Abstract
Unculturable polycyclic aromatic hydrocarbon (PAH)-degrading bacteria are a significant reservoir of the microbial potential to catabolize low-molecular-weight PAHs. The population of these bacteria is larger than the population of nah-like bacteria that are the dominant organisms in culture-based studies. We used the recently described phn genes of Burkholderia sp. strain RP007, which feature only rarely in culture-based studies, as an alternative genotype for naphthalene and phenanthrene degradation and compared this genotype with the genotypically distinct but ubiquitous nah-like class in different soils. Competitive PCR quantification of phnAc and nahAc, which encode the iron sulfur protein large (α) subunits of PAH dioxygenases in nah-like and phn catabolic operons, revealed that the phn genotype can have a greater ecological significance than the nah-like genotype.
It could mistakenly be inferred from available nucleotide sequence data that highly conserved nah-like gene clusters, which are isolated from polycyclic aromatic hydrocarbon (PAH)-degrading pseudomonads obtained from diverse geographic areas, are the dominant gene clusters involved in degradation of the low-molecular-weight PAHs naphthalene and phenanthrene. The results of probing and PCR amplification of nah sequences from contaminated soils and sediments also indicate that these sequences are ubiquitous (7, 17). Although PAH degraders with nah genotypes are easily isolated, it has been acknowledged that the nah catabolic cluster and closely related homologues may be present in only a small part of the PAH-degrading bacterial population (1, 4, 12, 20). Recently, the phn genes of Burkholderia sp. strain RP007 provided evidence that there is a different genotype that exhibits a low level of sequence homology with nah and has a different gene order yet encodes enzymes for an identical PAH degradation pathway (9, 10).
As increasingly diverse genes that encode enzymes for PAH catabolism are characterized (4–6, 14, 20), it is important not only to understand the function of these genes but also to determine their ecological significance in the context of environmental pollution. The objective of this study was, therefore, to compare the prevalence in aromatic hydrocarbon-contaminated soils of two distinct PAH catabolic genotypes. These genotypes were the divergent phn genes (9, 10), which are difficult to isolate by conventional microbiological methods (12), and the easily isolated nah-like genes (1, 7, 17). It has been shown that culture-based methods overemphasize nah-like genes and fail to detect bacteria with phn genotypes in contaminated soils in which both nahAc and phnAc are detected by PCR amplification and DNA hybridization (12). A molecular biological approach was required to overcome the disparity in the ease of culturing of host bacteria harboring these two genotypes. Highly specific primer combinations, which targeted genes that encode the iron sulfur protein large (α) subunits of the nah-like and phn PAH initial dioxygenases, allowed us to determine the number of copies of phnAc and nahAc present in soil by a quantitative competitive PCR (QC-PCR) technique (3, 11, 19). Although this approach did not allow us to determine relative activity or fluxes through nah-like and phn pathways, it did reveal the relative enrichment of populations after they were exposed to PAHs and allowed us to demonstrate that the phn genotype was enriched in response to the selective pressures exerted by specific PAHs in a range of soils.
MATERIALS AND METHODS
Soil samples and analysis.
Two soils from the Waikato region of New Zealand (38°S, 175°E) containing different levels of PAHs were selected for analysis. One soil (soil A) was obtained from the site of a former town gas-generating plant and was severely contaminated with PAHs; the other soil (soil B) was contaminated with petroleum fuels. The PAH content of each sample was determined commercially by using U.S. Environmental Protection Agency methods 3545, 3540, and 3630. The number of culturable heterotrophs present in each sample was determined on R2A plates (Difco Laboratories, Detroit, Mich.) by spreading dilutions of a soil suspension that was prepared by shaking 10 g of soil in 90 ml of 0.1% pyrophosphate along with 30 g of glass beads for 1 h at 25°C. The R2A plates were incubated at 25°C for 48 h. Noncontaminated (pristine) soils from central Siberia (61°N, 89°E), Ross Island in the Antarctic (77°S, 166°E), and a native New Zealand forest (38°S, 175°E) were used to assess the ubiquity of the phn genes in different environments.
Development of QC-PCR protocol.
The PCR primers used for specific probing of the nahAc and phnAc sequences were designed during a previous study (12). Primer nahAcfor (5′-TGGCGATGAAGAACTTTTCC) and primer nahAcrev (5′-AACGTACGCTGAACCGAGTC) amplify a 992-bp region encompassing nucleotides 63 to 1055 of Pseudomonas putida G7 nahAc (GenBank accession no. M83949). Primer P8073 (5′-TTCGAGCTGGAATGTGAGC) and primer P9047 (5′-AATAACCGGCGATTCCAAAC) amplify a 993-bp region encompassing nucleotides 82 to 1075 of Burkholderia sp. strain RP007 phnAc (GenBank accession no. AF061751). Since in this study we relied on accurate relative quantification of specific DNA templates, it was important that only homologous sequences were amplified. Therefore, an annealing temperature of 65°C was used; this temperature was determined empirically to be near the maximum optimum annealing temperature for the two primer sets. PCR amplification was carried out in 50-μl reaction mixtures that contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.25 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, 2.5 U of PLATINUM Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.), 0.2 μM forward primer, 0.2 μM reverse primer, and 0.1 μg of template DNA. The following PCR cycling conditions (Techne Cyclogene thermal cycler) were used: 94°C for 5 min; 35 cycles consisting of 94°C for 45 s, 65°C for 30 s, and 72°C for 60 s; 72°C for 10 min; and finally, cooling to 4°C. Maximum ramp rates were used throughout.
Each QC-PCR titration was performed by using six dilutions of standard DNA (prepared as described below), which resulted in a series of reaction mixtures that contained 10, 1, 0.1, 0.01, 0.001, and 0.0001 amol of pNΔSTD and pPΔSTD (equivalent to 6 × 106 to 60 copies of standard DNA template per reaction mixture). To each reaction mixture we added a known volume of target DNA that was extracted from 0.5 g of soil by the method of Berthelet et al. (2). Each QC-PCR preparation contained 25 to 125 ng of DNA based on spectrophotometric determination of absorbance at 260 nm. The DNA extract volume, which included the aqueous phase added plus the soil moisture, was recorded in order to obtain an accurate dilution factor for subsequent calculation of the gene copy number per gram (dry weight) in each soil. Standard and target amplicons generated during the QC-PCR experiment were quantified by electrophoresing 10 μl of each reaction mixture through a 1% agarose gel in Tris-borate-EDTA buffer. After electrophoresis, the DNA was stained for 3 h with SYBR Gold (FMC Bioproducts, Rockland, Maine), and the gel was photographed. The intensities of the target and standard amplicons were then determined by processing images of scanned photographs with Scion Image software (Scion Corp., Frederick, Md.). A regression analysis, based on the amplicon intensities and sizes of target and standard DNAs and the initial concentration of standard DNA, was used to calculate the concentration of nahAc or phnAc genes present in each soil DNA extract; this concentration was subsequently adjusted to obtain a gene copy value on a soil dry weight basis. QC-PCR titrations for the contaminated soil samples were replicated with aliquots of each soil DNA extract; mean values and standard errors are reported.
Construction of standard templates.
Deletion derivatives of the G7 nahAc and RP007 phnAc genes were constructed and used as standards for QC-PCR. Klenow fragment-treated nahAc and phnAc amplicons were ligated into the SmaI site of pUC18 to form pNTGT and pPTGT, and restriction fragments situated between the primer binding sites were then removed before religation of constructs following S1 nuclease treatment. Thus, a 351-bp HpaI-NsiI deletion of the G7 nahAc amplicon was used to generate pNΔSTD, and a 324-bp NsiI-ApaI deletion of the RP007 phnAc amplicon was used to generate pPΔSTD. The construct pNΔSTD yielded a 637-bp PCR product with primers nahAcfor and nahAcrev, and pPΔSTD yielded a 665-bp PCR product with primers P8073 and P9047. Each primer set exhibited identical amplification kinetics with the specific target and standard DNAs; therefore, an equimolar ratio of target and standard DNAs was used to calculate the QC-PCR titration. This ratio was determined by comparing the concentrations of the two PCR products in aliquots taken from PCR over a number of cycles (data not shown). Accurate standard solutions of pPΔSTD and pNΔSTD were prepared, and equimolar amounts based on a molecular mass of 660 Da/bp (15) were combined and diluted to obtain a stock preparation which contained 20 amol of both standard templates per μl. Tenfold dilutions of this mixture were used as standards in the QC-PCR experiments. Mixing the phn and nah templates ensured consistent dilution and did not interfere with subsequent PCR amplification.
Establishment of microcosms.
Enrichment for nahAc and phnAc was evaluated in microcosms containing pristine soil amended with different PAHs. The soil was collected from a native New Zealand forest. Ten-gram samples of soil and 1-ml portions of basal salts [4 g of Na2HPO4 per liter, 2 g of KH2PO4 per liter, 1 g of (NH4)2SO4 per liter] were placed in 100-ml glass flasks. Preparations were subjected to the following four treatments before they were incubated in the dark at 20°C for 60 days: no additional carbon source (blank); naphthalene supplied as a vapor from a vapor tube suspended above the soil; phenanthrene supplied as a vapor (as routinely used in our laboratory [9, 10, 12]); and 50 mg of fluoranthene added directly to the soil. Soil samples obtained from Siberia and Antarctica were also enriched in microcosms by using naphthalene and phenanthrene as supplementary carbon sources.
Sequencing of phnAc amplicons.
Amplicons of phnAc derived from PAH- and petroleum-contaminated soils were cloned into the SmaI site of pUC18 by using a SureClone ligation kit (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). The nucleotide sequences of selected amplification products were determined in both orientations by using the universal primers and a PRISM Ready Reaction DNA terminator cycle sequencing kit (Perkin-Elmer, Inc., Wellesley, Mass.). The Reaction mixtures were resolved by using an ABI model 377 sequencer at the Waikato DNA Sequencing Facility, and the sequences were analyzed by using Omiga 1.0 sequence analysis software (Oxford Molecular Group Ltd., Oxford, United Kingdom).
RESULTS AND DISCUSSION
QC-PCR analysis of phnAc and nahAc from contaminated soils.
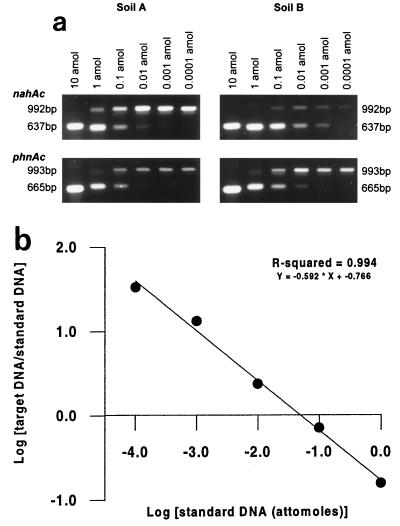
The two contaminated New Zealand soils used in this study were selected to represent high and low levels of PAH contamination and are described in Table 1. Figure 1 shows representative photographs of QC-PCR titration of phnAc and nahAc for the two soils (Fig. 1a), as well as an example of a QC-PCR calibration curve for phnAc (Fig. 1b). The lower limit of the QC-PCR titration series was 0.0001 amol of standard template (approximately 60 copies), which was equivalent to a detection limit of 2 × 105 copies of a given gene per gram (dry weight) of soil. By using PCR primers that were specific for a gene that encodes the iron sulfur protein large (α) subunit of the PAH dioxygenase from the recently described phn type of catabolic operon (10), we found that cells harboring phn genes are not as rare as microbiological culture techniques might lead us to believe (10, 12).
TABLE 1.
Soil properties
| Soil | Type of contamination | Concn (mg kg [dry wt]−1) of:
|
Concn of culturable heterotrophs (no. of cells g [dry wt]−1) | Concn (no. of copies g [dry wt]−1) of:
|
|||
|---|---|---|---|---|---|---|---|
| 16 PAHs | Naphthalene | Phenanthrene | phnAc | nahAc | |||
| A | Town gas site PAHs | 428 | 21.6 | 36.9 | 9.2 × 106 | 1.0 × 108 ± 0.3 × 108a | 2.0 × 108 ± 0.9 × 108 |
| B | Petrochemical | 2.7 | 0.29 | 0.86 | 1.3 × 107 | 6.2 × 107 ± 1.3 × 107 | 1.3 × 106 ± 0.9 × 106 |
Mean ± standard deviation (n = 3).
FIG. 1.
(a) QC-PCR of phnAc and nahAc amplified from two contaminated soils. Soil A was derived from a former town gas-generating site and contained high levels of PAHs. Soil B was contaminated with petrochemicals and contained low levels of PAHs. The upper band in each QC-PCR titration series contained the target amplicon (unknown); the lower band consisted of amplicons derived from the standard DNA. The amounts of standard DNA used (10 to 0.0001 amol) are indicated above the lanes. (b) Representative plot of the ratio of target DNA intensity to standard DNA intensity plotted against the initial concentration of standard DNA on logarithmic scales. Band intensities were quantified by processing images of Polaroid photographs of SYBR Gold-stained gels. The plot is not a representation of panel a.
Replicate QC-PCR experiments (n = 3) revealed that the PAH-contaminated soil (soil A) contained 1.0 × 108 ± 0.3 × 108 copies of phnAc per g of soil and 2.0 × 108 ± 0.9 × 108 copies of nahAc per g of soil, while the petroleum-contaminated soil (soil B) contained 6.2 × 107 ± 1.3 × 107 copies of phnAc per g of soil and 1.3 × 106 ± 0.9 × 106 copies of nahAc per g of soil. The numbers of copies of both phnAc and nahAc in these contaminated soils were significantly greater than the numbers of copies in a pristine soil, which did not contain detectable levels of either gene (i.e., less than 2 × 105 copies per g). The greater numbers of copies in the PAH-contaminated soil than in the petroleum-contaminated soil may reflect higher naphthalene and phenanthrene levels. Similar observations have been made in previous studies of the molecular ecology of nah-like genotypes in PAH-contaminated soil (16, 17). For the two contaminated New Zealand soils that we analyzed, the phn and nah-like genotypes were present (assuming that there was one copy of each gene per cell) at levels that were greater than or equal to the total numbers of heterotrophs determined for the same soil samples by culture techniques (Table 1). Although the presence of phnAc or nahAc genes in a soil does not a priori indicate that PAH-degrading activity is present, our data revealed elevated levels of the phnAc and nahAc genes in soils in which it can reasonably be expected that selection pressures exerted by PAHs would enrich populations able to degrade these substrates.
Diversity of phnAc amplicons.
Previous studies have shown that primers based on nah-like genes target a group of sequences that exhibit more than 75% sequence homology and form the phylogenetically distinct nah-like and dnt-ntd groups (7, 12, 18). We sequenced both strands of phnAc amplicons to determine the diversity of homologous sequences targeted in soil DNA by primers P8073 and P9047. The phn primers P8073 and P9047 amplified a very highly conserved group of phnAc genes that exhibit more than 98% sequence identity with the RP007 phnAc sequence. In this study two sequences derived from petroleum-contaminated soil and one sequence derived from PAH-contaminated soil were identical to the RP007 phnAc sequence, while an additional three sequences derived from PAH-contaminated soil samples differed at only 2, 12, and 15 nucleotide positions. Similarly high levels of sequence conservation were also found for six phnAc amplicons derived from two other contaminated soils in a previous study, which also exhibited more than 98% nucleotide homology with the RP007 phnAc sequence (9). Sequence errors due to Taq DNA polymerase or PCR amplicon contamination during cloning were statistically improbable since the majority of base substitutions (24 of 29 substitutions; >80%) resulted in synonymous (silent) substitutions. In addition, the Taq DNA polymerase error rate, <0.05%, as determined by sequencing two cloned phnAc amplicons derived from the Burkholderia sp. wild-type strain RP007, was also too low to account for these substitutions.
The highly conserved nature of the phnAc amplicons obtained with primers P8073 and P9047 may be an artifact of the high degree of primer specificity and stringent amplification conditions. We have not determined whether this observation is a true reflection of the diversity of phn genes in the environment, which would require the use of degenerate primers allied with less stringent annealing conditions during PCR amplification. What is certain is that the original isolation of the phn genes was fortuitous since bacteria harboring this genotype appear to be very difficult to isolate yet are ubiquitously distributed and may be present in relatively high numbers in PAH-contaminated soils.
Distribution of phn genes.
Having determined that phn genes were enriched in PAH-contaminated New Zealand soils, we were also interested in evaluating whether the phn genes are ubiquitous. Noncontaminated (pristine) soils obtained from central Siberia (61°N, 89°E), Ross Island in the Antarctic (77°S, 166°E), and a native New Zealand forest (38°S, 175°E) were used to assess the ubiquity of the phn genes in different environments. Enrichment of the PAH-degrading populations in these uncontaminated soils was necessary before we screened for the phnAc genotype since the levels of analogues of phnAc were initially below the limits of detection (i.e., less than 2 × 105 copies per g of soil) in these soils. Soil microcosms were therefore established, and PAH-degrading bacteria were enriched by using the low-molecular-weight PAHs naphthalene and phenanthrene. The broad geographic distribution of the phn genotype was confirmed when phnAc was amplified from a central Siberian soil, from an Antarctic soil from Ross Island, and from various New Zealand soils. The phn genes were enriched in soil microcosms that were incubated for 1 week in the presence of either naphthalene or phenanthrene, and after enrichment the levels were above our limit of detection (2 × 105 copies of phnAc per g [dry weight] of soil). Again it is particularly interesting that we detected phn genes in soils from areas as far afield as Siberia, Antarctica, and New Zealand and yet to our knowledge only one confirmed strain with a phn genotype has been isolated from the environment (9, 10, 12).
In situ specificity of the phn genes.
We also examined the effects of the selection pressures exerted by different low-molecular-weight PAHs on the phnAc and nahAc genotypes in an uncontaminated New Zealand soil. The levels of both nahAc and phnAc were initially below the detection limit (2 × 106 copies of phnAc per g [dry weight] of soil) in subsamples of this soil. This detection limit was higher than the detection limit for the contaminated soils due to the higher humic content of this soil, which required greater dilution of target DNA to a level which did not inhibit PCR amplification. As expected, selection pressures within the microcosms enriched for a PAH-degrading phenotype (8, 13) but interestingly favored the phn genotype and not the nah-like genotype, which we were not able to detect in any microcosm (Table 2). These findings imply that the nah-like genotype is not always ecologically dominant and confirm that it is not always realistic to represent PAH degradation by one genotype, as this genotype may not be present at a detectable (or significant) level in all environments (1, 12).
TABLE 2.
Microcosm enrichment
| Microcosm enrichment | Concn (no. of copies g of soil−1) of:
|
|
|---|---|---|
| phnAc | nahAc | |
| Preincubationa | NDb | ND |
| Control incubation | 2.0 × 108 | ND |
| Naphthalene | 1.8 × 109 | ND |
| Phenanthrene | 5.5 × 108 | ND |
| Fluoranthene | 4.9 × 108 | ND |
Soil sample obtained before microcosms were established.
ND, not detected (<2 × 106 copies per g [dry weight] of soil).
Enrichment of phnAc from uncontaminated soils appeared to be greatest with naphthalene (nine times greater than enrichment in the control microcosm), which has the greatest volatility, solubility, and, hence, availability of the three PAH substrates used for enrichment. Since previous studies have shown that the phn genes are induced by both naphthalene and phenanthrene (10), it is likely that phenanthrene also enriches for the phn genotype. Although the QC-PCR data for phenanthrene enrichment of phnAc may not be conclusive in the New Zealand soil microcosm (the level was only two to three times the level observed in the control microcosm), we have demonstrated that phnAc was enriched in both Siberian and Antarctic soils when they were exposed to phenanthrene. It remains to be established whether fluoranthene is degraded in situ by phn gene products or is able to induce transcription of the phn genes, since the evidence for enrichment in response to fluoranthene was not conclusive. Whether the increase observed in the control microcosm was real or due to variability and biases inherent in using small soil samples was not determined. A real increase may have occurred in response to natural soil substrates as a consequence of moisture, temperature, and inorganic nutrient level adjustments made while the control microcosm was being established.
ACKNOWLEDGMENTS
This work was supported by grant C09810 from the Foundation for Research Science and Technology, New Zealand.
We thank R. Fraser and D. W. F. Hunter for technical assistance and colleagues at Landcare Research for providing soil samples.
REFERENCES
- 1.Ahn Y, Sanseverino J, Sayler G S. Analyses of polycyclic aromatic hydrocarbon-degrading bacteria isolated from contaminated soils. Biodegradation. 1999;10:149–157. doi: 10.1023/a:1008369905161. [DOI] [PubMed] [Google Scholar]
- 2.Berthelet M, Whyte L G, Greer C W. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol Lett. 1996;138:17–22. doi: 10.1111/j.1574-6968.1996.tb08128.x. [DOI] [PubMed] [Google Scholar]
- 3.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goyal A K, Zylstra G J. Molecular cloning of novel genes for polycyclic aromatic hydrocarbon degradation from Comamonas testosteroni GZ39. Appl Environ Microbiol. 1996;62:230–236. doi: 10.1128/aem.62.1.230-236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyal A K, Zylstra G J. Genetics of naphthalene and phenanthrene degradation by Comamonas testosteroni. J Ind Microbiol Biotechnol. 1997;19:401–407. doi: 10.1038/sj.jim.2900476. [DOI] [PubMed] [Google Scholar]
- 6.Hedlund B P, Geiselbrecht A D, Bair T J, Staley J T. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl Environ Microbiol. 1999;65:251–259. doi: 10.1128/aem.65.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langworthy D E, Stapleton R D, Sayler G S, Findlay R H. Genotypic and phenotypic responses of a riverine microbial community to polycyclic aromatic hydrocarbon contamination. Appl Environ Microbiol. 1998;64:3422–3428. doi: 10.1128/aem.64.9.3422-3428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurie A D. Ph.D. thesis. Hamilton, New Zealand: University of Waikato; 1998. [Google Scholar]
- 10.Laurie A D, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181:531–540. doi: 10.1128/jb.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of abundance of an uncultured soil bacterium strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd-Jones G, Laurie A D, Hunter D W F, Fraser R. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol Ecol. 1999;29:69–79. [Google Scholar]
- 13.Mueller J G, Devereux R, Santavy D L, Lantz S E, Willis S G, Pritchard P H. Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie Leeuwenhoek. 1997;71:329–343. doi: 10.1023/a:1000277008064. [DOI] [PubMed] [Google Scholar]
- 14.Saito A, Iwabuchi T, Harayama S. Characterization of genes for enzymes involved in the phenanthrene degradation in Nocardioides sp. KP7. Chemosphere. 1999;38:1331–1337. doi: 10.1016/s0045-6535(98)00534-7. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- 16.Sanseverino J, Werner C, Fleming J, Applegate B, King J M, Sayler G S. Molecular diagnostics of polycyclic aromatic hydrocarbon biodegradation in manufactured gas plant soils. Biodegradation. 1993;4:303–321. doi: 10.1007/BF00695976. [DOI] [PubMed] [Google Scholar]
- 17.Stapleton R D, Sayler G S. Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microb Ecol. 1998;36:349–361. doi: 10.1007/s002489900121. [DOI] [PubMed] [Google Scholar]
- 18.Wilson M S, Bakermans C, Madsen E L. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl Environ Microbiol. 1999;65:80–87. doi: 10.1128/aem.65.1.80-87.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zachar V, Thomas R A, Goustin A S. Absolute quantification of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993;8:2017–2018. doi: 10.1093/nar/21.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zylstra G J, Kim E, Goyal A K. Comparative molecular analysis of genes for polycyclic aromatic hydrocarbon degradation. Genet Eng. 1997;19:257–269. doi: 10.1007/978-1-4615-5925-2_14. [DOI] [PubMed] [Google Scholar]