Abstract
Infective Cryptosporidium parvum oocysts were detected in mussels (Mytilus galloprovincialis) and cockles (Cerastoderma edule) from a shellfish-producing region (Gallaecia, northwest Spain, bounded by the Atlantic Ocean) that accounts for the majority of European shellfish production. Shellfish were collected from bay sites with different degrees of organic pollution. Shellfish harboring C. parvum oocysts were recovered only from areas located near the mouths of rivers with a high density of grazing ruminants on their banks. An approximation of the parasite load of shellfish collected in positive sites indicated that each shellfish transported more than 103 oocysts. Recovered oocysts were infectious for neonatal mice, and PCR-restriction fragment length polymorphism analysis demonstrated a profile similar to that described for genotype C or 2 of the parasite. These results demonstrate that mussels and cockles could act as a reservoir of C. parvum infection for humans. Moreover, estuarine shellfish could be used as an indicator of river water contamination.
Cryptosporidium parvum is a cause of diarrheal disease in humans and farm animals and is a major cause of diarrhea in children and neonatal ruminants (9). Moreover, in immunocompromised subjects, this disease can be life threatening. Transmission of C. parvum occurs mainly by ingestion of oocysts either by fecal-oral contact or through contaminated food or drinking water. Localized epidemics of food-borne cryptosporidiosis have been associated with uncooked sausage, offal, raw milk, apple cider, or foodstuffs, but waterborne transmission seems to play a more prominent role and is implicated in most outbreaks of human cryptosporidiosis (16). The presence of Cryptosporidium oocysts in drinking water supplies has been well documented since 1984, and waterborne epidemics of cryptosporidiosis have been reported frequently in the United States, United Kingdom, and Japan, among other countries (25). The potential for water contamination by cryptosporidial oocysts is high in areas where dumping of raw sewage is a common practice (25). In addition, the presence of waterborne C. parvum oocysts of animal origin needs to be considered, since a single neonatal ruminant can shed up to 1010 oocysts during the course of infection (21).
The presence of oocysts in river waters may also be a source of contamination of the marine environment. Rivers polluted by anthropogenic and livestock fecal discharges could play a major role in contamination by oocysts of shellfish in estuaries and coastal environments. Experimental data show that C. parvum oocysts can survive in seawater up to 30 ppt for as long as 1 month (11, 24). Oocysts also have been detected in seawater in Hawaii near a sewage discharge site (18). Moreover, oocysts with a size, shape, and appearance consistent with those of C. parvum have been detected in mussels (Mytilus edulis) from western Ireland (6) and in bent mussels (Ischadium recurvum) from Chesapeake Bay in the United States (14). Laboratory data show that the eastern oyster (Crassostrea virginica), the freshwater benthic clam (Carbicula fluminea), and different species of mussel (M. edulis and Mytilus galloprovincialis) can remove C. parvum oocysts from water and accumulate them on the gills and inside hemocytes in the hemolymph (6, 10, 11, 13, 27). Whether these or other marine animals actually act as reservoirs, retaining oocysts that initiate infection, remains to be clarified.
The present study was conducted to determine the potential role of mussels (M. galloprovincialis) and cockles (Cerastoderma edule) as reservoirs of C. parvum oocysts and their value as biological indicators of the presence of the parasite in water. We selected a shellfish-producing area on the coast of Gallaecia in northwest Spain that accounts for the majority of European mussel production, with an average of 250,000 metric tons/year, corresponding to 25% worldwide production (29). Shellfish samples were recovered from sites with different levels of anthropogenic and livestock fecal contamination. Moreover, the species, genotype, and infectivity of the oocysts detected were also determined.
MATERIALS AND METHODS
Experimental design and sample collection.
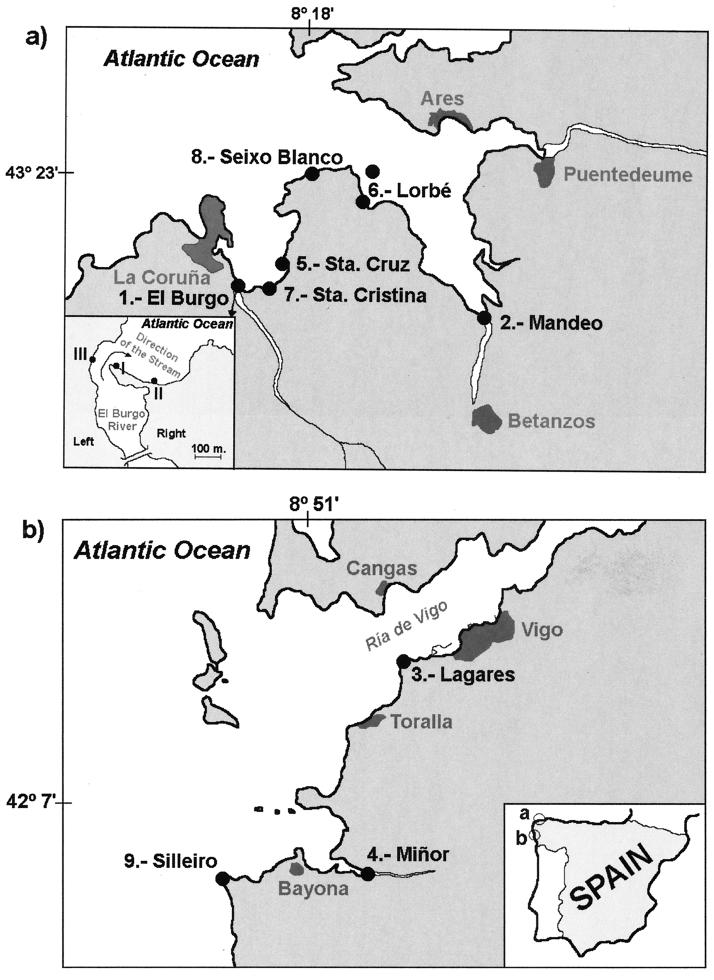
Marine blue mussels (M. galloprovincialis) about 4 cm long were collected in February 1999 from different points on the Gallaecian coast. These sites were selected based upon the criteria of degree of organic pollution, proximity to the river mouth, hydrography, oceanography, and the possible drag by the stream in the estuaries and adjacent shores. As shown in Fig. 1, four sample sites were located near (approximately 250 m away) the river mouths (sites 1, 2, 3, and 4), three sites were located inside the bay (sites 5, 6, and 7), and two sites were on the shores outside the bay (sites 8 and 9). The site characteristics of special relevance for this work are briefly described as follows. (i) Sites 1, 2, 3, and 4 are estuaries with a complex mixture of seawater and river freshwater; Santa Cruz (site 5) and Santa Cristina (site 7) are sites inside bays; Lorbé (site 6) has platforms for intensive mussel production; and Seixo Blanco (site 8) and Silleiro (site 9) are sites on shores outside bays (17). (ii) The basins of the El Burgo (site 1) and Mandeo (site 2) rivers support higher ruminant stocking densities than the Lagares (site 3) and Miñor (site 4) river basins (19). (iii) The estuaries of El Burgo (site 1), Mandeo (site 2), and Lagares (site 3) have higher levels of organic pollution than the estuary of Miñor (site 4) (15).
FIG. 1.
Sample sites at the Gallaecian coast (northwest Spain, bounded by the Atlantic Ocean). (a) La Coruña Bay. (b) Ría de Vigo.
Mussels were collected mainly from colonies growing naturally on the rocky substrate of the intertidal zone, and only one sample (site 6) was from either mussels growing naturally on seashore rocks or cultured mussels growing on floating platforms situated 1,000 m from the shore.
From each of the nine sites, up to 25 mussels were collected and transported in seawater to the laboratory. The mussels were placed in aquaria containing ASPM artificial seawater (Sigma-Aldrich Co., Saint Louis, Mo.) with f/2 salt mixture enrichment (Sigma-Aldrich Co.) at 18 ± 1°C and with a 12-h light-dark photocycle. They were fed ad libitum with a microalgal mixture of 40% Dunaliella tertiolecta, 30% Tetraselmis suecica, and 30% Rhodomonas baltica at 2.5 × 103 cells/ml (7). To determine the presence of Cryptosporidium oocysts in the material filtered by the mussels, the entire water content from each aquarium was collected and analyzed for oocysts at 24-h intervals as described below. Mussels were moved to new aquaria at 24, 48, and 72 h after collection under the same conditions as described above. After 72 h in aquaria, the mussels were dissected to determine the presence of oocysts in the tissues.
From the estuary of El Burgo (site 1 in Fig. 1a), one of the sites where Cryptosporidium oocysts were detected in mussels in February 1999, samples were collected again in March 1999. Mussels were collected from three sites, taking into account the possible drag by the stream (see Fig. 1a for details). Site I is an area of intense sedimentation of river materials; in contrast, sites II and III are under the effects of erosion (17). Cockles were also collected from site I. On this occasion, shellfish were cleaned externally by mechanical brushing immediately after collection until the elimination of epiphytes and were placed in an aquarium with f/2 isotonic medium (Sigma-Aldrich Co.) for 24 h. Afterward, they were transferred to a new clean aquarium and kept for a further 24 h under the same conditions as those described for mussels collected in February.
Oocyst detection and identification.
Water samples recovered at 24-h intervals from the aquaria and mussel tissues were processed as follows. Water samples were centrifuged at 1,300 × g for 10 min, and the pellets were resuspended in phosphate-buffered saline (PBS) and allowed to settle for 20 s. An amount corresponding to 10% (1.5 ml) of supernatant from material filtered by mussels was centrifuged again at 650 × g for 15 min, and the pellet was analyzed for oocysts.
For shellfish, the epiphytes were removed as aseptically as possible, and the shells were opened with a sterile scalpel. The abductor muscles were cut, and the flesh from each mussel was homogenized in a sterile food blender (Omni-mixer 2000; Omni). Aliquots of 300 μl of whole-tissue homogenate were analyzed for oocysts as described below.
Oocysts were detected in the samples described above by an indirect fluorescent-antibody test (IFAT). Samples were placed on glass microscope slides, air dried for 1 h, and fixed in cold acetone for 10 min. Polyclonal rabbit antiserum to C. parvum, diluted 1:40 with PBS (pH 7.4), was used (20). The slides were incubated at 37°C for 30 min, rinsed three times with PBS, and incubated at 37°C for 30 min with 1:160 fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G (Sigma-Aldrich Co.). After this second incubation, the slides were washed with PBS three times, mounted under coverslips with permanent resin mounting medium (Fluoprep; BioMerieux, Marcy l'Etoile, France), and examined by epifluorescence microscopy at a magnification of ×400. Oocysts were identified by positive staining (fluorescent apple green), size (3 to 5 μm), shape (spherical, often slightly irregular), and surface features (some appeared dented or flattened, and some appeared split open, with a pie slice-shaped piece missing).
IFAT-negative samples were processed by a direct immunomagnetic technique to increase the sensitivity of the test. Oocysts isolated from mussels and cockles for the infectivity assay (see below) and PCR-restriction fragment length polymorphism (RFLP) analysis (see below) were also immunomagnetically separated from debris. A commercially available immunomagnetic kit was used in both cases (anti-Cryptosporidium Dynabeads; Dynal A.S., Oslo, Norway). The procedure was performed as recommended by the manufacturer. Briefly, samples were placed in screw-cap vials, and 1 ml of 10× SL buffer B (Dynal A.S.) and 100 μl of bead conjugate were added. Each sample then was rotated through 360° for 1 h at room temperature. The beads were resuspended in 1 ml of 1× SL buffer A (Dynal A.S.), transferred to an Eppendorf tube, and separated using a magnetic particle concentrator (Dynal MPC-M); the supernatant was removed and discarded. The sediment was examined for oocysts by phase-contrast microscopy and by IFAT. In positive samples, oocysts were counted using a cell counting chamber (Neubauer Improved; Brand, Wertheim, Germany).
Oocyst infectivity assay.
The infectivity of the oocysts recovered from mussels was tested in a neonatal mouse model (2-day-old ICR mice). Four pups were orally inoculated with 103 oocysts/mouse from samples collected in February, and eight pups were inoculated with 104 oocysts/mouse from samples collected in March. Oocysts were resuspended in 25 μl of PBS and orally inoculated using a 24-gauge needle. Ten mice inoculated only with the same volume of PBS were used as negative controls. Eight mice inoculated with the same dose of C. parvum oocysts (from a calf isolate) were used as positive controls. The calf isolate was 2 weeks old and had been passaged in lambs. From each group, half of the mice were euthanatized on day 9 and the rest were euthanatized on day 12 postadministration (p.a.) by overexposure to ether, and the intestines were processed for oocyst detection as follows. From each mouse, the intestine was isolated, placed in a tube with 1 ml of PBS, and homogenized using a sterile food blender (Omni-mixer 2000). Whole-tissue homogenates were measured to calculate the parasite load as Cryptosporidium oocysts per mouse. A volume of 10 μl of each homogenate, diluted 1/5 in 1% sodium dodecyl sulfate–malachite green, was placed in a cell counting chamber (Neubauer Improved), and oocysts were counted by conventional microscopy at a magnification of ×400. All negative homogenates were processed for oocysts with anti-Cryptosporidium immunomagnetic beads by the same methodology as that used for mussel homogenates.
DNA isolation, PCR, and RFLP analysis.
Approximately 104 oocysts from mussels recovered at both the Mandeo and the El Burgo locations and 6 × 103 oocysts from cockles (El Burgo) were processed for PCR-RFLP characterization. As a positive control, 106 oocysts from the calf isolate maintained by passage in lambs were used. C. parvum genomic DNA was extracted following a previously described method (3, 22). Briefly, 100 μl of water containing 104 oocysts was mixed with 900 μl of lysis buffer (10 M guanidinium thyiocyanate, 0.1 M Tris-HCl [pH 6.4], 35 mM EDTA [pH 8], 2% [wt/vol] Triton X-100), and 0.3 g of 0.5-mm-diameter glass beads (Biospec Products, Inc., Bartlesville, Okla.) was added. The mixture was vortexed for 2 min and kept at room temperature for 5 min. The tubes were centrifuged at 12,000 × g for 15 s. The supernatant was recovered, 100 μl of coarse activated silica suspension (Sigma-Aldrich Co.) (3) was added to the supernatant, and the mixture was gently agitated at room temperature for 10 min. After centrifugation, the pellet was washed twice in 200 μl of washing buffer (10 mM guanidinium thyiocyanate, 0.1 M Tris-HCl) and once in 200 μl of acetone and then dried at 55°C. The pellet was resuspended in 100 μl of distilled water and centrifuged. The supernatant was recovered and used for PCR amplification.
The PCR oligonucleotide primers for the COWP (cry15 [5′-GTAGATAATGGAAGAGATTGTG-3′] and cry9 [5′-GGACTGAAATACAGGCATTATCTTG-3′]) and TRAP-C1 (Cp.E [5′-GGATGGGTATCAGGTAATAAGAA-3′] and Cp.W [5′-CAATTCTCTCCCTTTACTTC-3′]) loci have been previously described (26). Five microliters of DNA was added to a PCR mixture which contained PCR buffer with 2 mM MgCl2, 75 mM Tris-HCl, 50 mM KCl, 20 mM (NH4)2SO4, 0.001% bovine serum albumin (Biotools, Biotechnological & Medical Laboratories S.A., Madrid, Spain), 0.2 mM each deoxynucleoside triphosphate, 0.2 μM each primer, and 1.25 U of recombinant Thermus thermophilus DNA polymerase. PCR amplification was performed with a PCR thermal cycler (Mastercycler gradient; Eppendorf, Hamburg, Germany) for 40 cycles (92°C for 1 min, 55°C for 1 min, and 72°C for 1 min). Endonuclease treatment was carried out with RsaI (Amersham Life Science Inc., Cleveland, Ohio) in 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, and 5 U of RsaI for 4 h at 37°C. PCR products and restriction fragments were resolved on 1.5% agarose (Sigma-Aldrich Co.) and 4% high-resolution agarose (Ecogen SRL, Madrid, Spain) gels, respectively, and results were visualized by ethidium bromide staining.
RESULTS
Oocyst detection in mussels and cockles.
Cryptosporidium oocysts were detected only in mussels recovered from sites 1 and 2 (Table 1). None of the mussels from the other sites sampled in La Coruña Bay (Fig. 1a) nor any from the three sites sampled in Ría de Vigo (Fig. 1b) were positive. Only mussels from sites located near the mouths of rivers with basins supporting a high density of ruminants (Mandeo and El Burgo) were positive.
TABLE 1.
Results of detection of Cryptosporidium oocysts in shellfish collected at the Gallaecian coast
| Site code | Site name | No. of:
|
|
|---|---|---|---|
| Shellfisha | Cryptosporidium oocystsb | ||
| 1 | El Burgo | 17 | 1.0 × 105 |
| I | 14 | 8 × 104 | |
| 6 cockles | 3 × 104 | ||
| II | 11 | ND | |
| III | 18 | ND | |
| 2 | Mandeo | 5 | 1.5 × 104 |
| 3 | Lagares | 11 | ND |
| 4 | Miñor | 15 | ND |
| 5 | Santa Cruz | 20 | ND |
| 6 | Lorbé | 18 cultured mussels, 8 wild mussels | ND |
| 7 | Seixo Blanco | 25 | ND |
| 8 | Santa Cristina | 7 | ND |
| 9 | Silleiro | 11 | ND |
Mussels, unless otherwise indicated.
Oocysts recovered from water filtered during the first 24 h in aquaria. ND, none detected.
Of the mussels collected at sites 1 and 2, oocysts were recovered only during the first 48 h after collection. Most of the oocysts were detected during the first 24 h (Table 1), and small quantities of oocysts (under the quantification limit) were found in water filtered between 24 and 48 h after sampling. Oocysts were not detected in samples collected from aquaria after 72 h. Mussel tissue homogenates were also negative.
The results of a second, more detailed sampling of site 1, the El Burgo estuary, are shown in Table 1. Oocysts were detected in shellfish (mussels and cockles) collected from an area of intense sedimentation of river materials (site I in Fig. 1a) but not in mussels collected in sites under the effects of erosion (sites II and III in Fig. 1a). Shellfish washing medium used for external cleaning during the first 24 h after collection was also negative.
Morphological characteristics of oocysts (size, shape, surface features, and stain color [fluorescent apple green]) were consistent with those of C. parvum. Both the water filtered by mussels collected at sites 3 through 9 and their tissue homogenates were negative for oocysts.
Infectivity assay.
All mice orally inoculated with oocysts recovered from mussels were positive for C. parvum oocysts on days 9 and 12 p.a. The average numbers of oocysts per gut detected in mice inoculated with 103 oocysts were 5 × 103 oocysts/gut on day 9 p.a. and 104 oocysts/gut on day 12 p.a.; an average of 105 oocysts/gut was detected on day 9 p.a. in mice in the positive control group. Mice inoculated with 104 oocysts had loads of 7 × 104 oocysts/gut on day 9 p.a. and 5 × 104 oocysts/gut on day 12 p.a.; means of 9 × 106 oocysts/gut on day 9 p.a. and 3 × 105 oocysts/gut on day 12 p.a. were detected in positive control mice. Mice inoculated with PBS were negative on both days (9 and 12 p.a.).
PCR-RFLP analysis.
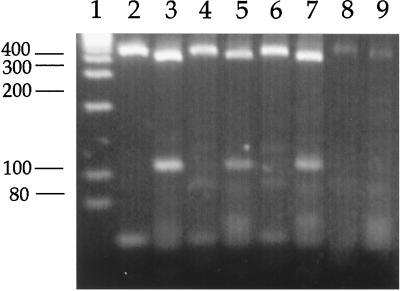
C. parvum isolates obtained from mussels (El Burgo and Mandeo locations) and cockles (El Burgo) had a PCR-RFLP electrophoretic profile similar to that of the calf isolate from our laboratory for the COWP (410 and 106 bp) and TRAP-C1 (455 and 51 bp) loci (Fig. 2), consistent with that described for the bovine or C genotype of the parasite.
FIG. 2.
PCR-RFLP analysis for the COWP and TRAP-C1 primers of C. parvum isolates from bovines (lanes 2 and 3), mussels (lanes 4 and 5 [El Burgo location]; lanes 6 and 7 [Mandeo location]), and cockles (lanes 8 and 9). Lane 1, molecular weight markers.
DISCUSSION
In this study, the presence of infective oocysts of C. parvum in environmental samples of blue mussels and cockles was verified using (i) immunofluorescence, (ii) an oocyst infectivity assay, and (iii) PCR-RFLP analysis. To our knowledge, viable oocysts of C. parvum were detected for the first time in these bivalves frequently consumed by humans and obtained from a region which accounts for the majority of European shellfish production. As suggested previously, mussels and cockles could represent an important reservoir of C. parvum infection for humans.
Apparently, C. parvum oocysts are able to survive in seawater as well as in marine freshwater shellfish. Previous studies (11, 24, 27) have demonstrated the survival of C. parvum oocysts in artificial seawater for at least 1 year under moderate oxygenation. Viable C. parvum oocysts have been recovered from experimentally infected freshwater clams (C. fluminea), marine eastern oysters (C. virginica), and mussels (M. edulis and M. galloprovincialis) (6, 10, 11, 13, 27). Oocysts of Cryptosporidium spp. were detected in naturally infected mussels from Ireland (6) and oysters and bent mussels from Chesapeake Bay (11, 14). The oocysts recovered from naturally infected oysters were infectious for mice and represented genotype C (11). Unfortunately, the authors (6, 14) did not confirm the species and infectivity of the oocysts detected in mussels.
Species differentiation of Cryptosporidium is difficult because (i) the size ranges of oocysts of various species, such as C. parvum and Cryptosporidium baileyi, overlap; (ii) the morphological characteristics of possible species of Cryptosporidium from marine hosts are not completely known; and (iii) no species-specific monoclonal antibodies are available. To date, molecular characterization and experimental infection in a mammalian model (such as neonatal mice infectivity), as used in this work, are two specific techniques for the identification of C. parvum detected in food and water samples.
The presence of oocysts in filtered material but not in tissue homogenate corroborated that oocysts had been harbored and then released by mussels and cockles but that endogenous infection and development of the parasite life cycle had not occurred. Previous studies have shown that under experimental conditions, infective oocysts of C. parvum accumulate in the hemolymph, gills, and intestinal tract of mussels (27), eastern oysters (10), and freshwater clams (13). All these data suggest that oocysts are not simply filtered out of the water by the gills but are also ingested and transported through the digestive tube. Oocysts remain infective for a mammalian host, implying that bivalves sold for human consumption should be tested for the presence of this parasite.
Moreover, the high average number of oocysts detected either in mussels or in cockles enhances the potential risk of infection of humans. An approximation of the parasite load of shellfish collected in El Burgo indicates that each shellfish transported about 5 × 103 oocysts. The assay of infectivity in neonatal mice indicated that many of these oocysts were infectious, since the dose applied was near the minimum infective dose for neonatal mice (12). These findings are very important in light of previous transmission studies showing that immunocompetent adult humans can be infected by as few as 30 oocysts and that 130 oocysts is the mean infective dose (8).
Shellfish such as mussels and cockles can harbor environmentally derived pathogenic microorganisms as a result of filtering large volumes of water and concentrating the recovered particles (28). Consequently, shellfish are usually depurated (prior to consumption), and standards regulating the quality of the waters used in the culture and depuration process are usually legislated in most developed countries. In this respect, the Spanish regulations (a copy of the EU regulations) are complex and cumbersome (1) and, to our knowledge, depuration rules are regulated only with respect to coliform contamination. Preventive measures for C. parvum are not taken into account, even in the most recent regulations (BOE, 9 April 1999 directive). However, depuration of shellfish could be easily improved, since results show that oocysts taken up by mussels are released within 48 h. Apparently, a depuration process of 72 h could be adopted to remove all the C. parvum oocysts from mussels.
PCR-RFLP studies, together with experimental infection studies, indicate the existence of two genetically distinct C. parvum populations (genotype H or 1 and genotype C or 2) (2, 4, 23, 26, 31). Genotype H or 1 is found exclusively in human infections, whereas genotype C or 2 is found in humans as well as in domestic ruminants. This finding suggests that C. parvum may not be a uniform species and that two independent transmission cycles may exist (5, 26). Molecular markers used in the present study demonstrated that C. parvum oocysts isolated from mussels and cockles in the study area were of genotype C or 2, corresponding to the lineage involved in the zoonotic cycle of transmission of human cryptosporidiosis (5) and possibly related to the high stocking rates of domestic ruminants on the banks of rivers supplying the bays.
Apparently, livestock is the main source of this C. parvum shellfish contamination. The density of grazing ruminants, farm practices, and rainfall play an important role in the runoff of oocysts into water (30). Consequently, the hydrography of river mouths, estuaries, and adjacent shores can greatly affect the distribution of C. parvum oocysts at the seashore. As expected, oocysts were detected only in mussels and cockles collected at the seashore near rivers mouths with very high potential contamination. The absence of contamination either in wild mussels or in cultured mussels recovered not far from these positive sites indicates that the risk for C. parvum in seawater could be limited to a narrow area near the outlet of sewer systems (human settlements) and near the mouths of rivers whose banks have many grazing ruminants. The river stream is able to transport the oocysts to the sea, but only the seashore areas affected by the extension of the river stream seem to be contaminated. Our results and those obtained for oysters and mussels from Chesapeake Bay (11, 14) demonstrated that shellfish, by their ability to recover and concentrate waterborne oocysts, are good indicators of river water contamination.
It is difficult to evaluate the implications of this finding for human health, but it seems that there is a risk of acquiring cryptosporidiosis by the consumption of shellfish eaten raw or insufficiently cooked and not depurated. Wild shellfish from natural colonies are consumed scarcely mainly by children and visitors. Moreover, the presence of self-limiting diarrheal problems of unknown etiology in children has been observed in primary day care centers in this area (J. Ucieda, personal communication). The epidemiological links between the contamination of wild shellfish with C. parvum and diarrheal disease in the human population need to be assessed.
ACKNOWLEDGMENTS
We thank Elena Carrillo and Mar Fernandez for technical support and Jesus Ucieda for assistance in collecting shellfish samples.
We also thank Ronald Fayer for critical comments on the manuscript.
REFERENCES
- 1.Alvarez Cobelas M, Cabrera Capitan F. La calidad de las aguas españolas. Mundo Científico. 1997;177:253–255. [Google Scholar]
- 2.Bonnin A, Fourmax M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygauthier-Tobas D, Gabriel-Pospisil F, Naciri M, Camerlynk P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansens C L, Wertheim-van Dillen P M E, Van der Noordaa J. Rapid and simple method for purification of nuclei acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casemore D P, Wright S E, Coop R L. Cryptosporidiosis: human and animal epidemiology. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 65–92. [Google Scholar]
- 6.Chalmers R M, Sturdee A P, Mellors P, Nicholson V, Lawlor F, Kenny F, Timpson P. Cryptosporidium parvum in environmental samples in the Sligo area, Republic of Ireland: a preliminary report. Lett Appl Microbiol. 1997;25:380–384. doi: 10.1046/j.1472-765x.1997.00248.x. [DOI] [PubMed] [Google Scholar]
- 7.Costas E. Genetic variability in growth rates of marine dinoflagellates. Genetica. 1990;83:99–102. [Google Scholar]
- 8.Dupont H L, Chapell C L, Sterling C R, Okhuysen P C, Rose J B, Jakubowski W. The infectivity of Cryptosporidium parvum in healthy volunteers. N Engl J Med. 1995;332:855–859. doi: 10.1056/NEJM199503303321304. [DOI] [PubMed] [Google Scholar]
- 9.Fayer R. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 1–42. [Google Scholar]
- 10.Fayer R, Farley C A, Lewis E J, Trout J M, Graczyk T K. Potential role of the Eastern oyster, Cassostrea virginica, in the epidemiology of Cryptosporidium parvum. Appl Environ Microbiol. 1997;63:2086–2088. doi: 10.1128/aem.63.5.2086-2088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fayer R, Graczyk T K, Lewis E J, Trout J M, Austin Farley C. Survival of infectious Cryptosporidium parvum oocysts in seawater and eastern oyster (Cassostrea virginica) in the Chesapeake Bay. Appl Environ Microbiol. 1998;64:1070–1074. doi: 10.1128/aem.64.3.1070-1074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finch G R, Daniels C W, Black E K, Schaefer F W, Belosevic M. Dose response of Cryptosporidium parvum in outbred neonatal CD-1 mice. Appl Environ Microbiol. 1993;59:3661–3665. doi: 10.1128/aem.59.11.3661-3665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graczyk T K, Fayer R, Cranfield M R, Bruce Conn D. Recovery of waterborne Cryptosporidium parvum oocysts by freshwater benthic clams (Corbicula fluminea) Appl Environ Microbiol. 1998;64:427–430. doi: 10.1128/aem.64.2.427-430.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graczyk T K, Fayer R, Trout J M, Farley C A. Cryptosporidium oocysts in Bent mussels (Ischadium recurvum) in the Chesapeake Bay. Parasitol Res. 1999;85:518–521. doi: 10.1007/s004360050590. [DOI] [PubMed] [Google Scholar]
- 15.Graña J, Macias F. Contaminación orgánica de las rías gallegas. Cuadernos Marisqueros Publ Tec. 1987;12:747–752. [Google Scholar]
- 16.Griffiths J K. Human cryptosporidiosis: epidemiology, transmission, treatment and diagnosis. Adv Parasitol. 1998;40:37–85. doi: 10.1016/s0065-308x(08)60117-7. [DOI] [PubMed] [Google Scholar]
- 17.Instituto Hidrográfico de la Marina. Derrotero de la costa NW de España. No. 2, Tomo 1. Cadiz, Spain: Servicio de Publicaciones de la Armada; 1993. [Google Scholar]
- 18.Johnson D C, Reynolds K A, Gerba C P, Pepper I L, Rose J B. Detection of Giardia and Cryptosporidium in marine waters. Water Sci Technol. 1995;31:439–442. [Google Scholar]
- 19.Ministerio de Agricultura Pesca y Alimentación. Anuario de estadística agraria. Madrid, Spain: Secretaría General Técnica; 1997. [Google Scholar]
- 20.Ortega-Mora L M, Troncoso J M, Rojo-Vazquez F A, Gomez-Bautista M. Cross-reactivity of polyclonal serum antibodies generated against Cryptosporidium parvum oocysts. Infect Immun. 1992;60:3442–3445. doi: 10.1128/iai.60.8.3442-3445.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortega Mora L M, Wright S E. Age-related resistance in ovine cryptosporidiosis: patterns of infection and humoral immune response. Infect Immun. 1994;62:5003–5009. doi: 10.1128/iai.62.11.5003-5009.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel S, Pedraza-Diaz S, McLauchlin J M, Casemore D. Molecular characterization of Cryptosporidium parvum from two large suspected waterborne outbreaks. Communicable Dis Public Health. 1998;1:231–233. [PubMed] [Google Scholar]
- 23.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C. Genetic polymorphisms among Cryptosporidium isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson L J, Campbell A T, Smith H V. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl Environ Microbiol. 1992;58:3494–3500. doi: 10.1128/aem.58.11.3494-3500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith H V, Rose J B. Waterborne cryptosporidiosis: current status. Parasitol Today. 1998;14:14–22. doi: 10.1016/s0169-4758(97)01150-2. [DOI] [PubMed] [Google Scholar]
- 26.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blanq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamburrini A, Pozio E. Long term survival of Cryptosporidium parvum oocysts in seawater and in experimentally infected mussels (Mytilus galloprovincialis) Int J Parasitol. 1999;29:711–715. doi: 10.1016/s0020-7519(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 28.Trollope D R. Use of mollusks to monitor bacteria in water. In: Grainger J M, Lynch J M, editors. Microbiological methods for environmental biotechnology. Society for Applied Bacteriology technical series 19. London, England: Academic Press Ltd.; 1984. pp. 393–409. [Google Scholar]
- 29.Varona M. La acuicultura se afianza como opción de futuro. Mar. 1997;2:28–30. [Google Scholar]
- 30.Walker F R, Stedinger J R. Fate and transport model of Cryptosporidium. J Environ Eng. 1999;125:325–333. [Google Scholar]
- 31.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]