Abstract
The ability to restrict gene delivery and expression to particular cell types is of paramount importance for many types of gene therapy, especially in the lung. The alveolar epithelial type I (ATI) cell, in particular, is an attractive cell type to target, as it comprises 95% of the internal surface area of the lung. We demonstrate, through microinjection of fluorescently labeled plasmids, that a DNA sequence within the rat T1α promoter was able to mediate ATI cell-specific plasmid DNA nuclear import due to the binding of ATI-enriched transcription factors. Promoter deletion analysis and site-directed mutagenesis of specific transcription-factor-binding sites within the +101 to − 200 bp region of the T1α promoter identified HNF3 and TTF-1 as critical transcription factors for import. To test for nuclear import in vivo, plasmids expressing GFP from the CMV promoter were delivered into the lungs of mice by electroporation and evaluated immunohistochemically 48 h later. Plasmids carrying the 1.3 kbp T1α sequence resulted in GFP expression almost exclusively in ATI cells. This represents a new and highly efficient way to target a specific lung epithelial cell type both in vitro and in vivo based on the restriction of DNA nuclear import.
INTRODUCTION
Cell-specific targeting remains a major limitation in the advancement of most gene delivery and gene therapy techniques. To date, three approaches have been used to achieve cell selectivity: direct delivery to anatomical locations, use of specific cell surface receptors to promote cell entry and use of cell-specific promoters to restrict transcription. Our laboratory has developed a method to target plasmid DNA to specific cell types by exploiting the natural nuclear import machinery within cells.1-3 We and others have identified several DNA sequences that target plasmids into the nuclei of non-dividing cells in either a general or cell-specific manner, depending on the DNA sequence.2,4-8 The common feature to these sequences (typically promoters or enhancers) is that they contain multiple binding sites for transcription factors. Under normal circumstances, a typical transcription factor would be transported into the nucleus either after synthesis or upon activation due to the presence of a nuclear localization sequence (NLS) in the protein, bind to its unique DNA target sequence present in various promoters and modulate transcription. However, if a plasmid containing the transcription-factor-binding site is present in the cytoplasm, the transcription factor may bind to this site before nuclear import. The NLS import machinery will then bind to the DNA-bound transcription factors and translocate the DNA–protein complex into the nucleus. We have shown in cultured cells and in animal models that this process can occur in either a general manner in all cells due to the binding of ubiquitous transcription factors (for example, the SV40 DTS), or a cell-specific manner due to the binding of cell-specific transcription factors to specific DNA nuclear targeting sequences (DTSs) (for example, SMGA DTS for smooth muscle cells or the SP-C DTS for alveolar epithelial type II (ATII) cells).5,9
The ability to target genes to specific cell types of the lung is highly attractive in terms of the development of therapies for lung disease, as well as for the enhancement of our understanding of how various cell types function. In particular, alveolar epithelial type I (ATI) cells cover more than 95% of the internal surface area of the lung and are involved in fluid homeostasis as evidenced by the cells’ high osmotic water permeability, expression of transport proteins and ability to transport ions.10-13 However, research on these is lacking due to the relative inability to isolate primary cells, poor cell models and the absence of suitable cell lines. Several recent papers, including those utilizing gene arrays and transgenic mice, have offered some new insights into this cell type, but the development of new approaches to target these cells would allow us to study them further.14,15
In order to identify a DTS active in ATI cells only, we examined the nuclear import activity of the promoters of several ATI-specific genes. Here we demonstrate that the +101 to − 1251 bp region of the T1α promoter functions as an ATI-specific DTS, and the +101 to − 200 bp region of this promoter acts as DTS in both ATI and ATII cells. This activity relies upon the binding sites for certain transcription factors, which differs depending upon the cell type. Moreover, both sequences restrict gene expression to the same cell types in vivo in electroporated mouse lungs as seen in microinjection studies in cultured cells. This is the first demonstration of the ability to target plasmid DNA specifically to ATI cells for gene therapy.
RESULTS
Characterization of ATI-like cells
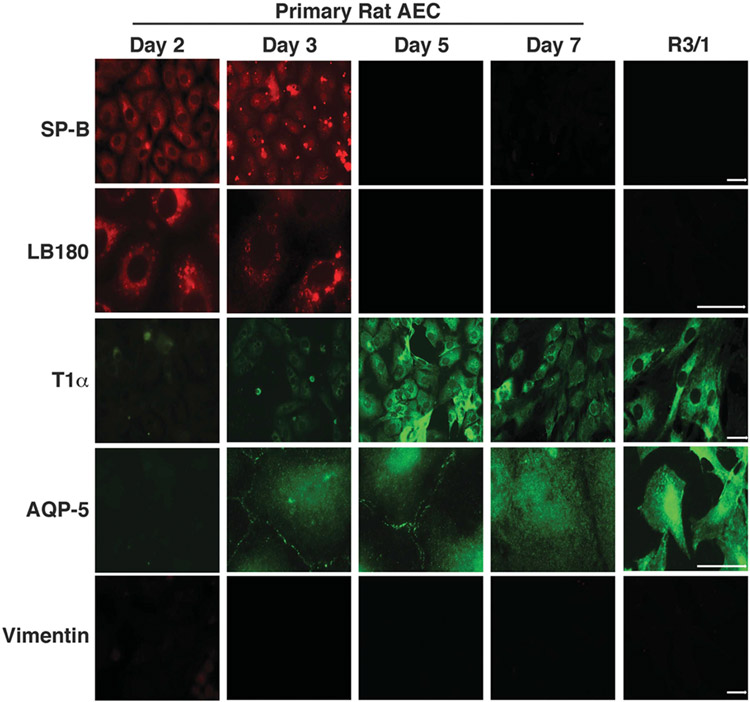
The study of ATI cells has been challenging due to the difficulty involved in isolating primary type I pneumocytes from animals as well as a limited selection of cell lines displaying type I characteristics. It has previously been demonstrated that primary ATII cells cultured on plastic begin to take on a type I cell phenotype after several days in culture.16-20 We characterized this process in our system by demonstrating that primary rat ATII cells exhibit a type I expression pattern by day 5 (D5) in culture, as evidenced by the increase in expression of T1α and Aquaporin-5 as well as the loss of Surfactant Protein B (SP-B) and lamellar body 180 (LB180) expression (Figure 1). We also obtained a rat cell line, R3/1, which has been previously reported to resemble ATI cells.21 In our hands, we demonstrated a highly type I cell phenotype (Figure 1). Reverse transcriptase PCR for transcripts of all of these genes confirmed the immunofluorescence data (data not shown). In contrast, a distinct type II phenotype was evident in D2 cells cultured on Matrigel, which helps maintain the type II phenotype while also making the cells adhere better for microinjection experiments, as well as D3 cells on glass (Figure 1). Therefore, all experiments were performed using R3/1 and D5 cells as models for ATI cells and D2 cells cultured on Matrigel as an ATII cell model.
Figure 1.
Analysis of alveolar epithelial cell phenotypes in culture. Primary rat alveolar epithelial type II cells were isolated and cultured on glass coverslips alone or coverslips precoated with Matrigel (day 2 only) for either 2, 3, 5 or 7 days. Immunofluorescence was then performed using antibodies specific to the ATII markers, SP-B and LB180, as well as the ATI markers T1α and AQP-5. The rat cell line, R3/1, was also analyzed. Staining for the fibroblast marker, vimentin, was used to verify purity of the cultures.
The T1α promoter mediates plasmid DNA nuclear import in type I-like alveolar epithelial cells
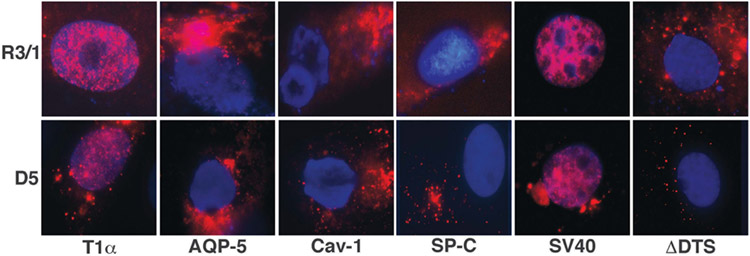
In an effort to identify a DNA nuclear import sequence that is active in type I alveolar epithelial cells, we selected the promoter regions of genes which, based on the literature, are expressed in a ATI cell selective manner. These promoters included the T1α, Aquaporin-5 and Caveolin-1 promoters.22 The Aquaporin-5 and Caveolin-1 promoters were PCR amplified from rat and mouse genomic DNA, respectively, and cloned into a plasmid containing tandem repeats of peptide nucleic acid (PNA)-binding sites for fluorescent labeling of the plasmid. Plasmid DNA nuclear import was assessed by cytoplasmic injection of these Cy3-PNA-labeled plasmids into either R3/1 cells or primary rat D5 cells. Cells were fixed at 5 h post-injection and the DNA was visualized by fluorescence microscopy for the Cy3-PNA-labeled plasmids (Figure 2). As expected, when plasmids containing the SV40 DTS, which supports DNA nuclear import in all cell types, were microinjected into the cytoplasm of both cell types, greater than 80% of the cells exhibited plasmid DNA within the nucleus at 5 h post-injection. In contrast, the plasmid backbone of these constructs (ΔDTS), which does not contain a DTS, showed no nuclear import (< 5%) after the incubation period, demonstrating that these cells, like all other mammalian cells tested to date, display sequence-specific DNA nuclear import.4 Plasmids carrying the T1α promoter sequence demonstrated robust DNA nuclear import in both R3/1 and D5 primary type I-like cells within 5 h. By contrast, plasmids carrying the Caveolin-1 promoter did not display any nuclear import in either cell type. The Aquaporin-5 promoter, however, produced variable results. In R3/1 cells, we observed a very low but reproducible level of nuclear import but no import was observed in D5 primary cells. Because of this weak and inconsistent activity, further evaluation of the AQP-5 promoter was not carried out. To demonstrate the selectivity of these ATI-like cells for DNA nuclear import, plasmids containing the SP-C DTS, an ATII-specific DTS sequence,9 were also microinjected into D5 and R3/1 cells, and showed no plasmid nuclear import in either cell type. These results suggest that the T1α promoter contains a DTS which is able to mediate plasmid nuclear import in ATI-like cells.
Figure 2.
The T1α promoter mediates plasmid nuclear import in ATI cells. Plasmids (0.5 mg ml−1) carrying the promoter regions of T1α, AQP-5, caveolin-1, SP-C, SV40 DTS or no DTS were labeled with Cy3-PNA and cytoplasmically microinjected into both D5 primary ATI cells or R3/1 cells. After 5 h, the cellular localization of the plasmids was analyzed by fluorescence microscopy.
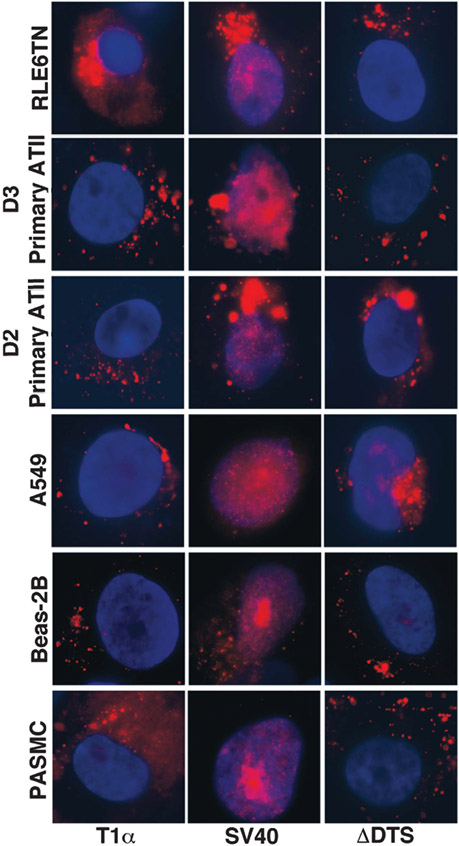
The T1α promoter does not mediate plasmid nuclear import in non-ATI cells
In order to determine if the nuclear import activity of the T1α DTS was restricted to type I cells, this plasmid was microinjected into the cytoplasm of RLE6TN cells, a type II cell line, D3 primary rat type II cells grown on glass, D2 rat primary type II cell cultured on Matrigel for 2 days, pulmonary artery smooth muscle cells (PASMC), A549, another ATII-like cell and Beas-2B (human bronchial epithelial cells) (Figure 3). All of these cells were also injected with plasmid containing the SV40 DTS to demonstrate that these cells are able to import DNA in a sequence-specific manner. Five hours post-injection, the cells were fixed and plasmid location was analyzed by visualization of the Cy3-PNA signal. As shown in Figure 3, plasmids carrying the T1α promoter failed to be imported in any of these cell types. This was also true for plasmids containing the promoterless plasmid (ΔDTS). By contrast, all showed nuclear localization of the ubiquitously active SV40 DTS. These data suggest that the T1α promoter nuclear import activity is likely restricted to type I cells.
Figure 3.
T1α promoter plasmids do not localize to the nucleus in non-ATI cells. A panel of different cell types were cytoplasmically microinjected with equal amounts of either T1α promoter plasmid, SV40 DTS plasmid or no DTS-containing plasmid. After 5 h, the location of the DNA was visualized by fluorescence microscopy.
Both general alveolar epithelial cell-specific nuclear import sequences as well as ATI-specific nuclear import sequences exist within the T1α promoter
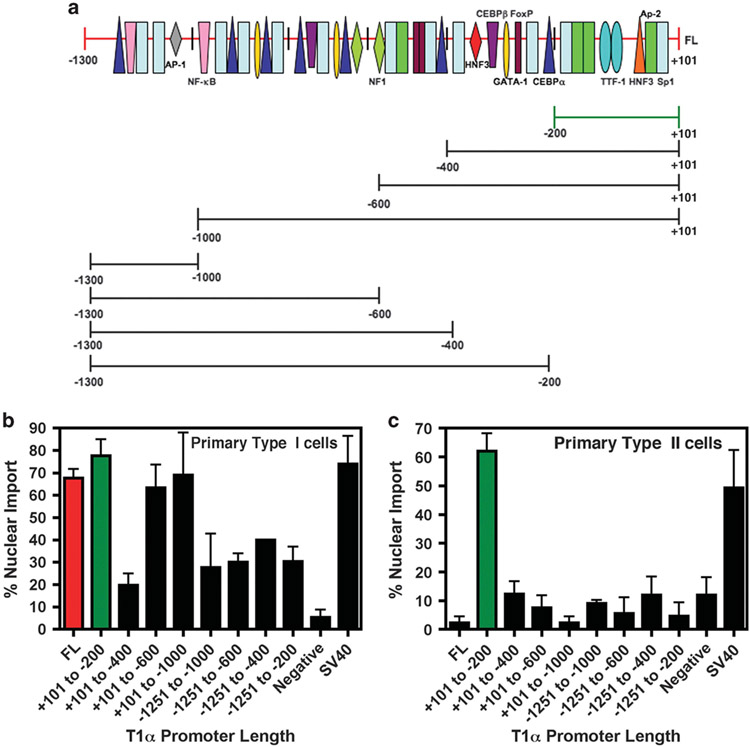
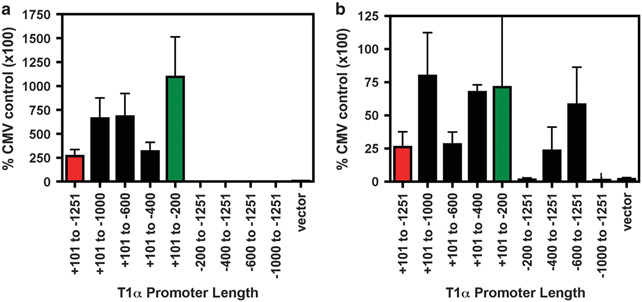
We next sought to analyze which sequences within the rat T1α promoter were required for ATI-specific nuclear import. Plasmids containing truncations of the full-length promoter were generated and tested for import activity following microinjection (Figure 4a). Both the 1.3 kb and 200 bp proximal T1α promoter show similar levels of nuclear import activity in both D5 ATII-like cells and the R3/1 cell line (data not shown and Figure 4b). This activity was not seen in the airway epithelial cell line, Beas-2B (data not shown). The other promoter fragments tested exhibited varying levels of nuclear import activity. However, the − 200 bp promoter fragment appears essential for maximal nuclear import activity since its removal destroyed all DNA nuclear import activity in type I cells. As seen in Figures 3 and 4c, the 1.3 kb promoter does not contain any nuclear import activity in primary ATII cells or the type II cell line, A549 (data not shown). Surprisingly, the 200-bp proximal T1α promoter did demonstrate nuclear import activity in both of these cell types (Figures 4b and c, green bar). This activity was then lost with promoter fragments containing more 5′ sequences of the promoter. These results suggest that sequences within 200-bp T1α promoter can mediate plasmid nuclear import in both ATI and ATII cells, whereas upstream sequences are required for ATI cell-specific nuclear import.
Figure 4.
The +101 to − 200 bp T1α promoter region is essential for maximal nuclear import activity. Plasmids containing either the wild-type 1.3. kb T1α promoter or various promoter truncations are shown (a) and were labeled with Cy3-PNA and cytoplasmically microinjected into either D5 primary ATI cells (b) or D3 primary ATII cells (c). Five hours post-microinjection, the cellular location of the plasmid was determined by fluorescence microscopy. The experiment was performed at least three times (mean % nuclear import±s.e.m.) with at least 50 cells counted.
Specific transcription-factor-binding sites within the 1.3 kb and 200 bp T1α promoter are important for T1α promoter-mediated nuclear import in ATI cells
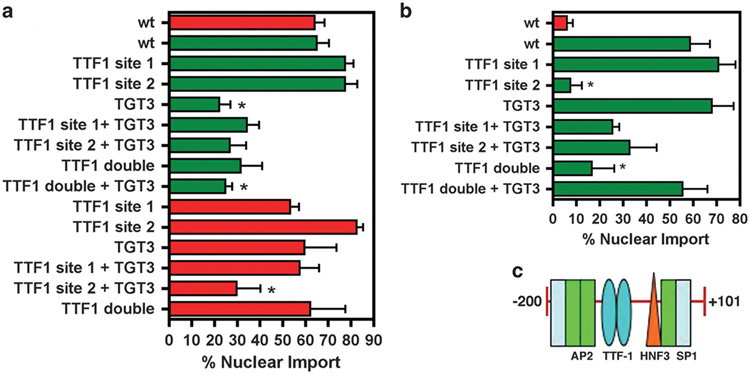
Previous studies have identified a set of transcription-factor-binding sites as important for T1α promoter transcriptional activity, including those for TGT3/HNF3 and thyroid transcription factor 1 (TTF-1).23 These binding sites reside at the − 106 to − 101 bp and − 128 to − 110 bp regions of the promoter, respectively23 (Figures 4a and 5c). In addition, a putative TTF-1-binding site exists in the − 139 to − 132 bp region23 (Figure 5c). Thus, in order to identify transcription factors potentially involved in ATI cell-specific DNA nuclear import, we mutated these binding sites within the 200 bp T1α promoter as well as the 1.3 kb promoter and subsequently microinjected these mutated plasmids into D5 cells. As shown in Figure 5a, we found that mutation of the TGT3/HNF3-binding site in the 200 bp promoter reduced nuclear import activity to < 30% in type I cells. Mutations in either TTF-1-binding site retain DNA nuclear import at normal levels in type I cells, suggesting that these sites do not function alone. However, a TTF-1 double mutation reduced DNA nuclear import to < 30% of the wild-type 200 bp fragment. In comparison, mutation of the TGT3/HNF3 site alone has no effect on import of the 1.3 kb promoter, but mutation of both the TGT3/HNF3 site and the second TTF-1 site reduced nuclear import activity to < 30% in the context of the full-length promoter. Overall, these results point to the importance of the TGT3/HNF3-binding site for promotion of ATI cell DNA nuclear import, with TTF-1 playing an interactive role with this site as well as itself.
Figure 5.
Transcription-factor-binding site requirements for T1α promoter nuclear import in ATI and ATII cells. (a) Plasmids containing the wild-type 1.3 kb T1α promoter, the +101 to − 200 bp truncated promoter and promoters containing the indicated binding site mutations were cytoplasmically microinjected into D5 primary ATI cells. The localization of the plasmid was visualized by fluorescence microscopy after 5 h. The experiment was performed at least three times (mean % nuclear import±s.e.m.) with at least 50 cells counted for each condition. (b) Plasmids containing the wild-type +101 to − 200 bp truncated promoter and truncated promoters containing the indicated binding site mutations were cytoplasmically microinjected into D2 primary ATII cells. The localization of the plasmid was visualized by fluorescence microscopy after 5 h. The experiment was performed at least three times (mean % nuclear import±s.e.m.) with at least 50 cells counted for each condition. (c) The +101 to − 200 bp sequence of the T1α promoter containing binding sites for TTF-1 and HNF3. *P < 0.05 compared with full-length wild type promoter in the respective cell type.
Specific transcription-factor-binding sites within 200 bp T1α promoter are important for T1α promoter-mediated nuclear import in ATII cells
We next sought to determine if similar binding sites were involved in the nuclear import activity of the − 200 bp proximal promoter in ATII cells. Upon microinjection into D2 ATII cells, the TGT3/HNF3 site mutation has no impact on nuclear import activity of the 200 bp promoter fragment (Figure 5b). TTF-1, in contrast, appeared to play a significant role as mutation of binding site 2 reduced activity to < 10%. A similar effect was seen when both site 1 and site 2 were mutated. These data are in contrast to that in ATI cells where the TGT3/HNF3-binding site alone is important for T1α promoter-mediated DNA nuclear import, in addition to an interaction between this site and that of TTF-1. In comparison, in ATII cells, the TTF-1 sites alone appear to be involved in plasmid DNA nuclear import in the context of the 200 bp proximal promoter.
Interaction of the first 200 bp of the T1α promoter with upstream promoter sequences is required for transcriptional activity
Given the fact that most studies of the transcriptional activity of the T1α promoter have been performed in ATII cell types,23,24 we sought to analyze the transcriptional ability of various regions of the promoter in relation to its DNA nuclear import activity. Owing to the difficulty of transfection of primary alveolar epithelial cells, we used R3/1 cells as an ATI model. As shown in Figure 6, the 200 bp fragment of the promoter produces the majority of detectable luciferase activity. Transcriptional activity decreases, however, with the addition of more 5′ sequences. Constructs lacking the first 200 bp exhibit undetectable transcriptional activity. This highlights the importance of the interaction of the 200 bp region with upstream sequences, which we also observe in the differences between nuclear import activities of the 200 bp fragment vs the 1.3 kb fragment. Similar studies in the ATII cell line, A549, demonstrated no detectable T1α promoter-driven luciferase activity as compared with R3/1 cells when normalized to percent of a CMV-driven luciferase control (Figure 6).
Figure 6.
T1α promoter transcriptional activity in ATI vs ATII cells. R3/1 (a) and A549 (b) cells were transfected with plasmids carrying the indicated promoter truncations upstream of luciferase using Lipofectamine 2000. Forty-eight hours post-transfection, luciferase activity was measured and normalized to total protein levels and the activity of CMV-driven luciferase plasmid in order to account for large differences in transfection efficiency between the two cell types.
Both the 1.3 kb and 200 bp T1α promoter act as DTSs to drive ATI cell-specific gene delivery in mice
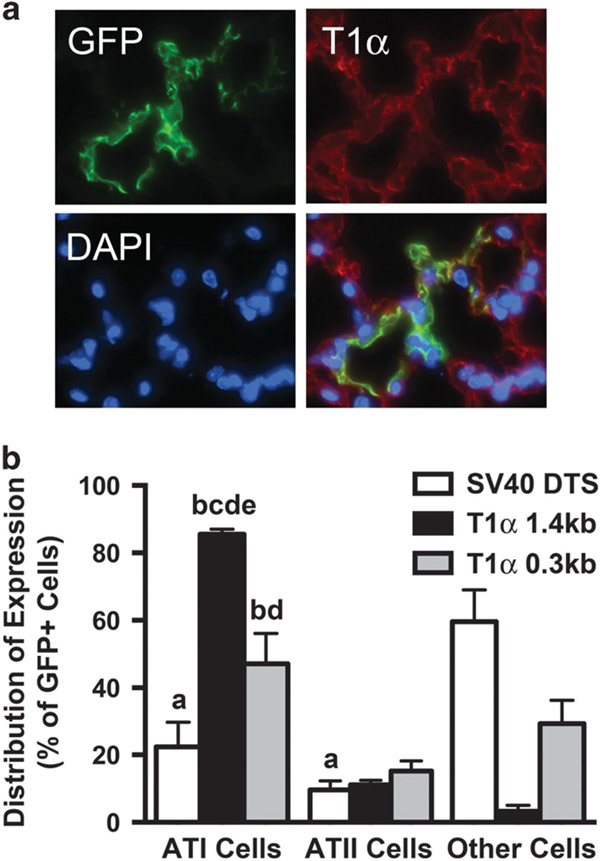
To test whether the T1α promoter acted as an ATI cell-specific DTS in vivo, we delivered CMV promoter-driven GFP expression plasmids that carried either the 1.3 kb T1α DTS, the 0.3 kb T1α DTS or the SV40 DTS downstream of the GFP gene to the lungs of mice using transthoracic electroporation. These plasmids express similar levels of GFP from the CMV promoter when delivered to dividing cells, but in the case of non-dividing cells, only those that enter the nucleus are able to express high levels of GFP. Gene delivery and expression were analyzed in the lungs of the mice 2 days later following perfusion and paraffin sectioning. Multiple sections from the distal, mid- and apical portions of the lungs from each animal were analyzed for GFP expression and the cellular distribution of expression was determined by co-staining with markers for either ATI (podoplanin) or ATII (SPC) cells (Figure 7).
Figure 7.
The T1α DTS promotes alveolar type I cell-specific gene delivery and expression in the lungs of mice. Plasmids expressing GFP from the CMV promoter and also carrying the SV40 DTS or either the 1.3 kb or 0.3 kb T1α promoter downstream of the GFP gene were electroporated into murine lungs (n = 5). Forty-eight hours later, the lungs were perfused, paraffin-embedded, sectioned and immunofluorescence was performed using antibodies for GFP to detect transgene expression. The sections were also co-stained with antibodies against T1α and SPC to visualize alveolar type I and type II cells, respectively. Four sections from each the top third, middle third or bottom third of the lungs were stained for each animal and the percentages of type I, type II or other cells expressing GFP was determined (mean±s.e.m.). (a) Representative photos (GFP, green; T1a, red; nucleus, blue). (b) Quantitative analysis of the in vivo data is shown. (a) P < 0.001 vs SV40 other cell types; (b) P < 0.0001 SV40 DTS ATI cells; (c) P < 0.0001 vs T1α 0.3 kb ATI cells; (d) P < 0.001 vs ATII cells (all plasmids); and (e) P < 0.001 vs SV40 DTS other cell types.
Expression from the SV40 DTS-containing plasmid was detected in both ATI and ATII cells (22.4 ± 7.4 and 9.6 ± 2.7%, respectively) as well as in non-alveolar cells (59.6 ± 9.4%), at levels that are roughly equivalent to the relative cell distribution of the lung.25 By contrast, over 85.6 ± 1.5% of the GFP-expressing cells following delivery of the 1.3 kb T1α DTS plasmid were ATI cells, whereas 11.1 ± 1.3% were ATII cells. The shorter 0.3 kb T1α DTS plasmid also showed the greatest expression in ATI cells (47.1 ± 9.0%), although at levels less than that seen with the full-length T1α promoter. Further, in contrast to results in cultured cells following microinjection, this short fragment did not support equal targeting to ATII cells, but rather showed expression at levels similar to, or only slightly higher than, that seen for the SV40 or 1.3 kb T1α promoter. Taken together, these results suggest that the T1α promoter/DTS enhances gene delivery and expression preferentially in ATI cells.
DISCUSSION
The ability to restrict gene expression to specific cell types is a necessary requirement for the further development of safe gene therapy methods. This, however, is a challenge. We have previously shown that DNA delivery can be restricted to specific cell types through the use of cell-specific nuclear import sequences that are not actively dividing. In this scenario, while DNA may be delivered to the cytoplasm of multiple cell types, it is only able to enter the nucleus of the cell type of interest. In the present study, we demonstrate that the rat T1α promoter functions as a cell-specific DTS in ATI cells. In our attempt to identify the minimal promoter sequence needed for nuclear import activity, we revealed that while the +101 to − 1251 bp (1.3 kb) promoter mediates nuclear import in an ATI-specific manner, a smaller fragment, +101 to − 200 bp, promotes import in both ATI and ATII cells. Mutagenesis studies revealed that multiple transcription-factor-binding sites are involved in nuclear import, with a significant role being played by TTF-1 and TGT3/HNF3. Moreover, when tested in the lungs of living mice, these sequences drive greatly restricted gene delivery and expression in type I cells.
The three promoters that we tested for type I cell DNA nuclear import are preferentially expressed in type I pneumocytes and have been shown to contain many of the same transcriptional control elements in their sequences. Transcription factors identified both by the presence of their binding sites in the DNA and by gel shift or mutational analyses of the promoters include both general and cell-specific factors such as AP1, AP2, SP1, NF1, NF-kB, C/EBP, GATA-1, GATA-6, HNF3 and TTF-1 among others. Surprisingly, however, despite the fact that each of these promoters contains multiple binding sites for many of these same factors, only the T1α promoter was able to support plasmid nuclear import, while the Aquaporin-5 and caveolin-1 promoters did not. This cannot be due to the presence or absence of a specific factor-binding site, since both the AQP-5 and T1α promoters bind to the same cohort of transcription factors, yet both have differing import activities. Indeed, the aquaporin-5 promoter actually contains more consensus binding sites for most of the same transcription factors than does the T1α promoter, hence it is unlikely to be a simple matter of abundance of bound transcription factors mediating DNA nuclear import. It is also unlikely that binding of certain cell-specific transcription factors, most notably TTF-1 or GATA proteins, causes differential nuclear import since AQP-5 binds TTF-1 and GATA-6 as does the SP-C promoter, yet the SPC promoter allows DNA nuclear import in type II cells while the AQP-5 promoter does not. Thus, it is possible that the organization of transcription-factor-binding sites on the promoter is an important factor in mediating DNA nuclear import, since although the tested promoters contain most of the same transcription-factor-binding sites, they are organized quite differently on the promoters, leading to the possibility of the formation of very different three-dimensional complexes on the DNA and it could be these complexes that control ability of the DNA to support nuclear import.
Our finding that the full-length T1α promoter mediates DNA nuclear import in an ATI-specific manner whereas the small fragment results in both ATI and ATII cell nuclear import reveals that there exists key transcriptional machinery differences between these two cell types. Since DNA nuclear import relies on the presence of transcription factors binding to promoter sequences, it is possible that the full-length T1α promoter sequence contains potential inhibitory sequences or binding sites that restrict import to ATI cells only. Also, the transcription factors themselves can interact with one another to form either active or inhibitory complexes that may affect nuclear import. For example, it has been demonstrated that TTF-1 is able to physically interact with HNF3α in a DNA-independent manner to prevent its binding to DNA.26 This may provide a method of control over transcription as well as DNA nuclear import. In terms of the specific proteins involved in nuclear import, the fact that we have found that both TTF-1 and TGT3/HNF3 sites are involved in ATI-specific nuclear import is not entirely surprising as mutation of both of these binding sites has been shown to result in a synergistic decrease in the transcriptional activity of the T1α promoter.23 Thus, these proteins are clearly present and playing a functional role within ATI cells. Interestingly, it has been shown that mutation of the TGT3/HNF3 site alone has a larger impact on promoter activity as compared with mutation of TTF-1 alone.23 This may suggest a greater role for this site in the T1α promoter, which is consistent with our observations that mutation of the TGT3 site alone significantly reduced nuclear import activity whereas mutation of TTF-1 did not.
It was rather surprising to find that the +101 to − 200 bp promoter fragment, which contains the binding sites for these proteins, supported nuclear import in ATII cells as well as ATI cells. It is established that ATII cells express TTF-1 and HNF3 proteins, and not only are these transcription factors important for T1α promoter activity, as previously mentioned, but also for the activity of ATII-specific genes SP-A, SP-B, SP-C and SP-D.27-30 Thus, given that both ATI and ATII cells express and utilize both TTF-1 and HNF3 proteins, the differences we observed in specificity of the 1.3 kb promoter vs the 200 bp promoter fragment may be due to differences in the relative ratios of TTF-1 and HNF3 in ATI and ATII cells. Our laboratory has shown that the levels of mRNA for TTF-1, HNF3α and HNF3β change as primary ATII cells transition to ATI cells in vitro (data not shown), and previous studies in human alveolar epithelial cell differentiation have demonstrated decreases in TTF-1 protein during ATII to ATI transition and concomitant increases in HNF3α expression.31 Therefore, the combination of proteins available for binding may determine the import activity of the promoter sequence.
Another surprising finding was the while the +101 to − 200 bp promoter fragment supported high level plasmid nuclear import in ATI cells, as did the +101 to − 600 bp, +101 to − 1000 bp and +101 to − 1251 bp (full-length promoter), the +101 to − 400 bp fragment showed greatly reduced nuclear import in these cells. This could suggest that there are sequences in this region (−200 to − 400) that are inhibitory to forming complexes competent for DNA nuclear entry. Such a sequence could act by causing retention of the DNA–protein complex in the cytoplasm, perhaps bound to microtubules or other cytoskeletal elements. Alternatively, this sequence could bind one or more proteins that prevent formation of a productive DNA nuclear import complex on the promoter proximal +101 to − 200 bp region. Future studies aimed at determining the bound protein constituents to this fragment compared with the minimal and full-length promoter could elucidate the mechanism.
In order for the plasmid DNA to be imported into the nucleus of a cell, the proteins bound to the plasmid must be oriented in such a way that their nuclear localization signals (NLSs) are exposed and free to interact with the nuclear import machinery. In the case of TTF-1 and HNF3β, the NLSs overlap with the DNA-binding domains of these proteins.32,33 Therefore, cell-specific import may involve the presence or absence of certain adaptors that bind to these transcription factors with their own NLSs exposed. For example, TAZ and SMAD3 have both been shown to interact with TTF-1 or TTF-1 and HNF3, respectively, to regulate transcription factor function.34,35 Thus, it is likely that, despite similarities in the organization of transcription-factor-binding sites within ATI and ATII-specific promoters, there is a complex set of interactions which determine transcriptional as well as nuclear import specificity. Previous work from our laboratory highlights the likely importance of this in that nuclear import of plasmid DNA carrying the SP-C promoter requires TTF-1, but TTF-1 alone is not the sole contributor for import as surfactant protein A and surfactant protein B promoters, which also contain binding sites for TTF-1, do not support nuclear import.9 Thus, specific protein–protein interactions are likely highly involved in cell-specific nuclear import, such as those with GATA-6 and NFI proteins in the context of SP-C promoter-driven nuclear import.9 Interestingly, in contrast to the T1α promoter, the SP-C promoter does not contain a binding site for HNF3α or HNF3β. Given that TTF-1 and HNF3 function and interact together in both positive and negative manners, this highlights a potential key difference between nuclear import in ATI vs ATII cells.26
While the ability to direct cell-specific nuclear import of plasmids in cultured type I and type II cells is interesting, the real utility of the approach is for use in the living lung. When plasmids carrying the SV40 DTS were electroporated into the lungs of mice, GFP expression from the ubiquitously active CMV promoter was relatively evenly distributed throughout cell types in relative accordance with the cells' abundance in the lung. Indeed, ATI and II cells make up approximately 8 and 15%, respectively, of the rat and human lung parenchyma,25,36 accounting for the < 30% of expression seen in these cells using the SV40 DTS-containing construct. That there are slightly more ATI-expressing cells than would be predicted from their abundance may be due to the fact that these cells make up 95% of the surface area of the alveoli. By contrast to the SV40 sequence, almost 90% of gene transfer and expression of plasmids carrying the 1.3 kb T1α promoter as a DTS was restricted to alveolar type I cells. Similarly, almost 50% of the gene transfer and expression seen with the shorter 0.3 kb T1α sequence was in type I cells as predicted from the microinjection experiments. However, there was very little if any specific targeting to type II cells in vivo, contrary to that seen in cultured cells. This may reflect differences between transcription factor and other gene expression in the cultured cell models and endogenous alveolar epithelial cells in the living lung or that the cultured type II cells are less stringent in their transcriptional machinery expression. These results demonstrate that the T1α promoter clearly functions as an ATI cell-specific DTS and can be used to drive type I cell-specific gene delivery in vivo.
MATERIALS AND METHODS
Plasmids
Plasmid containing the rat T1α promoter was a generous gift from Dr Maria Ramirez.37 The promoters for rat AQP-5 (−1702 to +123) and mouse Caveolin-1 (−828 to +50) were amplified by PCR from rat and mouse genomic DNA, respectively. pCMV-DTS and pSP-C have been described previously.9 Amplified sequences were cloned into the pCRII-TOPO plasmid (Invitrogen, San Diego, CA, USA). Plasmids used for microinjection were generated by amplification of the promoter regions out of originating plasmids using primers containing attB recombination sequences. In parallel, we designed an ‘acceptor’ vector containing the ccdB gene sequence flanked by attP sites and containing a GFP expression cassette driven by the CMV promoter as well as a PNA-binding site for fluorescent labeling of individual plasmids.38 A recombination event between the attB and attP sites within the PCR product and the vector was achieved using the BP clonase enzyme and resulted in the insertion of the promoter sequence between the attP sites. All plasmids were verified by DNA sequencing and then labeled with CY3-PNA for visualization by fluorescence.38 T1a promoter truncations were made by using attB primers specific for internal regions of the promoter followed by recombination into our acceptor vector. Plasmids containing site mutations within the T1α promoter were generated using the PCR-based Quickchange Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA).
Cell culture and microinjection
The A549 (CCL-185) and Beas-2B (CRL-9609) cell lines were obtained from the American Type Culture Collection (Washington, DC, USA). The R3/1 and RLE6TN cell lines were a gift from Dr Jacob Finkelstein (University of Rochester, Rochester, NY, USA).21,39 The Pulmonary Artery Smooth Muscle Cells were obtained from Dr Paul Babal (University of South Alabama, Mobile, AL USA). Primary rat alveolar epithelial cells were isolated as previously described.20 ATII cells were collected and cultured on glass coverslips either with or without pre-treatment with 50 μg ml−1 Matrigel (BD Biosciences, San Jose, CA, USA). All cells were maintained in Dulbecco’s modification of Eagle’s medium containing 10% fetal bovine serum and supplemented with antibiotics and antimycotics (Gibco, Carlsbad, CA, USA), except for R3/1 cells, which were grown in RPMI 1640 containing 10% fetal bovine serum and supplemented with antibiotics and antimycotics (Gibco). All cells were cytoplasmically microinjected using an Eppendorf Femtojet system (Hamburg, Germany) as previously described.40 Purified protein-free DNA was suspended in phosphate-buffered saline and injected at a concentration of 0.5 mg ml−1, which corresponds to roughly 20 000 plasmids per injected cell.
Immunofluorescence
Cells were cultured on coverslips and fixed with 4% formaldehyde for 10 min, followed by thorough washing in phosphate-buffered saline. For intracellular staining, the cell membranes were permeabilized for 10 min with 0.1% Triton X-100 in phosphate-buffered saline. The cells were treated with 10% fetal bovine serum for 60 min (to eliminate nonspecific binding of the antibodies) followed by overnight incubation with the following primary antibodies: anti-T1α (Developmental Studies Hybridoma Bank), anti-Lamellar Body 180 (Covance, Princeton, NJ, USA), anti-Aquaporin-5 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Vimentin (BD Pharmingen, San Jose, CA, USA) and anti-Surfactant Protein B (Chemicon, Billerica, MA, USA). The cells were next washed thoroughly in phosphate-buffered saline and incubated with the appropriate (Molecular Probes, Carlsbad, CA, USA) secondary antibodies for 60 min. After washing in phosphate-buffered saline, coverslips were mounted and immunofluorescence was observed by fluorescence microscopy.
Quantitative real-time PCR
Total RNA was isolated from cells using a Qiagen RNeasy plus mini kit (Qiagen, Valencia, CA, USA). cDNA was then synthesized from 1 μg of total RNA using the Promega Reverse Transcription System. Real-time PCR was performed on the samples using IQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA).
Luciferase assay
R3/1 and A549 cells were transfected with luciferase reporter plasmids using Lipofectamine 2000 according to the manufacturers instructions (Promega, Madison, WI, USA). Forty-eight hours post-transfection, the cells were lysed in 1 × Reporter Lysis Buffer (Promega), and luciferase activity was measured on a TD 20/20 Luminometer (Promega).
In vivo gene transfer and expression
Male C57BL/6 mice (9–11 weeks) were anesthetized with isoflurane and 100 μg of pCMV-GFP-T1α (1.4 kb), pCMV-GFP-T1α (0.32 kb) or pCMV-GFP-SV40 were delivered in 50 μl of 10 mm Tris-HCl (pH 8.0), 1 mm EDTA and 140 mm NaCl, to mouse lungs by aspiration. Eight, 10 ms square wave pulses at a field strength of 200 V cm−1 were immediately applied using cutaneous electrophysiology electrodes (Medtronic, Redmond, WA, USA) placed on the mouse chest with an ECM830 electroporator (BTX, Harvard Apparatus, Holliston, MA, USA). Two days later, lungs were perfused and inflated with 10% (vol/vol) buffered formalin immediately after mice were killed, and used for paraffin-embedded sections. Sections (5 μm) were rehydrated and immunostained with FITC conjugated anti-GFP antibody (Abcam, Cambridge, MA, USA) and hamster anti-T1α antibody as a marker for epithelial type I cells (Developmental Studies Hybridoma Bank at the University of Iowa, Iowa city, IA, USA) or rabbit anti-SPC antibody as a marker for epithelial type II cells (Santa Cruz, Santa Cruz, CA, USA). The immune complexes were detected using Alexa 546 anti-hamster or anti-rabbit secondary antibodies (Invitrogen, Grand Island, NY, USA) before sections were counterstained with 4',6-Diamidino-2-phenylindole, Dihydrochloride. Stained sections were visualized using a Leica DM RXA2 microscope (Leica, Wetzlar, Germany). All experimental procedures were performed according to institutional guidelines for the care and use of laboratory animals in an American Association for the Accreditation of Laboratory Animal Care-approved facility.
ACKNOWLEDGEMENTS
This work was supported by a postdoctoral fellowship from the American Heart Association (to LG) and by the National Institutes of Health grants HL81148, HL120521 and GM94228 (to DD), and T32 HL66988 (to XL). We thank Jacob Finkelstein (University of Rochester, Rochester, NY, USA) for R3/1 cells.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Degiulio JV, Kaufman CD, Dean DA. The SP-C promoter facilitates alveolar type II epithelial cell-specific plasmid nuclear import and gene expression. Gene Therapy 2010; 17: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp Cell Res 1997; 230: 293–302. [DOI] [PubMed] [Google Scholar]
- 3.Miller AM, Dean DA. Cell-specific nuclear import of plasmid DNA in smooth muscle requires tissue-specific transcription factors and DNA sequences. Gene Therapy 2008; 15: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear import. Exp Cell Res 1999; 253: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vacik J, Dean BS, Zimmer WE, Dean DA. Cell-specific nuclear import of plasmid DNA. Gene Therapy 1999; 6: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean DA, Byrd JN Jr, Dean BS. Nuclear targeting of plasmid DNA in human corneal cells. Curr Eye Res 1999; 19: 66–75. [DOI] [PubMed] [Google Scholar]
- 7.Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A et al. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J Virol 1998; 72: 6181–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mesika A, Grigoreva I, Zohar M, Reich Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol Ther 2001; 3: 653–657. [DOI] [PubMed] [Google Scholar]
- 9.DeGiulio J, Kaufman CD, Dean DA. The SP-C promoter facilitates alveolar type II epithelial cell-specific plasmid nuclear import and gene expression. Gene Therapy 2009; 17: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borok Z, Liebler JM, Lubman RL, Foster MJ, Zhou B, Li X et al. Na transport proteins are expressed by rat alveolar epithelial type I cells. Am J Physiol Lung Cell Mol Physiol 2002; 282: L599–L608. [DOI] [PubMed] [Google Scholar]
- 11.Dobbs LG, Gonzalez R, Matthay MA, Carter EP, Allen L, Verkman AS. Highly water-permeable type I alveolar epithelial cells confer high water permeability between the airspace and vasculature in rat lung. Proc Natl Acad Sci USA 1998; 95: 2991–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson MD, Bao HF, Helms MN, Chen XJ, Tigue Z, Jain L et al. Functional ion channels in pulmonary alveolar type I cells support a role for type I cells in lung ion transport. Proc Natl Acad Sci USA 2006; 103: 4964–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson MD, Widdicombe JH, Allen L, Barbry P, Dobbs LG. Alveolar epithelial type I cells contain transport proteins and transport sodium, supporting an active role for type I cells in regulation of lung liquid homeostasis. Proc Natl Acad Sci USA 2002; 99: 1966–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanderbilt JN, Allen L, Gonzalez RF, Tigue Z, Edmondson J, Ansaldi D et al. Directed expression of transgenes to alveolar type I cells in the mouse. Am J Respir Cell Mol Biol 2008; 39: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlin K, Mager EM, Allen L, Tigue Z, Goodglick L, Wadehra M et al. Identification of genes differentially expressed in rat alveolar type I cells. Am J Respir Cell Mol Biol 2004; 31: 309–316. [DOI] [PubMed] [Google Scholar]
- 16.Borok Z, Danto SI, Lubman RL, Cao Y, Williams MC, Crandall ED. Modulation of t1alpha expression with alveolar epithelial cell phenotype in vitro. Am J Physiol 1998; 275: L155–L164. [DOI] [PubMed] [Google Scholar]
- 17.Borok Z, Lubman RL, Danto SI, Zhang XL, Zabski SM, King LS et al. Keratinocyte growth factor modulates alveolar epithelial cell phenotype in vitro: expression of aquaporin 5. Am J Respir Cell Mol Biol 1998; 18: 554–561. [DOI] [PubMed] [Google Scholar]
- 18.Campbell L, Hollins AJ, Al-Eid A, Newman GR, von Ruhland C, Gumbleton M. Caveolin-1 expression and caveolae biogenesis during cell transdifferentiation in lung alveolar epithelial primary cultures. Biochem Biophys Res Commun 1999; 262: 744–751. [DOI] [PubMed] [Google Scholar]
- 19.Dobbs LG, Williams MC, Brandt AE. Changes in biochemical characteristics and pattern of lectin binding of alveolar type II cells with time in culture. Biochim Biophys Acta 1985; 846: 155–166. [DOI] [PubMed] [Google Scholar]
- 20.Dobbs LG, Williams MC, Gonzalez R. Monoclonal antibodies specific to apical surfaces of rat alveolar type I cells bind to surfaces of cultured, but not freshly isolated, type II cells. Biochim Biophys Acta 1988; 970: 146–156. [DOI] [PubMed] [Google Scholar]
- 21.Koslowski R, Barth K, Augstein A, Tschernig T, Bargsten G, Aufderheide M et al. A new rat type I-like alveolar epithelial cell line R3/1: bleomycin effects on caveolin expression. Histochem Cell Biol 2004; 121: 509–519. [DOI] [PubMed] [Google Scholar]
- 22.Williams MC. Alveolar type I cells: molecular phenotype and development. Annu Rev Physiol 2003; 65: 669–695. [DOI] [PubMed] [Google Scholar]
- 23.Ramirez MI, Rishi AK, Cao YX, Williams MC. TGT3, thyroid transcription factor I, and Sp1 elements regulate transcriptional activity of the 1.3-kilobase pair promoter of T1alpha, a lung alveolar type I cell gene. J Biol Chem 1997; 272: 26285–26294. [DOI] [PubMed] [Google Scholar]
- 24.Vanderbilt JN, Dobbs LG. Characterization of the gene and promoter for RTI40, a differentiation marker of type I alveolar epithelial cells. Am J Respir Cell Mol Biol 1998; 19: 662–671. [DOI] [PubMed] [Google Scholar]
- 25.Hyde DM, Magliano DJ, Plopper CG. Morphometric assessment of pulmonary toxicity in the rodent lung. Toxicol Pathol 1991; 19: 428–446. [DOI] [PubMed] [Google Scholar]
- 26.Minoo P, Hu L, Xing Y, Zhu NL, Chen H, Li M et al. Physical and functional interactions between homeodomain NKX2.1 and winged helix/forkhead FOXA1 in lung epithelial cells. Mol Cell Biol 2007; 27: 2155–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi M, Tong GX, Murry B, Mendelson CR. Role of CBP/p300 and SRC-1 in transcriptional regulation of the pulmonary surfactant protein-A (SP-A) gene by thyroid transcription factor-1 (TTF-1). J Biol Chem 2002; 277: 2997–3005. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Crouch EC, Rust K, Spaite E, Brody SL. Proximal promoter of the surfactant protein D gene: regulatory roles of AP-1, forkhead box, and GT box binding proteins. J Biol Chem 2000; 275: 31051–31060. [DOI] [PubMed] [Google Scholar]
- 29.Kelly SE, Bachurski CJ, Burhans MS, Glasser SW. Transcription of the lung-specific surfactant protein C gene is mediated by thyroid transcription factor 1. J Biol Chem 1996; 271: 6881–6888. [DOI] [PubMed] [Google Scholar]
- 30.Margana RK, Boggaram V. Functional analysis of surfactant protein B (SP-B) promoter. Sp1, Sp3, TTF-1, and HNF-3alpha transcription factors are necessary for lung cell-specific activation of SP-B gene transcription. J Biol Chem 1997; 272: 3083–3090. [DOI] [PubMed] [Google Scholar]
- 31.Apparao KB, Newman DR, Zhang H, Khosla J, Randell SH, Sannes PL. Temporal changes in expression of FoxA1 and Wnt7A in isolated adult human alveolar epithelial cells enhanced by heparin. Anat Rec (Hoboken) 2010; 293: 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian X, Costa RH. Analysis of hepatocyte nuclear factor-3 beta protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res 1995; 23: 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christophe-Hobertus C, Duquesne V, Pichon B, Roger PP, Christophe D. Critical residues of the homeodomain involved in contacting DNA bases also specify the nuclear accumulation of thyroid transcription factor-1. Eur J Biochem 1999; 265: 491–497. [DOI] [PubMed] [Google Scholar]
- 34.Park KS, Whitsett JA, Di Palma T, Hong JH, Yaffe MB, Zannini M. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J Biol Chem 2004; 279: 17384–17390. [DOI] [PubMed] [Google Scholar]
- 35.Li C, Zhu NL, Tan RC, Ballard PL, Derynck R, Minoo P. Transforming growth factor-beta inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors. J Biol Chem 2002; 277: 38399–38408. [DOI] [PubMed] [Google Scholar]
- 36.Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell number and cell characteristics of the normal human lung. Am Rev Respir Dis 1982; 126: 332–337. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez MI, Cao YX, Williams MC. 1.3 kilobases of the lung type I cell T1alpha gene promoter mimics endogenous gene expression patterns during development but lacks sequences to enhance expression in perinatal and adult lung. Dev Dyn 1999; 215: 319–331. [DOI] [PubMed] [Google Scholar]
- 38.Hillery E, Munkonge FM, Xenariou S, Dean DA, Alton EW. Nondisruptive, sequence-specific coupling of fluorochromes to plasmid DNA. Anal Biochem 2006; 352: 169–175. [DOI] [PubMed] [Google Scholar]
- 39.Driscoll KE, Carter JM, Iype PT, Kumari HL, Crosby LL, Aardema MJ et al. Establishment of immortalized alveolar type II epithelial cell lines from adult rats. In Vitro Cell Dev Biol Anim 1995; 31: 516–527. [DOI] [PubMed] [Google Scholar]
- 40.Gasiorowski JZ, Dean DA. Postmitotic nuclear retention of episomal plasmids is altered by DNA labeling and detection methods. Mol Ther 2005; 12: 460–467. [DOI] [PMC free article] [PubMed] [Google Scholar]