Abstract
Insulin-like growth factor 1 receptor (IGF-1R) gene is the main effector of insulin-like growth factor (IGF), which plays an important role in growth, development and reproduction of the animal organism. This study aimed to investigate the association of IGF-1R gene single nucleotide polymorphisms (SNPs) with egg quality and carcass traits of quail by direct sequencing. In this study, genomic DNA was extracted from quail blood samples of 46 Chinese yellow (CY) quail, 49 Beijing white (BW) quail and 48 Korean (KO) quail strains. Egg quality and carcass traits were measured and used for IGF-1R gene analysis in 3 quail strains. The results showed that 2 SNPs (A57G and A72T) of the IGF-1R gene were detected in 3 quail strains. The A57G was significantly associated with yolk width (YWI) in BW strain (P < 0.05). Whereas A72T was significantly associated with egg shell thickness (EST) in BW strain (P < 0.05), and significantly associated with egg weight (EW), egg long (EL), and egg short (ES) in KO strain (P < 0.05). Haplotypes based on 2 SNPs showed significant effect on EST in 3 quail strains (P < 0.05), it also has a significant effect on EW in KO strain (P < 0.05). Meanwhile, A72T was significantly associated with liver weight (LW) and dressing percentage (DP) in 3 strains (P < 0.05). Haplotypes showed significant effect on LW (P < 0.05). Therefore, the IGF-1R gene may be a molecular genetic marker to improve egg quality and carcass traits in quails.
Key words: IGF-1R, polymorphism, egg quality, carcass traits, quail
INTRODUCTION
Quails are small economic poultry widely farmed in China, which has characteristics such as short growth cycle, high reproductive ability, high egg production performance, high nutritional value, low investment and high economic benefits (Bai et al., 2021; Wang et al., 2023). Quails have been loved by consumers around the world in recent years because of its rich minerals, high protein, beneficial fatty acids and other nutrients, and the amount of quail farming has increased rapidly. However, there is little research on quail breeding at home and abroad, especially at the molecular level, which is still in its infancy (Minvielle, 2004; Priti and Satish, 2014). In recent years, marker-assisted selection has replaced traditional breeding methods as the new breeding option used to improve the animal economic traits (Zhang et al., 2014). Studies have revealed that many candidate genes are related to economic traits, among which IGF2, LEPR, and GnRH genes are well-accepted candidate genes that affect growth and production performance in livestock (Ali et al., 2021; El-Tarabany et al., 2022; Wang et al., 2023).
The insulin-like growth factor 1 receptor (IGF-1R) gene was the main effector of insulin-like growth factor (IGF). Studies had found that the IGF-1R gene played an important role in the animal organism, which not only regulated the vitality of IGFs, but also regulated the cell's own growth cycle, metabolism, proliferation and differentiation, immune regulation and important processes of adulthood have regulatory effects (Cardoso et al., 2021). There are numerous studies indicate that IGF-1R gene affects economic traits of animals, different mutation sites and genotypes have different effects on animal growth, carcass and reproduction traits. El-Magd et al. (2017) have proven that 2 SNPs (G64A and G280A) of the IGF-1 gene had a significant impact on growth traits of buffalo. Szewczuk et al. (2013) purported that the polymorphism of the IGF-1R gene was significantly associated with growth-related traits weight in Angus cattle. Ding et al. (2022) found 10 SNPs of the IGF-1 gene were detected in Hulun Buir sheep. Ma et al. (2019) found the IGF-1R CNV was significantly associated with body weight and body height of Jinnan cattle and was significantly associated with body height and hucklebone width of Qinchuan cattle. Wu et al. (2017) analyzed the correlation between the IGF-1 gene of Bian chickens’ growth traits, and found that the gene was highly correlated with chicken body weight (P < 0.05). Thus, the IGF-1R gene has been widely used to evaluate genetic diversity in many regions with different breeds.
Previous genetic studies showed that polymorphisms in the candidate gene were significantly associated with growth, egg production, and meat quality traits in many species (El-Magd et al., 2017; Wu et al., 2017; Ali et al., 2021). However, fewer studies have been conducted on association of polymorphisms with the IGF-1R gene with economic traits in poultry (Lei et al., 2008; Pu et al., 2016; Yang et al., 2022). Egg quality and carcass trait are important economic traits in poultry breeding. The objective of this study is to identify the IGF-1R gene polymorphism with the egg quality and carcass traits in Chinese yellow (CY) quail, Beijing white (BW) quail, and Korean (KO) quail strains by PCR sequencing, and to provide reference values for the future research of quail breeding.
MATERIALS AND METHODS
Ethics Statement
All experiment procedures in this study were approved by the Institutional Animal Care and Use Committee of College Animal Science, Henan University of Science and Technology (Luoyang, China), and animal experimentation in this study was conducted in strict accordance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China).
Experimental Animals, Housing Condition, and Phenotypic Measurements
A top of 46 female CY strain, 49 female BW strain, and 48 female KO strain were randomly selected from a commercial hatchery (Henan University of Science and Technology Quail Breeding Co. Ltd., Luoyang, China). All quail individuals were healthy and fed in single cages at the experimental farm of Henan University of Science and Technology under the same conditions. During the whole investigation, all quails were allowed to feed and drink ad libitum. Supplemental heaters were provided first 2 wk of growth. The daily lighting schedule was lights on from 5:00 am to 7:00 pm until 140 d. All 3 strains were fed a diet with 2,900 kcal/Kg of ME and 24% CP from d 1 to 49 (growth periods), ME and CP levels were 2,800 kcal/kg and 20% from d 50 to 140 (laying periods), respectively. One egg from each individual within each quail strain was selected for egg quality measurements at 7 wk of age. The egg quality included egg weight (EW), egg long (EL), egg short (ES), egg shape index (ESI), yolk height (YH), yolk width (YWI), yolk index (YI), yolk weight (YWE), eggshell thickness (EST), and albumen height (AH). The carcass traits included body weight (BOW), dressed carcass weight (DCW), whole net carcass weight (WNCW), heart weight (HW), liver weight (LW), breast muscle weight (BMW), leg muscle weight (LMW), dressing percentage (DP), whole net carcass rate (WNCR), heart rate (HR), liver rate (LR), breast muscle rate (BMR), and leg muscle rate (LMR) were measured at 20 wk of age in 3 quail strains.
DNA Samples, Primer Designing, PCR Amplification, and DNA Sequencing
Blood samples (5 mL) were taken from the wings of 143 quails (46 CY, 49 BW, and 48 KO) into a syringe containing 2% EDTA used as an anticoagulant and stored at −80°C for further experiment. Genomic DNA was isolated from venous blood samples using a poultry whole DNA extraction kit (Dingguo Changsheng Biotechnology Company, Beijing, China). Based on the potential SNPs of IGF-1R gene published in the NCBI database (https://www.ncbi.nlm.nih.gov/), the primer pairs were designed using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA), which were F-AACGCCTGGAGAACTGTACG and R-ATCGCTGAGGCTTTCCAAG. The primer specificity was verified by BLAST at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The expected amplified segment size was 155 bp. PCR was performed in a total volume of 20 μL, which included 10 μL of the 2 × Taq PCR Master Mix, 0.7 μL of each primer, 1 μL genomic DNA, and 7.6 μL double-distilled water. The reaction conditions were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 40 s, annealing for 56°C for 45 s, extension at 72°C for 35 s, and a final extension at 72°C for 10 min. The reaction system was stored under 4°C (Lei et al., 2008). Then, the amplified samples of the IGF-1R gene were sent to Beijing Tsingke Biological Co., Ltd. for sequencing.
Statistical Analysis
Sequence alignment and SNP identification were conducted via MegAlign program (version 5.0; DNAstar, Madison, WI). Chromas software (version 2.2.2; Technelysium, Queensland, Australia) was used to conduct sequence analyses. Genotypes and alleles were recorded using Excel (version 2016; Microsoft, Redmond, WA). The population genetic information was statistically analyzed using the Popgene version 1.32 (Yeh and Boyle, 1997). Finally, association analysis of polymorphisms was accomplished with the measured egg quality and carcass traits using Duncan's multiple range test in SPSS (version 26.0; IBM Corp., Armonk, NY) and expressed as means ± standard error (SE). Differences were considered highly significant or significant at P ≤ 0.01 or P ≤ 0.05, respectively. The association analysis model of egg quality was as follows:
| (1) |
Yij is the phenotype value, μ is the total mean value, Gi is the effect of genotype, and eij is the random error. The association analysis model of carcass traits was as follows:
| (2) |
Yijk is the phenotype value, μ is the total mean value, Si is the effect of strain, Gj is the effect of genotype, and eijk is the random error.
RESULTS AND DISCUSSION
Polymorphisms of IGF-1R Gene in Quail
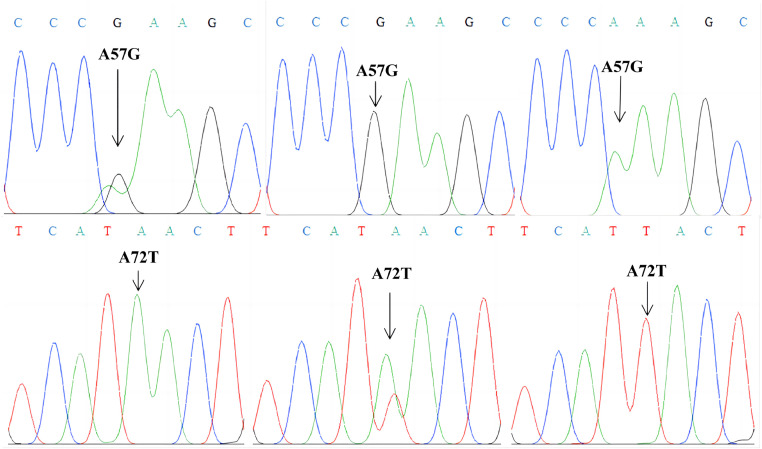
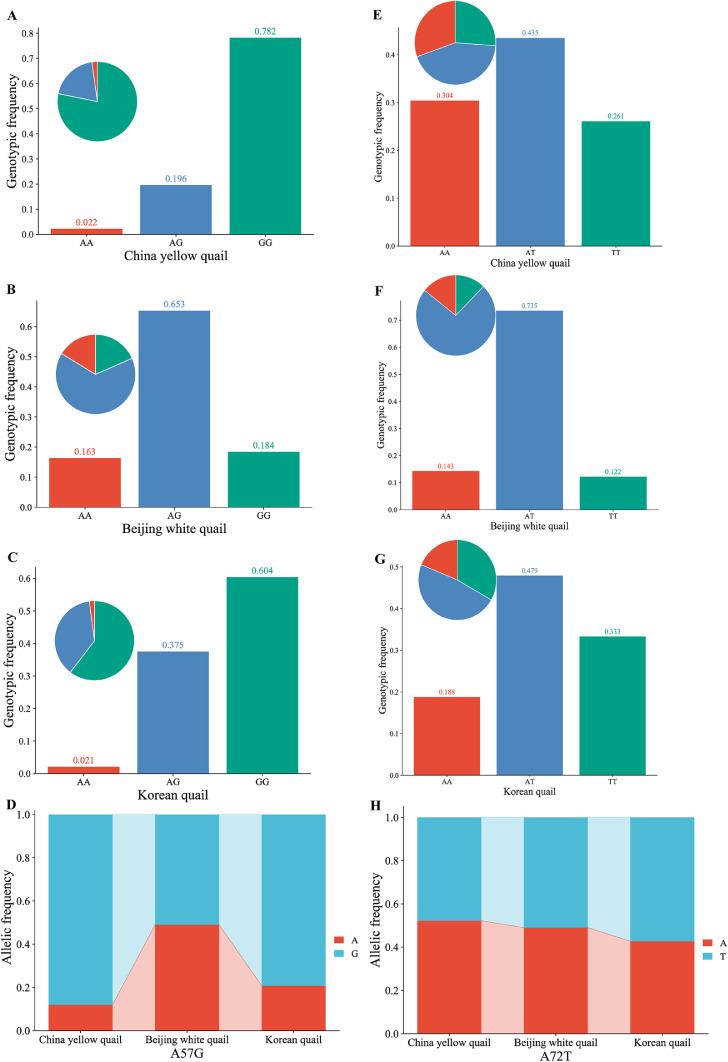
The egg quality traits and carcass traits are important economic traits in poultry breeding, which are controlled by genetic, environmental, and nutritional factors (Ye et al., 2017). The IGF-1R gene is well known to play an important role in economic traits of animals. It is reported that 10 SNPs of the IGF-1 gene were detected in Hulun Buir sheep. SNP8, haplotype combinations H5H5 and H5H6 of the IGF-1R gene showed superior growth traits during the early stage (Ding et al., 2022). In this study, we have detected polymorphisms of the IGF-1R gene in quails. Two SNPs (A57G and A72T) identified in the 3 quail strains of IGF-1R gene were genotyped by sequencing technology (Figure 1). It can be seen from Table 1 that 3 genotypes (AA, AG, and GG) were detected at the A57G site. Three genotypes (AA, AT, and TT) were detected at the A72T site. The AA, AG, and GG genotype frequencies were 2.2, 19.6, and 78.2% at the A57G site in CY strain, respectively (Figure 2A). The AA, AG, and GG genotype frequencies were 16.3, 65.3, and 18.4% at the A57G site in BW strain, respectively (Figure 2B). The AA, AG, and GG genotype frequencies were 2.1, 37.5, and 60.4% at the A57G site in KO strain, respectively (Figure 2C). Furthermore, allele G was the dominant gene in 3 strains (Figure 2D). The AA, AT, and TT genotype frequencies were 30.4, 43.5, and 26.1% at the A72TG site in CY strain, respectively (Figure 2E). The AA, AT, and TT genotype frequencies were 14.3, 73.5, and 12.2% at the A72TG site in BW strain, respectively (Figure 2F). The AA, AT, and TT genotype frequencies were 18.8, 47.9, and 33.3% at the A72TG site in KO strain, respectively (Figure 2G). Furthermore, allele A was the dominant gene of CY and BW strains, while allele T was the dominant gene of KO strain (Figure 2H). The PIC analysis results showed that all SNPs were in moderate polymorphism (0.25 < PIC < 0.50) except A57G site of CY quail that showed a low polymorphism (PIC < 0.25). The A57G and A72TG of CY and KO strains were in Hardy-Weinberg equilibrium (HWE) based on the chi-square test (P > 0.05). The A57G site of BW strain has deviated from the HWE (P < 0.05), the A72T site of BW strain has significantly deviated from the HWE (P < 0.01), which was not statistically significant (Table 1).
Figure 1.
Sequencing results of A57G and A72T site of IGF-1R gene.
Table 1.
Genotype frequency, allele frequency, and Hardy-Weinberg's law data of SNPs of IGF-1R gene in quail.
| Allelic frequency |
HWE3 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 | S2 | Genotypic frequency | Major | Minor | Chi-square | P | Ho4 | He5 | PIC6 | Ne7 | ||
| A57G | CY(46) | 0.022(AA) | 0.196(AG) | 0.782(GG) | 0.880 | 0.120 | 0.230 | 0.632 | 0.789 | 0.211 | 0.188 | 1.267 |
| BW(49) | 0.163(AA) | 0.653(AG) | 0.184(GG) | 0.510 | 0.490 | 4.608 | 0.032 | 0.500 | 0.500 | 0.375 | 1.999 | |
| KO(48) | 0.021(AA) | 0.375(AG) | 0.604(GG) | 0.792 | 0.208 | 0.899 | 0.343 | 0.670 | 0.330 | 0.275 | 1.492 | |
| A72T | CY(46) | 0.304(AA) | 0.435(AT) | 0.261(TT) | 0.522 | 0.478 | 0.763 | 0.382 | 0.501 | 0.499 | 0.375 | 1.996 |
| BW(49) | 0.143(AA) | 0.735(AT) | 0.122(TT) | 0.510 | 0.490 | 10.824 | 0.001 | 0.500 | 0.500 | 0.375 | 1.999 | |
| KO(48) | 0.188(AA) | 0.479(AT) | 0.333(TT) | 0.573 | 0.427 | 0.021 | 0.885 | 0.511 | 0.489 | 0.370 | 1.958 | |
Abbreviations: BW, Beijing white quail; CY, Chinese yellow quail; He5, heterozygosity; Ho4, homozygosity; HWE3, Hardy-Weinberg equilibrium test; KO, Korean quail; Ne7, effective allele numbers; PIC6, polymorphism information content; S2, strain; SNP1, single nucleotide polymorphism.
Figure 2.
The number of genotypes and frequencies of allelotypes in IGF-1R gene.
Association Analysis of IGF-1R Gene With Egg Quality in Quail
The present study was conducted to correlate the egg quality of quail at 7 wk of age (Table 2). The results showed that there was no significant association between the A57G site of IGF-1R gene with egg quality in CY strain (P > 0.05). The A57G site was significantly associated with the YWI in BW strain, and individuals with the GG genotype had significantly higher YWI than that of AG genotype (P < 0.05). In addition, there was no significant association between A57G site with egg quality in KO strain (P > 0.05). The results showed that A57G site had less effect on the egg quality in CY and KO quail strains.
Table 2.
Association analysis of A57G and A72T site with egg quality of quail.
| A57G (mean ± SE) |
A72T (mean ± SE) |
||||||
|---|---|---|---|---|---|---|---|
| S | EQ | AA | AG | GG | AA | AT | TT |
| CY(46) | EW | 10.900 ± 0.112 | 10.733 ± 0.131 | 10.706 ± 0.141 | 10.929 ± 0.211 | 10.568 ± 0.173 | 10.700 ± 0.203 |
| EL | 32.490 ± 0.172 | 32.142 ± 0.314 | 31.987 ± 0.207 | 32.391 ± 0.307 | 32.007 ± 0.287 | 31.642 ± 0.27 | |
| ES | 24.950 ± 0.094 | 24.914 ± 0.105 | 25.009 ± 0.118 | 25.121 ± 0.160 | 24.822 ± 0.149 | 25.098 ± 0.182 | |
| ESI | 1.302 ± 0.007 | 1.290 ± 0.014 | 1.279 ± 0.008 | 1.289 ± 0.009 | 1.290 ± 0.012 | 1.261 ± 0.011 | |
| YH | 8.000 ± 0.103 | 8.067 ± 0.133 | 7.991 ± 0.129 | 7.943 ± 0.174 | 8.011 ± 0.146 | 8.075 ± 0.250 | |
| YWI | 25.870 ± 0.199 | 24.437 ± 0.463 | 24.611 ± 0.227 | 24.486 ± 0.360 | 24.431 ± 0.312 | 25.018 ± 0.380 | |
| YI | 0.309 ± 0.005 | 0.331 ± 0.009 | 0.326 ± 0.007 | 0.326 ± 0.010 | 0.329 ± 0.008 | 0.324 ± 0.012 | |
| YWE | 3.600 ± 0.041 | 3.478 ± 0.072 | 3.429 ± 0.049 | 3.407 ± 0.062 | 3.389 ± 0.056 | 3.567 ± 0.099 | |
| EST | 0.208 ± 0.002 | 0.175 ± 0.004 | 0.176 ± 0.002 | 0.170 ± 0.004 | 0.179 ± 0.003 | 0.179 ± 0.004 | |
| AH | 1.570 ± 0.073 | 1.754 ± 0.156 | 1.725 ± 0.086 | 1.704 ± 0.105 | 1.663 ± 0.138 | 1.855 ± 0.113 | |
| BW(49) | EW | 10.514 ± 0.222 | 10.410 ± 0.163 | 10.757 ± 0.360 | 10.400 ± 0.497 | 10.432 ± 0.146 | 10.833 ± 0.246 |
| EL | 31.499 ± 0.241 | 31.684 ± 0.286 | 32.211 ± 0.695 | 31.565 ± 0.802 | 31.639 ± 0.266 | 32.435 ± 0.295 | |
| ES | 25.094 ± 0.322 | 24.807 ± 0.133 | 25.033 ± 0.266 | 24.773 ± 0.404 | 24.839 ± 0.122 | 25.273 ± 0.284 | |
| ESI | 1.257 ± 0.022 | 1.277 ± 0.010 | 1.287 ± 0.027 | 1.275 ± 0.032 | 1.274 ± 0.010 | 1.284 ± 0.020 | |
| YH | 7.886 ± 0.240 | 7.924 ± 0.130 | 7.814 ± 0.202 | 7.683 ± 0.294 | 7.935 ± 0.122 | 7.933 ± 0.198 | |
| YWI | 25.224 ± 0.393ab | 24.569 ± 0.227b | 26.250 ± 0.892a | 24.030 ± 0.792 | 25.065 ± 0.270 | 25.270 ± 0.431 | |
| YI | 0.313 ± 0.011 | 0.312 ± 0.013 | 0.301 ± 0.018 | 0.322 ± 0.019 | 0.307 ± 0.012 | 0.315 ± 0.011 | |
| YWE | 3.486 ± 0.055 | 3.362 ± 0.058 | 3.543 ± 0.181 | 3.267 ± 0.201 | 3.426 ± 0.057 | 3.483 ± 0.065 | |
| EST | 0.167 ± 0.006 | 0.159 ± 0.002 | 0.160 ± 0.005 | 0.151 ± 0.007b | 0.164 ± 0.002a | 0.153 ± 0.005ab | |
| AH | 1.997 ± 0.199 | 1.883 ± 0.077 | 1.683 ± 0.133 | 1.778 ± 0.193 | 1.870 ± 0.071 | 1.955 ± 0.237 | |
| KO(48) | EW | 10.307 ± 0.301 | 10.417 ± 0.172 | 10.380 ± 0.151 | 10.100 ± 0.287b | 10.045 ± 0.218b | 10.993 ± 0.225a |
| EL | 31.913 ± 0.457 | 31.839 ± 0.240 | 31.864 ± 0.219 | 31.457 ± 0.511b | 31.489 ± 0.281b | 32.609 ± 0.373a | |
| ES | 24.771 ± 0.224 | 24.834 ± 0.145 | 24.813 ± 0.121 | 24.626 ± 0.225b | 24.542 ± 0.167b | 25.287 ± 0.198a | |
| ESI | 1.288 ± 0.014 | 1.282 ± 0.008 | 1.284 ± 0.007 | 1.277 ± 0.016 | 1.283 ± 0.009 | 1.290 ± 0.015 | |
| YH | 6.593 ± 0.239 | 6.428 ± 0.105 | 6.484 ± 0.106 | 6.767 ± 0.112 | 6.380 ± 0.153 | 6.453 ± 0.224 | |
| YWI | 26.339 ± 0.466 | 26.876 ± 0.342 | 26.693 ± 0.276 | 26.446 ± 0.820 | 26.274 ± 0.406 | 27.399 ± 0.318 | |
| YI | 0.252 ± 0.012 | 0.241 ± 0.006 | 0.245 ± 0.006 | 0.258 ± 0.010 | 0.245 ± 0.009 | 0.237 ± 0.010 | |
| YWE | 3.440 ± 0.144 | 3.576 ± 0.073 | 3.530 ± 0.069 | 3.456 ± 0.176 | 3.450 ± 0.106 | 3.680 ± 0.095 | |
| EST | 0.169 ± 0.007 | 0.180 ± 0.003 | 0.176 ± 0.003 | 0.190 ± 0.006 | 0.172 ± 0.005 | 0.174 ± 0.004 | |
| AH | 1.375 ± 0.110 | 1.382 ± 0.088 | 1.380 ± 0.068 | 1.430 ± 0.134 | 1.334 ± 0.103 | 1.411 ± 0.127 | |
The difference between genotypes with different lowercase letters were significant (P < 0.05).
Abbreviations: AH, albumen height; BW, Beijing white quail; CY, Chinese yellow quail; EL, egg long; EQ, egg quality; ES, egg short; ESI, egg shape index; EST, egg shell thickness; EW, egg weight; KO, Korean quail; S, Strain; YH, yolk height; YI, yolk index; YWE, yolk weight; YWI, yolk width.
For the A72TG site, there was no significant association with egg quality in CY strain (P > 0.05). However, there was significantly associated with the EST in BW strain, and individuals with the AT genotype had significantly higher EST than that of AA genotype (P < 0.05). In addition, the A72TG site was significantly associated with the EW, ES, and EL in KO strain (P < 0.05), and individuals with the TT genotype had better performance on EW, ES, and EL (P < 0.05). The results showed that A72T site had less effect on the egg quality in CY quail strain.
In the linkage between 2 SNPs (A57G and A72T), there were 6, 8, and 6 haplotype combinations (combinations with the number of individuals higher than or equal to 3) in CY, BW, and KO quail strains, respectively (Table 3). The result showed that haplotype combination AAAA had significantly higher EST than AGAA in CY strain (P < 0.05). AAAA and AAAT combinations had significantly higher EST than AATT, AGAA and GGAA in BW strain (P < 0.05). AGTT and GGTT combinations had significantly higher EW than AGAT in KO strain (P < 0.05). AGAT combination had significantly lower EST than the other 5 haplotype combinations in KO strain (P < 0.05). Similar to this study, a previous study in Japanese quail showed that a SNP (c.2293G>A) of the IGF-1R gene was significantly associated with growth traits (Moe et al., 2007). Jin et al. (2014) showed that based a haplotype comprising of 3-SNP (rs14011783, rs14011780, and rs14011776) of IGF-1R gene was significantly associated with BW49, BW70, and FCR (P < 0.05), which similar to this study. It can be seen that there was a significant correlation between IGF-1R gene and economic traits of animals.
Table 3.
Correlation analysis of IGF-1R gene haplotype combinations with egg quality of quail.
| Traits (mean ± SE) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S | D | EW | EL | ES | ESI | YH | YWI | YI | YWE | EST | AH |
| CY | |||||||||||
| (46) | AAAA | 10.900 ± 0.112 | 32.490 ± 0.172 | 24.950 ± 0.094 | 1.302 ± 0.007 | 8.000 ± 0.103 | 25.870 ± 0.199 | 0.309 ± 0.005 | 3.600 ± 0.041 | 0.208 ± 0.002a | 1.570 ± 0.073 |
| AGAA | 10.850 ± 0.250 | 32.255 ± 0.025 | 25.160 ± 0.180 | 1.282 ± 0.010 | 7.650 ± 0.050 | 23.925 ± 0.145 | 0.320 ± 0.000 | 3.400 ± 0.000 | 0.169 ± 0.011b | 1.615 ± 0.285 | |
| AGAT | 10.717 ± 0.189 | 32.220 ± 0.467 | 24.758 ± 0.093 | 1.301 ± 0.019 | 8.200 ± 0.165 | 24.172 ± 0.504 | 0.340 ± 0.011 | 3.450 ± 0.092 | 0.175 ± 0.005ab | 1.815 ± 0.225 | |
| AGTT | 10.600 ± 0.112 | 31.450 ± 0.172 | 25.360 ± 0.094 | 1.240 ± 0.007 | 8.100 ± 0.103 | 27.050 ± 0.199 | 0.299 ± 0.005 | 3.800 ± 0.041 | 0.186 ± 0.002ab | 1.670 ± 0.073 | |
| GGAT | 10.500 ± 0.240 | 31.908 ± 0.369 | 24.852 ± 0.216 | 1.285 ± 0.016 | 7.923 ± 0.199 | 24.551 ± 0.401 | 0.324 ± 0.010 | 3.362 ± 0.071 | 0.182 ± 0.004ab | 1.593 ± 0.175 | |
| GGTT | 10.709 ± 0.223 | 31.659 ± 0.295 | 25.074 ± 0.198 | 1.263 ± 0.012 | 8.073 ± 0.274 | 24.833 ± 0.364 | 0.326 ± 0.013 | 3.545 ± 0.106 | 0.178 ± 0.004ab | 1.872 ± 0.123 | |
| BW(49) | AAAA | 11.100 ± 0.128 | 30.290 ± 0.225 | 26.160 ± 0.111 | 1.158 ± 0.009 | 8.300 ± 0.100 | 23.930 ± 0.233 | 0.347 ± 0.009 | 3.500 ± 0.050 | 0.182 ± 0.002a | 1.870 ± 0.065 |
| AAAT | 10.075 ± 0.085 | 31.628 ± 0.237 | 24.495 ± 0.229 | 1.292 ± 0.020 | 7.650 ± 0.393 | 25.050 ± 0.474 | 0.306 ± 0.017 | 3.425 ± 0.085 | 0.173 ± 0.004a | 1.820 ± 0.233 | |
| AATT | 11.100 ± 0.300 | 31.845 ± 0.005 | 25.760 ± 0.370 | 1.236 ± 0.018 | 8.150 ± 0.150 | 26.220 ± 0.080 | 0.311 ± 0.007 | 3.600 ± 0.000 | 0.147 ± 0.002b | 2.415 ± 0.515 | |
| AGAA | 9.900 ± 0.351 | 31.660 ± 0.802 | 24.173 ± 0.192 | 1.310 ± 0.033 | 7.433 ± 0.484 | 23.527 ± 0.477 | 0.317 ± 0.025 | 3.100 ± 0.100 | 0.142 ± 0.004b | 1.777 ± 0.384 | |
| AGAT | 10.427 ± 0.199 | 31.497 ± 0.348 | 24.853 ± 0.156 | 1.267 ± 0.011 | 8.009 ± 0.150 | 24.670 ± 0.271 | 0.311 ± 0.017 | 3.386 ± 0.072 | 0.162 ± 0.002ab | 1.926 ± 0.083 | |
| AGTT | 10.700 ± 0.344 | 32.730 ± 0.362 | 25.030 ± 0.346 | 1.308 ± 0.019 | 7.825 ± 0.287 | 24.795 ± 0.488 | 0.317 ± 0.017 | 3.425 ± 0.085 | 0.156 ± 0.007ab | 1.725 ± 0.208 | |
| GGAA | 10.800 ± 1.600 | 32.060 ± 2.580 | 24.980 ± 0.890 | 1.281 ± 0.058 | 7.750 ± 0.550 | 24.835 ± 2.775 | 0.319 ± 0.058 | 3.400 ± 0.700 | 0.148 ± 0.005b | 1.735 ± 0.335 | |
| GGAT | 10.740 ± 0.125 | 32.272 ± 0.589 | 25.054 ± 0.262 | 1.289 ± 0.034 | 7.840 ± 0.234 | 26.816 ± 0.787 | 0.294 ± 0.018 | 3.600 ± 0.130 | 0.165 ± 0.005ab | 1.662 ± 0.160 | |
| KO | |||||||||||
| (48) | AGAA | 9.933 ± 0.865ab | 30.987 ± 1.435 | 24.507 ± 0.598 | 1.263 ± 0.032 | 6.800 ± 0.153 | 26.270 ± 0.822 | 0.259 ± 0.002 | 3.467 ± 0.406 | 0.186 ± 0.006a | 1.200 ± 0.058 |
| AGAT | 9.640 ± 0.491b | 31.134 ± 0.607 | 24.316 ± 0.377 | 1.281 ± 0.027 | 6.320 ± 0.437 | 25.404 ± 1.102 | 0.253 ± 0.027 | 3.060 ± 0.252 | 0.150 ± 0.015b | 1.240 ± 0.084 | |
| AGTT | 10.943 ± 0.316a | 32.866 ± 0.540 | 25.210 ± 0.267 | 1.304 ± 0.021 | 6.700 ± 0.421 | 27.036 ± 0.474 | 0.249 ± 0.018 | 3.700 ± 0.148 | 0.175 ± 0.006a | 1.547 ± 0.216 | |
| GGAA | 10.183 ± 0.209ab | 31.692 ± 0.426 | 24.685 ± 0.220 | 1.284 ± 0.019 | 6.750 ± 0.159 | 26.533 ± 1.214 | 0.258 ± 0.016 | 3.450 ± 0.205 | 0.192 ± 0.009a | 1.545 ± 0.186 | |
| GGAT | 10.180 ± 0.240ab | 31.607 ± 0.323 | 24.617 ± 0.188 | 1.284 ± 0.009 | 6.400 ± 0.156 | 26.563 ± 0.398 | 0.242 ± 0.009 | 3.580 ± 0.096 | 0.179 ± 0.004a | 1.365 ± 0.135 | |
| GGTT | 11.038 ± 0.338a | 32.384 ± 0.536 | 25.354 ± 0.304 | 1.278 ± 0.023 | 6.238 ± 0.201 | 27.718 ± 0.423 | 0.226 ± 0.009 | 3.663 ± 0.132 | 0.173 ± 0.006a | 1.291 ± 0.146 | |
The difference between genotypes with different lowercase letters were significant (P < 0.05).
Abbreviations: AH, albumen height; BW, Beijing white quail; CY, Chinese yellow quail; D, diplotype; EL, egg long; EQ, egg quality; ES, egg short; ESI, egg shape index; EST, egg shell thickness; EW, egg weight; KO, Korean quail; S, strain; YH, yolk height; YI, yolk index; YWE, yolk weight; YWI, yolk width.
Association Analysis of IGF-1R Gene With Carcass Traits in Quail
The present study was conducted to correlate the carcass traits of quail at 20 wk of age (Table 4). The result showed that A57G site was not significantly related to carcass traits in 3 quail strains (P > 0.05). The A72T site was significantly associated with the LW in 3 quail strains, and individuals with the TT genotype had significantly higher LW than that of AT genotype (P < 0.05). The A72T site was significantly associated with the DP in 3 quail strains, and individuals with the TT genotype had significantly higher DP than that of AA and AT genotype (P < 0.05). In the linkage between 2 SNPs (A57G and A72T), 9 haplotype combinations (combinations with the number of individuals higher than or equal to 3) were formed in 3 strains (Table 5). The result showed that haplotype combination AATT, AGAA and AGTT had significantly higher LR than of AAAA and AAAT combination (P < 0.05). Similar to this study, a previous study in chicken showed that A17299834G SNP of the IGF-1R gene was significantly associated with carcass traits, and a haplotype based on 2 SNPs (A17299834G and C17293932T) showed significant correlation with most of the early growth traits and carcass traits (Lei et al., 2008). The IGF-1R gene may be used as the major gene affecting quail carcass traits. These studies suggest that the associations of the SNP or haplotype with economic traits in the present study were reliable.
Table 4.
Association of between A57G and A72T site with carcass traits of quail.
| A57G (mean ± SE) |
A72T (mean ± SE) |
|||||
|---|---|---|---|---|---|---|
| CT | AA | AG | GG | AA | AT | TT |
| BOW | 142.643 ± 3.615 | 140.546 ± 2.002 | 140.963 ± 2.146 | 141.493 ± 2.982 | 139.635 ± 1.921 | 143.461 ± 2.646 |
| DCW | 137.186 ± 3.516 | 135.524 ± 2.024 | 136.341 ± 2.164 | 136.367 ± 2.971 | 134.565 ± 1.909 | 139.283 ± 2.747 |
| WNCW | 86.086 ± 3.417 | 87.130 ± 1.566 | 90.574 ± 1.646 | 90.147 ± 2.579 | 87.706 ± 1.489 | 90.430 ± 2.104 |
| HW | 1.071 ± 0.102 | 1.054 ± 0.033 | 1.070 ± 0.028 | 1.160 ± 0.052 | 1.031 ± 0.023 | 1.074 ± 0.052 |
| LW | 4.157 ± 0.532 | 4.165 ± 0.125 | 3.943 ± 0.145 | 4.073 ± 0.288ab | 3.879 ± 0.087b | 4.426 ± 0.264a |
| BMW | 27.886 ± 1.648 | 27.789 ± 0.655 | 28.635 ± 0.650 | 28.427 ± 1.213 | 27.844 ± 0.563 | 28.97 ± 0.896 |
| LMW | 7.600 ± 0.307 | 7.324 ± 0.138 | 7.517 ± 0.145 | 7.600 ± 0.228 | 7.340 ± 0.140 | 7.578 ± 0.137 |
| DP | 96.179 ± 0.457 | 96.399 ± 0.153 | 96.682 ± 0.128 | 96.358 ± 0.145b | 96.348 ± 0.134b | 97.039 ± 0.179a |
| WNCR | 60.318 ± 1.624 | 62.005 ± 0.714 | 64.239 ± 0.538 | 63.669 ± 1.173 | 62.815 ± 0.564 | 63.044 ± 0.897 |
| HR | 1.263 ± 0.138 | 1.214 ± 0.034 | 1.189 ± 0.030 | 1.295 ± 0.055 | 1.185 ± 0.028 | 1.190 ± 0.052 |
| LR | 4.969 ± 0.802 | 4.816 ± 0.149 | 4.377 ± 0.149 | 4.589 ± 0.359 | 4.467 ± 0.110 | 4.922 ± 0.298 |
| BMR | 32.338 ± 1.198 | 31.895 ± 0.495 | 31.606 ± 0.423 | 31.482 ± 0.892 | 31.773 ± 0.392 | 31.997 ± 0.602 |
| LMR | 17.703 ± 0.573 | 16.850 ± 0.219 | 16.626 ± 0.190 | 16.917 ± 0.395 | 16.744 ± 0.176 | 16.857 ± 0.297 |
The difference between genotypes with different lowercase letters were significant (P < 0.05).
Abbreviations: BMR, breast muscle rate; BMW, breast muscle weight; BOW, body weight; CT, carcass trait; DCW, dressed carcass weight; DP, dressing percentage; HR, heart rate; HW, heart weight; LMR, leg muscle rate; LMW, leg muscle weight; LR, liver rate; LW, liver weight; WNCR, whole net carcass rate; WNCW, whole net carcass weight.
Table 5.
Correlation analysis of IGF-1R gene haplotype combinations with carcass traits of quail.
| Traits | AAAA | AAAT | AATT | AGAA | AGAT | AGTT | GGAA | GGAT | GGTT |
|---|---|---|---|---|---|---|---|---|---|
| BOW | 138.200 ± 1.388 | 148.167 ± 6.996 | 138.600 ± 3.980 | 139.833 ± 13.471 | 139.092 ± 2.221 | 145.538 ± 3.744 | 142.245 ± 2.618 | 139.135 ± 3.460 | 143.292 ± 4.401 |
| DCW | 134.300 ± 1.400 | 141.567 ± 7.626 | 133.767 ± 3.398 | 134.633 ± 13.165 | 133.831 ± 2.203 | 141.363 ± 4.033 | 137.027 ± 2.690 | 134.483 ± 3.443 | 139.275 ± 4.534 |
| WNCW | 92.300 ± 1.098 | 90.267 ± 6.636 | 79.833 ± 1.729 | 81.633 ± 11.341 | 87.027 ± 1.617 | 89.525 ± 3.427 | 92.273 ± 1.791 | 88.139 ± 2.768 | 93.683 ± 2.879 |
| HW | 1.200 ± 0.021 | 0.967 ± 0.088 | 1.133 ± 0.233 | 1.233 ± 0.186 | 1.046 ± 0.035 | 1.013 ± 0.072 | 1.136 ± 0.054 | 1.022 ± 0.033 | 1.100 ± 0.072 |
| LW | 3.200 ± 0.098 | 3.500 ± 0.265 | 5.133 ± 1.033 | 4.800 ± 0.458 | 3.969 ± 0.131 | 4.563 ± 0.298 | 3.955 ± 0.352 | 3.826 ± 0.122 | 4.158 ± 0.401 |
| BMW | 27.600 ± 0.444 | 29.433 ± 3.147 | 26.433 ± 2.617 | 23.600 ± 3.143 | 27.538 ± 0.702 | 30.175 ± 1.285 | 29.818 ± 1.198 | 27.983 ± 0.943 | 28.800 ± 1.353 |
| LMW | 7.900 ± 0.096 | 8.033 ± 0.371 | 7.067 ± 0.521 | 6.833 ± 0.696 | 7.358 ± 0.172 | 7.400 ± 0.208 | 7.782 ± 0.230 | 7.230 ± 0.245 | 7.825 ± 0.166 |
| DP | 97.178 ± 0.098 | 95.489 ± 0.641 | 96.536 ± 0.764 | 96.258 ± 0.183 | 96.204 ± 0.182 | 97.085 ± 0.291 | 96.311 ± 0.177 | 96.623 ± 0.195 | 97.134 ± 0.231 |
| WNCR | 66.787 ± 0.440 | 60.835 ± 2.523 | 57.644 ± 1.192 | 57.981 ± 2.711 | 62.642 ± 0.845 | 61.443 ± 1.394 | 64.938 ± 1.045 | 63.268 ± 0.799 | 65.462 ± 0.921 |
| HR | 1.300 ± 0.023 | 1.097 ± 0.175 | 1.416 ± 0.278 | 1.507 ± 0.035 | 1.206 ± 0.040 | 1.129 ± 0.060 | 1.237 ± 0.064 | 1.173 ± 0.040 | 1.174 ± 0.063 |
| LR | 3.467 ± 0.116b | 3.947 ± 0.547b | 6.492 ± 1.464a | 5.946 ± 0.256a | 4.596 ± 0.169ab | 5.108 ± 0.293a | 4.322 ± 0.417ab | 4.388 ± 0.144ab | 4.406 ± 0.354ab |
| BMR | 29.903 ± 0.308 | 32.490 ± 1.391 | 32.998 ± 2.625 | 28.975 ± 0.929 | 31.679 ± 0.614 | 33.691 ± 0.643 | 32.309 ± 1.099 | 31.785 ± 0.544 | 30.618 ± 0.727 |
| LMR | 17.118 ± 0.141 | 17.879 ± 0.634 | 17.723 ± 1.351 | 16.947 ± 1.163 | 16.915 ± 0.272 | 16.603 ± 0.366 | 16.891 ± 0.472 | 16.403 ± 0.218 | 16.811 ± 0.422 |
The difference between genotypes with different lowercase letters were significant (P < 0.05).
Abbreviations: BMR, breast muscle rate; BMW, breast muscle weight; BOW, body weight; DCW, dressed carcass weight; DP, dressing percentage; HR, heart rate; HW, heart weight; LMR, leg muscle rate; LMW, leg muscle weight; LR, liver rate; LW, liver weight; WNCR, whole net carcass rate; WNCW, whole net carcass weight.
In conclusion, 2 SNPs (A57G and A72T) or haplotype combination of IGF-1R gene which were significantly correlated to egg quality and carcass traits. Therefore, IGF-1R gene could be a molecular genetic marker to improve economic traits with quail breeding. However, due to limitation of the number of quail population, further studies in large populations with different quail strains are required to further assess associations of the IGF-1R gene polymorphisms with egg quality, carcass traits, and other economic traits.
ACKNOWLEDGMENTS
This research was supported by grants from National Natural Science Foundation of China (No. 31201777).
I declare that research on live animals is in line with the guidelines approved by the institutional animal care and use Committee (IACUC) through the use of appropriate management and laboratory techniques to avoid unnecessary discomfort of animals.
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- Ali D.A., Al-Shuhaib M.B.S., Farhadi G., Al-Kafajy F.R., Al-Thuwaini T.M., Esmailizadeh A. Detection of a novel single nucleotide polymorphism in IGF2 gene with a negative impact on egg production and body weight in Japanese quail (Coturnix japonica) J. Genet. Eng. Biotechnol. 2021;19:170. doi: 10.1186/s43141-021-00271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J.Y., Dong Z.H., Lei Y., Yang Y.B., Jia X.P., Li J.Y. Association analysis between polymorphism of gonadotrophin releasing hormone genes and growth traits of quail (Coturnix Coturnix) Braz. J. Poult. Sci. 2021;23 eRBCA-2020-1314. [Google Scholar]
- Cardoso S., López I.P., Piñeiro-Hermida S., Pichel J.G., Moreira P.I. IGF1R deficiency modulates brain signaling pathways and disturbs mitochondria and redox homeostasis. Biomedicines. 2021;9:158. doi: 10.3390/biomedicines9020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N., Tian D., Li X., Zhang Z., Tian F., Liu S., Han B., Liu D., Zhao K. Genetic polymorphisms of IGF1 and IGF1R genes and their effects on growth traits in Hulun Buir sheep. Genes. 2022;13:666. doi: 10.3390/genes13040666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Magd M.A., Saleh A.A., Nafeaa A.A., El-Komy S.M., Afifi M.A. Polymorphisms of the IGF1 gene and their association with growth traits, serum concentration and expression rate of IGF1 and IGF1R in buffalo. J. Zhejiang. Univ. Sci. B. 2017;18:1064–1074. doi: 10.1631/jzus.B1600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tarabany M.S., Saleh A.A., El-Araby I.E., El-Magd M.A. Association of LEPR polymorphisms with egg production and growth performance in female Japanese quails. Anim. Biotechnol. 2022;33:599–611. doi: 10.1080/10495398.2020.1812617. [DOI] [PubMed] [Google Scholar]
- Jin S., Chen S., Li H., Lu Y., Xu G., Yang N. Associations of polymorphisms in GHRL, GHSR, and IGF1R genes with feed efficiency in chickens. Mol. Biol. Rep. 2014;41:3973–3979. doi: 10.1007/s11033-014-3265-8. [DOI] [PubMed] [Google Scholar]
- Lei M., Peng X., Zhou M., Luo C., Nie Q., Zhang X. Polymorphisms of the IGF1R gene and their genetic effects on chicken early growth and carcass traits. BMC Genet. 2008;9:70. doi: 10.1186/1471-2156-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.L., Wen Y.F., Cao X.K., Cheng J., Huang Y.Z., Ma Y., Hu L.Y., Lei C.Z., Qi X.L., Cao H., Chen H. Copy number variation (CNV) in the IGF1R gene across four cattle breeds and its association with economic traits. Arch. Anim. Breed. 2019;62:171–179. doi: 10.5194/aab-62-171-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle F. The future of Japanese quail for research and production. World's Poult. Sci. J. 2004;60:500–507. [Google Scholar]
- Moe H.H., Shimogiri T., Kamihiraguma W., Isobe H., Kawabe K., Okamoto S., Minvielle F., Maeda Y. Analysis of polymorphisms in the insulin-like growth factor 1 receptor (IGF1R) gene from Japanese quail selected for body weight. Anim. Genet. 2007;38:659–661. doi: 10.1111/j.1365-2052.2007.01653.x. [DOI] [PubMed] [Google Scholar]
- Priti M., Satish S. Quail farming: an introduction. Int. J. Life Sci. 2014;2:190–193. [Google Scholar]
- Pu Y.J., Wu Y., Xu X.J., Du J.P., Gong Y.Z. Association of VIPR-1 gene polymorphisms and haplotypes with egg production in laying quails. J. Zhejiang Univ. Sci. B. 2016;17:591–596. doi: 10.1631/jzus.B1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczuk M., Zych S., Wójcik J., Czerniawska-Piątkowska E. Association of two SNPs in the coding region of the insulin-like growth factor 1 receptor (IGF1R) gene with growth-related traits in Angus cattle. J. Appl. Genet. 2013;54:305–308. doi: 10.1007/s13353-013-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.L., Li J.Y., Bai J.Y., Wang L.W., Fan H.D., Chen M.K., Zeng F.L., Lu X.N., He Y.H. Research Note: Polymorphisms of gonadotrophin-releasing hormone gene and their association with growth traits in quail (Coturnix Coturnix) Poult. Sci. 2023;102 doi: 10.1016/j.psj.2022.102439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.F., Wang D., Jin C.F., Zhang X.Q., Wu H.Q., Zhang L., Ding F.X., Xie K.Z., Zhang G.X. Polymorphisms of AluI and Hin1I loci of the IGF-1R gene and their genetic effects on growth traits in Bian chickens. Genet. Mol. Res. 2017;16 doi: 10.4238/gmr16029619. 10.4238. [DOI] [PubMed] [Google Scholar]
- Yang C., Teng J., Ning C., Wang W., Liu S., Zhang Q., Wang D., Tang H. Effects of growth-related genes on body measurement traits in Wenshang barred chickens. J. Poult. Sci. 2022;59:323–327. doi: 10.2141/jpsa.0210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M., Ye F., He L., Liu Y., Zhao X., Yin H., Li D., Xu H., Zhu Q., Wang Y. Molecular cloning, expression profiling, and marker validation of the chicken Myoz3 gene. Biomed. Res. Int. 2017;2017 doi: 10.1155/2017/5930918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh F.C., Boyle T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997;129:157. [Google Scholar]
- Zhang Y.R., Li Y.K., Fu C.Z., Wang J.L., Wang H.B., Zan L.S. Effects of bovine SMO gene polymorphisms on the body measurement and meat quality traits of Qinchuan cattle. Genet. Mol. Res. 2014;13:8105–8117. doi: 10.4238/2014.October.7.5. [DOI] [PubMed] [Google Scholar]