Summary
Background
DNA methylation (DNAm) in cord blood has been associated with various prenatal factors and birth outcomes. This study sought to fill an important knowledge gap: the link of cord DNAm with child postnatal growth trajectories from birth to age 18 years (y).
Methods
Using data from a US predominantly urban, low-income, multi-ethnic birth cohort (N = 831), we first applied non-parametric methods to identify body-mass-index percentile (BMIPCT) trajectories from birth to age 18 y (the outcome); then, conducted epigenome-wide association study (EWAS) of the outcome, interrogating over 700,000 CpG sites profiled by the Illumina Infinium MethylationEPIC BeadChip. Multivariate linear regression models and likelihood ratio tests (LRT) were applied to examine the DNAm-outcome association in the overall sample and sex strata.
Findings
We identified four distinct patterns of BMIPCT trajectories: normal weight (NW), Early overweight or obesity (OWO), Late OWO, and normal to very late OWO. DNAm at CpG18582997 annotated to TPGS1, CpG15241084 of TLR7, and cg24350936 of RAB31 were associated with BMIPCT at birth-to-3 y, 10 y, and 14 y, respectively (LRT FDR < 0.05 for all).
Interpretation
In this prospective birth cohort study, we identified 4 distinct and robust patterns of growth trajectories from birth to 18 y, which were associated with variations in cord blood DNAm at genes implicated in inflammation induction pathways. These findings, if further replicated, raise the possibility that these DNAm markers along with early assessment of BMIPCT trajectories may help identify young children at high-risk for obesity later in life.
Funding
Detailed in the Acknowledgements section.
Keywords: BMI percentiles, Critical periods, Childhood obesity, Epigenetics
Research in context.
Evidence before this study
Epigenetic signatures such as DNA methylation in cord blood might mirror prenatal nutritional differences and metabolic programming that may affect future risk of overweight or obesity (OWO). However, data are very limited on the link between cord epigenetic marks and child postnatal body mass index (BMI) longitudinal trajectories.
Added value of this study
This longitudinal birth cohort study offers an opportunity to investigate the prospective association between cord DNA methylation and postnatal BMI trajectory and avoid reverse causation. Using methods for unsupervised learning clustering and repeated BMI measurements, we defined four distinct BMI trajectory groups from birth to age 18 years. We performed epigenome-wide association study (EWAS) to uncover differentially methylated CpG sites between the BMI trajectory groups.
Implications of all the available evidence
This study showed significant link between cord DNA methylation marks with postnatal BMI trajectories. These findings warrant additional investigation and raise the possibility that combining DNA methylation markers with early assessment of BMI trajectories may help identify young children at high-risk for obesity later in life, thus offers the window of opportunity for precision early risk assessment and targeted prevention.
Introduction
Overweight or obesity (OWO) in childhood is classified by body-mass-index percentiles (BMIPCT) calculated according to age and sex-specific growth charts. The Center for Disease Control (CDC) and the World Health Organization (WHO) growth charts1,2 indicate the cut-off for overweight at >85th percentile, with >95th (CDC) or 97th (WHO) considered obesity.3 Low birth weight (<2500 g)4 or early development of obesity as early as the age of 2 years predicted obesity 30 years later,5 especially in children with severe obesity.6 Within the period from birth to young adulthood (age of 18 years), several obesity-related critical periods were highlighted7; for example, obesity at 3 years is a predictor for OWO in adolescence. Of adolescents with obesity, about half were OWO at the age of 5 years; The greatest acceleration in body mass index (BMI) occurs between 2 and 6 years of age. This acceleration is associated with the risk of OWO in adolescents. In contrast, most adolescents with normal weight had a normal weight from birth to 14 years. OWO children that become OWO adults have an increased risk of cardiometabolic diseases8 and cardiovascular mortality.9
Epigenetic modifications include changes to the genome that can alter gene expression without changing the DNA sequence,10 with DNA methylation (DNAm) as one of the mechanisms that underlie epigenetic regulation.11 Epigenetic alterations can be induced by several environmental factors, such as environmental carcinogens, alcohol, tobacco, exercise, and diet, and can occur during all life periods, from the embryonic and fetal stages to adulthood.12 Genotype and lifestyle are known risk factors for obesity, and increasing evidence indicates that these influences may start as early as the fetal and post-natal periods, altering the epigenetic regulation of specific genes central to this process.13
Cord DNAm represents the newborn epigenetic state during pregnancy and is potentially affected by maternal exposures and maternal and newborn genetics; Cord DNAm was associated with maternal exposures as pre-pregnancy weight and weight gain during pregnancy,14,15 maternal smoking,16 depression,17 and prenatal arsenic exposure.18 On the other hand, previous studies have linked cord DNAm with a child's weight in different life periods,19 body size,20 adiposity,21 and blood pressure percentiles.15 Yet, data regarding the association of cord DNAm with repeated and frequent weight measurements throughout years from birth to adolescence is limited but critically needed.
For this analysis, we used data from the ongoing, prospective Boston Birth Cohort (BBC), which has followed the study's children from birth up to age 21 years with a collection of repeated measurements of height and weight over time, offering the opportunity to define BMI longitudinal trajectory. Our study objectives were to examine the association between cord DNAm and child BMI trajectory across different stages of child growth and development, from birth to 1 year, 2 years, 3 years, 6 years, 10 years, and 14 years, and the entire paediatric age birth to 18 years. We further examined the association between cord DNAm and BMI trajectory among boys and girls, respectively, to identify potential sex differences.
Methods
This study included 831 mother–newborn pairs from the BBC (registered in ClinicalTrial.Gov NCT03228875), a US predominantly urban, low-income, multi-ethnic population. The BBC was initiated in 1998 with rolling enrollment at the Boston Medical Center in Boston, MA, as detailed elsewhere.22,23 In brief, mothers who delivered a singleton live birth at the Boston Medical Center were invited to participate 24–72 h after delivery. The BBC is enriched by preterm (<37 weeks of gestation) and low birthweight (<2500 g) births by design of over-sampling PTB at enrollment. Pregnancies resulting from in vitro fertilisation, multiple gestations (e.g., twins, triplets), fetal chromosomal abnormalities, major birth defects, or preterm birth due to maternal trauma were excluded. After mothers provided written informed consent, research assistants (RAs) administered a standardised questionnaire interview on maternal socio-demographic characteristics, lifestyle, including smoking and alcohol consumption, diet, and reproductive and medical history. Maternal and newborn clinical information, including birth outcomes, was abstracted from the medical records.
DNAm profiling in cord blood and quality control steps
Cord blood was obtained by the trained nursing staff of the labour and delivery service; the quality of the DNA samples has been demonstrated in our previous studies using the Illumina BeadChip.24 963 cord DNA samples (plus 21 replicates) were sent to the University of Minnesota Genomics Center for genome-wide DNAm profiling using the Illumina Infinium MethylationEPIC BeadChip (850K). With this platform, DNAm profiles for a total of 865,859 CpG sites (at locations of cytosines followed by guanine) were successfully generated, and a β-value for each CpG site was examined and reported, ranging from 0 to 1.0, to signify the percentage of DNAm at each CpG site. Quality control steps were detailed before.16 Briefly, systematic quality control steps were performed using existing analytic pipelines with the R/Bioconductor package ‘minfi’,25 and the distribution of each cell type (CD4+, CD8+, T cells, B cells, monocytes, granulocytes, natural killer cells, and nucleated red blood cells) was estimated.26 We excluded 7 sex mixed-up samples, 2 samples with call rate <98% methylation sites, 12 samples with mean log2 intensity <10, and 2 samples with logistic error. At the locus level, 4193 CpG sites with a detection p value >0.01 in more than 5% of the samples were removed. An additional 140,271 CpG sites were removed for the following reasons: having an annotated SNP at the measured or extension site or that overlapped with the probe and/or that potentially cross-hybridised to other genomic locations. We then performed the single-sample Noob (ssNoob) methods for background and dye bias correction27 and quantile normalisation to normalise type I and type II probes. These steps resulted in quality DNAm data for 721,395 CpG sites in 940 samples for subsequent analyses.
Sample selection: obesity groups and obesity-related critical period calculations
Out of the data available for the BBC, we selected 3029 children with height and weight measured in the follow-up visits. We calculated BMIPCT using WHO (birth to 2 years old1) and CDC growth charts (age 2 years and up2) for these children. As the frequencies of a child's standard well-care visits decreases by age, we defined the time windows of the BMIPCT data into 32 time points (Fig. S1): monthly measurements in the first year, quarterly measurements in the second, and yearly measurements from month 36 (3rd year) to the 216th month. We selected to focus on seven overlapping age periods representing different developmental phases. These included the BMIPCT trajectories from visit 1 after birth to the end of each age period: birth to 1 year, birth to 2, birth to 3, birth to 6, birth to 10, birth to 14, and birth to 18 years. To complete missing data (for the primary analysis), we fitted a LOWESS (locally weighted scatterplot smoothing) curve,28 as we demonstrated.29 Briefly, we adopted a locally weighted scatterplot smoothing curve within a time-limit aware scheme (LOWESS-TLS). Each missing BMIPCT data was predicted using a regression curve (LOWESS) of the available observations within the reasonable time frame around the missing time point. For the missing monthly data from birth to 1 year old, observations no more than 1 year apart were used; for missing quarterly data between 1 and 2 years old, imputation was allowed no more than one year away; for subsequent missing annual data, imputation was allowed from information no more than two years apart. The range of time limit at different ages was determined by our paediatrician epidemiologist based on the density of data availability and relevance to child growth. Based on the first age period in this analysis (birth to 1) and availability of children with DNAm analysis, we selected children that included at least one BMIPCT measurement at the first age period and refined the similar sample size at each period, thus resulting in a sample size of 831 for each age period allowing us to follow the observed or discovered trajectory and examine changes between OWO groups over time (A flow diagram of the sample selection for the primary analysis is presented in Fig. S2). The OWO groups, which represent the growth trajectory for seven age periods using BMIPCT of the observed and imputed data, were constructed separately for each period, as follows: first, we applied k-means clustering with k = 2 for each matrix containing the BMIPCT measurements (seven matrices, one for each age period). In each period, applying the k-means clustering identified two major groups. Next, we used Principal Component Analysis (PCA) to find the 1st and 2nd principal components. When examining the relationship between k-means clustering and PCA plot, we observed that PC1 drove the k-means clustering groups. A larger k corresponded to a more refined grouping based on the first principal component. To further obtain informative grouping, we chose to use the second principal component to further divide the two groups into 4 groups (Fig. S3), as previously demonstrated.30 The OWO groups resulting from this procedure represent four distinctive trajectories named retrospectively after calculating and plotting all BMIPCT trajectories: 1. Early OWO: children with early onset OWO who demonstrated a consistently high BMIPCT from birth to the end of each age period; 2. Late OWO: late onset OWO children that were NW at birth but experienced a rapid weight increase in the first months of life to become OWO by year 1; 3. NW to very late OWO: children distinguished from the late OWO by maintaining NW at early ages but becoming OWO by year 6; 4. NW children consistently kept NW from birth to the end of each age period. Table S1 presents the characteristics of the 831 children with calculated OWO groups and DNAm data used in this study compared to 2198 without OWO calculation.
Sample selection: time window analysis
For examining the association between cord DNAm and BMI at different ages (point prevalence), we used the available BMI data for the following 21 “time windows”: month 2 ± 1 to month 18 ± 1, month 20–24, month 36–48, month 48–60, month 60–72, month 72–84, month 84–96, month 96–108, month 108–120, month 120–132, month 132–144, month 144–158, and month 168 to the maximum age with available BMI measurement (if more than one measurement available we picked the latest one). We further stratified the data according to the reported sex of the child and repeated the association analysis of BMI with DNAm for each time window.
Definition of main covariates
For background characteristics and adjustment of the statistical models, we used the following data: maternal age at delivery, parity (Nulliparous or Multiparous), maternal education (below college or college and higher), maternal self-reported race (Black/African American, White, and Hispanic), maternal pre-pregnancy BMI, maternal diabetes (non, gestational diabetes or pre-existing DNAm), child's sex (female vs male; abstracted from the medical records, and verified during the DNAm data quality control), maternal smoking, birth weight (as continuous and binary with below 2500 g defined as the low birth weight (LBW)4), and gestational age (GEAA) at birth as continuous and binary with above/below week 37 defined as “preterm”.4 The estimation of GEAA was detailed before22 and was performed using an established algorithm based on both the last menstrual period and the result of early ultrasound (<20 weeks gestation). Standardised birth weight (SBWT) was defined as birth weight standardised by the mean and variance in the stratum of the corresponding ethnic group, sex, and gestational week in the reference population, as detailed before.31
Statistical analysis
The primary aim of this report is to examine the association between cord DNAm and BMIPCT growth trajectories across several age periods during childhood. Growth trajectories were classified into OWO groups, as detailed above. The secondary aims included examining the association of cord DNAm with birth weight/GEAA ratio and BMI at multiple selected time windows for the entire available cohort and stratified by sex. Weight and BMI data for the secondary analysis were not imputed. Prenatal and perinatal characteristics across subgroups of child BMI longitudinal trajectories from birth to 18 years were examined using the chi-square test for categorical variables and ANOVA for continuous variables. All statistical analyses were performed using R (version 4.1; R Foundation for Statistical Computing). For the regional plots, we used the coMET package.32 The “CMplot”33 and “ggplot2”34 were also used to output the main figures.
Identification of significantly associated CpG sites
To minimise the effect of outliers and ensure normality, we applied an inverse normal transformation to obtain inversely normalised β-values for each methylation site. We examined differential cord DNAm between OWO groups using the lmFit function in the “limma”35 package. The potential batch effect was estimated and accounted for using the “SmartSVA” package,36 with models additionally adjusted with calculated surrogate variables (SVs) for each of the linear models in this study. The linear regression models in the primary analysis included the inversely-normalised β-value at each CpG site as the outcome and the OWO groups (separately for each of the seven age periods) as the predictor, adjusting for potential confounders (“main model”): child's sex, maternal education, race, parity, GEAA, maternal diabetes, low birth weight (as binary outcome due to co-linearity with GEAA) and SVs. Since using “SmartSVA,” we did not adjust for cell-type composition, which was represented by some of the SVs.37 We performed sensitivity analyses for the primary outcome of the association between cord DNAm and OWO groups, including adding either folic acid or estimated cord blood cell composition to the main model and omitting LBW from the main model. The false discovery rate (FDR)38 was applied to correct for multiple testing, with FDR <0.05 as the genome-wide significance cutoff. A Likelihood Ratio Test (LRT) was applied to examine the goodness-of-fit between a full model, including the OWO groups, and a reduced model without. The LRT included the same covariates as the “limma” model, detailed above. We extracted the significant CpG sites resulting from the LRT and considered them significant associations with the OWO groups. For the secondary analysis, examining the associations of cord DNAm with SBWT and BMI at the range of ages, we used LRTs, with adjustments for potential confounders similar to the main model and detailed along with the results. We examined the BMI∗sex interactions using a joint model (joint testing the BMI main effect and BMI∗sex interaction term). The association between cord DNAm and BMI was also analysed using the LRT in stratification for sex.
Ethics
Written informed consent was obtained from all the study mothers. Institutional Review Boards of the Boston Medical Center and the Johns Hopkins Bloomberg School of Public Health approved the study. The study is also registered on ClinicalTrials.gov (NCT03228875).
Role of funders
The funder of the study had no role in study design, data collection, data analysis, data interpretation, the writing of the report, or the submission for publication.
Results
Population characteristics
Maternal and child characteristics across birth to 18 OWO groups are presented in Table 1. We further stratified these characteristics by sex and OWO group (Table S2). Significant differences between the OWO groups were observed for mothers with no previous childbirths (nulliparous) vs multiparous (p = 0.005; chi-square test), with the highest prevalence of nulliparous observed in the late OWO group, and the highest prevalence of multiparous was observed in the early OWO group. Differences in OWO prevalence in children of mothers with diabetes (either gestational or pregestational diabetes) vs children of a mother without any type of diabetes were observed (0.011; chi-square test), with most mothers with gestational diabetes in the NW group and most mothers with pregestational diabetes in the NW to very late OWO group. The early OWO group had the highest birth weight (p = 8.5e-06 vs late OWO; p = 2.5e-10 vs NW to very late OWO; p = 2.0e-08 vs NW; all ANOVA posthoc test Bonferroni adjusted) and had a later birth week compared with the late OWO and the NW to very late OWO (p = 0.011 and p = 0.009, respectively; ANOVA test). Maternal pre-pregnancy BMI was lowest in the NW group and significantly differed from the late OWO group (p = 4.3e-04; ANOVA test). The mean and median BMIPCT across OWO groups for the measurements included in the seven age periods calculation are presented in Table S3. The early OWO group showed a stable high BMIPCT during the first year, with a median BMIPCT of 77.6 [inter-quartile-range (IQR) = 27.6] in the first month and a median of 76.1 (IQR = 27.4). The late OWO had a median BMIPCT of 33.2 (IQR = 39.8) in the first month and gradually increased their BMIPCT to a median of 87.6 (IQR = 21.8) by the end of the first year of life. The other two groups remain at the lower BMIPCT, with the NW to very late OWO group having a median BMIPCT of 6.6 (first month; IQR = 16.6) and 37.2 (end of year 1; IQR = 33.5), and the NW group had a median BMIPCT of 48.6 (IQR = 38.0), that decreased to 28.2 (IQR = 30.0).
Table 1.
Prenatal and perinatal characteristics across subgroups of child BMI longitudinal trajectories from birth to 18 years.a
| Entire (N = 831) | Early OWO (N = 274) | Late OWO (N = 258) | NW to very late OWO (N = 140) | NW (N = 159) | p-valueb | |
|---|---|---|---|---|---|---|
| Maternal age at delivery (years) | 0.057 | |||||
| Mean (SD) | 28.4 (6.54) | 29.1 (6.46) | 28.5 (6.42) | 27.8 (6.80) | 27.4 (6.54) | |
| Maternal pre-pregnancy BMI (kg/m2) | 0.001 | |||||
| Mean (SD) | 26.9 (6.41) | 26.8 (6.37) | 28.0 (6.51) | 26.6 (5.9) | 25.4 (6.45) | |
| Gestational age at delivery (weeks) | <0.001 | |||||
| Mean (SD) | 38.6 (2.5) | 39.0 (1.87) | 38.4 (2.76) | 38.2 (2.80) | 38.5 (2.63) | |
| Parity (n (%)) | 0.005 | |||||
| Nulliparous | 374 (45.0%) | 102 (37.2%) | 134 (51.9%) | 69 (49.3%) | 69 (43.4%) | |
| Multiparous | 457 (55.0%) | 172 (62.8%) | 124 (48.1%) | 71 (50.7%) | 90 (56.6%) | |
| Maternal race (n (%)) | 0.134 | |||||
| Black/African American | 602 (72.4%) | 188 (68.6%) | 193 (74.8%) | 112 (80.0%) | 109 (68.6%) | |
| White | 44 (5.3%) | 17 (6.2%) | 9 (3.5%) | 7 (5.0%) | 11 (6.9%) | |
| Hispanic | 185 (22.3%) | 69 (25.2%) | 56 (21.7%) | 21 (15.0%) | 39 (24.5%) | |
| Maternal diabetes (n (%))c | 0.011 | |||||
| No | 763 (92.5%) | 255 (93.1%) | 230 (89.1%) | 128 (91.4%) | 150 (94.3%) | |
| Gestational diabetes | 35 (4.2%) | 13 (4.7%) | 10 (3.9%) | 3 (2.1%) | 9 (5.7%) | |
| Pregestational diabetes | 27 (3.3%) | 5 (1.8%) | 13 (5.0%) | 9 (6.4%) | 0 (0%) | |
| Maternal education (n (%)) | 0.493 | |||||
| Below college | 551 (66.3%) | 188 (68.6%) | 169 (65.5%) | 86 (61.4%) | 108 (67.9%) | |
| College and higher | 280 (33.7%) | 86 (41.4%) | 89 (34.5%) | 54 (38.6%) | 51 (32.1%) | |
| Maternal smoking (n (%)) | 0.973 | |||||
| Never smoked | 619 (74.5%) | 178 (77.7%) | 155 (71.8%) | 124 (66.3%) | 162 (81.4%) | |
| Ever smoked | 212 (25.5%) | 51 (22.3%) | 61 (28.2%) | 63 (33.7%) | 37 (18.6%) | |
| Maternal smoking during pregnancy (n (%)) | 0.252 | |||||
| Never smoked | 686 (82.6%) | 223 (81.4%) | 207 (80.2%) | 123 (87.9%) | 133 (83.6%) | |
| Ever smoked | 145 (17.4%) | 51 (18.6%) | 51 (19.8%) | 17 (12.1%) | 26 (16.4%) | |
| Baby's sex (n (%)) | ||||||
| Female | 396 (47.7%) | 145 (52.9%) | 120 (46.5%) | 67 (47.9%) | 64 (40.3%) | 0.083 |
| Male | 435 (52.3%) | 129 (47.1%) | 138 (53.5%) | 83 (52.1%) | 95 (59.7%) | |
| Child's birth weight (g) | ||||||
| Mean (SD) | 3120 (667) | 3350 (572) | 3080 (718) | 2910 (666) | 2970 (615) | <0.001 |
Bold values denote statistical significance at the p < 0.05 level.
BMI, body mass index; NW, normal weight; OWO, overweight or obesity.
BMI trajectories were defined using longitudinal BMI percentile data from birth to 18 years of age. Early OWO: children with consistently high BMIPCT; Late OWO: children with BMIPCT increased to OWO by the end of the first year; NW to very late OWO: children with NW at early life that was increased to OWO by the 6th year; NW: children with consistently normal BMIPCT.
Tested using ANOVA or chi-square tests.
Data available for N = 825.
Growth trajectories according to repeated BMIPCT measurements
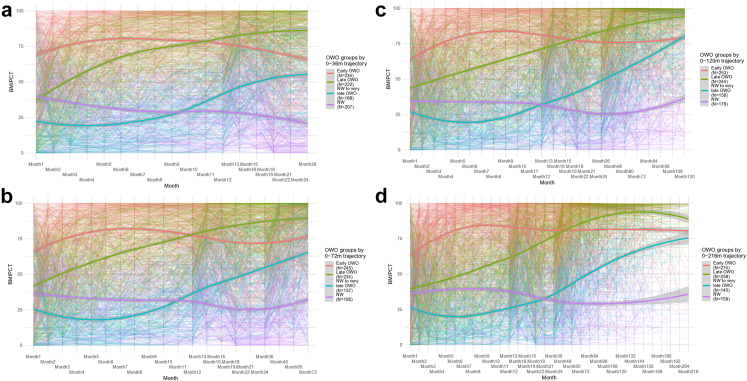
BMIPCT individual trajectories for selected age periods are presented in Fig. 1. Although the groups included two NW groups at earlier age periods (Fig. 1a), by the age of 6 years old (Fig. 1b), one NW group tended to become OWO (“NW to very late OWO”) and kept the OWO trajectory over the following periods (Fig. 1c and d).
Fig. 1.
Illustration of children with distinctive patterns of BMIPCT trajectories over selected age periods. BMI percentile trajectories of selected four age periods: (a) birth to 3y (36m), (b) 6y (72m), (c) 10y (120m), and (d) 18y (216m). Each plot presents individual BMIPCT trajectories and a smoothing curve for each OWO group. Early OWO: children with consistently high BMIPCT; Late OWO: children with BMIPCT increased to OWO by the end of the first year; NW to very late OWO: children with NW in early life that was increased to OWO by the 6th year; NW: children with consistently normal BMIPCT. BMI, body-mass-index; BMIPCT, BMI percentiles; m, months; NW, normal weight; OWO, overweight or obesity; y, years.
Cord DNAm and BMIPCT-based OWO groups
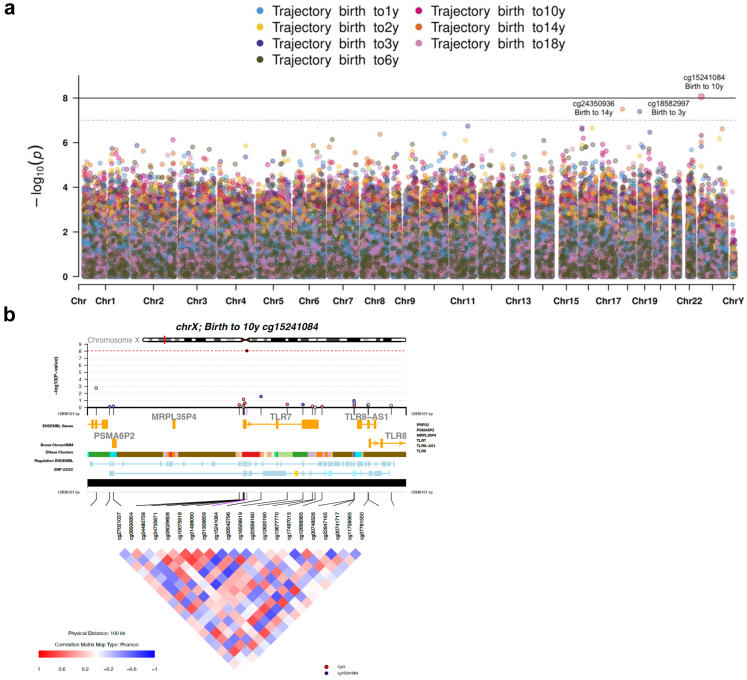
Overall, we observed 3 significant associations (FDR < 0.05) and five marginal associations (FDR < 0.1) in the LRT of DNAm and OWO groups across different age periods (Fig. 2a; Fig. S4; Table S4). We further examined the functional annotation and pattern of co-methylation using the coMET plot32 describing the genomic regions (±50 kb from each side of the leading CpG) of the leading CpG sites: cg15241084 (birth to 10 years; p = 8.71e-09; FDR = 0.006; Fig. 2b), cg18582997 (birth to 3 years; p = 4.10e-08; FDR = 0.029; Fig. S5a), and cg24350936 (birth to 14 years; p = 3.31e-08, FDR = 0.022; Fig. S5b).
Fig. 2.
EWAS results for the seven age periods and chromosome visualisation for the leading CpG sites. Likelihood Ratio Test results. (a) Full model: OWO group, maternal smoke, maternal education, gestational age, maternal diabetes, child's sex, race, parity, low birth weight, and batch effect variables. Reduced model: maternal smoke, maternal education, gestational age, maternal diabetes, child's sex, race, parity, low birth weight, and batch effect variables. (b) coMET plot describing the genomic region of birth to 10y LRT results for the association between OWO groups and cord DNA methylation, along with functional annotation of the region and pattern of co-methylation at the ±50 kb CpG sites of the leading CpG of critical period birth to 10y. CpG, cytosine phosphate guanine; EWAS, epigenome-wide association study; OWO, overweight or obesity; y, years.
We also examined the differential DNAm between the 4 OWO groups (a summary of the linear models for epigenome-wide DNAm associations is presented in Table 2) using linear models for microarray data, with overall 7 significant associations (FDR < 0.05), mainly between groups 1 and 4, highlighting the significant CpG sites from the LRT analysis: Toll like receptor 7 (TLR7) cg15241084 (at multiple age periods), Tubulin Polyglutamylase Complex Subunit 1 (TPGS1) cg18582997, and RAB31, Member RAS Oncogene Family (RAB31) cg24350936. Sensitivity analysis excluding birth weight (included in the main model as a binary outcome) or adding either folic acid or cell type to the models resulted in similar results (Tables S5–S7).
Table 2.
A comparison of the methylation difference between subgroups of BMI longitudinal trajectories by age periods.
| Age periods | Comparison groupsa | Gene | Chromosome | CpG | Association with OWO groupsb |
|||
|---|---|---|---|---|---|---|---|---|
| logFC | SE | p | q (FDR) | |||||
| Birth to 1y | 1 vs 4 | DHX32 | 10 | cg03786842 | 0.309 | 0.056 | 4.95e-08 | 0.035 |
| Birth to 2y | 3 vs 4 | PSMB10 | 16 | cg11451801 | −0.515 | 0.09 | 3.719e-08 | 0.0268 |
| Birth to 3y | 2 vs 3 | TPGS1 | 19 | cg18582997 | −0.38 | 0.068 | 4.08e-08 | 0.027 |
| Birth to 6y | 1 vs 4 | TLR7 | X | cg15241084 | −0.321 | 0.059 | 6.85e-08 | 0.048 |
| 1 vs 4 | - | 7 | cg09287864 | 0.252 | 0.047 | 1.34e-07 | 0.048 | |
| Birth to 10y | 1 vs 4 | TLR7 | X | cg15241084 | −0.359 | 0.06 | 3.87e-9 | 0.0027 |
| 2 vs 4 | KIAA0430 | 16 | cg16997622 | −0.378 | 0.071 | 7.32e-08 | 0.052 | |
| Birth to 14y | 2 vs 3 | RAB31 | 18 | cg24350936 | −0.386 | 0.066 | 7.937e-09 | 0.0057 |
NW, normal weight; OWO, overweight or obesity; y, years.
1 = early OWO; 2 = late OWO; 3 = NW to very late OWO; 4 = NW.
Model adjusted for maternal smoking, maternal education, gestational age, maternal diabetes, child's sex, race, parity, low birth weight, and batch effect variables. No associations for critical periods birth to 14y and birth to 18y were observed.
We repeated this analysis among the largest race/ethnic subgroup, non-Hispanic Black (N = 602). The significant associations observed for the entire cohort were attenuated [Birth to 1: cg03786842 (p = 6.64e-05, FDR = 0.871); Birth to 2: cg11451801 (p = 1.78e-06, FDR = 0.660); Birth to 3: cg18582997 (p = 3.13e-04, FDR = 0.999); Birth to 6: cg15241084 (p = 1.45e-05, FDR = 0.554) cg09287864 (p = 4.79e-06, FDR = 0.481); and Birth to 10: cg15241084 (p = 1.58e-05, FDR = 0.519), cg16997622 (p = 3.72e-05, FDR = 0.737) A differential methylation between late OWO and the group defined as NW to very late OWO in the age period of birth to 10 years was observed for cg04199164 annotated to Ubiquitin Specific Peptidase 40 (USP40; estimate (SE): 0.469 (0.083), p = 2.72e-08, FDR = 0.019). In the age period of birth to 14 years, a differential DNAm of Ras-related protein Rab-31 (RAB31) cg24350936 was observed (late OWO vs NW to very late OWO; estimate (SE): −0.463 (0.078), p = 4.83e-09, FDR = 0.003).
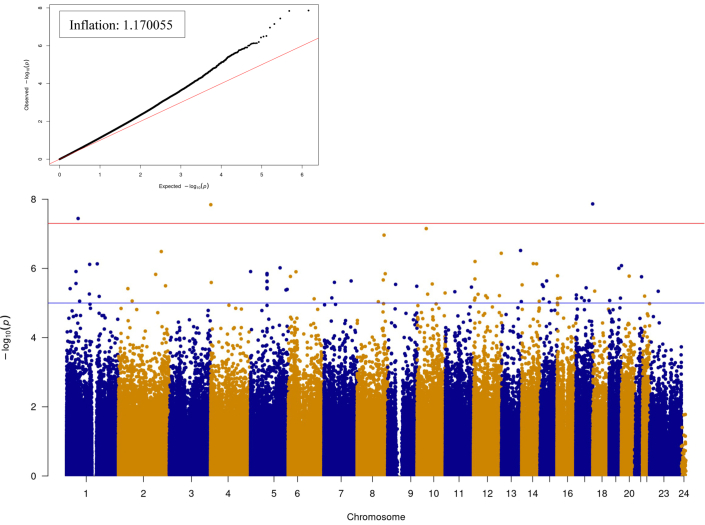
Cord DNAm and standardised birth weight
We examined the association between cord DNAm and SBWT for the entire available cohort (Fig. 3). Using the LRT test (with the same covariates as the main model, replacing LBW and GEAA with the independent variable SBWT), we detected a total of 27 significant associations (multivariate model; FDR < 0.05), with the following top 5 associations: cg04356233 of Major facilitator superfamily domain containing 7 (MFSD7; p = 1.43e-08, FDR = 0.005), cg04837676of Casein kinase I isoform delta (CSNK1D; p = 1.36e-08, FDR = 0.005), and cg27168858 of 24-Dehydrocholesterol Reductase (DHCR24; p = 3.36e-08, FDR = 0.008), cg23295629 of Pvt1 oncogene (PVT1; p = 1.09e-07, FDR = 0.015), and one unannotated site: cg20671781 (p = 7.06e-8, FDR = 0.012). As a sensitivity analysis, we added cell type composition to the model (Fig. S6), resulting in 169 significant associations (LRT FDR < 0.05), and found cg04837676, cg04356233, cg20671781, cg12723026, and cg05993265 as the top 5 significant CpG sites (FDR = 0.0003, FDR = 0.0004, FDR = 0.0006, FDR = 0.0015, and FDR = 0.003, respectively).
Fig. 3.
The association between cord DNA methylation and standardised birth weight; LRT results. Quantile–quantile (QQ) and Manhattan plots. Full model: INT CpGs ∼ SBWT + maternal smoking + Maternal education + maternal diabetes + race + sex of the child + parity + batch effect variable (50 SVs). For the reduced model, we excluded SBWT. The number of significant associations with FDR <0.05: 27. FDR, false discovery rate; LRT, Likelihood Ratio Test; SBWT, standardised birth weight.
Cord DNAm and repeated BMI measurements
Next, we examined the association between cord DNAm and BMI at different ages, starting at month 2 (±1 month), up to the maximum measurement available, with a total of 21 “time windows,” using LRT, in a multivariate model. For the entire sample available at each time window, we found significant CpG sites at the ages 12 months (N = 1), 20–24 months (N = 2), year 3 (N = 1), year 4 (N = 2), year 9 (N = 2), year 13 (N = 19), year 14 (N = 8), and at the maximum age available (N = 15; FDR < 0.05 for all; Fig. 4a) with the sex interaction analysis shown a noticeable increase in the number of significant CpG sites at month 12, month 20–24 to year 10, and from year 12 to maximum age examined. In girls (Fig. 4b), the increase occurred at the 2nd, 7th, and from the age of 12, with multiple significant CpG sites at month 4, month 12, month 15, month 18, month 20–24, year 3 to year 10, and year 12 to maximum. Among boys (Fig. 4b), the increase was smaller and later, compared with the girls, with fewer but significant CpG sites detected at year 7, year 8, and year 12 to maximum age (multivariate models excluding sex covariate; FDR < 0.05 for all). All FDR <0.05 (for each time window) associated CpG sites from the LRTs are available in Table S8 (top 10 CpG sites) and Table S9 (for associations with more than 10 significant hits per time window). The following top CpG sites overlapped between the time windows: ADAMTS2 cg17173355 in girls at year 3 and 4, SFT2D3 cg01900048 in girls at years 3 and 6, cg05646827 (unannotated) in boys at 12 and 13 years, and DDAH1 cg08654262 at years 13 and maximum age in boys. Fig. S7a and b shows significant CpG sites with a more conservative FDR, demonstrating significant associations' trajectory trends at certain ages.
Fig. 4.
Time window analysis: The association between cord DNA methylation and BMI (LRT results). Presented for the entire cohort and stratified by sex. Models adjusted for maternal smoking, education, gestational age, maternal diabetes, child's sex, race, parity, low birth weight, and batch effect (model a; stratification by sex using model a without sex covariate). (a) LRT results for reduced model excluding BMI and a joint interaction test (excluding BMI and BMI∗sex). (b) LRT results were stratified by sex. BMI, body mass index; FDR, false discovery rate; LRT, likelihood ratio test.
Discussion
In this multi-ethnic cohort with repeated BMIPCT measurements across the paediatric age periods from birth to 18 years, we identified 4 distinct and robust patterns of BMIPCT trajectories: early OWO, Late OWO, NW to very late OWO, and those who were consistently NW through the years from birth to adolescence. We found specific cord DNAm CpG sites associated with these patterns, some overlapping, in different age periods. A secondary analysis showed sex-specific DNAm patterns associated with BMI at different ages, from birth to adolescence, with DNAm mirroring sex differences in early development.
Our primary analysis used four OWO groups as the main outcome to associate with cord DNAm. Although previous studies have shown the association of cord DNAm with birth weight or BMI in different ages from early to late childhood,19,39 they mainly described point-prevalence BMI measurements up to 10 years old. We found three significant CpG sites annotated to TPGS1, also known as Chromosome 19 Open Reading Frame 20 (C19orf20), RAB31, and TLR7. Data on the methylation of these CpG sites is limited. In mice, C19orf20 was associated with obesity.40,41 In humans, methylation of cg18582997 annotated to C19orf20 was examined in an EWAS of repeated whole blood DNAm measurements from birth to late adolescence and found a decrease in cg18582997 methylation by −8.9e-4 per year.42 While there are no previous EWAS on cg24350936, methylation on other sites annotated to RAB31 gene were mostly associated with age42 and gestational age.43 TLR proteins are families of DNA sensors regulating the innate immune response by interacting with unmethylated CpG-DNA to activate a proinflammatory response to microbial DNA.44 In obesity and metabolic syndrome, TLRs may have a central role in inflammation induction, with an increase in TLRs expression in different tissues among obese individuals.45 We observed a significant association with OWO groups in several age periods for TLR7 cg15241084.46 Whole blood methylation of this CpG site was also associated with weekly alcohol intake.46 There is a need for further studies to explore the long-term obesity/BMI - methylation associations in these genes to map specific cord-blood CpGs to feature or distinguish OWO groups.
Excess calories and the reduction in energy expenditure leads to obesity and a loss of homeostasis in the adipose tissue that might promote chronic low-grade inflammation.47 Our results highlighted the methylation on the TLR7 gene, first-line essential immune receptors inducing inflammation,48 found to be associated with the OWO groups in two obesity-related critical periods age 6, and 10 years– and specifically between the extreme OWO groups – the NW and the early OWO. This may be explained by the adipose tissue volume and content differences reflected by excess weight. In the first year of life, the dominant adipose tissue is subcutaneous, which later decreases during the second year of life; The next increase in subcutaneous adipose tissue and the total body fat content occurs between the 8th and the 10th years of life, just before puberty.49 In obese children, there are early signs of adipose tissue dysfunction, reflected by the secretion of adipokines and adipocyte hypertrophy.50 A recent publication found differential gene expression between lean and obese children (ages 10 and 13 years, respectively) in genes involved in inflammation in subcutaneous adipose tissue.51 These adiposity-related time windows and obesity-related adipose tissue dysfunction can explain our observation of the differential methylation between the NW and early OWO groups. Our observations from this analysis may promote an understanding of the epigenetic role in adipose tissue distribution in childhood.
DNAm is an essential mechanism for regulating gene expression.52 Hypo-methylated CpG regions are commonly associated with gene activation, while hyper-methylation in a CpG region is usually associated with lower expressed or silenced genes53; however, the effect of lower and higher methylation depends on the location of the CpGs. High-density DNAm, especially in the promoter region of a gene, can repress long-term mRNA expression, as methylation near gene promoters is correlated with low or no transcription.54 Transcriptional activation of some genes is associated with hypomethylation of enhancer DNA, allowing transcription factor binding.55 In our study, cg15241084, located in the promotor, according to the functional annotation track, showed a strong correlation with the surrounding CpG sites. A previous study reported that in 9 years old children, the umbilical cord tissue methylation of specific CpG sites located in the promotor of chromosomes 7 and 9 of endothelial nitric oxide synthase (eNOS) and retinoid X receptor-α (RXRA) genes, respectively, were associated with body fat.56 Another study examining DNAm in the blood of 355 young individuals (N = 130 ages 14–16 years; N = 225 ages 18–34 years) for known obesity-related single nucleotide polymorphisms, found that out of 107 CpG sites, about a third were located in genes promotors.57 We also observed differential methylation of cg18582997 and cg24350936 (both late OWO vs NW to very late OWO), located in the enhancer and promoter with weak and moderate correlation between the surrounding CpG sites. Although DNAm cannot solely explain gene expression, exploring the region of the DNAm promotes the understanding of this mechanism, functionality, and, in this study context, the effect on long-term children's obesity trajectories.
Obesity in adulthood and childhood obesity have some similar complications, such as elevated blood pressure, type 2 diabetes, cardiovascular diseases, and asthma58; However, obesity among children may also lead to alterations in development, and with early puberty in girls, but delayed in boys.59 In our two EWAS models exploring the association of DNAm with SBWT, we observed many significant associations, with 3 CpG sites repeating in the top 5 of both analyses: cg04837676, cg20671781, and cg04356233. Previous studies found no association between cg04837676 and cg04356233 with any phenotypes or exposures. However, the methylation of cg20671781 was associated with birth weight60 and prenatal maternal smoking.61 Furthermore, in the EWAS of repeated whole blood DNAm measurements from birth to late adolescence mentioned above,42 cg20671781 was associated with weight at the age of 6 years. The inflations for both EWASs of SBWT were ∼1.2, suggesting some systematic bias to be considered while interpreting these results. Our “time window” analysis allowed us to examine methylation patterns across the entire cohort and stratified by sex. We observed a higher number of significant findings in girls, with the number of significant sex interactions decreasing at the age of six, with the boys having less significant associations, compared with the girls, who demonstrated an increase in the number of significant associations during the following years. This pattern smilingly mirrors the biological catch-up of boys, legging the girls’ pre-puberty period. According to the GWAS catalogue,62 there are currently 49 known genes associated with childhood obesity. Concordantly, our time window analysis showed significant methylation in previously recognised childhood obesity-related genes, such as and Brain-Derived Neurotrophic Factor (BDNF) (Table S9). The period-overlapping cg01900048 measured in cord blood was associated with prenatal phthalate exposure63 in a cohort examining altered patterns of cord blood DNAm in 336 Mexican-American newborns. The other identified CpGs were not previously associated with any traits when measured in cord blood. Our analysis resulted in many significant associations in some of the time windows and the birthweight/GEAA EWAS; thus, it should be interpreted with caution and considered exploratory.
This work has some limitations. Our sample size is not adequately powered to in more refined subgroup analyses in the future. Nevertheless, to our knowledge, the present study is by far the largest with well-characterised longitudinal BMI trajectories from birth to 18 years. The unique strength of the BBC, namely, dense repeated measurements of BMIPCT in a traditionally understudied US minority birth cohort, also limited our ability to find appropriate existing sample to replicate our study results. However, we anticipate that a US national birth cohort, such as ECHO, will offer many opportunities to replicate our study findings and to have a greater sample size and power to conduct more refined subgroup analyses in the future. Nevertheless, to the best of our knowledge, this is the first and largest prospective study to demonstrate distinct long-term OWO trajectory groups associations with cord blood DNAm. BMIPCT was calculated using two different growth charts: WHO and CDC; however, this allowed us to explore the trajectory from birth to 1 and 2 years old. This study is based on a US, mostly urban, low-income, African American/Black population; thus, the generalizability of the results should be carefully examined. Yet, this might also be an advantage as previously published studies were mostly European ancestry samples. We did not adjust the linear models to cell type composition; However, a previous publication has shown that adjusting for batch effect, including cell mixture by SmartSVA, is sufficient.37 Nevertheless, we added sensitivity analyses showing that adjusting for cell type did not affect the main findings. In our primary analysis, we could not find any previously reported CpG site associated with BMIPCT in children; This might be because we examined the association of cord DNAm with obesity groups and not with BMIPCT as a continuous variable or due to our more frequent visits available for this cohort that resulted in a higher density of BMI measurements. Our secondary findings support this assumption The DNAm in cord blood could reflect prenatal influences on epigenetic programming during its most sensitive period that may have a long-lasting impact on child postnatal growth. The longitudinal nature of our study offers an opportunity to investigate the prospective association between cord DNAm and postnatal BMI trajectory and avoid reverse causation. We acknowledge that our findings are preliminary, and the biological mechanism underlying the link between cord DNAm and BMI trajectory remains to be explored. In this context, we hope this present study will stimulate and inform future studies along the same line. Our findings, if further confirmed by future studies, will provide new insight into the developmental origins of childhood obesity and will also improve our ability in early risk assessment and early prevention of childhood obesity. The strengths of this study include the use of EPIC array, allowing us to detect a high number of CpG sites with more CpGs at methylation dynamic sites according to the design of the chip, and a high density of measurements from birth to adolescence used to create obesity/growth patterns and associate them with cord DNAm. Multiple visits also reduce measurement errors, as a non-differential misclassification can occur when dealing with real-life data, even when following the same protocol. Finally, we examined the longitudinal BMI data in two ways: by using age periods, we highlighted the weight/weight gain trajectory and the association with birth methylation and by using point-prevalence BMI measurements and later stratifying by sex, to explore further which ages during the follow-up time were more associated with DNAm. This allowed us to uncover novel CpGs associated with obesity and replicate previous methylation findings in CpGs annotated to BMI-related genes.
In conclusion, we found that cord blood CpG sites DNAm were associated with child BMI longitudinal trajectories and revealed potential sex differences in the associations. These findings lent further support for the developmental origins of OWO and epigenetic alterations as a potential underlying mechanism. These findings, if further replicated, raise the possibility that these DNAm markers, along with an early assessment of BMIPCT trajectories, may help identify young children at high risk for obesity later in life, thus offering the window of opportunity for precision early risk assessment and targeted prevention.
Contributors
A.Y.M., X.H., X.W., and L.L. were responsible for study conception. C.P., W.G.A., and X.W. supervised collection of phenotypic data and biospecimens. A.Y.M. and L.L. verified the underlying data. A.Y.M. was responsible for drafting of the manuscript. X.H., G.W., L.L., and X.W. supervised DNA methylation data generation. X.H. performed DNA methylation quality control and data cleaning. A.Y.M. performed most of the statistical analyses under the guidance and technical support of L.L. W.H. performed imputation of the BMIPCT data. T.C. derived the BMIPCT trajectory outcome. X.W. was responsible for overseeing acquisition of the epidemiological and clinical data as well as biospecimens. All the authors were responsible for critical review and revision of the manuscript and contributed to data interpretations. All authors read and approved the final version of the manuscript.
Data sharing statement
The data, data dictionary, and analytical programs for this manuscript are not currently available to the public. However, they can be made available upon reasonable request and after the review and approval of the institutional review board. The source code to replicate the results upon request will be provided.
Declaration of interests
A.Y.M., W.H., T.C., X.H., C.P., W.G.A., G.W., X.W., L.L. declare no competing interests.
Acknowledgement
The authors wish to thank the study participants in the BBC, the nursing staff at labour and delivery of the Boston Medical Center, as well as the field team for their contributions to the Boston Birth Cohort. The Boston Birth Cohort (the parent study) was supported in part by the National Institutes of Health (NIH) grants (2R01HD041702, R01HD086013, R01HD098232, R21AI154233, R01ES031272, R03HD096136, U01 ES034983, and R01ES031521); and by the Health Resources and Services Administration (HRSA) of the U.S. Department of Health and Human Services (HHS) (UT7MC45949). Dr Yaskolka Meir is supported by the Council for Higher Education- Zuckerman support program for outstanding postdoctoral female researchers. This information or content and conclusions are those of the author and should not be construed as the official position or policy of, nor should any endorsements be inferred by the funding agencies.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104550.
Contributor Information
Xiaobin Wang, Email: xwang82@jhu.edu.
Liming Liang, Email: lliang@hsph.harvard.edu.
Appendix A. Supplementary data
References
- 1.Body mass index-for-age (BMI-for-age). OMS 2021:1–6. https://www.who.int/toolkits/child-growth-standards/standards/body-mass-index-for-age-bmi-for-age
- 2.Growth charts - individual growth charts n.d. https://www.cdc.gov/growthcharts/charts.htm
- 3.Tyson N., Frank M. Childhood and adolescent obesity definitions as related to BMI, evaluation and management options. Best Pract Res Clin Obstet Gynaecol. 2018;48:158–164. doi: 10.1016/J.BPOBGYN.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Organization W.H. Vol. 3. World Health Organization; 2004. (International statistical classification of diseases and related health problems: alphabetical index). [Google Scholar]
- 5.Jornayvaz F.R., Vollenweider P., Bochud M., Mooser V., Waeber G., Marques-Vidal P. Low birth weight leads to obesity, diabetes and increased leptin levels in adults: the CoLaus study. Cardiovasc Diabetol. 2016;15:73. doi: 10.1186/S12933-016-0389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward Z.J., Long M.W., Resch S.C., Giles C.M., Cradock A.L., Gortmaker S.L. Simulation of growth trajectories of childhood obesity into adulthood. N Engl J Med. 2017;377:2145–2153. doi: 10.1056/NEJMOA1703860/SUPPL_FILE/NEJMOA1703860_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geserick M., Vogel M., Gausche R., et al. Acceleration of BMI in early childhood and risk of sustained obesity. N Engl J Med. 2018;379:1303–1312. doi: 10.1056/NEJMoa1803527. [DOI] [PubMed] [Google Scholar]
- 8.Juonala M., Magnussen C.G., Berenson G.S., et al. Childhood adiposity, adult adiposity, and cardiovascular risk factors. N Engl J Med. 2011;365:1876–1885. doi: 10.1056/NEJMoa1010112. [DOI] [PubMed] [Google Scholar]
- 9.Sommer A., Twig G. The impact of childhood and adolescent obesity on cardiovascular risk in adulthood: a systematic review. Curr Diab Rep. 2018;18:1–6. doi: 10.1007/s11892-018-1062-9. [DOI] [PubMed] [Google Scholar]
- 10.McKay J.A., Mathers J.C. Diet induced epigenetic changes and their implications for health. Acta Physiol. 2011;202:103–118. doi: 10.1111/j.1748-1716.2011.02278.x. [DOI] [PubMed] [Google Scholar]
- 11.Teven C.M., Liu X., Hu N., et al. Epigenetic regulation of mesenchymal stem cells: a focus on osteogenic and adipogenic differentiation. Stem Cells Int. 2011;2011:201371. doi: 10.4061/2011/201371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tammen S.A., Friso S., Choi S.-W. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013;34:753–764. doi: 10.1016/j.mam.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lillycrop K.A., Burdge G.C. Epigenetic changes in early life and future risk of obesity. Int J Obes. 2011;35:72–83. doi: 10.1038/ijo.2010.122. [DOI] [PubMed] [Google Scholar]
- 14.Morales E., Groom A., Lawlor D.A., Relton C.L. DNA methylation signatures in cord blood associated with maternal gestational weight gain: results from the ALSPAC cohort. BMC Res Notes. 2014;7:1–10. doi: 10.1186/1756-0500-7-278/TABLES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin C.L., Jima D., Sharp G.C., et al. Maternal pre-pregnancy obesity, offspring cord blood DNA methylation, and offspring cardiometabolic health in early childhood: an epigenome-wide association study. Epigenetics. 2019;14:325. doi: 10.1080/15592294.2019.1581594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R., Hong X., Zhang B., et al. DNA methylation mediates the effect of maternal smoking on offspring birthweight: a birth cohort study of multi-ethnic US mother–newborn pairs. Clin Epigenetics. 2021;13:1–13. doi: 10.1186/s13148-021-01032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemoda Z., Massart R., Suderman M., et al. Maternal depression is associated with DNA methylation changes in cord blood T lymphocytes and adult hippocampi. Transl Psychiatry. 2015;5(4) doi: 10.1038/tp.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kile M.L., Andres Houseman E., Baccarelli A.A., et al. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics. 2014;9:774–782. doi: 10.4161/EPI.28153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vehmeijer F.O.L., Küpers L.K., Sharp G.C., et al. DNA methylation and body mass index from birth to adolescence: meta-analyses of epigenome-wide association studies. Genome Med. 2020;12:1–15. doi: 10.1186/s13073-020-00810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Relton C.L., Groom A., St Pourcain B., et al. DNA methylation patterns in cord blood DNA and body size in childhood. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kresovich J.K., Zheng Y., Cardenas A., et al. Cord blood DNA methylation and adiposity measures in early and mid-childhood. Clin Epigenetics. 2017;9:86. doi: 10.1186/s13148-017-0384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Zuckerman B., Pearson C., et al. Maternal cigarette smoking, metabolic gene polymorphism, and infant birth weight. JAMA. 2002;287:195–202. doi: 10.1001/jama.287.2.195. [DOI] [PubMed] [Google Scholar]
- 23.Pearson C., Bartell T., Wang G., et al. Boston Birth Cohort profile: rationale and study design. Precis Nutr. 2022;1 doi: 10.1097/PN9.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong X., Hao K., Ji H., et al. Genome-wide approach identifies a novel gene-maternal pre-pregnancy BMI interaction on preterm birth. Nat Commun. 2017;8:1–10. doi: 10.1038/ncomms15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aryee M.J., Jaffe A.E., Corrada-Bravo H., et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakulski K.M., Feinberg J.I., Andrews S.V., et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11:354–362. doi: 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortin J.-P., Triche T.J., Jr., Hansen K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleveland W.S. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74:829–836. [Google Scholar]
- 29.Huang W., Yaskolka Meir A., Olapeju B., et al. 2022. Defining longitudinal trajectory of body mass index percentile and predicting childhood obesity: methodologies and findings in the Boston birth cohort. Precision Nutrition. PMC Manuscript ID: NIHMS1886758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao T., Zhao J., Hong X., et al. Cord blood metabolome and BMI trajectory from birth to adolescence: a prospective birth cohort study on early life biomarkers of persistent obesity. Metabolites. 2021;11:739. doi: 10.3390/metabo11110739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L., Wang X., Laird N., Zuckerman B., Stubblefield P., Xu X. Polymorphism in maternal LRP8 gene is associated with fetal growth. Am J Hum Genet. 2006;78:770. doi: 10.1086/503712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin T.C., Yet I., Tsai P.-C., Bell J.T. coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics. 2015;16:1–5. doi: 10.1186/s12859-015-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin L., Zhang H., Tang Z., et al. rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics Proteomics Bioinformatics. 2021;19(4):619–628. doi: 10.1016/j.gpb.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickham H. Springer; 2016. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 35.Ritchie M.E., Phipson B., Wu D.I., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J., Behnam E., Huang J., et al. Fast and robust adjustment of cell mixtures in epigenome-wide association studies with SmartSVA. BMC Genomics. 2017;18:1–13. doi: 10.1186/s12864-017-3808-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- 39.Mao L.L., Xiao X.H., Zhang Q., et al. DNA methylation and birth weight: a genome-wide analysis. Biomed Environ Sci. 2017;30:667–670. doi: 10.3967/bes2017.088. [DOI] [PubMed] [Google Scholar]
- 40.Campbell P.K., Waymire K.G., Heier R.L., et al. Mutation of a novel gene results in abnormal development of spermatid flagella, loss of intermale aggression and reduced body fat in mice. Genetics. 2002;162:307–320. doi: 10.1093/genetics/162.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérusse L., Rankinen T., Zuberi A., et al. The human obesity gene map: the 2004 update. Obes Res. 2005;13:381–490. doi: 10.1038/oby.2005.50. [DOI] [PubMed] [Google Scholar]
- 42.Mulder R.H., Neumann A., Cecil C.A.M., et al. Epigenome-wide change and variation in DNA methylation in childhood: trajectories from birth to late adolescence. Hum Mol Genet. 2021;30:119–134. doi: 10.1093/HMG/DDAA280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohlin J., Håberg S.E., Magnus P., et al. Prediction of gestational age based on genome-wide differentially methylated regions. Genome Biol. 2016;17:1–9. doi: 10.1186/s13059-016-1063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Briard B., Place D.E., Kanneganti T.-D. DNA sensing in the innate immune response. Physiology. 2020;35:112–124. doi: 10.1152/physiol.00022.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jialal I., Kaur H., Devaraj S. Toll-like receptor status in obesity and metabolic syndrome: a translational perspective. J Clin Endocrinol Metab. 2014;99:39–48. doi: 10.1210/jc.2013-3092. [DOI] [PubMed] [Google Scholar]
- 46.Dugué P., Wilson R., Lehne B., et al. Alcohol consumption is associated with widespread changes in blood DNA methylation: analysis of cross-sectional and longitudinal data. Addict Biol. 2021;26 doi: 10.1111/adb.12855. [DOI] [PubMed] [Google Scholar]
- 47.Tsigalou C., Vallianou N., Dalamaga M. Autoantibody production in obesity: is there evidence for a link between obesity and autoimmunity? Curr Obes Rep. 2020;9:245–254. doi: 10.1007/S13679-020-00397-8/FIGURES/1. [DOI] [PubMed] [Google Scholar]
- 48.Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/ANNUREV.IMMUNOL.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 49.Körner A., Kiess W., Landgraf K. Pediatric Obesity. Springer; 2018. White adipose tissue accumulation and dysfunction in children with obesity; pp. 95–115. [Google Scholar]
- 50.Landgraf K., Rockstroh D., Wagner I.V., et al. Evidence of early alterations in adipose tissue biology and function and its association with obesity-related inflammation and insulin resistance in children. Diabetes. 2015;64:1249–1261. doi: 10.2337/db14-0744. [DOI] [PubMed] [Google Scholar]
- 51.Landgraf K., Kühnapfel A., Schlanstein M., et al. Transcriptome analyses of adipose tissue samples identify EGFL6 as a candidate gene involved in obesity-related adipose tissue dysfunction in children. Int J Mol Sci. 2022;23:4349. doi: 10.3390/ijms23084349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unnikrishnan A., Jackson J., Matyi S.A., et al. Role of DNA methylation in the dietary restriction mediated cellular memory. Geroscience. 2017;39(3):331–345. doi: 10.1007/s11357-017-9976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 55.Saluz H.P., Jiricny J., Jost J.P. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci U S A. 1986;83:7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Godfrey K.M., Sheppard A., Gluckman P.D., et al. Epigenetic gene promoter methylation at birth is associated with child's later adiposity. Diabetes. 2011;60:1528–1534. doi: 10.2337/db10-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voisin S., Almén M.S., Zheleznyakova G.Y., et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:1–16. doi: 10.1186/s13073-015-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kinlen D., Cody D., O'Shea D. Complications of obesity. QJM. 2018;111:437–443. doi: 10.1093/QJMED/HCX152. [DOI] [PubMed] [Google Scholar]
- 59.Burt Solorzano C.M., McCartney C.R. Obesity and the pubertal transition in girls and boys. Reproduction. 2010;140:399. doi: 10.1530/REP-10-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Küpers L.K., Monnereau C., Sharp G.C., et al. Meta-analysis of epigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-09671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sikdar S., Joehanes R., Joubert B.R., et al. Comparison of smoking-related DNA methylation between newborns from prenatal exposure and adults from personal smoking. Epigenomics. 2019;11:1487–1500. doi: 10.2217/EPI-2019-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buniello A., MacArthur J.A.L., Cerezo M., et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2018;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Solomon O., Yousefi P., Huen K., et al. Prenatal phthalate exposure and altered patterns of DNA methylation in cord blood. Environ Mol Mutagen. 2017;58:398–410. doi: 10.1002/EM.22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.