Abstract
Objective
Comprehensive and reliable genome-wide variant analysis of a small number of cells has been challenging due to genome coverage bias, PCR over-cycling, and the requirement of expensive technologies. To comprehensively identify genome alterations in single colon crypts that reflect genome heterogeneity of stem cells, we developed a method to construct whole-genome sequencing libraries from single colon crypts without DNA extraction, whole-genome amplification, or increased PCR enrichment cycles.
Results
We present post-alignment statistics of 81 single-crypts (each contains four- to eight-fold less DNA than the requirement of conventional methods) and 16 bulk-tissue libraries to demonstrate the consistent success in obtaining reliable coverage, both in depth (≥ 30X) and breadth (≥ 92% of the genome covered at ≥ 10X depth), of the human genome. These single-crypt libraries are of comparable quality as libraries generated with the conventional method using high quality and quantities of purified DNA. Conceivably, our method can be applied to small biopsy samples from many tissues and can be combined with single cell targeted sequencing to comprehensively profile cancer genomes and their evolution. The broad potential application of this method offers expanded possibilities in cost-effectively examining genome heterogeneity in small numbers of cells at high resolution.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-023-06333-y.
Keywords: Next generation sequencing, High coverage whole-genome sequencing, Sequencing library construction without DNA extraction
Introduction
Whole-genome sequencing (WGS) library preparation generally requires > 50 ng of purified human DNA input, equivalent to 8,333 diploid cells (not considering losses during DNA extraction), to achieve 30X average depth and > 90% breadth coverage of the genome for a comprehensive DNA variant analysis. Using tissue culture and whole-genome amplification (WGA) with limited starting material can introduce mutations and genome coverage bias. These problems persist despite the recent development of single-cell sequencing technology; therefore, comprehensive variant analysis without sequencing multiple cells cannot be achieved [1–4].
Our goal is to use the colon crypt, which consists of 1,000 to 2,000 cells all derived from a single stem cell [5–8], as a model system to identify mutations occurring in the single stem cells to understand genome heterogeneity. To achieve this goal, we modified the conventional WGS library preparation steps to generate high-quality sequencing libraries reaching a balanced ≥ 30X depth post-alignment coverage consistently from 81 single human colon crypts without WGA or DNA purification. This method will empower researchers to reliably generate streamlined high-quality libraries from tissues containing small numbers of cells.
Materials and methods
Tissue collection and crypt isolation
A small piece of colon is collected from individuals who have undergone surgery to remove part of the colon under the standard of care at either Keck Hospital of USC or Children’s Hospital Los Angeles through the Norris Comprehensive Cancer Center Translational Pathology Core.
A 5 mm x 5 mm colon specimen is cut into smaller pieces, washed with 10 ml 1X Phosphate Buffered Saline with 9 mM EDTA (1XPBS/EDTA) three times, and incubated in 1XPBS/EDTA for 20 min at room temperature. The liquid is decanted after incubation and 2 ml of 1XPBS (without EDTA) is added. After 10-second vortexing at high speed, 20 to 30 individual crypts are identified and transferred under an inverted microscope into separate low-binding microfuge tubes. The presence of one single colon crypt in each tube is confirmed under the microscope before storing at -80oC. The remaining crypts in the suspension are spun down for bulk DNA extraction using the phenol/chloroform extraction and ethanol precipitation method.
Whole-genome sequencing library construction
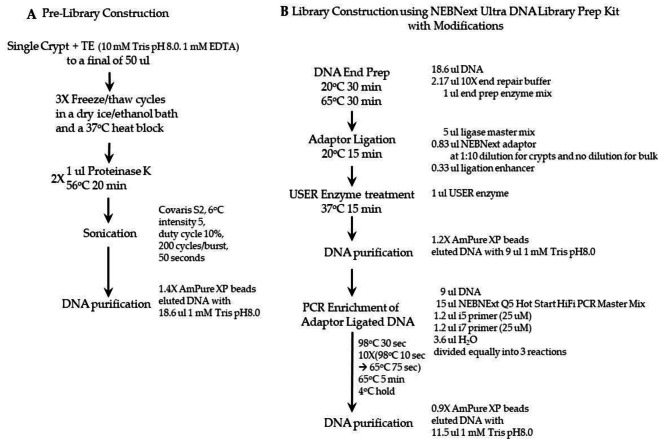
The detailed workflow of single colon crypt sequencing library construction is illustrated in Fig. 1. The entire workflow through the enrichment step must be carried out without delay between each step to minimize DNA loss, and low-binding tubes are preferred. Specifically, reagents are pre-aliquoted or ready to be aliquoted just prior to each step, and no more than eight libraries are processed in each session. After freeze-and-thaw cycles, each colon crypt sample is treated with proteinase K, transferred to a Covaris microtube, and immediately sonicated and transferred into a fresh microtube containing 70 ul of pre-aliquoted AmPure XP beads (Backman Coulter). Throughout the experiment, the bead purification is done according to the manufacturer’s instruction, and the DNA is eluted with 1 mM Tris pH 8.0 preheated to 55oC.
Fig. 1.
Whole-genome sequencing library construction from single colon crypts without DNA extraction. (A) Preparation and fragmentation of single colon crypt for library construction. (B) Workflow of library construction
The NEBNext Ultra DNA Library Prep Kit (NEB #E7370) is used according to the manufacturer’s instructions with modifications. Only 1/3 of the recommended amount for all reagents is used in the NEBNext end prep and the adaptor ligation steps with a 1:10 dilution of the adaptor. A 30 ul PCR mixture (instead of the 50 ul reaction recommended) is divided into three PCR tubes for 10 cycles of amplification for the enrichment step.
The bulk tissue library construction is essentially the same as single crypt library construction except for 50 ng of purified bulk tissue DNA and an undiluted adaptor in the adaptor ligation reaction are used.
Assessment and assembly of sequencing library
Detailed steps of assessing and assembling each colon crypt library and pooling of multiple libraries for each sequencing flow cell are summarized in Supplemental Fig. 1. Only one of the three PCR reactions is analyzed by BioAnalyzer (Model 2100, Agilent, Santa Clara, CA), and all three PCR reactions are quantitated by Qubit assay (Qubit 2.0 Fluorometer, Thermo Fisher Scientific). Each crypt-sequencing library is assembled by combining an equal quantity of library DNA from each of the three PCR tubes. The remaining DNA from each PCR tube is stored separately for variant verification in the future. The bulk tissue sequencing library is assembled by combining PCR products from all three tubes after BioAnalyzer analysis. All final sequencing libraries are purified with AmPure XP beads and evaluated by BioAnalyzer and Qubit analyses post-assembly.
A total of 81 crypt libraries and 16 bulk libraries are constructed and sequenced in six pools of up to 20 libraries per pool. Each pool is quality controlled by BioAnalyzer and qPCR analyses and shallow sequenced by MiSeq to verify adequate pooling.
Sequencing and post-run quality assessment
Each library pool is diluted to a 0.7 nM final concentration and sequenced (150 bp paired-end) on an S4 flow cell using NovaSeq 6000 (Illumina, San Diego) S4 300 cycles reagent kit (v1.5) in the Keck Genomics Platform Core facility at USC. Post-sequencing, the read quality of sequencing reads is assessed by FastQC using BCL2FASTQ (v1.8.4).
Sequencing reads are aligned to GRCh38 by BWA (v0.7.8-r455), followed by GATK’s Base Recalibrator (v3.5.0) to detect quality score errors. Next, Picard Tools (v1.128) merges aligned BAMs and marks duplicate reads. GATK’s IndelRealigner minimizes mismatches across local alignments; Picard Tools GC Bias determines coverage bias; and Picard HS Metrics determines hybrid-selection metrics. Picard MultiMetrics and Samtools Stats (v1.2) collect multiple classes of metrics. VCFtools(v0.1.17) [9], Plinkv1.9 (v1.90b6.7) [10], SnpSniffer (v.7.0.0) (https://github.com/tgen/snpSniffer), and a variation of global ancestry principal component analysis [11] were used to analyze genome concordance between the bulk and crypt libraries for allele biasing.
Results
All 97 libraries show a normal size distribution from 300 to 1000 bp with a peak of ~ 400 to 500 bp with no adaptor contamination and are of sufficient quantity when analyzed using BioAnalyzer (Supplemental Fig. 2). The yield of the 81 crypt libraries ranges from 69 ng to 685 ng with an average of 310 ng (Supplemental Fig. 3), within the 160 to 320 ng yield expected from the NEBNext Ultra kit for 6 ng DNA input with 10 PCR cycles of enrichment. NovaSeq 6000 (Illumina) generated > 120Gb of data per library. The libraries generated an average of 12 × 109 clusters and 25 × 109 reads at 91.6% >Q30 per run (Table 1). The overall reads generated from all 6 flow cells exceeded the Illumina recommended benchmark of a good quality sequencing run.
Table 1.
Whole Genome Sequencing Performance on s$v1.5 Illumina NovaSeq 6000 2 × 150 bp
| PF Clusters | Paired-End Reads (B) | Yield (Gb) | %>Q30 | Mean Quality | |
|---|---|---|---|---|---|
| Run 6 | 12,913,455,779 | 26 | 3,489 | 90.8 | 35.4 |
| Run 5 | 10,882,379,236 | 22 | 3,286 | 91.8 | 35.6 |
| Run 4 | 12,811,554,090 | 26 | 3,869 | 91.0 | 35.4 |
| Run 3 | 13,106,143,219 | 26 | 3,968 | 92.3 | 35.7 |
| Run 2 | 12,401,208,646 | 25 | 3,745 | 91.8 | 35.6 |
| Run 1 | 12,262,229,436 | 25 | 3,773 | 91.7 | 35.5 |
| Average | 12,396,161,734 | 25 | 3,688 | 91.6 | 35.5 |
| Illumina Recommendations | 16–20 | 2,400-3,000 | > 85% | ||
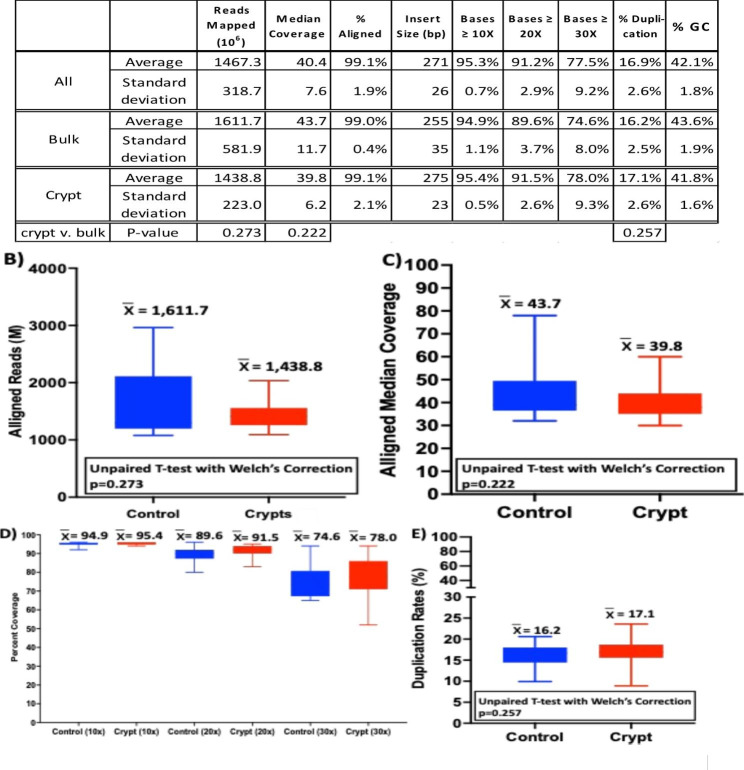
Next, the sequencing reads are aligned to the human genome and post-alignment statistics are collected (Supplemental Table 1). The performance of each set of crypt libraries is compared to a control library for the same patient. More than 1,100 × 106 reads are mapped for each library, and there is no significant difference (p = 0.273) between the average number of reads per library generated from crypts and bulk controls (Fig. 2A and B). The percentage of aligned reads is 99% for both groups (Fig. 2A). The high mapping rates and the library insert size (Fig. 2A) indicate that these libraries are free from contamination and of good mapping quality.
Fig. 2.
Post-alignment analysis of crypt and control (bulk) whole-genome libraries. (A) Summary of statistical analysis on all, bulk (control), and crypt libraries. Whisker plot presentation of (B) the number of aligned reads in millions (M); (C) median depth of coverage in multiples of genome; (D) percent of the genome covered with 10X, 20X, and 30X depth; and (E) duplication rates in percent of total aligned reads for bulk and crypt libraries. In B) through E), blue represents bulk libraries; red represents crypt libraries; the brackets indicate the range; and the average is shown above each bar
Deep post-alignment genome coverage depth and broad coverage overall and at ≥ 10, ≥20, and ≥ 30 depth observed here indicate uniformity of the libraries and more than adequate usable data for reliable variant calling. The average median depth is 44X and 40X for bulk and crypt libraries, respectively, with no significant difference between the two groups (p = 0.222) (Fig. 2A and C). A similar average and standard deviation for the percentage of coverage across 10X, 20X, and 30X depth indicates sequencing uniformity within each group as well as between the two groups (Fig. 2A and D).
A high percentage of duplicate reads indicates low library diversity that can be caused by sample degradation, suboptimal sonication, inadequate adaptor ligation, and high PCR bias. The average duplication rate is 16.2% and 17.1% for bulk and crypt libraries, respectively, with no significant difference between the two groups (p = 0.257) (Fig. 2A and E). These duplication rates are within a workable range when using a low quantity of DNA as inputs [12]. The GC content, which is 41% for the human reference genome, reflects the balance of representation in a sequencing library. In our study, the average GC content is 43.6% and 41.8% of the bulk and crypt libraries, respectively (Fig. 2A).
Further SNP-based genome concordance analyses indicate no allele biasing in the crypt libraries. Plinkv1.9 analysis of > 5,000 SNPs shows an average concordance of 0.997 (Supplemental Table 2). Of 300 known heterozygous SNPs analyzed using SnpSniffer, bulk and crypt libraries from the same individual show an average of 0.998 concordance (Supplemental Table 3). Furthermore, principal component analysis also showed high genotype concordance estimated from 45,679 high-quality SNP distributed across autosomes between crypt and bulk libraries (Supplemental Table 4).
The comparisons described above clearly demonstrate that all 97 libraries are of good quality, and crypt libraries are comparable to bulk (control) libraries with no evidence of allele biasing.
Discussion
We present a cost-effective method that consistently and reliably generates high-quality WGS libraries from as little as 1,000 to 2,000 cells, which is four- to 8-fold less than the purified DNA used in conventional sequencing library construction. Considering material loss during DNA extraction, our improvement is even more substantial. This improved method allows a comprehensive mutation analysis of a small number of cells and offers new possibilities for thorough genomic examination of material-limiting tissue sources. The key improvements are achieved by minimizing sample loss by constructing the library directly from the tissue without DNA extraction; optimizing the speed of completing each step; increasing reaction efficiency; and reducing PCR duplicates by dividing the enrichment PCR reaction into multiple reactions for 10 cycles of amplification.
We demonstrate that high-quality sequencing libraries could be made directly from < 2,000 cells without DNA extraction and that the quantity and size distribution of the library material generated are comparable to libraries constructed with ample purified DNA. All six sequencing runs outperformed Illumina’s recommended benchmark, and each library achieved greater than 30X genome coverage after removing duplicate reads post-alignment. Other studies successfully identified variants using microdissected colon crypts achieving a < 20X median depth coverage of the genome [8, 13]. All our crypt libraries, which exceed 31X median depth and cover > 90% of the genome at ≥ 15X depth, are more than sufficient for reliable variant calling. There is no significant difference in the depth of coverage and percentage of genome coverage between the 81 crypt libraries and the 16 control bulk libraries. Multiple analyses show no indication of allele biasing in the crypt libraries. We clearly show that the single crypt libraries are of comparable quality to libraries constructed from high quantities of purified DNA. Furthermore, our experimental design with three PCR reactions allows confirmation of whether any specific variant is an artifact from PCR or sequencing.
Our method can be applied to other tissue sources with minimal optimization and offers the possibility of interrogating small biopsy samples or limited tissue sources with depth and breadth of coverage equivalent to conventional WGS libraries. Our method can be used to profile variants in small numbers of cells, followed by targeted single-cell sequencing to obtain comprehensive cancer genome profiles and their evolution more cost-effectively and reliably than sequencing the whole genome of multiple single cells. An additional advantage of our method is that single-cell suspension is not required as the starting material; therefore, it can be applied to a broader selection of tissues. The potential broad applications of our method, without requiring any costly new equipment, would offer new possibilities in the comprehensive examination of genome heterogeneity in normal as well as diseased cells.
Limitations
Our method is reliable, easily streamlined, and cost-effective without additional expensive technologies. It has possible applications across various tissue types with some needed optimizations for samples with a high lipid content or connective tissue. NEBNext Ultra II DNA Library Prep Kit can be used for our method, but the NEBNext Ultra II FS Library Prep Kit cannot be used because the DNA fragmentation step requires purified DNA.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the Norris Comprehensive Cancer Center Translational Pathology Core for the sample collection. We would like to acknowledge the Norris Comprehensive Cancer Center Molecular Genomics Core and the Keck Genomics Platform at the University of Southern California for the sequencing work. We also want to thank Dr. Tracy Grikscheit for facilitating sample collection from CHLA.
Abbreviations
- WGS
Whole-genome sequencing
- WGA
whole-genome amplification
Author Contribution
ZM oversaw the whole-genome sequencing and analyzed the sequencing data post-sequencing runs. CLH designed the experiment and developed the library construction protocol. CLH and CYO isolated the colon crypts and constructed the sequencing libraries. JW assisted in specimen collection and provided advice on colon anatomy and crypt identification. YJ performed the SnpSniffer and principal component analysis. DVDB and MRL provided advice on experimental and study design. All authors read and approved the final manuscript.
Funding
This work is supported by funds from NIA R01 AG 067615 and the Catherine and Joseph Aresty Endowment.
Data Availability
All data supporting the results of this article are included in this article and the supplementary information. The original alignment stats from PicardTools generated from the 97 sequencing libraries in this study are available from the corresponding author upon request.
Declarations
Ethics approval and consent to participate
All samples were collected from patients who had undergone surgery to remove part of the colon for diseases under the standard of care at either Keck Hospital of USC or Children’s Hospital Los Angeles through the Norris Comprehensive Cancer Center Translational Pathology Core. The collection of tissue samples was approved by the Institutional Reviewer Board of the University of Southern California under an exempt protocol (HS-20-00531) in accordance with the Declaration of Helsinki and with informed consent from the patients or their legal guardians.
Consent for publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ruan Q, Ruan W, Lin X, Wang Y, Zou F, Zhou L, Zhu Z, Yang C. Digital-WGS: Automated, highly efficient whole-genome sequencing of single cells by digital microfluidics. Sci Adv. 2020;6(50):eabd6454. doi: 10.1126/sciadv.abd6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez-Pena V, Natarajan S, Xia Y, Klein D, Carter R, Pang Y, Shaner B, Annu K, Putnam D, Chen W, Connelly J, Pruett-Miller S, Chen X, Easton J, Gawad C. Accurate genomic variant detection in single cells with primary template-directed amplification. Proc Natl Acad Sci U S A. 2021;118(24):e2024176118. doi: 10.1073/pnas.2024176118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biezuner T, Raz O, Amir S, Milo L, Adar R, Fried Y, Ainbinder E, Shapiro E. Comparison of seven single cell whole genome amplification commercial kits using targeted sequencing. Sci Rep. 2021;11(1):17171. doi: 10.1038/s41598-021-96045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan X, Yang C, Li W, Bai X, Zhou X, Xie H, Wen L, Tang F. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021;22(1):195. doi: 10.1186/s13059-021-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolas P, Kim KM, Shibata D, Tavaré S. The stem cell population of the human colon crypt: analysis via methylation patterns. PLoS Comput Biol. 2007;3(3):e28. doi: 10.1371/journal.pcbi.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozar S, Morrissey E, Nicholson AM, van der Heijden M, Zecchini HI, Kemp R, Tavaré S, Vermeulen L, Winton DJ. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell. 2013;13(5):626–33. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Baker AM, Cereser B, Melton S, Fletcher AG, Rodriguez-Justo M, Tadrous PJ, Humphries A, Elia G, McDonald SA, Wright NA, Simons BD, Jansen M, Graham TA. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014;8(4):940–7. doi: 10.1016/j.celrep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, Georgakopoulos N, Torrente F, Noorani A, Goddard M, Robinson P, Coorens THH, O’Neill L, Alder C, Wang J, Fitzgerald RC, Zilbauer M, Coleman N, Saeb-Parsy K, Martincorena I, Campbell PJ, Stratton MR. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574(7779):532–7. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 9.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R. 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manojlovic Z, Christofferson A, Liang WS, Aldrich J, Washington M, Wong S, Rohrer D, Jewell S, Kittles RA, Derome M, Auclair S, Craig DW, Keats J, Carpten JD. Comprehensive molecular profiling of 718 Multiple Myelomas reveals significant differences in mutation frequencies between African and European descent cases. PLOS Genetics. 2017; 13(11): e1007087 NEB Technical Note Improved library preparation with the NEBNext® Ultra™ II DNA LibraryPrep Kit for Illumina Addressing lower input amounts and challenging sample types. https://www.neb-online.de/wp-content/uploads/2020/03/Improved-library-preparation-with-the-NEBNext-Ultra-II-DNA-Library-Prep-Kit-for-Illumina-E7645.pdf [DOI] [PMC free article] [PubMed]
- 12.NEB Technical Note Improved library preparation with the NEBNext® Ultra™ II DNA LibraryPrep Kit for Illumina Addressing lower input amounts and challenging sample types. https://www.neb-online.de/wp-content/uploads/2020/03/Improved-library-preparation-with-the-NEBNext-Ultra-II-DNA-Library-Prep-Kit-for-Illumina-E7645.pdf
- 13.Olafsson S, McIntyre RE, Coorens T, Butler T, Jung H, Robinson PS, Lee-Six H, Sanders MA, Arestang K, Dawson C, Tripathi M, Strongili K, Hooks Y, Stratton MR, Parkes M, Martincorena I, Raine T, Campbell PJ, Anderson CA. Somatic evolution in non-neoplastic IBD-Affected Colon. Cell. 2020;182(3):672–684e11. doi: 10.1016/j.cell.2020.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the results of this article are included in this article and the supplementary information. The original alignment stats from PicardTools generated from the 97 sequencing libraries in this study are available from the corresponding author upon request.