 M NaCl–T50 buffer, and strongly bound ones with 1 M NaCl–T50 buffer. Selected fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fractions containing the highest chromate reductase activity were pooled, concentrated with a Centricon YM-10 apparatus (Millipore Corp., Bedford, Mass.), dialyzed against 0.025 M Tris-acetic acid buffer (pH 8.3), and used in the chromatofocusing step.
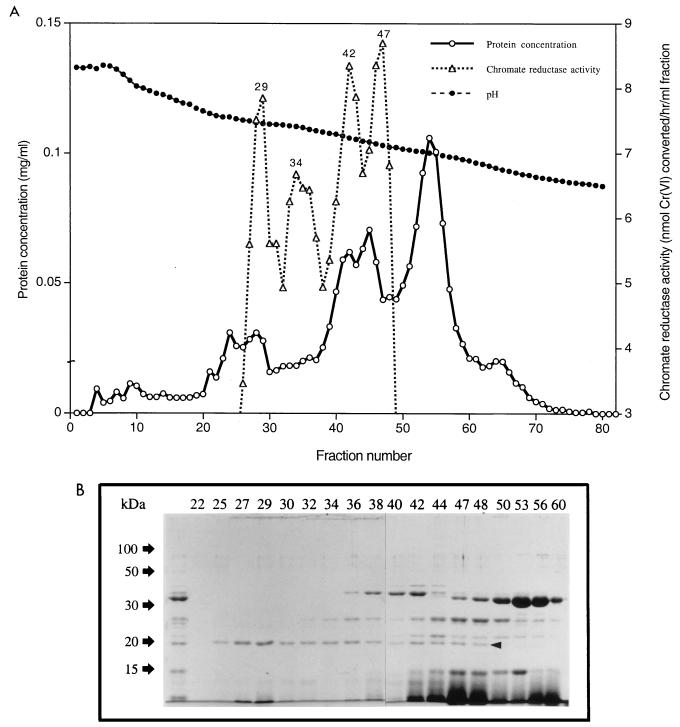
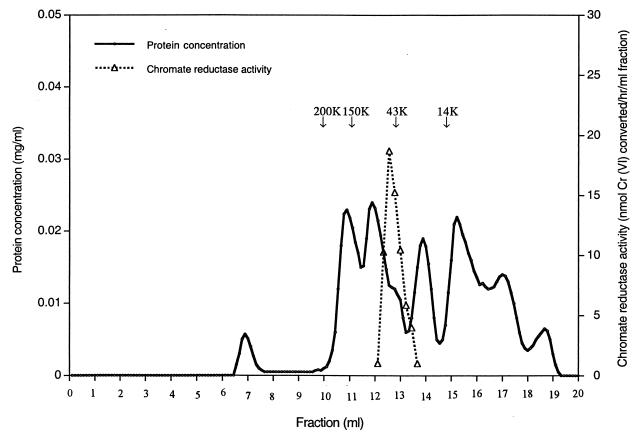
The chromatofocusing step employed a Polybuffer exchanger 94 (Pharmacia Biotech) column (1.5 by 44 cm) equilibrated with 0.025 M Tris-acetic acid buffer (pH 8.3). A 13-fold distilled-water dilution of Polybuffer 96-acetic acid (pH 6.0) served as the eluent. A 5-ml volume of the eluent was applied to the column before the sample to minimize exposure to pH extremes. Proteins were eluted at a flow rate of 20 ml/h until the fraction pH reached ca. 6.0. The chromate reductase activity and pH of each fraction were measured. The fractions showing high chromate reductase activity (fractions 27 to 31 [see Fig. 2A]) were concentrated using Centricon YM-10, dialyzed against 0.2 M NaCl-T50 buffer and used in gel filtration. This employed a Superose 12 HR 10/30 column, with 0.2 M NaCl–T50 buffer as the eluent (run at 15 ml/h) in a fast protein liquid chromatography (FPLC) system. The chromate reductase activity of each fraction was measured. Elution volumes of proteins of known molecular mass (β-amylase [200 kDa], alcohol dehydrogenase [150 kDa], ovalbumin [43 kDa], chymotrypsinogen A [25 kDa], and RNase A [13.7 kDa] dissolved in 0.2 M NaCl–T50 buffer) were measured and used as reference standards in chromate reductase native molecular mass determination.
FIG. 2.
(A) Chromatofocusing using a Polybuffer exchanger 94 column of fractions 23 to 31 containing the chromate reductase activity obtained from ion-exchange chromatography. The column was eluted as described in Materials and Methods, and the chromate reductase activity of each fraction was measured. (B) SDS-PAGE (12% polyacrylamide) analysis of fractions obtained from chromatofocusing (Fig. 2A). The fractions are identified by numbers above the lanes; the unnumbered lane represents the pooled active fractions obtained from anion-exchange chromatography (Fig. 1). A 100-μl volume of each fraction was concentrated 10-fold before loading. The arrowhead shows the position of the presumptive chromate reductase band.
Analytical techniques.
The protein concentration was measured by a Bio-Rad assay kit, using bovine serum albumin as the standard. In the FPLC column eluent, protein was monitored by A280 measurement. PAGE (7) under denaturing conditions was performed with 12 and 4% acrylamide resolving and stacking gels, respectively. A MiniProtean II electrophoresis cell (Bio-Rad) was used, and the gels were stained with Coomassie brilliant blue R-250 or with silver (3).
XANES spectroscopy.
X-ray absorption spectroscopy provides information on the electronic and structural state of an element (17). In the X-ray absorption near-edge-structure (XANES) spectrum, the stable oxidation states of chromium, Cr(VI) and Cr(III), can be distinguished by the pronounced pre-edge feature of the former (5). X-ray absorption spectra were collected on beamline 4-3 at the Stanford Synchrotron Radiation Laboratory run under dedicated conditions at 3 GeV. The ring intensity varied from ca. 100 mA directly after a fill to ca. 40 mA before a fill; energy selection was accomplished using a double monochromator composed of Si(220) crystals. The beam passing through the monochromator was detuned (75% at 6,000 eV) to minimize higher-order harmonics and had a vertical spread of 1 mm. Incident and transmitted X-ray intensities were recorded in 15-cm-long ionization chambers. The fluorescence intensity was used as the absorption measure for unknown samples and was established using a 13-element Ge solid-state detector (1). The spectrometer was calibrated to element Cr (foil), with the inflection point of the metal set to 5,989 eV. Standards of sodium chromate and Cr2O3 were used as reference for end-point oxidation states. Spectra were collected from −100 eV below the edge to 200 eV above, using 0.25-eV steps across the pre- and main-edge regions. For kinetic experiments, the total scan time was minimized so as to acquire successive spectra every 3.1 min. The enzyme reaction was started off-line by adding chromate (final concentration, 1.5 mM). The reaction mixture was then transferred to a 0.5-cm2 Teflon cell, which was sealed with Kapton film, and placed in the spectrometer. The lapse time between starting the reaction and performing the scan was 1.1 min.
Chemicals.
NADH and NADPH were purchased from Boehringer Mannheim (Indianapolis, Ind.). Tris, ammonium sulfate, 1,5-diphenylcarbazide, silver nitrate, β-mercaptoethanol, and Coomassie brilliant blue R-250 were obtained from Sigma (St. Louis, Mo.). The polyvinylidene difluoride membrane was purchased from Bio-Rad Laboratories (Hercules, Calif.). The column chromatography materials were obtained from Pharmacia Fine Chemicals (Uppsala, Sweden). All other chemicals, of analytical grade, were from standard suppliers.
RESULTS
Purification of chromate reductase.
The method and purification parameters are summarized in Table 1. The soluble extract had a 2.5-fold-higher chromate reductase specific activity than the crude extract did. Also, its total reductase activity was almost twice that of the crude extract (Table 1), suggesting that some crude extract component(s) inhibited the enzyme. That the crude extract contains activity-modifying compounds was also found by Suzuki et al. (18) for P. ambigua. The soluble extract was subjected to stepwise ammonium sulfate precipitation, and the highest reductase specific activity fraction (55 to 70% ammonium sulfate saturation) was applied to a DEAE Sepharose CL-6B column.
From this column, the reductase eluted as a sharp peak when the eluent was T50 buffer without NaCl (Fig. 1), indicating that the enzyme did not bind to the column at this pH and that therefore its isoelectric point (pI) is 7.0 or higher. An additional, evidently acidic, pI chromate reductase eluted as a broad peak of low activity at higher NaCl concentrations, which corresponded to approximately 50% of the total activity (data not shown); no further purification of this enzyme was attempted. Fractions of the higher-pI chromate reductase activity were analyzed by SDS-PAGE. A 20-kDa band correlated directly in intensity with the reductase activity of individual fractions (Fig. 1, inset), suggesting that this band represented the enzyme protein. Since little other protein eluted with the reductase (Fig. 1), we were able to achieve a 44-fold purification of the enzyme at the DEAE-Sepharose step, resulting in an overall purification of over 150-fold (Table 1); in replicate experiments, purification of up to 200-fold was attained at this stage.
FIG. 1.
Anion-exchange chromatography of the ammonium sulfate fraction (see Results) on a DEAE-Sepharose CL-6B column. The column was preequilibrated with 50 mM Tris-HCl buffer (pH 7.0). The inset shows an SDS-PAGE gel prepared from the specified fractions (numbers above the lanes). The arrowhead indicates the 20-kDa protein band whose intensity corresponds to chromate reductase activity in individual fractions.
Further purification of the higher-pI reductase (Fig. 1) was done by chromatofocusing. DEAE-Sepharose fractions containing the highest chromate reductase activity (fractions 23 to 31) were pooled, concentrated, applied to a Polybuffer exchanger 94 column, and eluted as described in Materials and Methods. Four closely spaced chromate reductase activity peaks corresponding to pI values of 7.46, 7.41, 7.24, and 7.13 were found in the eluate (Fig. 2A). A 0.1-ml sample of each fraction was concentrated and analyzed by SDS-PAGE (Fig. 2B). The 20-kDa band observed previously was again seen in the active fractions, and the band intensity correlated with the reductase activities in the fractions (Fig. 2), further indicating that it represented the chromate reductase protein. Fractions 27 to 31, which constituted the first peak of chromate reductase activity (Fig. 2A), were chosen for the next purification step, since they showed the highest enzyme specific activity. These were pooled, concentrated (see Materials and Methods), and used in the final stage of purification.
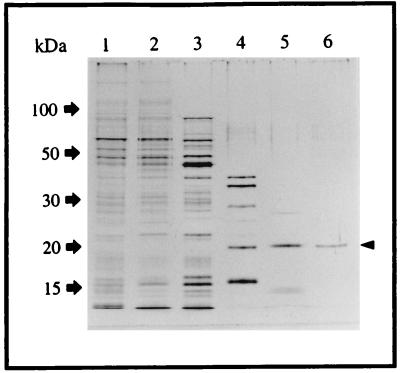
The final purification step employed the FPLC system. The sample was applied to the Superose 12 HR 10/30 column (see Materials and Methods). Chromate reductase eluted as a single peak of activity under these nondenaturing conditions (Fig. 3), which is consistent with the interpretation that a single protein, with a native molecular mass of ca. 50 kDa, is present in fractions 27 to 31 of Fig. 2A. Material from this peak showed only a single band of 20 kDa on a silver-stained SDS-PAGE gel (Fig. 4), further strengthening the conclusion that this peak represents pure chromate reductase activity. The way in which 20-kDa monomers (Fig. 1, 2B, and 4) may give rise to a 50-kDa native protein (Fig. 3) is considered in the Discussion. This stage of purification represents some 600-fold overall purification (Table 1).
FIG. 3.
FPLC of active fractions (fractions 27 to 31) from the chromatofocusing column (Fig. 2A) using a Superose 12 HR 10/30 column. These fractions were concentrated and analyzed. Note the presence of a single sharp peak of chromate reductase activity.
FIG. 4.
Silver stain of SDS-PAGE of proteins at various stages of purification. Lanes: 1, crude extract (0.5 μg of protein loaded on the gel); 2, soluble extract (0.5 μg); 3, ammonium sulfate fraction (0.5 μg); 4, DEAE-Sepharose active fractions (0.3 μg); 5, Polybuffer exchanger 94 active fractions (0.2 μg); 6, gel filtration active fractions (0.1 μg). The arrowhead indicates the position of the chromate reductase.
The other closely spaced peaks of activity seen in Fig. 2A were not further analyzed. Since they differ only slightly in their pI values, they may represent isoenzymes of the major peak of activity that was further purified or minor purification-induced modification of the enzyme protein.
Analysis of chromate reductase activity showing fractions at each purification step by silver staining of SDS-PAGE gels is shown in Fig. 4. The data confirm that the 20-kDa band does indeed represent chromate reductase activity, and since the resolving power of silver staining is <1 ng (3), they are consistent with the conclusion that the final step resulted in complete purification of the chromate reductase activity.
Optimum reaction conditions and stability.
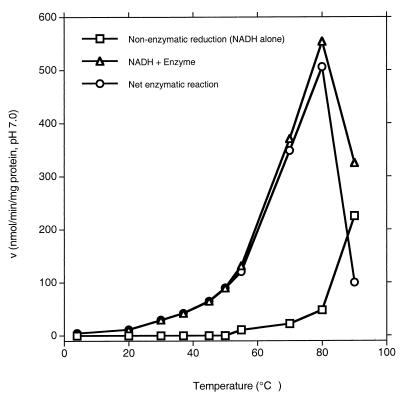
A ca. 200-fold-purified chromate reductase preparation (active fractions obtained at the DEAE-Sepharose CL-6B step [Table 1]) was used in these and subsequent studies. The enzyme absolutely required NADH or NADPH for activity, and both were equally effective (data not shown). The reductase became sharply more active as the reaction temperature increased up to 80°C and then showed a sharp decline as the temperature was increased further (Fig. 5). Nonenzymatic conversion of chromate became significant only above ca. 70°C.
FIG. 5.
Effect of reaction temperature on chromate reductase activity. Net enzymatic reaction rates were obtained by subtracting nonenzymatic reduction from reduction in the presence of enzyme and NADH. The assay was conducted at pH 5.0, using 50 mM citric acid–NaOH buffer (pH 5.0).
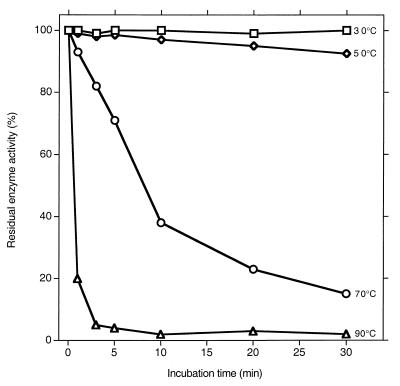
The linearity of the enzyme reaction rates appeared to last for shorter durations at higher temperatures (data not shown). Since one explanation for this is loss of enzyme activity, we measured enzyme stability at different temperatures. Following incubation of the enzyme for different periods at various temperatures, its residual activity was measured at 50°C. Little loss of activity was seen upon a 30-min exposure to 30 or 50°C, but appreciable inactivation occurred at higher temperatures (Fig. 6). For this reason, even though the activity at 50°C was only 20% of that at 80°C, the former temperature was used in further studies. The optimal pH was 5.0. That it was the pH and not the chemical composition of a given buffer that determined the activity is suggested by the fact that different buffers gave very similar activities at pH 6 and 7 (see Materials and Methods).
FIG. 6.
Residual enzyme activity after incubation at the indicated temperatures for the indicated duration. The assay was conducted at 50°C using 50 mM citric acid–NaOH buffer (pH 5.0).
Enzyme kinetics.
The apparent Km value obtained from the Lineweaver-Burk plot was 374 μM CrO42−; the Vmax was 1.72 μmol/min/mg of protein (data not shown). Inhibition of the enzyme activity by selected metals and anions was also determined. Cd2+ and Zn2+, when supplied as chloride salts, had little effect at concentrations up to 10 mM, and AsO43− was also not inhibitory (Table 2). CdSO4, however, produced significant inhibition at 1 mM and above, suggesting that the SO42− was inhibitory. Since CuSO4 was no more inhibitory than CdSO4, Cu2+ was also noninhibitory. Kinetic measurements, using CdSO4 concentrations of 0 to 9 mM (at 0.1 to 0.5 mM chromate concentrations), showed that sulfate inhibited the enzyme noncompetitively (Ki ≈ 11 mM [data not shown]).
TABLE 2.
Effect of selected inhibitors on chromate reductase activity
| Chemical and concn (mM) | Remaining activity (%) in:
|
|
|---|---|---|
| Buffer 1a | Buffer 2a | |
| CdCl2 | ||
| 1 | 97 | 113 |
| 3 | 92 | 107 |
| 10 | 86 | 89 |
| CdSO4 | ||
| 1 | 69 | 63 |
| 3 | 60 | 35 |
| 10 | 5 | 6 |
| CuSO4 | ||
| 5 | 31 | NDb |
| 10 | 15 | ND |
| 25 | 15 | ND |
| KH2AsO4 | ||
| 1 | 105 | ND |
| 5 | 98 | ND |
| 10 | 80 | ND |
| ZnCl2 | ||
| 0.1 | 98.6 | ND |
| 1 | 85.5 | ND |
| 5 | 85 | ND |
Buffer 1 is citric acid-NaOH buffer, and buffer 2 is sodium acetate buffer. Since the activities found in the two buffers were very similar, not all determinations were made using both the buffers.
ND, not determined.
End product of chromate conversion.
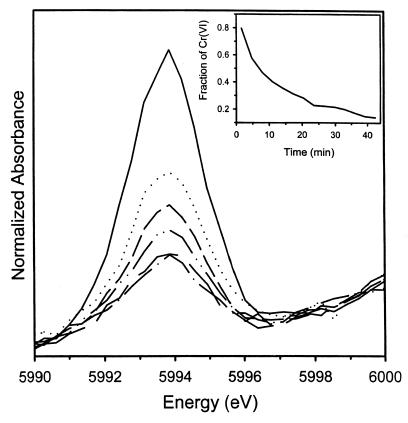
As stated in Materials and Methods, the end product was determined by XANES; chromium was speciated using the spectral analysis and calibration procedure of Patterson et al. (13). The fraction of Cr(VI) was calculated by dividing the height of the Cr(VI) pre-edge peak by the total atomic absorption; that of Cr(III) was calculated from the difference between the amount of chromium represented by the pre-edge peak and the total absorption jump (Fig. 7 and 8). Good agreement (r2 ≥ 0.988) was found between the known and calculated amounts of Cr(VI) in standards by using this method.
FIG. 7.
XANES spectra of chromium at successive times after chromate was reacted with chromate reductase. The reduction in the pre-edge intensity relative to the main-edge intensity with time denotes the progressive transformation of Cr(VI) to Cr(III).
FIG. 8.
The isolated pre-edge feature of the Cr-XANES spectra illustrating diminished intensity with increased reaction time. (Inset) Fraction of Cr(VI) in the sample as a function of time determined from the ratio of the pre-edge to main-edge intensity.
The relative contribution of the pre-edge feature in the XANES spectra diminished progressively with time during the reaction (Fig. 7 and 8), indicating a diminishing fraction of Cr(VI). At 2.7 min of the reaction, Cr(VI) constituted more than 90% of the total chromium in the sample, but by 40 min, it was depleted to less than 10% (inset, Fig. 8), with a proportionate increase in Cr(III).
Cr(V) also produces a pre-edge peak in the XANES spectrum, which is slightly shifted (ca. 1 eV) to lower energy. The sensitivity (>5%) of our assay was not high enough to permit a determination of whether Cr(V) accumulated during the transformation. Cr(V) is an unstable species, and if it was produced during the reaction, its accumulation is likely to be only transient.
DISCUSSION
Only a single peak of chromate reductase activity was detected at the final (FPLC) stage of purification, and the silver-stained gels of active fractions at this stage showed only a single band (Fig. 4). This is the first report of purification to homogeneity of a bacterial chromate reductase. Ishibashi et al. (4) could purify the enzyme from P. putida PRS 2000 only partially, while Suzuki et al. (18) achieved a 38-fold purification of chromate reductase from P. ambigua. From the N-terminal and internal amino acid sequence of our purified enzyme and the sequence data of P. putida obtained from The Institute of Genomic Research, we have determined the sequence of our chromate reductase-encoding gene, which confirms that the enzyme is indeed a novel protein and embodies features of other reductases (C. H. Park, M. Keyhan, and A. Matin, unpublished data).
That the chromate reductase purified in this study from P. putida MK1 is a new enzyme is supported by its properties. It is soluble, while that of Enterobacter cloacae (21) is membrane bound; although the P. putida PRS2000 and P. ambigua reductases are also soluble (4, 18), they exhibit different properties from that of the MK1 enzyme. For example, the MK1 enzyme is optimally active at 80°C, but the optimal reaction temperature of the P. ambigua enzyme is 50°C; the MK1 enzyme has a pH optimum of 5.0, while P. ambigua and PRS2000 enzymes have pH optima of 8.6 and between 6.5 and 7.5, respectively. Similarly, the Km values of the three enzymes are different: 370 μM for MK1 versus 13 and 40 μM for the P. ambigua and PRS2000 enzymes, respectively. The MK1 enzyme is inhibited by sulfate, while the PRS2000 enzyme is not. The elution patterns of the MK1 and PRS2000 enzymes from anion-exchange columns also point to differences between them, with the pI value of the former being higher than 7.0 (Fig. 1) and that of the latter being lower than 7.0 (4).
The MK1 enzyme gave a monomer molecular mass on SDS-PAGE of 20 kDa and a molecular mass for the native protein on gel filtration of ca. 50 kDa. Therefore, it is not known whether the enzyme is a dimer or a trimer. The P. ambigua chromate reductase exhibited a similar behavior, with a molecular mass of 25 kDa on SDS-PAGE and a mass of 65 kDa on gel filtration (18). Proteins can exhibit nonproportionate movement upon SDS-PAGE and gel filtration for several reasons. One possibility is that both reductases possess intrasubunit disulfide cross-linkages that, by producing altered conformation and Stokes radii, influence protein movement in gels differently (16).
The N-terminal amino acid of the MK1 enzyme is not methionine (Park et al., unpublished), raising the possibility that it is a periplasmic protein. Most periplasmic proteins are synthesized with a leader sequence that is cleaved off as they cross the inner membrane (14). Since chromate can easily pass through the membranes and in the cytoplasm is undoubtedly toxic to the bacterium (20), it would be advantageous to the cell to have chromate detoxification capability localized close to the cell surface. However, although the growth of MK1 is not affected by chromate up to a concentration of 0.8 mM (Park et al., unpublished), it remains to be determined if the reductase contributes to this resistance. Anaerobic growth of P. putida MK1 was not supported by chromate (Park et al., unpublished).
The question of the role that chromate reductases play in bacterial physiology has not been explored. The E. cloacae enzyme might be involved in anaerobic respiration, with chromate as the electron acceptor (21), but no definite role for the soluble reductases has been established. In Pseudomonas fluorescens, chromate reductase activity was equally strong in chromate-resistant and -sensitive strains, suggesting that this activity did not confer protection against chromate toxicity (12). Ishibashi et al. (4) suggested that chromate reduction might be a vicarious property of reductases with different primary roles. The fact that we found an additional peak of chromate reductase activity upon ion-exchange chromatography, as did Suzuki et al. (18), may be consistent with this possibility.
However, whether or not the soluble chromate reductases have an actual physiological purpose of detoxifying Cr(VI), such activities can still be useful in bioremediation. Indeed, cometabolic processes account for many successful bioremediation approaches. For example, methane monooxygenase of methanotrophs has been exploited to bioremediate trichloroethylene, even though the physiological role of this enzyme is to oxidize methane (10). The soluble chromate reductases can also be similarly useful in cometabolic chromate remediation. The fact that the optimal activity of our enzyme occurs at a high temperature does not detract from its potential usefulness in chromate remediation. It is not unusual for even essential enzymes to exhibit maximum activity in vitro under conditions different from the optimal growth conditions of an organism; possibly their maximal activity in vivo more closely matches the organism's optimal growth conditions (7a). Similarly, the P. putida enzyme studied here still exhibits significant activity at 30°C, even though it is less than 1/10 the maximal activity; this may be amenable to further enhancement by protein engineering. Further, the gene could be cloned into a thermophile, and in an immobilized enzyme-bioreactor-based remediation system, its thermotolerant product can have a decisive advantage. These considerations also apply to the chromate reductase purified by Suzuki et al. (18), which has a temperature optimum of 50°C; the optimal temperatures for other chromate reductases reported so far remain to be determined.
Systematic studies are needed to determine the real nature of activities so far identified as chromate reductases; it is pertinent to note in this connection that we have found chromate reductase activity in enzymes characterized in completely different contexts (Park et al., unpublished). A future communication will deal with the cloning and molecular regulation of the gene encoding the enzyme purified in this study, the effect of a knockout mutation of this gene on P. putida physiology, and comparative molecular features of the various enzymes that exhibit chromate reductase activity. This information will greatly facilitate the use of protein and genetic engineering to enhance the chromate remediation potential of P. putida.
ACKNOWLEDGMENT
This work was supported by Department of Energy grant DE-FG03-97ER-62494-A002 to A.M.
REFERENCES
- 1.Cramer S P, Tench O, Yocum M, George G N. A 13-element Ge detector for fluorescence EXAFS. Nuclear Instr Methods Phys. 1988;A266:586–591. [Google Scholar]
- 2.Greenberg A E, Connors J J, Jenkins D, Franson M A, editors. Standard methods for the examination of water and wastewater. 15th ed. Washington, D.C.: American Public Health Association; 1981. pp. 187–190. [Google Scholar]
- 3.Heukeshoven J, Dernick R. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis. 1985;6:103–112. [Google Scholar]
- 4.Ishibashi Y, Cervantes C, Silver S. Chromium reduction in Pseudomonas putida. Appl Environ Microbiol. 1990;56:2268–2270. doi: 10.1128/aem.56.7.2268-2270.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jardine, P. M., S. E. Fendorf, M. A. Mayes, I. L. Larsen, S. C. Brooks, and W. B. Bailey. Fate and transport of hexavalent chromium in undisturbed heterogeneous soil. Environ. Sci. Technol., in press.
- 6.Kim Y, Watrud L, Matin A. A carbon starvation survival gene of Pseudomonas putida is regulated by ς54. J Bacteriol. 1995;177:1850–1859. doi: 10.1128/jb.177.7.1850-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 7a.Matin A. Regulation of enzyme synthesis as studied in continuous culture. In: Calcott P H, editor. Continuous culture of cells. II. Boca Raton, Fla: CRC Press, Inc.; 1981. pp. 69–97. [Google Scholar]
- 8.Matin A. Starvation promoters of Escherichia coli: their function, regulation and use in bioprocessing and bioremediation. Recombinant DNA technology II. Ann N Y Acad Sci. 1994;721:277–291. doi: 10.1111/j.1749-6632.1994.tb47401.x. [DOI] [PubMed] [Google Scholar]
- 9.Matin A, Little C D, Fraley C D, Keyhan M. Use of starvation promoters to limit growth and selectively express trichloroethylene and phenol transformation activity in recombinant Escherichia coli. Appl Environ Microbiol. 1995;61:3323–3328. doi: 10.1128/aem.61.9.3323-3328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty P L. Ground-water treatment for chlorinated solvents. In: Matthews J E, editor. Handbook of bioremediation. Ann Arbor, Mich: Lewis Publishers; 1994. pp. 87–116. [Google Scholar]
- 11.Norris R D. In-situ bioremediation of soils and groundwater contaminated with petroleum hydrocarbons. In: Matthews J E, editor. Handbook of bioremediation. Ann Arbor, Mich: Lewis Publishers; 1994. pp. 17–37. [Google Scholar]
- 12.Ohtake H, Cervantes C, Silver S. Decreased chromate uptake in Pseudomonas fluorescens carrying a chromate resistance plasmid. J Bacteriol. 1987;169:3853–3856. doi: 10.1128/jb.169.8.3853-3856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson R R, Fendorf S E, Fendorf M J. Reduction of chromate by amorphous iron sulfide. Environ Sci Technol. 1997;31:2039–2044. [Google Scholar]
- 14.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley R G, Zachara J M, Wobber F J. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface science research. Report DOE/ER-0547T. U.S. Washington, D.C.: Department of Energy; 1992. [Google Scholar]
- 16.Smith B J. SDS polyacrylamide gel electrophoresis of proteins. Methods Mol Biol. 1994;32:23–34. doi: 10.1385/0-89603-268-X:23. [DOI] [PubMed] [Google Scholar]
- 17.Stohr J. NEXAFS spectroscopy. New York, N.Y: Springer-Verlag; 1992. [Google Scholar]
- 18.Suzuki T, Miyata N, Horitsu H, Kawai K, Takamizawa K, Tai Y, Okazaki M. NAD(P)H-dependent chromium(VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate is formed during the reduction of Cr(VI) to Cr(III) J Bacteriol. 1992;174:5340–5345. doi: 10.1128/jb.174.16.5340-5345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timmis K N, Steffan R J, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 20.Venitt S, Levy L S. Mutagenicity of chromate in bacteria and its relevance to chromate carcinogenesis. Nature. 1974;250:493–495. doi: 10.1038/250493a0. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Mori T, Komori K, Sasatsu M, Toda K, Ohtake H. Isolation and characterization of an Enterobacter cloacae strain that reduces hexavalent chromium under anaerobic conditions. Appl Environ Microbiol. 1989;55:1665–1669. doi: 10.1128/aem.55.7.1665-1669.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wetterhahn K E, Hamilton J W. Molecular basis of hexavalent chromium carcinogenicity: effect on gene expression. Sci Total Environ. 1989;86:113–129. doi: 10.1016/0048-9697(89)90199-x. [DOI] [PubMed] [Google Scholar]