Abstract
As the Urtica dioica L. whole plant’s essential oil has presented significant multiple activities, it was therefore evaluated using the GC–MS technique. This essential oil was investigated for its antioxidant, phytotoxic, and antibacterial activities in vitro. The GC–MS analysis data assisted in the identification of various constituents. The study of the essential oil of U. dioica showed potential antioxidant effects and antibacterial activity against the selected pathogens Escherichia coli -ATCC 9837 (E. coli), Bacillus subtilis-ATCC 6633 (B. subtilis), Staphylococcus aureus-ATCC6538 (S. aureus), Pseudomonas aeruginosa-ATCC 9027 (P. aeruginosa), and Salmonella typhi-ATCC 6539 (S. typhi). The library of 23 phytochemicals was docked by using MOE software, and three top virtual hits with peroxiredoxin protein [PDB ID: 1HD2] and potential target protein [PDB ID: 4TZK] were used; hence, the protein–ligand docking results estimated the best binding conformations and a significant correlation with the experimental analysis, in terms of the docking score and binding interactions with the key residues of the native active binding site. The essential oil in the silico pharmacokinetic profile explained the structure and activity relationships of the selected best hits, and their additional parameters provided insight for further clinical investigations. Therefore, it is concluded that the U. dioica essential oil could be a potent antioxidant and antibacterial agent for aromatherapy through its topical application, if further tested in a laboratory and validated.
Keywords: Urtica dioica L. essential oil, GC–MS, phytochemical analysis, antioxidant activities, antibacterial activities
1. Introduction
The genus Urtica is associated with the family “Urticaceae” in the division of Angiosperms, which present peculiar flowering characteristics which have been known for a long time, and are also utilized as traditional medicine and are significant as food [1]. The closest associates of this genus are the stinging nettle Urtica dioica L. (U. dioica) and the small nettle U. urens L., which are native to Asian, African, European, and North American areas [2]. U. dioica L., commonly known as the common nettle, is a persistent plant that grows in moderate and humid wilderness ranges across the world [3]. It possesses pointed leaves and grows from 1 to 1.4 m high. The isolation, purification, and characterization of antimicrobial plant-derived compounds remains a stimulating research field, particularly in the drug development field for multi-drug resistant bacteria [4,5].
U. dioica is extensively recycled as a traditional medicine for the management of allergies [6], stone formation, anemia, rashes [7], hypertension [8], antiarthritic and antirheumatic effects [9], antidandruff, galactagogue, hemostatic, and anti-ulcer activity, stomachache and liver dysfunction, and anti-inflammatory [10,11], antihyperglycemic [10], antioxidant, acute diuretic, natriuretic, and hypotensive effects [12]. It has been used in the traditional therapy of antifungal agents [13].
The chemical constituents of the U. dioica plant also exhibit anticancer and anti-diabeties mellitus (anti-DM) properties. Existing knowledge suggests that U. dioica possesses fatty acids, sterols, lignans, carotenoids, plastocyanins, glycoproteins, lectins, polysaccharides, terpenes, and flavonoids as its main phytoconstitutents [14]. The identification of essential oils is an interesting component of phytochemical investigation [15,16,17].
As the plant U. dioica in its crude form and its various constituents exhibited biological potential, we intended to perform a gas chromatography–mass spectrometry (GC–MS) analysis of U. dioica’s essential oil, and to evaluate its antioxidant, phytotoxic, and anti-bacterial potential. For the validation of the experimental results, we followed in silico techniques, such as molecular docking (MD) and ADMET, to highlight each ligand’s behavior with respect to specific selected proteins.
In this study, the U. dioica plant’s oil revealed potential antioxidant and antimicrobial activities, potentially for first time while using GC–MS-derived bioactive compounds from the U. dioica plant’s oil. The most significant outcome of this study was to reveal the phytotoxic effect of the oil for the first time. In this study, another objective was to accomplish the docking interaction of the compounds with proteins, using in silico methods and mutagenesis experiments, which are in the pipeline to be performed in upcoming studies. 1HD2 is an extensively studied antioxidant enzyme, and 4TZK is used in several studies to test antibacterial effects.
2. Materials and Methods
2.1. Sampling
Our research was conducted to collect samples in a suitable season from a specific area, where the plant growth is good and available. According to the sampling plans, the whole U. dioica plant was collected in March 2019 from the Dardarez Landidak regions of Bannu (Latitude: 32°59′9.9996″ N and Longitude: 70°36′14.9904″ E), KPK, Pakistan. The plant was identified by Dr. Faizan Ullah, Assistant Professor, Department of Botany, University of Science and Technology Bannu, Bannu, Pakistan.
2.2. Extract Preparation
The whole plant material was dried in a shady area, ground well, and then soaked in 80% aq. methanol (MeOH) for one week. The soaked material was evaporated by using a rotary evaporator to obtain a dark, gummy residue. The gummy material was initially extracted with n-hexane to remove the fatty materials. The defatted MeOH extract was suspended in water, and then the aqueous fraction was further fractionated with dichloromethane (DCM), ethyl acetate (EtOAc), and butanol. The DCM fraction was preceded further by column chromatography and the essential oil part was passed through gas chromatography (GC) and gas chromatography mass spectrum (GC–MS) instruments, and the components were identified.
2.3. Gas Chromatography–Mass (GC–MS) Spectrum Analysis
The essential oil component of the dichloromethane fraction of the U. dioica was analyzed through a GC–MS Agilent 6890N Network GC system, combined with an Agilent 5973 Network Mass Selective Detector (GC–MS) instrument, under computer control. The injector temperature was set at 220 °C for 5 min. At a split ratio of 1:10. 1 µL, a volume of 1000 ppm of the essential oil solution (GC Grade n, hexane, scharlau, chemia, Barcelona, Spain) was injected. Initially, the column was maintained at 50 °C for 2 min and then increased to 150 °C, at which it was held isothermal for 5 min, and a second rmp (200 per minutes) was applied to 220 °C and held isothermal for 10 min. The total run time was 120 min. Thus, it was maintained between 180 °C and 230 °C, respectively. The MS was performed in the electron ionization mode (70 eV) [18].
2.4. Antioxidant Activity
The antioxidant activity was estimated with a DPPH assay [19,20]. The DPPH solution was set via dissolving 3.2 mg in 100 mL of 82% methanol, and then 2.8 mL of the DPPH solution was added to the glass vial and monitored with the addition of 0.2 mL of the test sample solution, leading to the final concentration of 1 µg/mL, 5 µg/mL, 10 µg/mL, 25 µg/mL, 50 µg/mL, and 100 µg/mL. Ascorbic acid was used as a standard. The process was performed under dark conditions at fixed room temperature for 1 h, the discoloration was measured at 517 nm in triplicate by using a UV spectrophotometer (Deuterium lamp, Shimadzu, Japan), and the radical scavenging capacity was expressed as a percentage effect (E%) and estimated by using the following equations.
2.5. Phytotoxic Activity
The phytotoxic activity was estimated with the application of a modified protocol of Lemna minor L. [15]. Inorganic E-medium was prepared by mixing the appropriate inorganic constituents into 1 L of distilled water, and the pH was accustomed at 5.5–5.6 by adding KOH solution and autoclaved at 121 °C for 15 min. The essential oil served as a stock solution. A total of three sterilized flasks for each concentration were inoculated with 1000 µL, 100 µL, and 10 µL of the stock solution for 500, 50, and 5 ppm, respectively. Each flask, medium (20 mL), and essential oil, each encompassing a rosette of three fronds of the Lemna minor L., were added. In total, two supplemented flasks, one with the standard drug (Paraquat) and the other with the E-medium, were served as positive and negative controls, respectively. All the flasks were wrought with cotton and were reserved in the growth cabinet for seven days. The number of fronds per flask was calculated and noted at day seven. The results were interpreted by considering the growth regulation as a percentage and considered with reference to the negative control.
| (1) |
2.6. Antibacterial Activity
To understand the antibacterial effect, Microplate Alamar Blue Assay was used [21,22]. Mueller–Hinton medium was used for the growth of the organism. This is a non-selective, non-differential, microbiological growth medium. It contained beef extract, acid casein hydrolysate, starch, and agar. The beef extract and casein acid hydrolysate provided nitrogen, vitamins, carbon, amino acids, sulfur, and other essential nutrients. The inoculation adjustment was done to a 0.5 McFarland turbidity index. The essential oil samples were mixed in pure DMSO with a 1:1 ratio [23]. The media were poured into all the wells that did not contain any test samples. The reaction mixture was a total of 200 µL and 96-well microtiter plates were used. In the end, about 5 × 106 cells were added in each well of the controls, as well as the tests. The plate was wrapped with paraffin film and was incubated for about 20 h. After incubation, the suggested dye, Alamar Blue, was distributed in each well, and the plate was well shaken at 80 RPM in an incubator with a shaking mode for about 3 h. The plate was covered with aluminum and essential oil while the shaking was carried out in the shaking incubator. The color of the Alamar Blue dye changing from blue to pink showed the growth of certain bacterial strains such as Escherichia coli -ATCC 9837 (E. coli), Bacillus subtilis-ATCC 6633 (B. subtilis), Staphylococcus aureus-ATCC6538 (S. aureus), Pseudomonas aeruginosa-ATCC 9027 (P. aeruginosa), and Salmonella typhi-ATCC 6539 (S. typhi). The absorbance of each well was monitored in the ELISA reader at a 570–600 nm wavelength.
2.7. Computational Analysis
2.7.1. Selection of Protein Targets and Chemical Compounds
In pharmaceutical research and computer-aided drug design (CADD), one of the most significant computational techniques is called MD [24,25]. The fundamental requirement of protein–ligand MD protocols is to find the probable binding geometries of a suspected ligand with a known three-dimensional structure and a target protein [26]. This may be accomplished by comparing the structures of both the hypothetical ligand and the targeting protein. The 2D structures of the phytochemical dataset were sketched with the ChemDraw application for further analysis [27] (Table 1). In this study, we targeted the peroxiredoxin protein [PDB ID: 1HD2] [28] and the potential selected macromolecule [PDB ID: 4TZK] [29] for the MD analysis, in order to check its correlation with the experimental analysis of the whole Urtica dioica L. plant’s essential oil compounds.
Table 1.
Phytochemical compounds identified from U. dioica essential oil of DCM fraction.
| Name of Compound | Mass/RT (min) |
% Area | Molecular Formula |
Fragments Ions |
Structure |
|---|---|---|---|---|---|
| Nonanoic acid, 9-oxo-, ethyl ester | 200/33.69 | 2.32 | C11H20O3 | 41,55,88,29,43,101,83,60,157,155 |
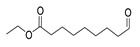
|
| Carophyllen oxide | 220/35.70 | 0.28 | C15H24O | 43,41,79,93,91,95,69,55,67,81 |
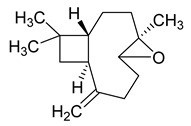
|
| Limonen-6-ol, pivalate | 236/36.06 | 0.23 | C15H24O2 | 57,41,43,93,55,107,109,91,85,119 |
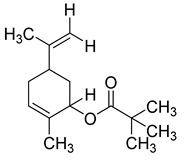
|
| β-Himachalenoxide | 220/36.39 | 0.45 | C15H24O | 110,220,95,192,43,41,69,109,151,93 |
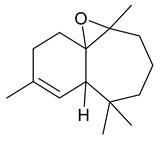
|
| 4-tert-Butyltoluene | 148/36.85 | 0.4 | C11H16 | 133,105,148,93,41,91,134,39,115,77 |
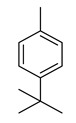
|
| α-Bisabolol | 222/37.27 | 1.25 | C15H26O | 109,119,69,43,93,41,95,121,67,71 |
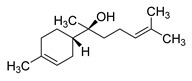
|
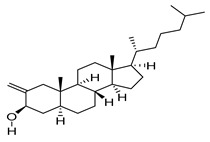
| Cholestan-3-ol, 2-methylene-, (3β, 5α)- | 400/36.32 | 0.18 | C28H48O | 69,81,71,95,67,83,79,93,97,105 |
|
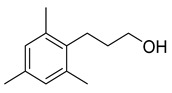
| Benzenepropanol, 2, 4, 6-trimethyl- | 178/39.84 | 0.17 | C12H18O | 133,178,134,120,119,91,145,105,117,115 |
|
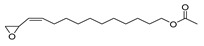
| Z-(13, 14-Epoxy) tetradec-11-en-1-ol acetate | 268/36.61 | 1.01 | C16H28O3 | 43,97,69,55,41,82,67,81,84,83 |
|
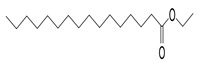
| Hexadecanoic acid, ethyl ester, | 284/40.11 | 0.47 | C18H36O2 | 88,101,43,55,41,57,69,73,71,70 |
|
| 3, 7, 11, 15-Tetramethyl-2-hexadecen-1-ol | 296/40.14 | 1.01 | C20H40O | 81,82,43,95,123,55,41,57,71,68 |
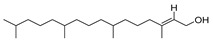
|
| 2-Pentadecanone, 6, 10, 14-trimethyl | 268/41.30 | 1.76 | C18H36O | 43,58,71,57,59,41,55,69,85,95 |
|
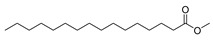
| Hexadecanoic acid, methyl ester | 270/43.47 | 0.66 | C17H34O2 | 74,87,43,41,55,75,29,57, 143 |
|
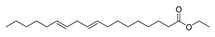
| 9, 12-Octadecadienoic acid, ethyl ester | 308/54.29 | 1.92 | C20H36O2 | 67,81,55,95,68,54,96,69 |
|
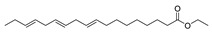
| Ethyl 9, 12, 15-octadecatrienoate | 306/54.76 | 3.33 | C20H34O2 | 79,67,95,93,81,55,8o,107,91 |

|
| 1, 2-Benzenedicarboxylic acid, mono (2-ethylhexyl) ester | 278/80.83 | 5.12 | C16H22O4 | 149,167,57,71,43,70 |
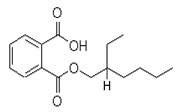
|
| 16-Hentriacontanone | 450/91.16 | 1.52 | C31H62O | 239,57,43,71,255,55,58,245,69,41 |
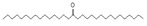
|
| Z-5-Methyl-6-heneicosen-11-one | 322/92.79 | 0.94 | C22H42O | 43,55,57,41,83,169,85,81,69 |
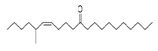
|
| 18-Pentatriacontanone | 506/96.41 | 1.161 | C35H70O | 71,267,69,85,83,283,97,82 |
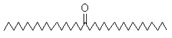
|
| E, E, Z-1, 3, 12-Nonadecatriene-5, 14-diol | 294/99.40 | 8.39 | C15H26O2 | 55,95,81,41,67,69,96,83,43,57 |
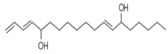
|
| 9-Octadecenoic acid (Z)-, 9-octadecenyl ester, (Z) | 532/103.82 | 1.00 | C36H68O2 | 55,69,83,97,82,96,57,95,67 |
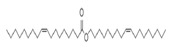
|
| Tricyclo[20.8.0.0(7,16)]triacontane, 1(22),7(16)-diepoxy | 444/104.24 | 0.42 | C30H52O2 | 55,67,95,81,69,41,43,83,79,109 |
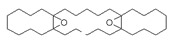
|
2.7.2. Molecular Docking (MD) and Interaction Analysis
MD is a suitable molecular modeling procedure, which is significantly based on the searching and ranking of poses to generate an MD score scheme, and the best binding configurations that are estimated from a best-docked complex, which help to explore the binding interactions of a protein–ligand; therefore, it could be useful to understand the molecular functions of a ligand that is confined within the active residual region of the selected enzyme [30,31]. For MD purposes, 5 compounds were used, and the selected proteins’ three-dimensional structures (PDB ID: 1HD2 and 4TZK), in .pdb format, were downloaded from the PDB and prepared in a molecular operating environment (MOE) software [32,33]. The water molecules from the ligand that was already bound to the selected enzyme molecule were detached, and for the heteroatoms, 3D protonation was performed to prepare its structure for the MD procedure. In each protein structure, an active site was acknowledged, and PRO40, THR44, PRO45, LEU116, PHE120, GLY46, CYS47, ARG127, and THR147 residues were selected for the active residual region of the 1HD2 protein, while GLY14, ILE15, ILE16, ALA22, PHE41, ALA190, ALA191, GLY192, PRO193, ILE194, MET147, ASP148, PHE149, MET155, VAL65, GLN66, THR17, SER20, ILE21, HIS93, SER94, ILE96, GLY97, PHE98, MET99, MET103, LEU63, ASP64, GLY104, ILE122, PRO156, ALA157, TYR158, MET161, LYS165, THR196, ALA198, MET199, ILE202, LEU207, ALA211, ILE215, and LEU218 residues were selected for the active site of the 4TZK protein. A structural optimization was also applied by notable estimations, such as the addition of hydrogen atoms and an Amber14 force field scheme, and a realistic scheme with chiral constraints was applied, as were geometrical constraints for extra energy control of the potent bounded conformers. The surfaces and maps panel module was used to manage the surface’s structural transparency and resulted in evidence of important amino acids in the selected region of the enzyme selected as the initial deigned conformer. A database of 23 phytochemicals that was retrieved from the experimental analysis was created in the MOE software, in order to perform MD simulations, and the database was saved with an .mdb extension for next-level analysis. The enhancement and estimations of binding-free energies (ΔG) were evaluated with the application of a scoring function (GBVI/WSA dg) to screen the top-ranked poses that were performed [34]. Pi, hydrogen, and hydrophobic interactions as the consistent scoring pattern were established in the form of an MD score of the correct binding poses [35]. The database of the docked complex, which was generated in the MOE, was visualized for a detailed understanding of the mode of the binding interactions of the ligands in the selected active site of the target protein.
2.7.3. Pharmacokinetic/ADMET Profile Estimation
The selected best-docked compounds were used for the estimations of the ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties in order to justify the drug-like assumptions, as this is considered to be an essential criterion for the drug-like screening of chemical libraries [24,36]. For the purpose of the ADMET profile estimation of the selected potential hits, SwissADME [37] and Datawarrior tools [38] were used.
3. Results and Discussion
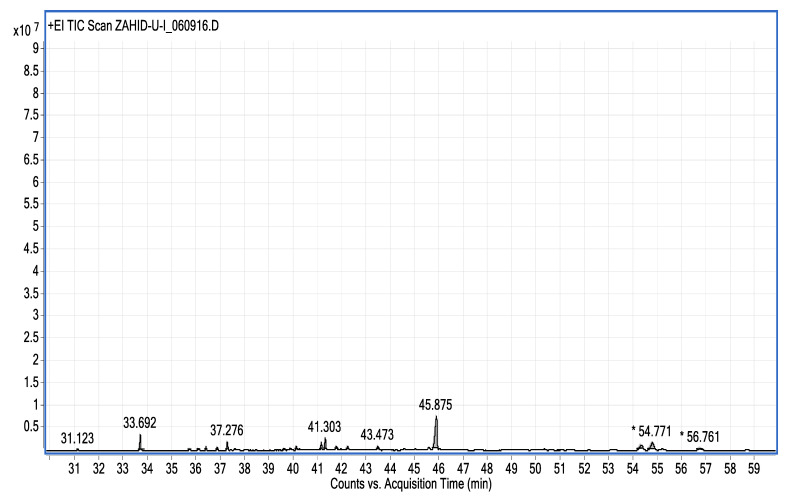
The study showed a GC–MS analysis of the isolated essential oils of U. dioica (Figure 1 and Figure 2). The GC–MS chromatogram showed various peaks, which, upon comparison with data from the literature, were identified as nonanoic acid, 9-oxo- ethyl ester, caryophyllene oxide, limonen-6-ol-pivalate, β-himachalenoxid, 4-tert-butyltoluene, α-bisabolol, cholestan-3-ol,2-methylene-, (3β, 5α), benzenepropanol 2,4,6-trimethyl, Z-(13,14-epoxy)tetradec-11-en-1-ol acetate, hexadecanoic acid, ethyl ester, 3,7,11,15-tetramethyl-2-hexadecen-1-ol, 2-pentadecanone, 6,10,14-trimethyl, hexadecanoic acid, methyl ester, 9,12-octadecadienoic acid, ethyl ester, ethyl 9,12,15-octadecatrienoate, 1,2-benzenedicarboxylic acid, mono(2-ethylhexyl) ester, 16-hentriacontanone, Z-5-methyl-6-heneicosen-11-one, 18-pentatriacontanone, E,E,Z-1,3,12-nonadecatriene-5,14-diol, 9-octadecenoic acid (Z), 9-octadecenyl ester, (Z), and tricyclo [20.8.0.0(7,16)] triacontane, 1(22),7(16)-diepoxy. The identification of these compounds was made by the comparison of their retention time and the mass spectra of those stored in the computer library and published literature. The mass, retention time, % area, molecular formula, fragment ions, and their structures, are hereby summarized in Table 1.
Figure 1.
GC–MS chromatogram showing peaks in the range of 31–59, * denoted high values.
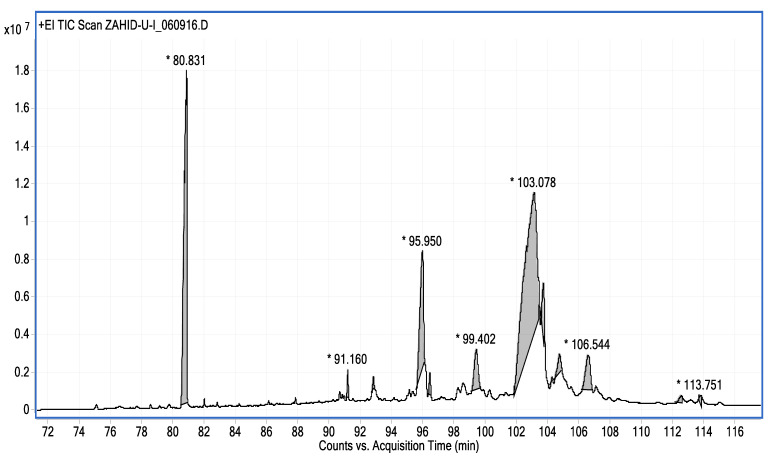
Figure 2.
GC–MS chromatogram showing peaks in the range of 72–116, * denoted high values.
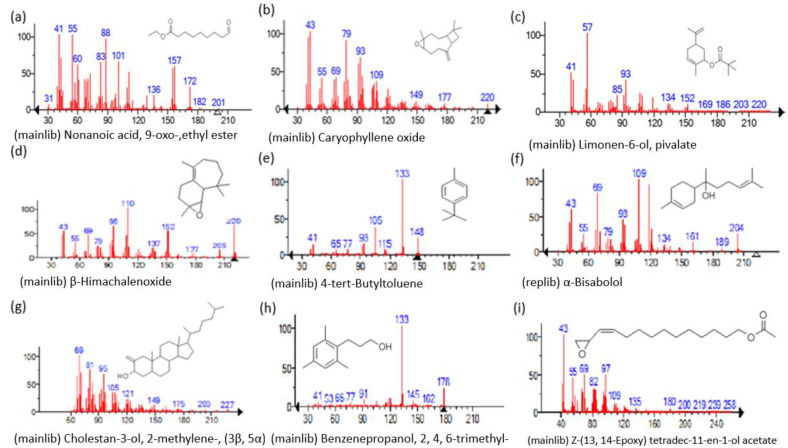
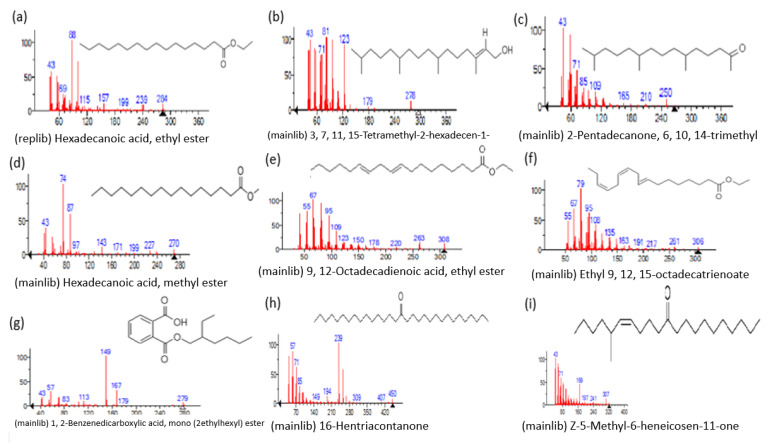
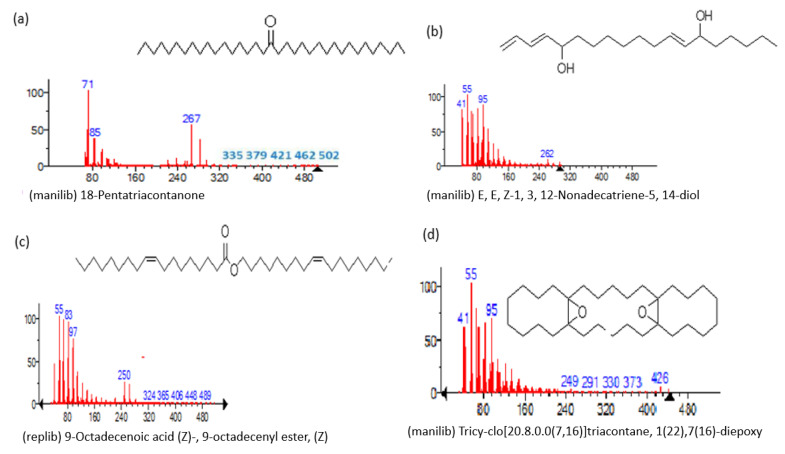
The mass spectrum of the identified compounds is presented in Figure 3, Figure 4 and Figure 5.
Figure 3.
Representing the mass spectrum of Nonanoic acid, 9-oxo-, ethyl ester with RT= 33.692 (a), Caryophyllene oxide with retention time (RT) =35.706 (b), Limonen-6-ol, pivalate with RT = 36.06 (c), β- Himachaleoxide with RT = 36.39 (d), 4-tert-Butyltoluene with RT = 36.853 (e), α-Bisabolol with RT = 37.276 (f), Cholestan-3-ol, 2-methylene-, (3β, 5α) with RT = 36.325 (g), Benzenepropanol, 2, 4, 6-trimethyl- with RT =36.942 (h), and Z-(13,14-Epoxy) tetradec-11-en-1-ol acetate with RT = 36.613 (i).
Figure 4.
Representing the mass spectrum of Hexadecanoic acid, ethyl ester with RT = 41.613 (a), 3, 7, 11, 15 -Tetramethyl-2-hexadecen-1-ol with RT = 41.140 (b), 2-Pentadecanone, 6, 10, 14-trimethyl- with RT = 41.303 (c), Hexadecanoic acid, methyl ester with RT = 43.470 (d), 9,12-Octa decadienoic acid, ethyl ester with RT = 54.297 (e), Ethyl 9,12,15-octadecatrienoate with RT = 54.767 (f), 1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester with RT = 80.831 (g), 16-Hentriacontanone with RT = 91.164 (h), and Z-5-Methyl-6-heneicosen-11-one with RT = 92.799 (i).
Figure 5.
Representing the mass spectrum of 18-Pentatriacontanone with RT = 96.417 (a), E, E, Z-1, 3, 12-Nonadecatriene-5,14-diol with RT = 99.402 (b), 9-Octadecenoic acid (Z)-, 9-octadecenyl ester, (Z) - with RT = 103.82 (c), and Tricyclo [20.8.0.0(7, 16)] triacontane, 1(22), 7(16)-diepoxy- with RT = 104.247 (d).
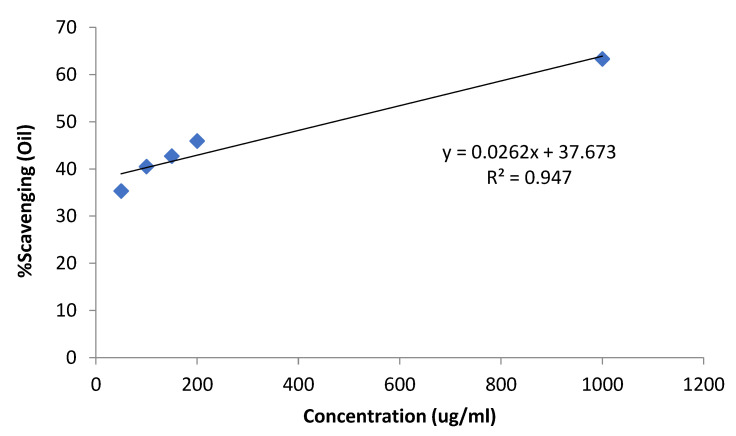
In the antioxidant assay (Table 2), concentrations were taken in (µg/mL), starting from 50, 100, 150, 200, and 1000 and shows the values in Figure 6. The values of the essential oil that were obtained are 39.3, 40.5, 42.7, 45.9, and 63.3, while the values of the standard drug (ascorbic acid) are 70.5, 73.7, 77.8, 79.4, and 87.4, based on increasing concentrations. The U. dioica essential oil showed significant DPPH scavenging activity, as compared to the standard’s ascorbic acid. The essential oil of U. dioica, at low concentration, showed no growth inhibition, while at higher concentration, it showed inhibition power. Similar activity was reported by Kukri et al., using crude extract [39]. In a recent study conducted by Chaqroune and Taleb (2022), the findings showed significant antioxidant effects of the essential oil. The methanol and ethanol were used as solvents for extraction [40,41]. These findings may correlate to this study, as essential oils have the potential to be used as antioxidant agents.
Table 2.
Antioxidant activity of U. dioica essential oil.
| Conc. (µg/mL) | %Scavenging (Essential Oil) | (Essential Oil) | %Scavenging (Ascorbic Acid) |
|---|---|---|---|
| 50 | 35.3 ± 1.8 | 70.5 ± 2.3 | |
| 100 | 40.5 ± 1.9 | 71.7 ± 3.1 | |
| 150 | 42.7 ± 2.1 | 470.4 | 72.9 ± 2.0 |
| 200 | 45.9 ± 1.7 | 73.4 ± 2.4 | |
| 1000 | 63.3 ± 1.8 | 87.4 ± 3.0 |
Figure 6.
Equation to determine the values of essential oil.
In the phytotoxic assay (Table 3), the concentrations were taken in µg/mL, starting from 10, 100, 250, 500 and 1000 µg/mL. The concentration of the standard drug (0.015 µg/mL) was used. The percentage of the growth inhibition of the essential oil showed 26% and 62.5% at the concentrations of 500 and 1000 µg/mL, respectively, while at other concentration levels, showed a 0% result.
Table 3.
Phytotoxic activity of U. dioica essential oil.
| Conc. (µg/mL) | Essential Oil | Control | %Growth Inhibition | Std. Drug Conc. (µg/mL) |
|---|---|---|---|---|
| 10 | 24 | 24 | 0 | 0.015 |
| 100 | 24 | 24 | 0 | |
| 250 | 24 | 24 | 0 | |
| 500 | 17 | 24 | 26 | |
| 1000 | 09 | 24 | 62.5 |
Table 4 contains the antibacterial results against five bacteria, E. coli, B. subtilis, S. aureus, S. aeruginosa, and S. typhi. Although the essential oil did not respond to the concentration of 250 μg/mL, it started to exhibit antibacterial activity against E. coli and B. subtilis at the concentration of 500 μg/mL, with an inhibition zone diameter of 10 ± 0.25 mm and 12 ± 0.32 mm. The antibacterial activity exhibited by the essential oil of U. dioica against the tested bacterial strains, E. coli, B. subtilis, S. aureus, S. aeruginosa, and S. typhi, showed good antibacterial activity (1000 μg/mL) as compared to the reference drug, ofloxacin (0.25 µg/mL). These findings concluded that the U. dioica essential oil could be used as a potential antibacterial agent for aromatherapy or topical applications against E. coli and B. subtilis. In addition, previous studies have also been reported on the antibacterial effects of U. dioica extract against selected pathogens [42,43]. The antibacterial activities of essential oils against the group of bacteria (E. coli, B. subtilis, and S. aureus) has been proven by Zeroual et al., 2021 [44]. Similarly, the current findings also showed a significant outcome against E. coli, B. subtilis, S. aureus, S. aeruginosa, and S. typhi bacteria.
Table 4.
Antibacterial activity of Urtica dioica essential oil against selected bacteria strains.
| Bacteria | U. dioica Essential Oil | Control (Ofloxacin) 0.25 µg/mL |
||
|---|---|---|---|---|
| 250 μg/mL | 500 μg/mL | 1000 μg/mL | ||
| Inhibition Zone Diameter (mm) | ||||
| E. coli | 0 | 10 ± 0.25 | 32 ± 1.4 | 92.47 ± 2.3 |
| B. subtilis | 0 | 12 ± 0.32 | 35 ± 1.3 | 91 ± 3.1 |
| S. aureus | 0 | 0 | 25 ± 1.2 | 94 ± 2.4 |
| P. aeruginosa | 0 | 0 | 26 ± 1.1 | 94 ± 3.1 |
| S. typhi | 0 | 0 | 20 ± 1.5 | 95 ± 2.3 |
Computational Analysis
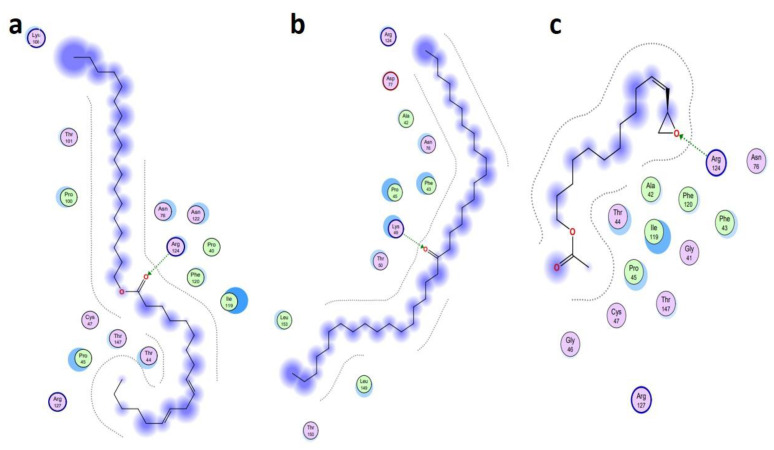
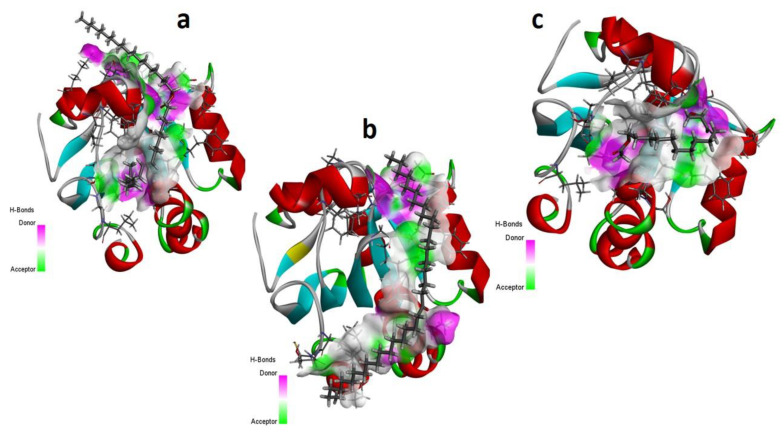
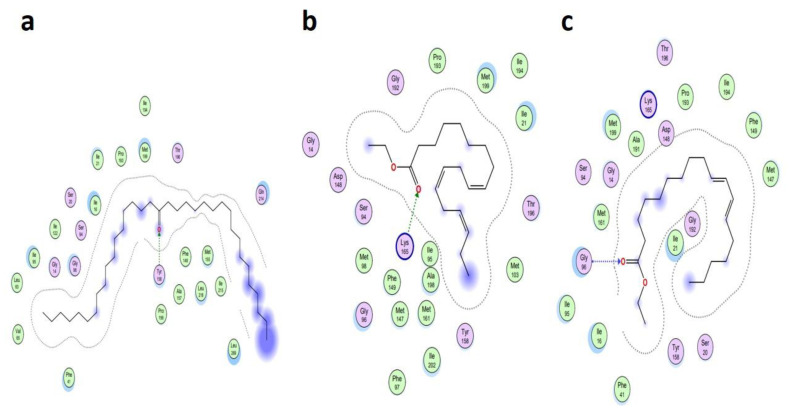
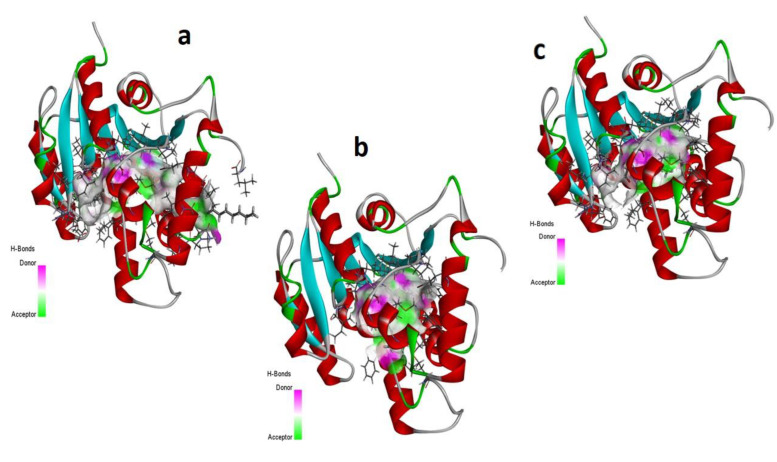
The MD investigations of the selected ligands with two target proteins were performed by MOE software. The binding energies of the selected ligands within the best binding pose were studied and it was observed that four ligands presented good results in terms of their binding interactions and binding energies in kcal/mol. A total of two best- bounded ligands in the vicinity of the active binding site of both the target proteins are shown in Figure 7, Figure 8, Figure 9 and Figure 10. The library of the 23 compounds was docked, and the three top virtual hits, with the antioxidant protein [PDB ID: 1HD2] [28] and antibacterial protein [PDB ID: 4TZK] [29] from the MD results, significantly presented a correlation with the experimental analysis of the whole Urtica dioica L. plant’s essential oil compounds. Hence, the two-dimensional interaction plots and three-dimensional presentation of the hydrogen binding pocket are shown in Figure 7, Figure 8, Figure 9 and Figure 10.
Figure 7.
Interaction plot of top three scored docked ligands CID_ 22287839 (a), CID_10440 (b), and CID_ 5363633 (c), within active site of antioxidant protein [PDB ID: 1HD2], in the vicinity of 4Å.
Figure 8.
Hydrogen bonding capacity of top three scored docked ligands CID_ 22287839 (a), CID_10440 (b), and CID_5363633 (c), within the active site of antioxidant protein [PDB ID: 1HD2]. Purple colored area presents the hydrogen bond donor capacity and the green colored area presents the hydrogen bond acceptor capacity.
Figure 9.
Interaction plot of top three scored docked ligands CID_10440 (a), CID_5367460 (b), and CID_22371644 (c), within active site of antibacterial protein [PDB ID: 4TZK], in the vicinity of 4Å.
Figure 10.
Hydrogen bonding capacity of top three scored docked ligands CID_10440 (a), CID_5367460 (b), and CID_22371644 (c), within the active site of antibacterial protein [PDB ID: 4TZK]. Purple colored area presents the hydrogen bond donor capacity and the green colored area presents the hydrogen bond acceptor capacity.
Table 5 presents the summary of the interaction analysis, the binding energies of the best-bounded conformation of the ligands with the target proteins in kcal/mol, and the interaction plot of top three best-docked ligands. 9-Octadecenoic acid (Z)-, 9-octadecenyl ester, (Z) [CID_22287839] presented the best-bounded conformation at −6.1991 kcal/mol, 18-Pentatriacontanone [CID_10440] presented the best-bounded conformation at −5.7512 kcal/mol, and Z-(13, 14-Epoxy) tetradec-11-en-1-ol acetate [CID_5363633] presented the best-bounded conformation at −5.2222 kcal/mol, within the active site of the antioxidant protein [PDB ID: 1HD2] and in the vicinity of 4Å (Figure 7 and Figure 8). Each color presents the type of interacting residues, and the hydrogen bonds are highlighted in each figure. In the interaction plot of the top three best-docked ligands, 18-Pentatriacontanone [CID_10440] presented the best-bounded conformation at −8.2366 kcal/mol, Ethyl 9, 12, 15-octadecatrienoate [CID_5367460] presented the best-bounded conformation at −7.8228 kcal/mol, and 9, 12-Octadecadienoic acid, ethyl ester [CID_22371644] presented the best-bounded conformation at −7.7674 kcal/mol, within the active site of the antibacterial protein [PDB ID: 4TZK] and in the vicinity of 4Å (Figure 9 and Figure 10).
Table 5.
Summary of interaction analysis of the two best virtual hits.
| Ligand Name | Binding Energy (Kcal/mol) | Binding Interaction | |||
|---|---|---|---|---|---|
| Interacting Residues | Interaction Type |
Bond Distance |
Bond Energy (Kcal/mol) | ||
| Antioxidant protein [PDB ID: 1HD2] | |||||
| 9-Octadecenoic acid (Z)-, 9-octadecenyl ester, (Z) [CID_22287839] |
−6.1991 | O2–NH2 ARG 124 (A) | H-acceptor | 3.15 | −2.0 |
| 18-Pentatriacontanone [CID_10440] |
−5.7512 | O1–NZ LYS 49 (A) | H-acceptor | 2.92 | −6.4 |
| Z-(13, 14-Epoxy) tetradec-11-en-1-ol acetate [CID_5363633] |
−5.2222 | O1–NH2 ARG 124 (A) | H-acceptor | 3.05 | −0.5 |
| Antibacterial protein [PDB ID: 4TZK] | |||||
| 18-Pentatriacontanone [CID_10440] |
−8.2366 | O1–OH TYR 158 (A) | H-acceptor | 2.93 | −2.3 |
| Ethyl 9, 12, 15-octadecatrienoate [CID_5367460] |
−7.8228 | O2–NZ LYS 165 (A) | H-acceptor | 3.16 | −1.1 |
| 9, 12-Octadecadienoic acid, ethyl ester [CID_22371644] |
−7.7674 | O2–N GLY 96 (A) | H-acceptor | 3.33 | −1.8 |
The reported in silico applied protocols highlighted the standing of the ADMET profile for the short-listing of large chemical libraries, in order to justify the potential drug-like hits that can be tolerable in the design and development of novel drugs [34]. The ADMET justifications with the SwissADME and Datawarrior tools validated the properties of the selected hits, such as molecular weight (MW), partition coefficient/lipophilic parameters (logP values), hydrogen bond acceptor (HBA), hydrogen bond donor (HDB), total polar surface area (TPSA), molar refractivity (MR), and rotatable bond (RB), which are vital drug-like characteristics that are considered for the selected hits. As preliminary drug discovery protocols, they endorse estimations of drug-likeness, water solubility, pharmacokinetics, and toxicity estimations, along with medicinal chemistry perceptions, as highlighted for the selected potential hits in Table 6.
Table 6.
Computational protocols applied for ADMET profile.
| Chemical Parameters | 9-Octadecenoic Acid (Z)-, 9-Octadecenyl Ester, (Z) | 18-Pentatriacontanone | Z-(13, 14-Epoxy) tetradec-11-en-1-ol Acetate | Ethyl 9, 12, 15-Octadecatrienoate | 9, 12-Octadecadienoic Acid, Ethyl Ester |
|---|---|---|---|---|---|
| Physicochemical Properties | |||||
| Molecular weight (MW) (g/mol) | 532.92 | 506 | 268.39 | 306.48 | 308.50 |
| Rotatable bonds | 32 | 32 | 13 | 15 | 16 |
| Hydrogen bond acceptors (HBA) | 2 | 1 | 3 | 2 | 2 |
| Hydrogen bond donors (HBD) | 0 | 0 | 0 | 0 | 0 |
| Molar Refractivity (MR) | 175.50 | 170.56 | 78.81 | 98.12 | 98.59 |
| Total polar surface area (TPSA) (Å) | 26.30 | 17.07 | 38.83 | 26.30 | 26.30 |
| Bioavailability Score | 0.17 | 0.17 | 0.55 | 0.55 | 0.55 |
| Lipophilicity | |||||
| Log Po/w (iLOGP) | 8.73 | 8.68 | 3.44 | 4.82 | 5.01 |
| Water Solubility | |||||
| Class | Insoluble | Insoluble | Soluble | Moderatley soluble | Moderatley soluble |
| Pharmacokinetics | |||||
| GI absorption | Low | Low | High | High | High |
| BBB permeant | No | No | Yes | No | No |
| P-gp substrate | Yes | Yes | No | No | No |
| CYP1A2 inhibitor | No | No | Yes | Yes | Yes |
| CYP2C19 Inhibitor | No | No | No | No | No |
| CYP2C9 inhibitor | No | No | Yes | Yes | Yes |
| CYP2D6 inhibitor | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | No | No |
| Log Kp (skin permeation) (cm/s) | 1.61 | 2.51 | −4.74 | −3.44 | −2.79 |
| Toxicity estimation | |||||
| Mutagenic | No | No | Yes | No | No |
| Tumorigenic | No | No | Yes | No | No |
| Reproductive effects | No | No | No | No | No |
| Irritant effects | No | No | Yes | No | No |
| Medicinal chemistry-related properties | |||||
| PAINS | No | No | No | No | No |
| Brenk | 1 t: isolated_alkene | No | 3: Three-membered_heterocycle, isolated_alkene | 1: isolated_alkene | 1: polyene |
| Synthetic accessibility | 5.29 | 4.57 | 3.60 | 3.26 | 3.53 |
Not all the physicochemical properties of the virtual hits are in the acceptable range, and have some violations of Lipinski [42] and Veber’s [43] theory of drug-likeness because of a large MW. The lipophilicity and water solubility class also presented very good outcomes for Z-(13, 14-Epoxy) tetradec-11-en-1-ol acetate, Ethyl 9, 12, 15-octadecatrienoate, and 9, 12-Octadecadienoic acid, ethyl ester. The gastrointestinal drug absorption (GI-DA) [44] and blood–brain barrier (BBB) permeability [44] were also calculated for the selected five hits. The CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4 inhibitory potential was estimated, and it was observed that Z-(13, 14-Epoxy) tetradec-11-en-1-ol acetate, Ethyl 9, 12, 15-octadecatrienoate, and 9, 12-Octadecadienoic acid, ethyl ester is CYP1A2 and CYP2C9 inhibitors, although both compounds, 9-Octadecenoic acid (Z)-, 9-octadecenyl ester, (Z) and 18-Pentatriacontanone are P-glycoprotein (P-gp) substrates. The Log Kp (skin permeation) values were high for all the hits, and the PAINS alert and Brenk alert that were supported by the medicinal chemistry parameter evaluation [30] showed minor violations, which provides suggestions to improve the structures’ functionality and activity before moving a drug to the next phase of development. Synthetically, all the hits were highly accessible, with high scores, although minor toxicity was presented for one hit, Z-(13, 14-Epoxy) tetradec-11-en-1-ol acetate. Hence, the selected hits presented moderate drug-like activities.
4. Conclusions
The present study was conducted on U. dioica essential oil, which resulted in the identification of 22 bioactive compounds. For this identification, a GC–MS analysis was carried out. However, this study was not conducted with an individual compound. The whole U. dioica plant’s essential oil revealed potential antioxidant and antimicrobial activities in this study, potentially for the first time using the GC–MS-derived bioactive compounds of the U. dioica plant’s essential oil. The most significant outcome of this study was to reveal the phytotoxic effect of these essential oils for the first time. Moreover, future research could surely contribute to the selection of an individual compound obtained from the essential oil of U. dioica. The MD investigations assisted the experimental studies with respect to the protein–ligand binding conformation representations, in terms of the binding affinity and docked score, and helps in understanding the mechanism, while the ADMET estimation helps to justify the drug-like characteristics. Hence, these hits are recommended for clinical investigations, and it is expected that, in drug development, our results on antioxidant and antibacterial hits could surely contribute to the selection of significant drug candidates obtained from the essential oil of U. dioica.
Author Contributions
Conceptualization M.Z.K., A.K.A. and S.J.; methodology, M.Z.K. and S.J.; software, M.Z.K., S.J. and S.B. validation, A.K.A. and S.B.; formal analysis, M.Z.K., S.J., M.S., A.K.A. and S.B.; investigation, M.Z.K., S.J. and S.B.; resources, M.Z.K., S.J. and A.M.S.A.M.; data curation, A.K.A. and A.M.S.A.M.; writing—original draft preparation, A.K.A., M.Z.K., S.J. and S.B.; review and editing, A.K.A., S.B., G.M.A., N.A.T.N., J.A.A. and M.M.A.-D.; visualization, A.K.A.; supervision, M.Z.K., S.J. and M.S.; project administration, M.Z.K. and S.J. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be available upon request. The data are not publicly available due to university research and management policy.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R30), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Motamedi H., Seyyednejad S.M., Bakhtiari A., Vafaei M. Introducing Urtica dioica, a native plant of Khuzestan, as an antibacterial medicinal plant. Jundishapur J. Nat. Pharm. Prod. 2014;9:e15904. doi: 10.17795/jjnpp-15904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kregiel D., Pawlikowska E., Antolak H. Urtica spp.: Ordinary plants with extraordinary properties. Molecules. 2018;23:1664. doi: 10.3390/molecules23071664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu L.-F., Monro A.K., Wen F., Xin Z.-B., Wei Y.-G., Zhang Z.-X. The rediscovery and delimitation of Elatostemasetulosum WT Wang (Urticaceae) PhytoKeys. 2019;126:79. doi: 10.3897/phytokeys.126.35707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahangarpour A., Mohammadian M., Dianat M. Antidiabetic effect of hydroalcholic Urtica dioica leaf extract in male rats with fructose-induced insulin resistance. Iran. J. Med. Sci. 2012;37:181. [PMC free article] [PubMed] [Google Scholar]
- 5.Christaki E., Bonos E., Giannenas I., Florou-Paneri P. Aromatic plants as a source of bioactive compounds. Agriculture. 2012;2:228–243. doi: 10.3390/agriculture2030228. [DOI] [Google Scholar]
- 6.Mittman P. Randomized, double-blind study of freeze-dried Urtica dioica in the treatment of allergic rhinitis. Planta Med. 1990;56:44–47. doi: 10.1055/s-2006-960881. [DOI] [PubMed] [Google Scholar]
- 7.Durak I., Biri H., Devrim E., Sözen S., Avcı A. Aqueous extract of Urtica dioica makes significant inhibition on adenosine deaminase activity in prostate tissue from patients with prostate cancer. Cancer Biol. Ther. 2004;3:855–857. doi: 10.4161/cbt.3.9.1038. [DOI] [PubMed] [Google Scholar]
- 8.Farzami B., Ahmadvand D., Vardasbi S., Majin F.J., Khaghani S. Induction of Insulin Secretion by a Component of Urtica dioica Leaves Extract in Perfused Islets of Langehans and its Vivo Effects in normal and Streptozotocin Diabetic Rats. J. Ethnopharmacol. 2003;89:47–53. doi: 10.1016/S0378-8741(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 9.Asghar N., Naqvi S.A.R., Hussain Z., Rasool N., Khan Z.A., Shahzad S.A., Sherazi T.A., Janjua M.R.S.A., Nagra S.A., Zia-Ul-Haq M., et al. Compositional difference in antioxidant and antibacterial activity of all parts of the Carica papaya using different solvents. Chem. Cent. J. 2016;10:5. doi: 10.1186/s13065-016-0149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riehemann K., Behnke B., Schulze-Osthoff K. Plant extracts from stinging nettle (Urtica dioica), an antirheumatic remedy, inhibit the proinflammatory transcription factor NF-κB. FEBS Lett. 1999;442:89–94. doi: 10.1016/S0014-5793(98)01622-6. [DOI] [PubMed] [Google Scholar]
- 11.Obertreis B., Giller K., Teucher T., Behnke B., Schmitz H. Anti-inflammatory effect of Urtica dioica folia extract in comparison to caffeic malic acid. Arzneimittelforschung. 1996;46:52–56. [PubMed] [Google Scholar]
- 12.Bnouham M., Merhfour F.-Z., Ziyyat A., Mekhfi H., Aziz M., Legssyer A. Antihyperglycemic activity of the aqueous extract of Urtica dioica. Fitoterapia. 2003;74:677–681. doi: 10.1016/S0367-326X(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 13.Tahri A., Yamani S., Legssyer A., Aziz M., Mekhfi H., Bnouham M., Ziyyat A. Acute diuretic, natriuretic and hypotensive effects of a continuous perfusion of aqueous extract of Urtica dioica in the rat. J. Ethnopharmacol. 2000;73:95–100. doi: 10.1016/S0378-8741(00)00270-1. [DOI] [PubMed] [Google Scholar]
- 14.Hadizadeh I., Peivastegan B., Kolahi M. Antifungal activity of nettle (Urtica dioica L.), colocynth (Citrullus colocynthis L. Schrad), oleander (Nerium oleander L.) and konar (Ziziphus spina-christi L.) extracts on plants pathogenic fungi. Pak. J. Biol. Sci. 2009;12:58–63. doi: 10.3923/pjbs.2009.58.63. [DOI] [PubMed] [Google Scholar]
- 15.Gül S., Demirci B., Başer K.H.C., Akpulat H.A., Aksu P. Chemical composition and in vitro cytotoxic, genotoxic effects of essential essential oil from Urtica dioica L. Bull. Environ. Contam. Toxicol. 2012;88:666–671. doi: 10.1007/s00128-012-0535-9. [DOI] [PubMed] [Google Scholar]
- 16.Fahad K., Hani S., Walid M., Fouad A.S. Efficient utilization of aquaculture effluents to maximize plant growth, yield, and essential essential oil s composition of Origanum majorana Annals of Agricultural cultivation. Sciences. 2021;66:1–7. [Google Scholar]
- 17.Huda J.A., Mohammed Y.H., Imad H.H. Phytochemical analysis of Urtica dioica leaves by fourier-transform infrared spectroscopy and gas chromatography-mass spectrometry. J. Pharm. Phytoth. 2015;7:238–252. [Google Scholar]
- 18.Gülçin I., Küfrevioǧlu Ö.I., Oktay M., Büyükokuroǧlu M.E. Antioxidant, antimicrobial, antiulcer and analgesic activities of nettle (Urtica dioica L.) J. Ethnopharmacol. 2004;90:205–215. doi: 10.1016/j.jep.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Mzid M., Ben Khedir S., Ben Salem M., Regaieg W., Rebai T. Antioxidant and antimicrobial activities of ethanol and aqueous extracts from Urtica urens. Pharm. Biol. 2017;55:775–781. doi: 10.1080/13880209.2016.1275025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Körpe D.A., İşerİ Ö.D., Sahin F.I., Cabi E., Haberal M. High-antibacterial activity of Urtica spp. seed extracts on food and plant pathogenic bacteria. Int. J. Food Sci. Nutr. 2013;64:355–362. doi: 10.3109/09637486.2012.734290. [DOI] [PubMed] [Google Scholar]
- 21.Ranjbar R., Shamami K.S. Evaluation of antimicrobial activity of Eucalyptus essential essential oil and Urtica alcoholic extract on Salmonella enteritidis and Shigella dysenteriae in vitro conditions. Bull. Environ. Pharmacol. Life Sci. 2015;4:56–59. [Google Scholar]
- 22.Salehzadeh A., Asadpour L., Naeemi A.S., Houshmand E. Antimicrobial activity of methanolic extracts of Sambucus ebulus and Urtica dioica against clinical isolates of methicillin resistant Staphylococcus aureus. Afr. J. Tradit. Complement. Altern. Med. 2014;11:38–40. doi: 10.4314/ajtcam.v11i5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bibi S., Hasan M.M., Wang Y.-B., Papadakos S.P., Yu H. Cordycepin as a Promising Inhibitor of SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) Curr. Med. Chem. 2022;29:152–162. doi: 10.2174/0929867328666210820114025. [DOI] [PubMed] [Google Scholar]
- 24.Khan M.S., Mehmood B., Yousafi Q., Bibi S., Fazal S., Saleem S., Sajid M.W., Ihsan A., Azhar M., Kamal M.A. Molecular docking studies reveal rhein from rhubarb (Rheum rhabarba-rum) as a putative inhibitor of atp-binding cassette super family g member 2. Med. Chem. 2021;17:273–288. doi: 10.2174/1573406416666191219143232. [DOI] [PubMed] [Google Scholar]
- 25.Saleem U., Shehzad A., Shah S., Raza Z., Shah M.A., Bibi S., Chauhdary Z., Ahmad B. Antiparkinsonian activity of Cucurbita pepo seeds along with possible underlying mechanism. Metab. Brain Dis. 2021;36:1231–1251. doi: 10.1007/s11011-021-00707-6. [DOI] [PubMed] [Google Scholar]
- 26.Mendelsohn L.D. ChemDraw 8 ultra, windows and macintosh versions. J. Chem. Inf. Comput. Sci. 2004;44:2225–2226. doi: 10.1021/ci040123t. [DOI] [Google Scholar]
- 27.Declercq J.-P., Evrard C., Clippe A., Vander Stricht D., Bernard A., Knoops B. Crystal structure of human peroxiredoxin 5, a novel type of mammalian peroxiredoxin at 1.5 Åresolution. J. Mol. Biol. 2001;311:751–759. doi: 10.1006/jmbi.2001.4853. [DOI] [PubMed] [Google Scholar]
- 28.He X., Alian A., Stroud R., de Montellano P.R. Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis. J. Med. Chem. 2006;49:6308–6323. doi: 10.1021/jm060715y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibi S., Sakata K. Current Status of Computer-Aided Drug Design for Type 2 Diabetes. Curr. Comput. Aided-Drug Des. 2016;12:167–177. doi: 10.2174/1573409912666160426120709. [DOI] [PubMed] [Google Scholar]
- 30.Bibi S., Sakata K. An Integrated Computational Approach for Plant-Based Protein Tyrosine Phosphatase Non-Receptor Type 1 Inhibitors. Curr. Comput. Aided Drug Des. 2017;13:319–335. doi: 10.2174/1573409913666170406145607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilar S., Cozza G., Moro S. Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery. Curr. Top. Med. Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 32.Saleem U., Bibi S., Shah M.A., Ahmad B., Saleem A., Chauhdary Z., Anwar F., Javaid N., Hira S., Akhtar M.F., et al. Anti-Parkinson’s evaluation of Brassica juncea leaf extract and underlying mechanism of its phytochemicals. Front. Biosci.-Landmark. 2021;26:1031–1051. doi: 10.52586/5007. [DOI] [PubMed] [Google Scholar]
- 33.Yousafi Q., Batool J., Khan M.S., Perveen T., Sajid M.W., Hussain A., Mehmood A., Saleem S. In Silico Evaluation of Food Derived Bioactive Peptides as Inhibitors of Angiotensin Converting Enzyme (ACE) Int. J. Pept. Res. Ther. 2021;27:341–349. doi: 10.1007/s10989-020-10090-y. [DOI] [Google Scholar]
- 34.Khan M., Patujo J., Mushtaq I., Ishtiaq A., Tahir M.N., Bibi S., Khan M.S., Mustafa G., Mirza B., Badshah A., et al. Anti-diabetic potential, crystal structure, molecular docking, DFT, and optical-electrochemical Studies of new Dimethyl and Diethyl Carbamoyl-N, N’-disubstituted based thioureas. J. Mol. Struct. 2021;1253:132207. doi: 10.1016/j.molstruc.2021.132207. [DOI] [Google Scholar]
- 35.Zeb M.A., Rahman T.U., Sajid M., Xiao W., Musharraf S.G., Bibi S., Akitsu T., Liaqat W. GC-MS Analysis and In Silico Approaches of Indigofera heterantha Root Essential oil Chemical Constituents. Compounds. 2021;1:116–124. doi: 10.3390/compounds1030010. [DOI] [Google Scholar]
- 36.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sander T., Freyss J., Von Korff M., Rufener C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015;55:460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 38.Kukrić Z.Z., Topalić-Trivunović L.N., Kukavica B.M., Matoš S.B., Pavičić S.S., Boroja M.M., Savić A. V Characterization of antioxidant and antimicrobial activities of nettle leaves (Urtica dioica L.) Acta Period. Technol. 2012;43:257–272. doi: 10.2298/APT1243257K. [DOI] [Google Scholar]
- 39.Chaqroune A., Taleb M. Effects of Extraction Technique and Solvent on Phytochemicals, Antioxidant, and Antimicrobial Activities of Cultivated and Wild Rosemary (Rosmarinus officinalis L.) from Taounate Region. Biointerface Res. Appl. Chem. 2022;12:8441–8452. [Google Scholar]
- 40.Gul H., Naseer R.D., Abbas I., Khan E.A., Rehman H.U., Nawaz A., Abdel-Daim M.M. The Therapeutic Application of Tamarix aphylla Extract Loaded Nanoemulsion Cream for Acid-Burn Wound Healing and Skin Regeneration. Medicina. 2022;59:34. doi: 10.3390/medicina59010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zenão S., Aires A., Dias C., Saavedra M.J., Fernandes C. Antibacterial potential of Urtica dioica and Lavandula angustifolia extracts against methicillin resistant Staphylococcus aureus isolated from diabetic foot ulcers. J. Herb. Med. 2017;10:53–58. doi: 10.1016/j.hermed.2017.05.003. [DOI] [Google Scholar]
- 42.Jyoti K., Baunthiyal M., Singh A. Characterization of silver nanoparticles synthesized using Urtica dioica Linn. leaves and their synergistic effects with antibiotics. J. Radiat. Res. Appl. Sci. 2016;9:217–227. doi: 10.1016/j.jrras.2015.10.002. [DOI] [Google Scholar]
- 43.Zeroual A., Sakar E.H., Eloutassi N., Mahjoubi F., Chaouch M., Chaqroune A. Wild chamomile [Cladanthus mixtus (L.) chevall.] collected from central-northern Morocco: Phytochemical profiling, antioxidant, and antimicrobial activities. Biointerface Res. Appl. Chem. 2021;11:11440–11457. [Google Scholar]
- 44.Khan A., Hussain S., Ahmad S., Suleman M., Bukhari I., Khan T., Wei D.Q. Computational modelling of potentially emerging SARS-CoV-2 spike protein RBDs mutations with higher binding affinity towards ACE2: A structural modelling study. Comput. Biol. Med. 2022;141:105163. doi: 10.1016/j.compbiomed.2021.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon request. The data are not publicly available due to university research and management policy.