Abstract
Evidence from the literature suggests an association between the microbiome and asthma development. Here, we aimed to identify the current evidence for the association between asthma and the upper airway, lower airway and/or the gut microbiome. An electronic systemic search of PubMed, EBSCO, Science Direct and Web of Science was conducted until February 2022 to identify the eligible studies. The Newcastle–Ottawa Scale and the Systematic Review Centre for Laboratory Animal Experimentation risk of the bias tools were used to assess quality of included studies. Twenty-five studies met the inclusion criteria. Proteobacteria and Firmicutes were identified as being significantly higher in the asthmatic children compared with the healthy controls. The high relative abundance of Veillonella, Prevotella and Haemophilus in the microbiome of the upper airway in early infancy was associated with a higher risk of asthma development later in life. The gut microbiome analyses indicated that a high relative abundance of Clostridium in early childhood might be associated with asthma development later in life. The findings reported here serve as potential microbiome signatures associated with the increased risk of asthma development. There is a need for large longitudinal studies to further identify high-risk infants, which will help in design strategies and prevention mechanisms to avoid asthma early in life.
Keywords: asthma, children, dysbiosis, gut microbiome, immunity, lung microbiome
1. Introduction
Asthma is a chronic inflammatory disease that affects the respiratory system and leads to significant morbidity and mortality [1]. Individuals suffering from asthma exhibit an array of symptoms, from wheezing and coughing to chest tightness and shortness of breath [2]. These manifestations vary in time of onset and intensity between asthmatic patients [2]. The common triggers that may lead to asthma exacerbation include, but are not limited to, viral respiratory infections, air pollution, tobacco smoke and exercise [3]. Allergies, genetics, respiratory infections during infancy and environmental features are risk factors for asthma development [3]. However, the exact aetiology of asthma is not well understood.
Evidence from the literature suggests that there is an association between the human microbiome and the development of asthma [4]. Both human studies and studies performed on experimental animal models have linked the dysbiosis of the early-life gut microbiome to a greater risk for the development of asthma in individuals who are genetically susceptible to this disease [4,5,6,7]. The gut microbiome has been shown to regulate the immune responses associated with chronic inflammatory diseases in humans and animal models [5,8]. The establishment of the human gut microbiome starts at birth and is influenced by many factors, including the mode of delivery, antibiotic use and the feeding method [9]. The human gut microbiome consists of not only bacteria but also fungi, protozoa and viruses [10]. The microbiome is dynamic and withstands changes due to age, dietary modifications and environmental and medical interventions, such as the use of antimicrobial agents, throughout an individual’s lifetime [10]. While most of the normal gut microbiome is composed of the phyla Firmicutes and Bacteroidetes, the less common phyla are Actinobacteria, Proteobacteria, Fusobacteria and Verrucomicrobia [10,11]. The most prevalent genera in the normal gut microbiome are Bacteroides, Faecalibacterium and Bifidobacterium [10,11]. The healthy fungal gut microbiome consists mainly of Saccharomyces cerevisiae, Malassezia restricta and Candida albicans [12].
On the other hand, data on the relationship between the lung microbiome and asthma remain limited. This is mainly due to the difficulty of sampling and the long-standing dogma about the lungs being a sterile environment [13]. However, studies have identified the normal lung microbiome, which includes the bacterial genera Prevotella, Streptococcus, Veillonella, Neisseria, Haemophilus and Fusobacterium [13]. Fungal microbiomes in healthy lungs include mainly Ascomycota (Aspergillus, Cladosporium, Eremothecium and Vanderwaltozyma) and Microsporidia (Systenostrema) [14,15].
In addition, emerging evidence confirms a crosstalk at what is termed the ‘gut–lung axis’, where changes in the gut microbiome may have an impact on the development of lung diseases and vice versa [16,17]. This occurs via the mesenteric lymph nodes, where elements of the microbiome and their metabolites are transported to and from the lungs [18]. Discrepancies in the gut–lung axis are associated with an increased emergence of asthma as well as other acute and chronic respiratory diseases [19].
This systematic review fills the knowledge gaps regarding the association between asthma and the upper airway, lower airway and/or gut microbiome, which has not been specifically addressed previously. In fact, the published systematic reviews have mostly investigated the association between the gut microbiota and asthma or allergic diseases without including the upper and lower airways. In 2018, Zimmerman and colleagues systematically reviewed the intestinal microbiota composition and the development of allergic diseases from birth to 20 years of age [20]. The authors reported that early-life gut microbial exposure indeed has a role in allergic disease development [20]. Melli and colleagues in 2015 examined the early literature (2007–2013) on the link between the gut microbiota and allergic diseases in children and reached a similar conclusion [21]. Nonetheless, the majority of the studies included in the above-mentioned reviews [20,21] utilised traditional bacterial cultures and polymerase chain reaction (PCR) techniques to characterise the gut microbiota composition and specifically studied the intestinal microbiota–allergy association. A more recent systematic review, in which the authors retrieved studies that utilised genomic sequencing to measure the microbiome composition and diversity, explored the link between the intestinal microbiome and respiratory diseases (including asthma) [22]. The authors highlighted that disruptions in gut microbiota composition alone might not directly lead to respiratory diseases and there is a need for large longitudinal studies [22]. The main objective of the current systematic review was therefore to identify the current evidence for the association between asthma and the upper airway, lower airway and/or gut microbiome in humans and in animals. This study intended to determine the upper airway, lower airway and gut microbiome characteristics commonly associated with asthma. Hence, the findings of this study might have an impact on our understanding of the potential role of the microbiome in asthma development.
2. Materials and Methods
We initially performed a non-systematic search within relevant journals for asthma and microbiomes to identify the existing systematic reviews related to these topics. However, the available systematic reviews were generally limited to upper airway or gut microbiome investigations in humans and paid little attention to the lower airway microbiome and animal-based studies. The current review was developed based on the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [23]. The study team consisted of researchers with experience in microbiology, immunology and respiratory care. It also included a researcher with experience in systematic reviews who was familiar with searching variable databases. The Covidence software from Veritas Health Innovation, Melbourne, Australia (available at https://www.covidence.org/ and accessed on 31 January 2023), was used to manage the retrieved studies, track the status of each study and update the PRISMA flow diagram.
2.1. Eligibility Criteria
The eligibility criteria consisted of original articles published in English between inception and February 2022 that addressed asthma diagnosis as an outcome among children up to 18 years old and investigated microbial communities in the upper airway, lower airway or the gut in humans or animals. Studies that addressed asthma diagnosis as a subgroup analysis were also included. The exclusion criteria consisted of studies that examined environmental and/or pollutant microbiomes and asthma or that reported asthma symptoms and/or atopic/allergy diseases without an asthma diagnosis.
2.2. Information Sources and Search Strategy
We comprehensively searched the following major electronic databases from 3 to 5 March 2022: PubMed, EBSCO, Science Direct and Web of Science. The search strategy was applied as appropriate for each database. The general search keywords used were: (asthma) AND (microbiome OR dysbiosis OR microbiota). The following filters were applied: age (up to 18 years), language (English) and literature type (original/academic journals). More details on the search strategy are provided in Supplementary File S1.
2.3. Selection and Data-Collection Process
All studies were imported to EndNote version X9 and then uploaded to Covidence software. After duplicates were removed, two stages of screening were conducted. First, two independent reviewers screened the titles and abstracts of the imported studies. Second, two independent reviewers conducted full-text screenings for the studies included during the first stage of screening. Finally, independent reviewers performed data extraction based on a data collection form designed specifically to address the objectives of this review (Supplementary Materials Table S1). Conflicts in the screening stages and the data collection process were resolved through regular discussion meetings with all authors.
2.4. Data Items
The data collection form (Supplementary Materials Table S1) included the following variables that were extracted from each study: the citation and title of the article, the country where the study was conducted, the study type (human or animal based), the study design, the sample size for each group, the age for each group, the microbiome environment (the upper airway, lower airway and/or the gut), the type of specimen collected for the microbiome analysis, the time of specimen collection (one time point or different time points), the microbiome detection method, the genomic DNA extraction method, the sequencing platform used, the microbial community diversity assessment (α-diversity, β-diversity, or both), the bioinformatics pipeline used and the study findings.
2.5. Risk of Bias Assessment
The quality of the included human non-randomised studies was assessed using Newcastle–Ottawa Scale (NOS) tools adapted for each study’s design. Three tools were used: (1) the NOS adapted for cross-sectional studies [24], (2) the NOS for case-control studies and (3) the NOS for cohort studies. The NOS tools were used to assess quality based on different items categorised into three domains (selection, comparability and exposure or outcome). Then, the quality of each study was rated as good, fair or poor by translating the results of the NOS to the Agency for Health Research and Quality standards, as described previously [22]. For animal intervention studies, the Systematic Review Centre for Laboratory Animal Experimentation (SYRCLE) risk of bias tool was used [25]. Details of the tools used are described in Supplementary Materials Table S2.
2.6. Synthesis Methods
Due to the nature of the present systematic review, the descriptive data were extracted using a data collection tool that was generated specifically to address the objective of this review (Supplementary Materials Table S1).
3. Results
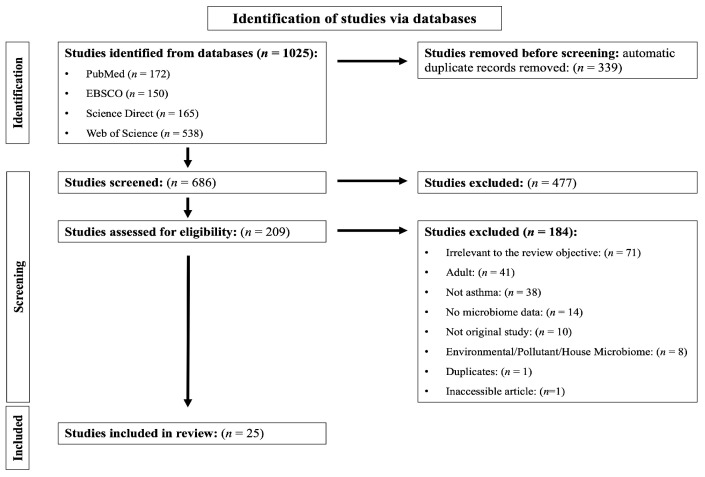
The literature search resulted in a total of 1025 studies, which were uploaded to Covidence. After the duplicates were automatically removed (n = 339), 686 studies remained. The titles and abstracts were screened, as a result of which 477 studies were considered irrelevant to the aim of the current review and excluded. The full text of the remaining 209 studies was examined for eligibility. As a result, 184 were excluded for the reasons detailed in Figure 1. The screening phase resulted in 25 studies that met the inclusion criteria and were identified as eligible for inclusion in the present review.
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flowchart describing the studies excluded and analysed in the current systematic review.
3.1. Quality of the Included Studies
Table 1, Table 2 and Table 3 show the quality assessment results of the included human studies (n = 22) based on the NOT criteria for case-control, cohort and cross-sectional studies, respectively. Sixteen human studies out of twenty-two were classified as good quality [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41], four were classified as fair quality [42,43,44,45] and only two were classified as poor quality [13,46]. The limitations were generally related to the potential selection bias. The quality evaluation for the animal intervention studies (three out of twenty-five) is described in Table 4. The three animal intervention studies [47,48,49] generally indicated the potential performance and detection bias in aspects specifically related to the blinding procedures.
Table 1.
Newcastle–Ottawa Scale for the human case-control studies.
| Citation | Selection | Comparability | Exposure | Total | Rate | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Is the Case (Asthma) Definition Adequate? | Representativeness of the Cases | Selection of Controls |
Definition of Controls |
Comparability of Cases and Controls on the Basis of the Design or Analysis |
Ascertainment of Exposure | Same Method of Ascertainment for Cases and Controls |
Non-Response Rate | |||
| [26] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | Good |
| [46] | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | Poor |
| [27] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | Good |
| [42] | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 6 | Fair |
| [28] | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 8 | Good |
| [43] | 1 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 6 | Fair |
| [44] | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 6 | Fair |
Table 2.
Newcastle–Ottawa Scale for the human cohort studies.
| Citation | Selection | Comparability | Outcome | Total | Rate | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Representativeness of the Exposed Cohort | Selection of the Non-Exposed Cohort | Ascertainment of Exposure to Implants | Demonstration That Outcome of Interest (Asthma) Was Not Present at Start of Study | Comparability of Cohorts on the Basis of the Design or Analysis | Assessment of Outcome | Was Follow Up Long Enough for Outcome to Occur | Adequacy of Follow-Up of Cohorts |
|||
| [29] | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 8 | Good |
| [30] | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 9 | Good |
| [31] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | Good |
| [32] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | Good |
| [45] | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 6 | Fair |
Table 3.
Newcastle–Ottawa Scale for human cross-sectional studies.
| Citation | Selection | Comparability | Outcome | Total | Rate | ||||
|---|---|---|---|---|---|---|---|---|---|
| Representativeness of the Sample | Sample Size | Non-Respondents | Ascertainment of the Exposure | The Subjects in Different Outcome Groups Are Comparable, Based on the Study Design or Analysis. Confounding Factors Are Controlled | Assessment of Outcome | Statistical Test | |||
| [33] | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 8 | Good |
| [34] | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 8 | Good |
| [35] | 1 | 0 | 1 | 2 | 2 | 1 | 1 | 8 | Good |
| [36] | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 7 | Good |
| [37] | 1 | 0 | 1 | 2 | 2 | 2 | 1 | 9 | Good |
| [38] | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 8 | Good |
| [13] | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Poor |
| [39] | 1 | 1 | 0 | 1 | 2 | 2 | 1 | 8 | Good |
| [40] | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 7 | Good |
| [41] | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 7 | Good |
Table 4.
The systematic review centre for the laboratory animal experimentation risk of the bias assessment tool for animal studies.
| Citation | Selection Bias | Performance Bias | Detection Bias | Attrition Bias | Reporting Bias | Other | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sequence Generation | Baseline Characteristics | Allocation Concealment | Random Housing | Blinding | Random Outcome Assessment | Blinding | Incomplete Outcome Data | Selective Outcome Reporting | Was the Study Apparently Free of Other Problems That Could Result in High Risk of Bias? | |
| [47] | Yes | Yes | Unclear | Yes | Unclear | Unclear | Unclear | No | No | No |
| [48] | Yes | Yes | Yes | Yes | Unclear | Yes | Unclear | No | No | No |
| [49] | Unclear | Yes | Unclear | Unclear | Unclear | Unclear | Unclear | No | No | Unclear |
3.2. Characteristics of the Included Studies
3.2.1. Clinical Studies
Twenty-two out of the twenty-five studies identified in this review were clinical and examined the upper airway, the lower airway and/or the gut microbiome in healthy controls and/or asthmatic children (Table 5). Ten studies (out of twenty-two) examined the upper airway microbiome [26,29,30,31,33,34,35,36,37,38], while only three studies investigated the lower airway microbiome [13,39,46]. One study analysed both the upper and lower airway microbiomes in healthy controls and children with severe persistent asthma [27]. In a study conducted in 2021, both the upper airway and gut microbiome investigations were performed in healthy controls and asthmatic children [42]. Seven studies (out of twenty-two) analysed faecal specimens to characterise the gut microbiome [28,32,40,41,43,44,45]. The specimen types used to examine the upper airway microbiome were nasal swab [27,29,35,42], nasal wash [33], hypopharyngeal aspirate [30], nasopharyngeal swab [34], nasopharyngeal wash [31], saliva [26] and throat swab [35,36,37,38]. In contrast, the specimen types used to study the lower airway were broncho-alveolar lavage (BAL) [13,27] and sputum [39,46], while faecal specimens were used to study the gut microbiome [28,32,40,41,42,43,44,45].
3.2.2. Animal Intervention Studies
Three out of the twenty-five identified studies were conducted using animal models (Table 6). All three studies used murine models consisting of BALB/c mice [47], Sprague–Dawley (SD) rats [48] and C57BL/6 mice [49]. Regarding asthma induction, for both the BALB/c mouse model [47] and the SD rat model [48], the animals were sensitised by intraperitoneal injections of ovalbumin (OVA) and then challenged by OVA aerosol inhalation. However, there were variations among the methods used in each study, including the frequency and dose schedule of OVA exposure. For the interleukin-13 (IL-13) transgenic (TG) C57BL/6 mouse model, asthma was induced by lung-specific IL-13 overexpression [49]. The first animal intervention study performed 16S rRNA sequencing on both the nasal lavage fluid and BAL to characterise the upper and lower airway microbiomes in mice with OVA-induced asthma [47]. The second study extracted the lung tissues from rats with allergic asthma to characterise the lower airway microbiome [48]. BAL, lung tissue and faecal specimens were collected from IL-13 transgenic mice simulating chronic asthma to examine both the lower airway and gut microbiomes [49].
Table 6.
Overview of the included animal-based studies that investigated the microbiome and asthma.
| Citation and Title of the Article | Country | Sample Size | Age | Type of Sample Collected | Time of Sample Collection | Microbiome Detection Method | Genomic DNA Extraction Kit | Sequencing Platform | Microbiome Diversity Assessment | Bioinformatics Pipeline Used | Study Findings |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Upper and lower airway microbiome | |||||||||||
| [47] ‘Respiratory Microbiota Profiles Associated with the Progression from Airway Inflammation to Remodelling in Mice With OVA-Induced Asthma’. |
China | Female BALB/c mice: n = 30 Control group: n = 6 Ovalbumin group: n = 24 |
4–6 weeks | Nasal lavage fluid and BAL |
Control group was sacrificed at the end of the experiment: n = 6 mice Experimental groups were sacrificed at different time points for sample collection as follows: 1 week: n = 6 mice 2 weeks: n = 6 mice 4 weeks: n = 6 mice 6 weeks: n = 6 mice |
16S rRNA gene sequencing (V3-V4 region) |
OMEGA soil DNA extraction kit | Illumina MiSeq | α-diversity: Shannon index and β-diversity: Weighted UniFrac distance |
QIIME 2 | Upper airway microbiome of the OVA induced mice had significantly higher α-diversity than control mice. Insignificant α-diversity difference in the lower airway microbiome of the OVA induced mice and control mice. Significant difference detected in β-diversity between the OVA-induced mice and control mice. The dominant respiratory microbiome in the acute inflammatory and airway remodelling stages were different. Acute inflammatory stage: ↑ Relative abundance of Pseudomonas spp. Airway remodelling stage: ↑ Relative abundance of Staphylococcus spp. and Cupriavidus spp. |
| Lower airway microbiome | |||||||||||
| [48] ‘High-throughput 16S rDNA sequencing of the pulmonary microbiome of rats with allergic asthma’ |
China | Normal control group: n = 4 Saline control group: n = 4 Allergic asthma group: n = 4 |
4–6 weeks | Lung tissues | 1 time point Normal control group: lung tissues on day 0 Saline control and allergic asthma groups: lung tissues on day 29 |
16S rRNA gene sequencing (V4−V5 region) |
No mention | Illumina high-throughput technology (Illumina PE250) | α-diversity: Chao index, coverage index, Shannon index, and Simpson index and β-diversity: Bray–Curtis |
Mothur | The α-diversity of the lower airway microbiome in the allergic asthma group increased. Significant difference between normal control group and allergic asthma group was detected. Normal control group: ↑ Proteobacteria. Allergic asthma group: ↑ Firmicutes. |
| Lower airway and gut microbiome | |||||||||||
| [49] ‘Alteration of Lung and Gut Microbiota in IL-13-Transgenic Mice Simulating Chronic Asthma’. |
Korea | IL-13 overexpressing transgenic (TG) mice: n = 30 C57BL/6 wild-type (WT) mice: n = 30 |
10-week-old mice for both groups | BAL, lung tissue and faecal | 1 time point | 16S rRNA gene sequencing (no mention of region) | FastDNA SPIN Kit | Illumina MiSeq | α-diversity: Shannon index, Chao1 index, and the Inverse Simpson’s diversity index and β-diversity: Weighted UniFrac distances |
QIIME | No significant difference in α-diversity was observed. Altered β-diversity in lung and gut microbiota in the IL-13 TG mice compared to the WT mice. IL-13 TG mice (lungs): ↑ Proportion of Proteobacteria and Cyanobacteria. ↓ Amount of Bacteroidetes IL-13 TG mice (gut): ↓ Firmicutes and Proteobacteria. |
Table 5.
Overview of the included clinical studies that investigated microbiome and asthma.
| Citation and Title of the Article | Country | Study Design | Sample Size | Age | Sample Collected | Time of Sample Collection | Microbiome Detection Method | Genomic DNA Extraction Method | Sequencing Platform | Microbiome Diversity Assessment | Bioinformatics Pipeline Used | Findings |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upper airway microbiome | ||||||||||||
| [29] ‘Longitudinal Changes in Early Nasal Microbiota and the Risk of Childhood Asthma’ |
Finland | Cohort | 2-month visit: n = 704 13-month visit: n = 665 24-month visit: n = 570 |
2-month visit: 2.5 (2.4–2.7) 13-month visit: 13.5 (13.1–13.9) 24-month visit: 25.0 (24.6–25.5) |
Nasal swabs | 3 time points: 2, 13 and 24 months | 16S rRNA gene sequencing (V4 region) | Automated MagNA Pure 96 System | Illumina MiSeq | α-diversity: Shannon index and β-diversity: Bray–Curtis |
UPARSE OUT clustering | Insignificant difference in α-diversity as well as β-diversity between children who developed asthma by age 7 years and those who did not. ↑ Relative abundance of Haemophilus over age 2 to 13 months was associated significantly with higher risk of asthma. ↑ Relative abundance of Lactobacillus at age 2 months was associated significantly with lower risk of asthma. |
| [30] ‘Infant airway microbiota and topical immune perturbations in the origins of childhood asthma’ |
Denmark | Cohort | 700 | The cohort was followed up from the age of 1 week until 6 years of life | Hypopharyngeal aspirates | Different time-points: Hypopharyngeal aspirates were obtained at ages 1 week, 1 month and 3 months |
16S rRNA gene sequencing (V4 region) |
PowerMag Soil DNA Isolation Kit | Illumina MiSeq | α-diversity: Shannon index and β-diversity: Bray–Curtis and UniFrac, weighted |
Mothur | At age 1 month: ↑ α-diversity and a difference in β-diversity in children who developed asthma in the first 6 years of life compared to those who did not. ↑ Relative abundance of Veillonella and Prevotella at age 1 month were associated significantly with asthma development by age 6 years. At ages 1 week and 3 months: Insignificant association between α- or β-diversity or any taxa and the development of asthma. |
| [33] ‘Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas’ |
USA | Cross-section | 163 children and adolescents | Age for all participants years (SE): 11.0 (0.3) |
Nasal washes | 205 nasal washes. 1 time point: 163 sample 2 time points: 42 samples (patients came back for an additional visit (5.5 to 6.5 months apart), and one additional sample was taken) |
16S rRNA gene sequencing (V4 region) | QIAGEN QIAamp DNA Kit | Illumina MiSeq | α-diversity: Shannon index, ACE indices, and Faith’s phylogenetic diversity index and β-diversity: UniFrac (unweighted and weighted), Bray-Curtis, and Jaccard distances |
Mothur | Operational taxonomic units of pathogenic Moraxella, Staphylococcus, Streptococcus and Haemophilus were present in 95% of nasal microbiotas in asthmatics. |
| [31] ‘Nasopharyngeal Microbiome Diversity Changes over Time in Children with Asthma’ |
USA | Cohort | 40 children and adolescents | 6–18 years; mean = 11 years | Nasopharyngeal washes | Two samples (5.5 to 6.5 months apart) | 16S rRNA gene sequencing (V4 region) |
QIAGEN QIAamp DNA Kit | Illumina MiSeq | α-diversity: Good’s coverage, Chao1, Shannon indices, and Faith’s phylogenetic diversity index and β-diversity: UniFrac (unweighted and weighted) |
Mothur | The nasopharyngeal core microbiome of asthmatics at the 95% level: Moraxella, Staphylococcus, Streptococcus, Haemophilus, Fusobacterium. 86% of the total reads in asthmatics were: Moraxella, Staphylococcus, Dolosigranulum, Corynebacterium, Prevotella, Streptococcus, Haemophilus, Fusobacterium and a Neisseriaceae. |
| [34] ‘Different functional genes of upper airway microbiome associated with natural course of childhood asthma’ |
Korea | Cross-section | Healthy children (controls), n = 31 Children with asthma, n = 30 Children with asthma in remission, n = 30 |
Years Healthy children (controls): 7.1 ± 1.1 Children with asthma: 8 ± 0.9 Children with asthma in remission: 7.6 ± 1.4 |
Nasopharyngeal swabs | 1 time point | 16S rRNA gene sequencing (V1-V3 region) |
PowerMag Microbiome RNA/DNA isolation kit (MP Biomedicals, Santa Ana, CA, USA) | Illumina TruSeq DNA | α-diversity: Shannon index and β-diversity: UniFrac (unweighted and weighted) |
No mention | Control group: ↑ Relative abundance of Haemophilus and Moraxella. Asthma and remission groups: ↑ Relative abundance of Streptococcus, Dolosigranulum, and Corynebacterium. Asthma group: ↑ Relative abundance of Staphylococcus. |
| [26] ‘Bacterial salivary microbiome associates with asthma among African American children and young adults’ |
USA | Case control | Asthma cases, n = 57 Healthy controls, n = 57 |
Asthma case: 15.6 ± 3.3 Healthy controls: 15 ± 3.9 |
Saliva | 1 time point | 16S rRNA gene sequencing (V4 region) |
Oragene DNA Discover OGR-500 self-collection kits | Illumina MiSeq | α-diversity: Shannon index | QIIME | Significant difference in α-diversity between asthma cases and healthy controls. Asthma cases: ↓ Relative abundance of Streptococcus. ↑ Relative abundance of Veillonella. Healthy controls: ↑ Relative abundance of Streptococcus. ↓ Relative abundance of Veillonella. |
| [35] ‘Bacterial microbiota of the upper respiratory tract and childhood asthma’ |
Europe | Cross-section | Throat swabs: Children with asthma, n = 125 Controls, n = 202 Nasal swabs: Children with asthma, n = 39 Controls, n = 29 |
6 to 12 years | Nasal and throat swabs | 1 time point | 16S rRNA gene sequencing (V3-V5 region) |
QIAmp DNA Mini Kit | Pyrosequencing, Roche 454-GS FLX Titanium | α-diversity: Shannon index and β-diversity: Unweighted UniFrac distances |
QIIME | Asthma was associated with alterations in nasal (not throat) microbiome. Asthmatic children versus controls: ↓ α- and β-diversity and lower abundance of Moraxella of nasal microbiome. |
| [36] ‘Integration of metagenomics-metabolomics reveals specific signatures and functions of airway microbiota in mite-sensitized childhood asthma’ |
China | Cross-section | Control: n = 28 Asthma: n = 27 |
Years Control: 4.54 ± 0.3 Asthma: 4.32 ± 0.85 |
Throat swabs | 1 time point. Asthma case: swabs were collected before inhaled or nasal administration of corticosteroids for regular daily treatment. Control: no mention. |
Shotgun metagenome sequencing | FastDNA SPIN Kit for Soil (MP Biomedical) | Illumina HiSeq | α-diversity: Shannon index and β-diversity: Bray–Curtis index |
Metagenome assembly by MEGAHIT and contig binning by MetaBAT |
No difference in α-diversity between asthma and control groups, but β-diversity difference was detected between the two groups. Asthma group: Predominance of Neisseria elongate. Control group: Significant enrichment of Eubacterium sulci, Leptotrichia wadei and Prevotella spp. |
| [37] ‘Integrated metabolic and microbial analysis reveals host-microbial interactions in IgE-mediated childhood asthma’ |
Taiwan | Cross-section | Asthma (non-atopic, lowly sensitize): n = 15 Asthma (non-atopic, highly sensitize): n = 13 Healthy controls: n = 25 |
Years Asthma (non-atopic, lowly sensitized): 3.7 ± 0.6 Asthma (non-atopic, highly sensitized): 3.5 ± 0.7 Healthy controls: 3.6 ± 0.7 |
Throat swabs | 1 time point, no time specified | 16S rRNA gene sequencing (V3-V4 region) |
FastDNA Spin Kit for Soil (MP Biomedical, Solon, OH, USA) | Illumina HiSeq 2500 | α-diversity: Shannon index and Chao1 index | QIIME | No statistically significant difference in airway taxa composition between asthma and healthy controls. Highly sensitized asthma children: ↓ Relative abundance of Dialister, Streptococcus, Prevotella, Tannerella, Atopobium and Ralstonia. |
| [38] ‘Comparison of Oropharyngeal Microbiota from Children with Asthma and Cystic Fibrosis’ |
Germany | Cross-sectional | Control children: n = 62 Children with asthma: n = 27 Children with cystic fibrosis (CF): n = 57 |
Years (min–max) Control: 10.1 (8–12) Asthma: 10 (8–12) CF: 10.61 (6–12) |
Throat swabs | 1 time point | 16S rRNA gene sequencing (V4 region) |
QIAamp Mini Kit | Illumina MiSeq system | α-diversity: Shannon index and Chao1 index and β-diversity: Morisita–Horn similarity index |
Mothur | High level of similarity was detected between control, asthma and CF groups. Core microbiome in healthy controls, children with asthma and CF: Prevotella, Streptococcus, Neisseria, Veillonella and Haemophilus. |
| Lower airway microbiome | ||||||||||||
| [13] ‘Disordered microbial communities in asthmatic airways’ |
Ireland | Cross-sectional | Difficult asthma, n = 13 Non-asthmatic controls, n = 7 |
Asthmatic children: 11.8 ± 2.8 years Controls: 11.3 ± 5.7 years |
Bronchoalveolar lavage (BAL) | 1 time point, time not specified | 16S rRNA gene sequencing (V3 region) and cloning | DNeasyn (Qiagen) | No mention | α-diversity: Chao1 index | DOTUR program | Asthmatic children: Significant increase in Proteobacteria Children with difficult asthma: ↑ Staphylococcus spp. Controls: ↑ Bacteroidetes (Prevotella spp.). |
| [46] ‘Altered respiratory microbiota composition and functionality associated with asthma early in life’ |
United Arab Emirates | Case control | Paediatric asthmatic: n = 11 Paediatric healthy: n = 9 |
Years, mean (SD, range) Paediatric asthmatic: 6.7 (4.1, 12) Paediatric healthy: 8 (3.1, 8) |
Sputum | 1 time point: Spontaneous coughed up sputum (expectorated phlegm/mucous) was the first preference of sample collection whenever possible in all subjects. |
16S rRNA gene sequencing for bacteria (V4 region) ITS2-gene based microbial profiling for fungi |
MoBio PowerMag Soil DNA Isolation | Illumina MiSeq | α-diversity: Shannon index and β-diversity: Bray-Curtis index |
Mothur | Asthmatic versus non-asthmatic controls: Significant difference of bacteria and fungi between the two groups. Significant difference in Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria. Paediatric asthma: ↑ Relative abundance in Streptococcus spp. and Moraxella spp. Difference in Ascomycota, Basidiomycota phyla and other unclassified fungi. ↓ Penicillium aethiopicum and Alternaria spp. |
| [39] ‘Gram-negative microbiota is related to acute exacerbation in children with asthma’ |
Korea | Cross-section | Total children, n = 95 Children with asthma exacerbation: n = 22 Children with stable asthma: n = 67 Controls: n = 6 |
Years Asthma exacerbation: 9.0 (6.4/10.9) Stable asthma: 8.0 (6.6/9.7) Controls: 13.2 (10.7/14.9) |
Sputum | 1 time point | 16S rRNA gene sequencing (V3-V4 region) |
FastDNA SPIN Kit for Soil (MP Biomedicals, USA) | Illumina MiSeq | α-diversity: ACE, Chao1, Jackknife, NPShannon, Shannon and Simpson and β-diversity: Jensen–Shannon, Bray–Curtis, Generalised UniFrac, and UniFrac indices |
No mention | No difference in α-diversity detected between asthma exacerbation and stable asthma children. Significant difference in β-diversity detected between asthma exacerbation and stable asthma children. Asthma exacerbation: Phylum level: ↑ Abundance of Proteobacteria. ↓ Abundance of Saccharibacteria and Actinobacteria. Genus level: ↑ Abundance of Veillonella, Neisseria, Haemophilus, Fusobacterium, Oribacterium, Campylobacter and Capnocytophaga ↓ Saccharimonas, Rothia, Porphyromonas, Gemella and Actinomyces. |
| Upper and lower airway microbiome | ||||||||||||
| [27] ‘Integrative study of the upper and lower airway microbiome and transcriptome in asthma’ |
USA | Case control | Children with severe persistent asthma: n = 27 Healthy controls: n = 27 |
Years Children with severe persistent asthma: 11, IQR 8 Healthy controls: 13, IQR 6 |
Nasal swabs BAL |
1 time point | 16S rRNA gene sequencing (V3-V4 region) |
Qiagen DNeasy Mini Kit | Illumina MiSeq | α-diversity: Shannon index and β-diversity: UniFrac distance index |
QIIME | α-diversity was higher in bronchial (BAL) versus nasal. Significant difference in β-diversity detected between bronchial (BAL) and nasal. Asthmatic children Nasal microbiome: Moraxella and Alloiococcus are hub genera. Bronchial microbiome: no hubs. Nasal Streptococcus was enriched in children with persistent asthma versus healthy controls. |
| Upper airway and gut microbiome | ||||||||||||
| [42] ‘Altered IgA Response to Gut Bacteria Is Associated with Childhood Asthma in Peru’ |
Peru | Case control | Asthma: n = 40 Control children: n = 40 |
Years Asthma: 14.6 ± 1.5 Controls: 13.3 ± 2.3 |
Nasal swabs and faecal specimens | 1 time point: Biospecimens samples (nasal swabs and faecal) were collected the same day of the home visit or during the same week | 16S rRNA gene sequencing (V4 region) |
Faecal specimens ethanol-based method Nasal swabs: no information was provided |
Illumina MiSeq | α-diversity: Shannon index and Renyi entropy and β-diversity: Bray–Curtis and UniFrac distances |
DADA 2 | α-and β-diversity of faecal as well as nasal swabs showed no difference between asthma and controls. |
| Gut microbiome | ||||||||||||
| [32] ‘Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age’ |
Denmark | Cohort | 411 infants | Full-term infants born at >36 week and were followed until 6 years | Faecal specimens | 2 time points: At 1 month old and 12 months old |
16S rRNA gene and denaturing gradient gel electrophoresis (V3 region) | QIAamp DNA stool Mini Kit (Qiagen, Hilden, Germany) | None | Band richness and principal component analysis | BioNumerics software 4.50 | No association between bacterial diversity of the infant’s gut microbiota and asthma in the first 6 years of life. |
| [28] ‘Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma’ |
Canada | Case control | Total 76. Asthmatic: n = 39 Healthy control: n = 37 |
Follow up from birth till 4 years. | Faecal specimens | 2 time points: 3 months and 1 year of age |
16S rRNA gene sequencing (V3 region) |
Mo Bio dry bead tubes (Mo Bio Laboratories) | Illumina Hi-Seq. 2000 | α-diversity: Shannon index and β-diversity |
Mothur | At 3 months asthmatic children: ↓ Abundance of Lachnospira. ↑ Abundance of Clostridium neonatale. Negative association between the ratio of Lachnospira and Clostridium neonatale and asthma risk by 4 years of age. |
| [43] ‘Early infancy microbial and metabolic alterations affect risk of childhood asthma’ |
Canada | Longitudinal nested case control | Control: n = 74 Atopy and wheeze: n = 22 Atopy only: n = 87 Wheeze only: n = 136 |
Baseline: 1 year of age Follow-up: 3 years of age |
Faecal specimens | 2 time points: at 3 months and 1 year |
16S rRNA gene sequencing (V3 region) |
Qiagen DNA Stool Mini Kit | Illumina HiSeq 2000 | α-diversity: Shannon index | Mothur | No significant difference in α-diversity among four groups. Children at risk of asthma: ↓ Relative abundance of Lachnospira, Veillonella, Faecalibacterium and Rothia. |
| [44] ‘Gut microbial-derived butyrate is inversely associated with IgE responses to allergens in childhood asthma’ |
Taiwan | Case control | Children with rhinitis: n = 27 Children with asthma: n = 34 Healthy controls, n = 24 |
Years Controls: 5.7 ± 0.8 Rhinitis: 6.0 ± 0.9 Asthma: 5.6 ± 0.9 |
Faecal specimens | 1 time point. Time not specified. | 16S rRNA gene sequencing (V3-V4 region) |
FastDNA Spin Kit for Faeces (MP Biomedical) | Illumina HiSeq 2500 | α-diversity: species richness | QIIME | Children with rhinitis and asthma versus healthy controls: ↓ Relative abundance of Firmicutes. ↓ Relative abundance of Faecalibacterium, Roseburia, SMB53 and Dialister. ↑ Relative abundance of Escherichia, Enterococcus and Clostridium. |
| [40] ‘Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies’ |
Taiwan | Cross-section | Controls: n = 26 Asthma: n = 35 Rhinitis: n = 28 |
Controls: 5.6 ± 0.8 Asthma: 5.5 ± 0.9 Rhinitis 5.9 ± 0.9 |
Faecal specimens | 1 time point | 16S rRNA gene sequencing (V3-V4 region) |
FastDNA Spin Kit for Faeces (MP Biomedical, Solon, OH, USA) | Illumina HiSeq 2500 | α-diversity: Shannon index and Chao 1 index and β-diversity: Bray–Curtis and Weighted UniFrac distance |
QIIME | Relatively lower α-diversity in allergic disease than control (insignificant). No significant difference in β-diversity in allergic airway disease. Children with asthma and allergic rhinitis versus healthy controls: ↓ Relative abundance of Firmicutes. ↓ Relative abundance of Dorea spp. ↑ Relative abundance of Clostridium spp. |
| [45] ‘Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation’ |
USA | Cohort | 1 month: n = 130 infants 6 months: n = 168 infants |
1 month and 6 month infants | Faecal specimens | 2 time points: 1 month and 6 months. | 16S rRNA gene sequencing (V4 region) (ITS)2 rRNA sequencing for fungi |
In-house kit: Modified cetyltrimethylammonium bromide buffer-based protocol |
Illumina MiSeq | α-diversity: Shannon index and β-diversity: Unweighted UniFrac distance and Bray–Curtis |
QIIME | The highest risk group: ↓ Relative abundance of Bifidobacterium, Akkermansia and Faecalibacterium. ↑ Relative abundance of Candida and Rhodotorula. |
| [41] ‘Correlations of Inflammatory Factors with Intestinal Flora and Gastrointestinal Incommensurate Symptoms in Children with Asthma’ |
China | Cross-section | Asthmatic group: n = 70 Control group: n = 25 |
Years Asthmatic group: 9.03 ± 2.01 Control group: 8.12 ± 2.13 |
Faecal specimens | 1 time point (exact time was not mentioned) | SYBR GREEN I fluorescence quantitative polymerase chain reaction | No mention | Not applicable | Total load of bacteria between observation group and control group | Not applicable | The total load of bacteria: asthmatic group > control group Asthmatic group: ↓ Load of Bifidobacterium and Lactobacillus. ↑ Load of Escherichia coli, Helicobacter pylori, Streptococcus and Staphylococcus. Control group: ↑ Load of Bifidobacterium and Lactobacillus. ↓ Load of Escherichia coli, Helicobacter pylori, Streptococcus and Staphylococcus. |
3.3. Microbiome Quantification
The 25 identified studies in the current review analysed the bacteriome (Table 5). Two studies investigated only the mycobiome in addition to the bacteriome [45,46]. However, none of the identified studies evaluated the virome. Of the 25 included studies, 23 (92.0%) utilised 16S rRNA gene sequencing to characterise bacterial communities in the upper airway, lower airway or faecal specimens, as shown in Table 5 and Table 6. These studies targeted different sequencing regions on 16S rRNA, consisting of region V3 (n = 3; 13.0%) [28,43], V4 (n = 9; 39.1%) [26,29,30,31,33,35,38,42,45,46], V1–V3 (n = 1; 4.3%) [34], V3–V4 (n = 6; 26.0%) [27,37,39,40,47], V4–V5 (n = 1; 4.3%) [48] and V3–V5 (n = 1; 4.3%) [35]. One study did not indicate the targeted sequencing region [49]. Additionally, a single study used 16S rRNA gene sequencing with cloning [13] and another study used the 16S rRNA gene and denaturing gradient gel electrophoresis [32]. In both studies, V3 was the targeted sequencing region [13,32]. In addition, one study used shotgun metagenome sequencing [36] and another study used the SYBR GREEN I fluorescence quantitative polymerase chain reaction method [41] to characterise the microbiome. In the two studies that characterised the mycobiome, the internal transcribed spacer region (ITS)2 of the rRNA gene was amplified and sequenced using the Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA) [45,46].
3.4. Diversity Assessments
As shown in Table 5 and Table 6, 18 out of the 25 identified studies (72.0%) assessed both the α- and β-diversity of the upper airway, lower airway and/or gut microbiome. These studies have reported contradictory findings related to α- and β-diversity. For instance, an insignificant difference was observed in both α- and β-diversity between asthmatic children and non-asthmatics [29,40,42]. On the contrary, a significant difference in α- and β-diversity of the upper airway, lower airway and/or gut microbiome was detected between asthmatic children and non-asthmatics [27,30,35,46,48]. Five studies evaluated only the α-diversity of the microbiome in the upper airway, lower airway and/or gut microbiome (20.0%) [13,26,37,43,44] and demonstrated conflicting data. For example, Espuela-Ortiz and colleagues (2019) reported a significant difference in the α-diversity of the upper airway microbiome between asthma cases and healthy controls [26]. Another study detected insignificant differences in the airway taxa composition between asthma patients and healthy controls [37]. However, Bisgaard and colleagues (2011) estimated band richness and conducted principal component analysis (PCA), which resulted in no association between the bacterial diversity of the infant’s gut microbiome and asthma development in the first 6 years of life [32]. The total load of bacteria for asthmatic children and healthy controls was calculated, and the authors reported a higher bacterial load in asthmatic children than in the healthy control group [41].
3.5. Microbiome Outcome
3.5.1. Human Studies
The data presented in Table 5 indicates that the microbiome in the upper airways of asthmatic children has a significantly high relative abundance of Moraxella, Staphylococcus, Streptococcus, Haemophilus, Fusobacterium, Dolosigranulum, Corynebacterium, Veillonella and Neisseria elongate [26,31,33,34,36]. However, a significantly low relative abundance of Streptococcus, Moraxella, Dialister, Prevotella, Tannerella, Atopobium and Ralstonia was identified in the upper airways of asthmatic children [26,35,37]. An increased relative abundance of Haemophiles in children aged 2 to 13 months was significantly associated with a higher risk of asthma development [29]. An additional study reported that a high relative abundance of Veillonella and Prevotella at age 1 month was significantly associated with asthma development by age 6 [30]. However, a significantly high abundance of Lactobacillus at age 2 months was associated with a lower risk of asthma development, suggesting that this bacterium plays a protective role [29].
The lower airway microbiome indicated a significant increase in Protobacteria in asthmatic children, particularly in asthma exacerbation cases [13,39], while a significant decrease in Saccharibacteria and Actinobacteria was detected [39]. Moreover, asthma exacerbation was associated with a high relative abundance of Veillonella, Neisseria, Haemophilus, Fusobacterium, Oribacterium, Campylobacter and Capnocytophaga in sputum [39]. However, Saccharimonas, Rothia, Porphyromonas, Gemella and Actinomyces were detected with low significant relative abundance in asthma exacerbation cases [39]. A high relative abundance of Streptococcus, Moraxella and Staphylococcus was identified in asthmatic children, with the latter detected in difficult asthma cases [13,46]. A mycobiome analysis revealed a significantly low abundance of Penicillium aethiopicum and Alternaria spp. in sputum specimens collected from asthmatic children [46].
The gut microbiome studies that examined the faecal specimens of asthmatic children revealed a significant increase in the relative abundance of Clostridium, Escherichia and Enterococcus [32,40,44]. In addition, a higher load of E. coli, Helicobacter pylori, Streptococcus and Staphylococcus was detected in the faecal specimens of asthmatic children [41]. A lower load of Bifidobacterium and Lactobacillus was detected in the faecal specimens of the same group, indicating that these bacteria play a protective role [41]. The mycobiome analysis of faecal specimens obtained from infants revealed a high relative abundance of Candida and Rhodotorula, which were associated with a high risk of developing asthma [45]. In contrast, the relative abundance of Lachnospira, Faecalibacterium, Roseburia, SMB53, Dialister and Dorea was significantly decreased in asthmatic children [28,40,44]. Lachnospira, Veillonella, Faecalibacterium, Rothia, Bifidobacterium and Akkermansia were significantly decreased in high-risk children [43,45].
3.5.2. Animal Intervention Studies
A respiratory microbiome analysis identified an increase in the relative abundance of Pseudomonas spp. during the acute inflammatory stage, while Staphylococcus spp. and Cupriavidus spp. increased during the airway remodelling stage in mice with OVA-induced asthma [47]. The bacterial phylum Firmicutes were detected at higher levels in the lower airway (lung tissues) microbiomes of rats with allergic asthma [48]. Proteobacteria and Cyanobacteria phyla were identified at higher levels in the lungs of IL-13 TG mice [49]. The microbiome analysis of faecal specimens extracted from IL-13 TG mice reflected a lower level of Firmicutes and Protobacteria, whereas the lung microbiome indicated a low level of Bacteroidetes [49].
4. Discussion
The aims of the current study were to examine the association between asthma and the upper airway, lower airway and/or gut microbiome in humans and animals and identify the characteristics of the upper airway, lower airway and the gut microbiome commonly associated with asthma.
The data presented in this review demonstrated that the clinical specimens collected from both the control and asthmatic children were mostly from the upper airway (i.e., a nasal swab, nasal wash, hypopharyngeal aspirate, nasopharyngeal swab, nasopharyngeal wash, throat swab and saliva). Only three studies collected specimens from the lower airway (BAL and sputum) [13,39,46], and one contained specimens from both the upper and lower airways [27]. The limited number of lower airway microbiome studies might contribute to the difficulty in collecting lower airway human specimens (specifically from healthy children) as it is more convenient to collect specimens from the upper airway.
Evidence of the association between asthma and changes in the upper and lower airways and/or gut microbiome was synthesized. The phyla Proteobacteria (Haemophilus, Moraxella, Neisseria, Campylobacter, Escherichia and Helicobacter) and Firmicutes (Veillonella, Staphylococcus, Streptococcus, Dolosigranulum, Oribacterium, Alloiococcus, Clostridium and Enterococcus) were identified as being significantly higher in the asthmatic children [13,39] compared with the healthy controls. These findings confirm the previous observations that Proteobacteria (Haemophilus, Moraxella and Neisseria) and Firmicutes (Staphylococcus and Streptococcus) were the most abundant bacteria in asthmatic children [50].
A previous literature review performed in 2019 reported that the most dominant genera in the upper airways of infants are Corynebacterium, Dolosigranulum, Haemophilus, Moraxella, Staphylococcus and Streptococcus [51]. However, in this study, we found that the upper airway microbiome in 1-month-old infants indicated an increase in the relative abundance of Veillonella and Prevotella, which were associated with asthma development later in life [30]. Both genera were considered normal flora of the upper respiratory system and their increased abundance in infants suggests their potential involvement in asthma development later in life [30]. Furthermore, the upper airway microbiome in infants ranging in age between 2 and 13 months indicated a higher abundance of Haemophilus, which was associated with a higher risk of asthma development later in life [29]. This substantiates the results of a previous review, which highlighted that dysregulated Haemophilus was common in asthmatic children [52].
As shown in Table 5, the upper airways of asthmatic children have a significant high relative abundance of Moraxella, Staphylococcus, Streptococcus, Haemophilus, Fusobacterium, Dolosigranulum, Corynebacterium, Veillonella and Neisseria elongate and a high relative abundance of Streptococcus, Moraxella and Staphylococcus was determined in their lower airways. The above-mentioned bacteria are known as normal human microbiota in the respiratory tract [53]. Furthermore, Staphylococcus, Streptococcus and Haemophilus, followed by Moraxella and Veillonella were the most frequently reported bacterial genera in the respiratory system of asthmatic children (Table 5). Previous studies have highlighted that the clinical characteristics of asthma patients and the type of immune response stimulated by aeroallergens influence airway microbiome composition [54]. For instance, Moraxella catarrhalis, a species of Haemophilus, and Streptococcus were the predominant respiratory tract bacteria in patients with severe asthma and corticosteroid resistance [54]. The literature points to a lack of metabolomic investigations of the association between the metabolic characteristics of these dysbiotic bacteria and asthma phenotypes and treatment prognosis. For instance, asthma patients with steroid resistance might have a higher abundance of airway microbial communities that can degrade steroids [55].
It has been established in the literature that the dysbiosis of the normal gut microbiome plays an important role in the development of immune disorders, including asthma [56,57]. This is explained by the key role of the gut microbiome in shaping the human immune system [55]. Differences in the gut microbiome in terms of composition and diversity were previously reported between healthy and asthmatic children [52]. In this study, the high relative abundance of the genus Clostridium was detected in faecal specimens collected from asthmatic children in three studies [28,40,44]. Previous studies have shown that the Clostridium species have an impact on the host’s immune system [7]. In addition, infant colonization with Clostridium species is associated with a higher risk of allergy development [7]. This substantiates the findings of the current review as a predominance of the Clostridium species was detected during early childhood and was associated with asthma development [28].
The studies analysed in this review lacked consistency in reporting their findings. Some of the studies on bacterial communities in airways and/or gut have identified most of the detected bacterial taxa at the phylum level [13,39,40,44,46], whereas the others have identified the detected taxa at the genus level [26,27,28,29,30,31,33,34,35,36,37,38,41,43,45] (Table 5). Due to this inconsistency, making an accurate comparison of these studies became challenging. Moreover, bacteria belonging to different genera under the same phylum might have different effects on a host. For instance, this review revealed that the genus Lactobacillus, which belongs to the phylum Firmicutes, is associated with a low risk of asthma development, suggesting that the bacteria under this phylum play a protective role in asthma. By contrast, other genera under the same phylum Firmicutes, such as Veillonella, are significantly associated with asthma development later in life, suggesting their contributory role in asthma development. Therefore, it has been recommended that the use of reporting guidelines (i.e., the Strengthening the Organization and Reporting of Microbiome Studies [STORMS] checklist) must be adopted in future human microbiome studies [58].
Contradictory findings on microbiome diversity were reported by the included clinical studies (Table 5). Of the 22 clinical studies, 15 determined both the α- and β-diversity of the upper airway, lower airway and/or gut microbiomes, but they reported conflicting findings on α- and β-diversity between the asthmatic and non-asthmatic children. As depicted in Table 5, the clinical studies were conducted in different geographic locations, including North America, Europe, Asia, and Middle East, and they analysed clinical specimens obtained from different ethnic groups. The literature highlighted that the gut microbiome composition is associated with ethnicity and geography [59]. Furthermore, the sample sizes in 16 clinical studies were heterogeneous, with minimum and maximum sample sizes of 20 [46] and 923 [29] children, respectively. This sample size variation might have contributed to the variations in the diversity metrics [60]. The clinical studies also varied with respect to technical protocols, next-generation sequencing platforms and bioinformatics pipelines, as described in Table 5, and these variations might have influenced the quality of the obtained microbiome data [61].
There is limited literature on the use of animal intervention studies to examine the association between asthma development and microbiomes. The criteria related to random housing, blinding and random outcome assessment may hinder the research on such studies as the validity might be compromised. The quality assessment of animal intervention studies included in this review [47,48,49] generally indicated the potential performance and detection bias in aspects related to the blinding procedures, which might influence the validity of the results of these studies [47,48,49]. Furthermore, the current review indicated a lack of microbiome data related to viruses, archaea and micro-eukaryotes (such as protozoa). The characterization of these rare microbiome components might have a valuable impact on our understanding of asthma development.
5. Conclusions
The phyla Proteobacteria and Firmicutes were identified as being significantly higher in the asthmatic children compared with the healthy controls. The high relative abundance of Veillonella, Prevotella and Haemophilus in the microbiome of the upper airway in early infancy was associated with a higher risk of asthma development later in life. Gut microbiome analyses indicated that a high relative abundance of the genus Clostridium in early childhood might be associated with asthma development later in life. The findings reported here serve as potential microbiome signatures associated with an increased risk of asthma development. There is a need for human studies targeting the lower airway as well as well-designed animal intervention studies to further identify high-risk infants, which will help in design strategies and prevention mechanisms to avoid asthma early in life.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11040939/s1, File S1: search strategy; Table S1: data collection form; Table S2: quality assessment tools.
Author Contributions
Conceptualization, M.G.A. and A.M.A.; methodology, M.G.A., A.M.A.-M., A.S.A., H.Y.A., N.A.A., R.A. and A.M.A.; validation, M.G.A., A.M.A.-M., A.S.A., H.Y.A., N.A.A., R.A. and A.M.A.; formal analysis, M.G.A., A.M.A.-M., A.S.A., H.Y.A., N.A.A., R.A. and A.M.A.; investigation, M.G.A., A.M.A.-M., A.S.A., H.Y.A., N.A.A., R.A. and A.M.A.; data curation, M.G.A., A.M.A.-M., A.S.A., H.Y.A., N.A.A., R.A. and A.M.A.; writing—original draft preparation, M.G.A., A.M.A.-M., A.S.A., H.Y.A., N.A.A., R.A. and A.M.A.; writing—review and editing, M.G.A. and R.A.; visualization, M.G.A., A.M.A.-M., A.S.A., H.Y.A., N.A.A., R.A. and A.M.A.; supervision, M.G.A.; project administration, M.G.A., A.M.A.-M. and A.M.A.; funding acquisition, M.G.A. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of King Abdullah International Medical Research Center (KAIMRC), Riyadh, Saudi Arabia (protocol code RYD-22-419812-169312 on 20 November 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The APC was funded by King Abdullah International Medical Research Center (KAIMRC), Riyadh, Saudi Arabia.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Asher M.I., Rutter C.E., Bissell K., Chiang C.-Y., El Sony A., Ellwood E., Ellwood P., García-Marcos L., Marks G.B., Morales E., et al. Worldwide trends in the burden of asthma symptoms in school-aged children: Global Asthma Network Phase I cross-sectional study. Lancet. 2021;398:1569–1580. doi: 10.1016/S0140-6736(21)01450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mims J.W. Asthma: Definitions and pathophysiology. Int. Forum Allergy Rhinol. 2015;5:S2–S6. doi: 10.1002/alr.21609. [DOI] [PubMed] [Google Scholar]
- 3.Pijnenburg M.W., Fleming L. Advances in understanding and reducing the burden of severe asthma in children. Lancet Respir. Med. 2020;8:1032–1044. doi: 10.1016/S2213-2600(20)30399-4. [DOI] [PubMed] [Google Scholar]
- 4.Barcik W., Boutin R.C.T., Sokolowska M., Finlay B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity. 2020;52:241–255. doi: 10.1016/j.immuni.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manfredo Vieira S., Hiltensperger M., Kumar V., Zegarra-Ruiz D., Dehner C., Khan N., Costa F.R.C., Tiniakou E., Greiling T., Ruff W., et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrahamsson T., Jakobsson H.E., Andersson A., Björkstén B., Engstrand L., Jenmalm M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy. 2014;44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 7.Van Nimwegen F.A., Penders J., Stobberingh E.E., Postma D.S., Koppelman G.H., Kerkhof M., Reijmerink N.E., Dompeling E., Brandt P.A.V.D., Ferreira I., et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 2011;128:948–955.e3. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 8.Doré E., Joly-Beauparlant C., Morozumi S., Mathieu A., Lévesque T., Allaeys I., Duchez A.-C., Cloutier N., Leclercq M., Bodein A., et al. The interaction of secreted phospholipase A2-IIA with the microbiota alters its lipidome and promotes inflammation. JCI Insight. 2022;7:e152638. doi: 10.1172/jci.insight.152638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasan N., Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G.A.D., Gasbarrini A., Mele M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7:14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., Fernandes G.R., Tap J., Bruls T., Batto J.M., et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nash A.K., Auchtung T.A., Wong M.C., Smith D.P., Gesell J.R., Ross M.C., Stewart C.J., Metcalf G.A., Muzny D.M., Gibbs R.A., et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilty M., Burke C., Pedro H., Cardenas P., Bush A., Bossley C., Davies J., Ervine A., Poulter L., Pachter L., et al. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen L.D.N., Viscogliosi E., Delhaes L. The lung mycobiome: An emerging field of the human respiratory microbiome. Front. Microbiol. 2015;6:89. doi: 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver D., Gago S., Bromley M., Bowyer P. The Human Lung Mycobiome in Chronic Respiratory Disease: Limitations of Methods and Our Current Understanding. Curr. Fungal Infect. Rep. 2019;13:109–119. doi: 10.1007/s12281-019-00347-5. [DOI] [Google Scholar]
- 16.Budden K.F., Gellatly S.L., Wood D.L.A., Cooper M.A., Morrison M., Hugenholtz P., Hansbro P.M. Emerging pathogenic links between microbiota and the gut–lung axis. Nat. Rev. Microbiol. 2016;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 17.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 18.Bingula R., Filaire M., Radosevic-Robin N., Bey M., Berthon J.-Y., Bernalier-Donadille A., Vasson M.-P., Filaire E. Desired Turbulence? Gut-Lung Axis, Immunity, and Lung Cancer. J. Oncol. 2017;2017:5035371. doi: 10.1155/2017/5035371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokholm J., Blaser M.J., Thorsen J., Rasmussen M.A., Waage J., Vinding R.K., Schoos A.-M.M., Kunøe A., Fink N.R., Chawes B.L., et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018;9:141. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann P., Messina N., Mohn W.W., Finlay B.B., Curtis N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: A systematic review. J. Allergy Clin. Immunol. 2018;143:467–485. doi: 10.1016/j.jaci.2018.09.025. [DOI] [PubMed] [Google Scholar]
- 21.Melli L., Carmo-Rodrigues M.D., Araújo-Filho H., Solé D., de Morais M.B. Intestinal microbiota and allergic diseases: A systematic review. Allergol. Immunopathol. 2015;44:177–188. doi: 10.1016/j.aller.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Alcazar C.G.-M., Paes V.M., Shao Y., Oesser C., Miltz A., Lawley T.D., Brocklehurst P., Rodger A., Field N. The association between early-life gut microbiota and childhood respiratory diseases: A systematic review. Lancet Microbe. 2022;3:e867–e880. doi: 10.1016/S2666-5247(22)00184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009;62:e1–e34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Moskalewicz A., Oremus M. No clear choice between Newcastle–Ottawa Scale and Appraisal Tool for Cross-Sectional Studies to assess methodological quality in cross-sectional studies of health-related quality of life and breast cancer. J. Clin. Epidemiol. 2020;120:94–103. doi: 10.1016/j.jclinepi.2019.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Hooijmans C.R., Rovers M.M., de Vries R.B.M., Leenaars M., Ritskes-Hoitinga M., Langendam M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014;14:43. doi: 10.1186/1471-2288-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Espuela-Ortiz A., Lorenzo-Diaz F., Baez-Ortega A., Eng C., Hernandez-Pacheco N., Oh S.S., Lenoir M., Burchard E.G., Flores C., Pino-Yanes M. Bacterial salivary microbiome associates with asthma among african american children and young adults. Pediatr. Pulmonol. 2019;54:1948–1956. doi: 10.1002/ppul.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chun Y., Do A., Grishina G., Grishin A., Fang G., Rose S., Spencer C., Vicencio A., Schadt E., Bunyavanich S. Integrative study of the upper and lower airway microbiome and transcriptome in asthma. JCI Insight. 2020;5:e133707. doi: 10.1172/jci.insight.133707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiemsma L.T., Arrieta M.-C., Dimitriu P.A., Cheng J., Thorson L., Lefebvre D.L., Azad M.B., Subbarao P., Mandhane P., Becker A., et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin. Sci. 2016;130:2199–2207. doi: 10.1042/CS20160349. [DOI] [PubMed] [Google Scholar]
- 29.Toivonen L., Karppinen S., Schuez-Havupalo L., Waris M., He Q., Hoffman K.L., Petrosino J.F., Dumas O., Camargo C.A., Hasegawa K., et al. Longitudinal Changes in Early Nasal Microbiota and the Risk of Childhood Asthma. Pediatrics. 2020;146:20200421. doi: 10.1542/peds.2020-0421. [DOI] [PubMed] [Google Scholar]
- 30.Thorsen J., Rasmussen M.A., Waage J., Mortensen M., Brejnrod A., Bønnelykke K., Chawes B.L., Brix S., Sørensen S.J., Stokholm J., et al. Infant airway microbiota and topical immune perturbations in the origins of childhood asthma. Nat. Commun. 2019;10:5001. doi: 10.1038/s41467-019-12989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pérez-Losada M., Alamri L., Crandall K.A., Freishtat R.J. Nasopharyngeal Microbiome Diversity Changes over Time in Children with Asthma. PLoS ONE. 2017;12:e0170543. doi: 10.1371/journal.pone.0170543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bisgaard H., Li N., Bonnelykke K., Chawes B.L.K., Skov T., Paludan-Müller G., Stokholm J., Smith B., Krogfelt K.A. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011;128:646–652.e5. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 33.Pérez-Losada M., Authelet K.J., Hoptay C.E., Kwak C., Crandall K.A., Freishtat R.J. Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. Microbiome. 2018;6:179. doi: 10.1186/s40168-018-0564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim B.-S., Lee E., Lee M.-J., Kang M.-J., Yoon J., Cho H.-J., Park J., Won S., Lee S.Y., Hong S.J. Different functional genes of upper airway microbiome associated with natural course of childhood asthma. Allergy. 2018;73:644–652. doi: 10.1111/all.13331. [DOI] [PubMed] [Google Scholar]
- 35.Depner M., Ege M.J., Cox M.J., Dwyer S., Walker A.W., Birzele L.T., Genuneit J., Horak E., Braun-Fahrländer C., Danielewicz H., et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J. Allergy Clin. Immunol. 2017;139:826–834.e13. doi: 10.1016/j.jaci.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 36.Chiu C., Chou H., Chang L., Fan W., Dinh M.C.V., Kuo Y., Chung W., Lai H., Hsieh W., Su S. Integration of metagenomics-metabolomics reveals specific signatures and functions of airway microbiota in mite-sensitized childhood asthma. Allergy. 2020;75:2846–2857. doi: 10.1111/all.14438. [DOI] [PubMed] [Google Scholar]
- 37.Chiu C.-Y., Cheng M.-L., Chiang M.-H., Wang C.-J., Tsai M.-H., Lin G. Integrated metabolic and microbial analysis reveals host–microbial interactions in IgE-mediated childhood asthma. Sci. Rep. 2021;11:23407. doi: 10.1038/s41598-021-02925-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boutin S., Depner M., Stahl M., Graeber S.Y., Dittrich S.A., Legatzki A., von Mutius E., Mall M., Dalpke A.H. Comparison of Oropharyngeal Microbiota from Children with Asthma and Cystic Fibrosis. Mediat. Inflamm. 2017;2017:5047403. doi: 10.1155/2017/5047403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y.H., Jang H., Kim S.Y., Jung J.H., Kim G.E., Park M.R., Hong J.Y., Na Kim M., Kim E.G., Kim M.J., et al. Gram-negative microbiota is related to acute exacerbation in children with asthma. Clin. Transl. Allergy. 2021;11:e12069. doi: 10.1002/clt2.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiu C.-Y., Chan Y.-L., Tsai M.-H., Wang C.-J., Chiang M.-H., Chiu C.-C. Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies. World Allergy Organ. J. 2019;12:100021. doi: 10.1016/j.waojou.2019.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Li T., Yuan H., Pan W., Dai Q. Correlations of Inflammatory Factors with Intestinal Flora and Gastrointestinal Incommensurate Symptoms in Children with Asthma. Med. Sci. Monit. 2018;24:7975–7979. doi: 10.12659/MSM.910854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh C.-S., Rengarajan S., Kau A., Tarazona-Meza C., Nicholson A., Checkley W., Romero K., Hansel N.N. Altered IgA Response to Gut Bacteria Is Associated with Childhood Asthma in Peru. J. Immunol. 2021;207:398–407. doi: 10.4049/jimmunol.2001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arrieta M.-C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., Kuzeljevic B., Gold M.J., Britton H.M., Lefebvre D.L., et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 44.Chiu C., Cheng M., Chiang M., Kuo Y., Tsai M., Chiu C., Lin G. Gut microbial-derived butyrate is inversely associated with IgE responses to allergens in childhood asthma. Pediatr. Allergy Immunol. 2019;30:689–697. doi: 10.1111/pai.13096. [DOI] [PubMed] [Google Scholar]
- 45.Fujimura K.E., Sitarik A.R., Havstad S., Lin D.L., LeVan S., Fadrosh D., Panzer A.R., LaMere B., Rackaityte E., Lukacs N.W., et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al Bataineh M.T., Hamoudi R.A., Dash N.R., Dash N.R., Ramakrishnan R.K., Almasalmeh M.A., Sharif H.A., Al-Hajjaj M.S., Hamid Q. Altered respiratory microbiota composition and functionality associated with asthma early in life. BMC Infect. Dis. 2020;20:697. doi: 10.1186/s12879-020-05427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng J., Wu Q., Zou Y., Wang M., He L., Guo S. Respiratory Microbiota Profiles Associated with the Progression From Airway Inflammation to Remodeling in Mice With OVA-Induced Asthma. Front. Microbiol. 2021;12:2372. doi: 10.3389/fmicb.2021.723152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong Y., Hu S., Zhou H., Zeng H., He X., Huang D., Li X. High-throughput 16S rDNA sequencing of the pulmonary microbiome of rats with allergic asthma. Genes Dis. 2020;7:272–282. doi: 10.1016/j.gendis.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sohn K.-H., Baek M.-G., Choi S.-M., Bae B., Kim R.Y., Kim Y.-C., Kim H.-Y., Yi H., Kang H.-R. Alteration of Lung and Gut Microbiota in IL-13-Transgenic Mice Simulating Chronic Asthma. J. Microbiol. Biotechnol. 2020;30:1819–1826. doi: 10.4014/jmb.2009.09019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Losol P., Park H.-S., Song W.-J., Hwang Y.-K., Kim S.-H., Holloway J.W., Chang Y.-S. Association of upper airway bacterial microbiota and asthma: Systematic review. Asia Pac. Allergy. 2022;12:e32. doi: 10.5415/apallergy.2022.12.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sbihi H., Boutin R., Cutler C., Suen M., Finlay B.B., Turvey S.E. Thinking bigger: How early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy Eur. J. Allergy Clin. Immunol. 2019;74:2103–2115. doi: 10.1111/all.13812. [DOI] [PubMed] [Google Scholar]
- 52.Pulvirenti G., Parisi G.F., Giallongo A., Papale M., Manti S., Savasta S., Licari A., Marseglia G.L., Leonardi S. Lower Airway Microbiota. Front. Pediatr. 2019;7:393. doi: 10.3389/fped.2019.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tille P. Bailey & Scott’s Diagnostic Microbiology. Elsevier Health Sciences; Amsterdam, The Netherlands: 2015. [Google Scholar]
- 54.Green B.J., Wiriyachaiporn S., Grainge C., Rogers G., Kehagia V., Lau L., Carroll M.P., Bruce K.D., Howarth P.H. Potentially Pathogenic Airway Bacteria and Neutrophilic Inflammation in Treatment Resistant Severe Asthma. PLoS ONE. 2014;9:e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivas M.N., Crother T.R., Arditi M. The microbiome in asthma. Curr. Opin. Pediatr. 2016;28:764–771. doi: 10.1097/MOP.0000000000000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalliomäki M., Kirjavainen P., Eerola E., Kero P., Salminen S., Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 57.Penders J., Thijs C., van den Brandt P.A., Kummeling I., Snijders B., Stelma F., Adams H., van Ree R., Stobberingh E.E. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mirzayi C., Renson A., Furlanello C., Sansone S.-A., Zohra F., Elsafoury S., Geistlinger L., Kasselman L.J., Eckenrode K., van de Wijgert J., et al. Reporting guidelines for human microbiome research: The STORMS checklist. Nat. Med. 2021;27:1885–1892. doi: 10.1038/s41591-021-01552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaulke C.A., Sharpton T.J. The influence of ethnicity and geography on human gut microbiome composition. Nat. Med. 2018;24:1495–1496. doi: 10.1038/s41591-018-0210-8. [DOI] [PubMed] [Google Scholar]
- 60.Kers J.G., Saccenti E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022;12:4366. doi: 10.3389/fmicb.2021.796025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allali I., Arnold J.W., Roach J., Cadenas M.B., Butz N., Hassan H.M., Koci M., Ballou A., Mendoza M., Ali R., et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017;17:194. doi: 10.1186/s12866-017-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.