Abstract
The most common biological control agents (BCAs) of the genus Trichoderma have been reported to be strains of Trichoderma virens, T. harzianum, and T. viride. Since Trichoderma BCAs use different mechanisms of biocontrol, it is very important to explore the synergistic effects expressed by different genotypes for their practical use in agriculture. Characterization of 16 biocontrol strains, previously identified as “Trichoderma harzianum” Rifai and one biocontrol strain recognized as T. viride, was carried out using several molecular techniques. A certain degree of polymorphism was detected in hybridizations using a probe of mitochondrial DNA. Sequencing of internal transcribed spacers 1 and 2 (ITS1 and ITS2) revealed three different ITS lengths and four different sequence types. Phylogenetic analysis based on ITS1 sequences, including type strains of different species, clustered the 17 biocontrol strains into four groups: T. harzianum-T. inhamatum complex, T. longibrachiatum, T. asperellum, and T. atroviride-T. koningii complex. ITS2 sequences were also useful for locating the biocontrol strains in T. atroviride within the complex T. atroviride-T. koningii. None of the biocontrol strains studied corresponded to biotypes Th2 or Th4 of T. harzianum, which cause mushroom green mold. Correlation between different genotypes and potential biocontrol activity was studied under dual culturing of 17 BCAs in the presence of the phytopathogenic fungi Phoma betae, Rosellinia necatrix, Botrytis cinerea, and Fusarium oxysporum f. sp. dianthi in three different media.
Trichoderma species have been investigated as biological control agents (BCAs) for over 70 years (14), but it is only recently that strains have become commercially available. This is largely a result of the change in public attitude toward the use of chemical pesticides and fumigants such as methyl bromide (44).
Knowledge concerning the behavior of these fungi as antagonists is essential for their effective use since they can act against target organisms in several ways (16). Strains of Trichoderma can produce extracellular enzymes (13) and antifungal antibiotics (11), but they may also be competitors to fungal pathogens (40), promote plant growth (15), and induce resistance in plants (5, 48). The commercial use of Trichoderma BCAs must be preceded by precise identification, adequate formulation, and studies about the synergistic effects of their mechanisms of biocontrol.
The most important BCAs against plant pathogenic fungi have been reported to be Trichoderma virens (24) von Arx, Beih, T. harzianum Rifai, and T. viride Pers.:Fr. (29, 36). In addition, there are isolates belonging to these species which are unable to act as effective BCAs. The fact that T. virens and T. harzianum were assigned to section Pachybasium (Sacc.) Bisset (3) and T. viride in section Trichoderma (2) is a demonstration of the taxonomical diversity of the BCAs included in Trichoderma, a genus in which the species concept is very wide (2, 33).
T. virens (formerly Gliocladium virens) is morphologically very similar to the anamorph of Hypocrea gelatinosa (32) and its internal transcribed spacer (ITS) sequences have revealed that it is close in proximity to T. harzianum (21).
T. harzianum is a species aggregate, grouped on the basis of conidiophore branching patterns with short side branches, short inflated phialides, and smooth and small conidia. T. harzianum has been divided in three, four, or five subspecific groups, depending on the strains and on the attributes considered (12). Four biotypes (Th1, Th2, Th3, and Th4) were originally proposed based on its pathogenicity on mushrooms (38). Molecular studies have confirmed the distribution of T. harzianum in the same four Seaby's biotypes, of which Th1 and Th3 seem to be nonpathogenic to mushrooms (28). Phylogenetic analysis based on ITS sequences (27) has revealed that Th1 is the most recent ancestor for aggressive mushroom colonizers which were included in Th2 (26) and Th4 (28) of T. harzianum. Th3 is coincident with T. atroviride Karsten, a previously described species with similarities in colony character to T. harzianum (21).
T. harzianum has been neotypified by Gams and Meyer (10) and redistributed, after molecular examination, into two major groups: T. harzianum sensu lato (s.l.) and T. viride-T. atroviride complex.
T. harzianum s.l. includes the most common strains used as BCAs (27) and comprises Th1 and T. inhamatum (20, 18). Colombian isolates of T. harzianum were initially characterized as T. inhamatum (45) due to their lack of sterile appendages to the conidiophores and globose conidia as main features. Th1 and T. inhamatum have been considered to be two different species not only because of their morphological features but also because of two base sequence differences in ITS1 (10). However, T. harzianum and T. inhamatum are also considered as cospecific (2), and their ITS1 sequence variability ranges within the divergences described for Th1 (20). Strains from Th2 and Th4 are not BCAs (27) and are also distinct from T. harzianum s.l. according to molecular information (10).
T. viride-T. atroviride complex is characterized by specific restriction fragment length polymorphism (RFLP) patterns and contains isolates with quickly darkening conidia (10). T. viride includes an aggregate of species with globose or subglobose to ellipsoidal warted conidia; most of them produce antibiotics and a typical coconut odor (6). The morphology of T. viride has not been clear for many years until two types of conidial ornamentation were described (23). Recently, it has been found that T. viride is a paraphyletic group and an integrated morphological and molecular approach has confirmed the redefinition of types I and II of T. viride in two species. Type I is the true T. viride species, which also includes the anamorph of Hypocrea rufa, and is grouped together with strains of T. atroviride and T. koningii (22). Type II represents the new species T. asperellum (37, 22), which has ovoidal rather than globose conidiation and also darker and faster conidiation.
Strains of Trichoderma used in this work were previously studied and selected as BCAs against 10 isolates of five different soilborne fungal plant pathogens (12). The distribution of these strains in physiological and biochemical groups was related to the biological activity against different plant pathogens, and a preliminary molecular characterization into ribosomal DNA (rDNA) groups was initiated in an earlier study (12). In the present study, we have followed the recent nomenclature applied by Gams and Meyer (10) to T. harzianum s.l. and by Samuels et al. (37) and Lieckfeldt et al. (22) to T. asperellum.
The present study was carried out to distinguish strains of Trichoderma by trying to demonstrate the influence of both the fungal target and the substrate composition under controlled conditions before exploring their potential capacity for biocontrol in natural environments. We have made use of ITS sequencing and polymorphisms originated by hybridization of total genomic DNA with a mitochondrial DNA (mtDNA) probe to study 16 biocontrol strains previously identified as T. harzianum and one biocontrol T. viride strain. The strain distribution in several genotypes could also support the idea of developing antifungal formulations in which different Trichoderma BCAs could be combined. We have also studied, in three different media, the behavior of these BCAs in dual cultures against Phoma betae, Rosellinia necatrix, Botrytis cinerea, and Fusarium oxysporum f. sp. dianthi, which are fungi pathogenic to sugar beet, avocado, vine, and carnation, respectively.
MATERIALS AND METHODS
Strains.
The strains of Trichoderma spp. utilized in this study are given in Table 1. Seventeen strains were tested in RFLP analysis of mtDNA and ITS sequencing and were also used in confrontation assays in vitro against phytopathogenic fungi. All of the sequences corresponding to strains represented in Table 1 were included in phylogenetic analysis based on the ITS1 sequence.
TABLE 1.
Strains used in this study
| Straina | Strain no. | Geographic origin | Sourceb | EMBL no. | Taxon label in sequencec |
|---|---|---|---|---|---|
| T. asperellum | U.S.d | TR 48 | AJ230669 | ||
| T. atroviride | U.S. | DAOM 165779 | Z48817 | ||
| T. hamatum (ex-neotype strain) | Canada | DAOM 167057 | Z48816 | ||
| T. harzianum* | 3 | France | IMI 20179 | AJ224020 | Asp |
| T. harzianum* | 11 | France | IMI 352941 | AJ224008 | Atr |
| T. harzianum* | 24 | Spain | IMI 352940 | AJ224006 | Inh |
| T. harzianum* | 260 | England | IMI 352939 | AJ224007 | Atr |
| T. harzianum* | 2923 | Zimbabwe | IMI 281112 | AJ224009 | Atr |
| T. harzianum* | 2924 | India | IMI 293168 | AJ224010 | Inh |
| T. harzianum* | 2925 | Colombia | IMI 296235 | AJ224011 | Har |
| T. harzianum* | 2926 | England | IMI 298371 | AJ224012 | Inh |
| T. harzianum* | 2927 | England | IMI 298372 | AJ224013 | Inh |
| T. harzianum* | 2928 | England | IMI 298373 | AJ224014 | Inh |
| T. harzianum* | 2929 | England | IMI 298374 | AJ224015 | Inh |
| T. harzianum* | 2930 | India | IMI 304056 | AJ224016 | Inh |
| T. harzianum* | 2931 | India | IMI 304057 | AJ224017 | Inh |
| T. harzianum* | 2932 | India | IMI 304058 | AJ224018 | Lon |
| T. harzianum* | 2933 | Sri Lanka | IMI 300082 | AJ224019 | Inh |
| T. harzianum* | ThVA | Spain | IMI 20268 | AJ224021 | Asp |
| T. harzianum | Germany | CBS 978.70 | AJ222723 | ||
| T. harzianum (ex-neotype strain) | England | CBS 226.95 | AJ222720 | ||
| T. harzianum | 6884 | Australia | CBS 819.68 | Z68189 | |
| T. harzianum | Th2I (63) | Northern Ireland | P. R. Mills | U78880 | |
| T. harzianum | Th4C (92) | Canada | D. L. Rinker | U78882 | |
| T. inhamatum (ex-type strain) | Colombia | CBS 273.78 | Z68187 | ||
| T. koningii | The Netherlands | CBS 457.96 | Z79628 | ||
| T. koningii | U.S. | GJS 91-7 | Z95923 | ||
| T. longibrachiatum (ex-type strain) | U.S. | CBS 816.68 | Z31019 | ||
| T. parceramosum (ex-type strain) | U.S. | CBS 259.85 | Z31015 | ||
| T. viride* | 25 | Colombia | IMI 296237 | AJ223773 | Asp |
| T. viride | England | CBS 240.63 | X93979 | ||
| T. viride | U.S. | TR 22 | AJ230678 | ||
| T. viride-T. atroviride | 5502 | Japan | CBS 470.94 | AJ222719 |
Biological control strains included in hybridization studies and dual-culture tests and sequenced in this work are indicated with an asterisk. ITS1 sequences of all strains were included in the phylogenetic analysis.
Abbreviations: CBS, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands; DAOM, Department of Agriculture (Mycology), Ottawa, Canada; GJS, collection of Gary J. Samuel, Betsville, Mass.; IMI, International Mycological Institute, Egham, United Kingdom; TR, collection of Earl Nelson (maintained at the USDA-ARS Beltsville Collection).
Asp, T. asperellum; Atr, T. atroviride; Inh, T. inhamatum; Har, T. harzianum; Lon, T. longibrachiatum.
U.S., United States.
Fungal growth conditions and DNA extraction.
Cultures were maintained on potato dextrose agar (PDA) (Difco) at 25°C, and mycelia for DNA extraction were grown in liquid cultures (120 rpm) at 25°C in potato dextrose broth (PDB) (Difco) for 48 h. Hyphae were collected on filter paper in a Buchner funnel, washed with distilled water, frozen, and lyophilized. Genomic DNA was extracted using the method of Raeder and Broda (31). DNA was resuspended in 50 μl of 10 mM Tris–1 mM EDTA (pH 8) and quantified by use of ethidium bromide fluorescence (35).
The mtDNA was isolated from T. harzianum 2924. Briefly, 20 g of frozen mycelium was ground in liquid nitrogen. Cell lysis was achieved in 0.33 M sucrose–1 mM EDTA–10 mM tricine (pH 7.5), and the mixture was centrifuged at 4,000 rpm for 10 min at 4°C. The supernatant was filtered through cheesecloth and centrifuged at 12,000 rpm for 60 min at 4°C. The supernatant was discarded, and the mitochondrial pellet was resuspended in 150 mM NaCl–1 mM EDTA–100 mM Tris (pH 8 buffer)–100 μg of proteinase K (Sigma, St. Louis, Mo.) per ml. Mitochondria were lysed by incubation for 40 min at 65°C in sodium dodecyl sulfate and Nonidet P-40 (Sigma) to final concentrations of 1% (wt/vol) and 4% (wt/vol), respectively. After incubation for 10 min on ice, any nonlysed material was removed by centrifugation at 10,000 rpm for 20 min at 4°C. The supernatant was decanted and purified by cesium chloride-bisbenzimide gradient centrifugation at 44,000 rpm for 48 h. The mtDNA band was removed and washed with butanol to extract the dye and the mtDNA was concentrated by ethanol precipitation.
PCR amplification.
Primers ITS1 and ITS4 described by White et al. (47) were synthesized by Boehringer Mannheim (Mannheim, Germany) and used to amplify a fragment of rDNA including ITS1 and ITS2 and the 5.8S rDNA gene. PCR amplifications were performed in a total volume of 50 μl by mixing 40 ng of the template DNA with 0.2 μM concentrations of each primer, 200 μM concentrations of each deoxynucleoside triphosphate, and 2.5 U of Taq DNA polymerase in GeneAmp 10× PCR Buffer II (100 mM Tris-HCl, pH 8.3; 500 mM KCl) (Perkin-Elmer). These reactions were subjected to an initial denaturation of 5 min at 95°C, followed by 35 cycles of 1.5 min at 94°C, 2 min at 55°C, and 3 min at 72°C, with a final extension of 5 min at 72°C in a Perkin-Elmer Cetus thermal cycler. Aliquots (4 μl) were analyzed by electrophoresis in 1.2% (wt/vol) agarose gel in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA [pH 8]), stained with ethidium bromide, and photographed over a transilluminator. The molecular size marker was φX174-HaeIII (Promega, Madison, Wis.).
Sequencing of amplification products.
Sequencing of the rDNA region, including the spacers ITS1 and ITS2 and 5.8S rDNA, was carried out by automated DNA sequencing with fluorescent terminators using ABI 377 Prism Sequencer (Applied Biosystems, Foster City, Calif.) at the Department of Molecular Biology, University of Alcalá de Henares, Madrid, Spain. Prior to sequencing, PCR products were purified with Centricon-100 concentrators (Amicon, Madison, Wis.) according to the manufacturer's specifications. Each Trichoderma isolate was sequenced in both directions using primers ITS1-ITS3 and ITS2-ITS4 (47) with a T7 DNA Polymerase Sequencing Kit (Pharmacia, Uppsala, Sweden).
Phylogenetic analysis.
Sequence alignments were conduced with the CLUSTAL W program (43). The alignment was then optimized manually. Nucleotide divergences were estimated using the Kimura's two-parameter method (17), excluding gaps and equivocal sites with the DNADIST program implemented in the PHYLIP package, version 3.5 (8). Phylogenetic inference was performed by the neighbor-joining (NJ) method (34) with the NEIGHBOR program from the PHYLIP package. Relative support for particular clades in the NJ tree was estimated by using 1,000 replications of the bootstrap procedure (7), performed with the SEQBOOT, DNADIST, and NEIGHBOR programs from the PHYLIP package. Fusarium sambucinum (accession number X65477), and Verticillium albo-atrum (accession number L19499), were used as outgroup when necessary.
Southern blotting and hybridization.
Genomic DNA (8 μg) was digested with EcoRI or EcoRV (Promega) restriction enzymes until completion at 37°C according to the manufacturer's instructions. Restriction fragments were separated in 0.8% (wt/vol) agarose gel in 1× TAE buffer at 0.5 V cm−1 overnight. HindIII/EcoRI-HindIII-digested λ DNA was used as the molecular size marker. The DNA was capillary transferred from the gel to a nylon membrane (Hybond N; Amersham-Buchler, Braunschweig, Germany) (41), immobilized by baking at 80°C for 90 min, and hybridized with an mtDNA probe, obtained after purification of mitochondria from T. harzianum 2924 and digestion with BamHI. The probe was labeled with digoxigenin using the DIG DNA Labeling Mix Kit (Boehringer Mannheim).
The filters were prehybridized in 6× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate; pH 7.0)–1× Denhardt's solution–5% SDS–denatured herring sperm DNA (5 mg/ml) for 2 h at 65°C. Hybridization was done overnight after the addition of 20 ng of denatured probe per ml according to the procedure described by Sambrook et al. (35). Filters were washed twice at 37°C with 2× SSC plus 0.1% SDS for 5 min, with two final high-stringency washes at 65°C with 0.1× SSC plus 0.1% SDS for 15 min each.
The blots were subjected to immunological detection using anti-digoxigenin antibody to alkaline phosphatase and CDP-Star (Boehringer Mannheim) according to the manufacturer's instructions. Chemiluminescent signals were detected by exposing the filters to an X-ray film (Fuji Photofilm Europe Gmbh, Düsseldorf, Germany) for up to 5 min.
Confrontation assays in vitro.
In vitro confrontations were studied between the Trichoderma isolates and the phytopathogenic fungi Fusarium oxysporum f. sp. dianthi (CECT 20270), isolated from the metula of carnation in Esperanza (Santa Fe, Argentina); Phoma betae 17 (CECT 20198), isolated from a sugar beet seedling in Salamanca (Spain); Botrytis cinerea B05.10 (CECT 20382), a monosporic line of an original strain isolated from vine in Italy; and Rosellinia necatrix 397 (CECT 20383), isolated from avocado root in Málaga (Spain). Agar plugs cut from the growing edge of a 4-day colony of each phytopathogenic fungus were placed 1 cm from the border of petri dishes containing PDA, malt agar (MA) (Difco) or corn meal agar (CMA) (Difco). P. betae and R. necatrix were allowed to grow at 25°C for 3 days before the sowing of Trichoderma strains at 1 cm from the border on the opposite side of the same petri plates where these target pathogens were grown. In the same way, F. oxysporum f. sp. dianthi was grown 1 day before the antagonists and B. cinerea, and the biocontrol strains were sown at the same time. Growth parameters in all dual cultures were read after 7 days. Morphological characteristics of the sporulation on the colony pathogen and the sporulation and production of a yellow pigment on the surface of the medium were recorded by using a 0 to 3 scale in which the values were coded as follows: 0, absence; 1, weak; 2, heavy; and 3, very heavy. In order to facilitate the application of the statistical treatment, these values were transformed into a scale in which 1 was the maximum rate. Mean values were registered in a StatView 4.01 program and analyzed by using a Fisher exact test. The assay was repeated twice.
Sequences corresponding to ITS1, ITS2, and the 5.8S rDNA gene from Trichoderma strains used in this study were deposited and are available in the EMBL database under the accession numbers given in Table 1.
RESULTS
PCR amplification.
A single product of approximately 560 to 600 bp was obtained from all the PCR amplifications with primers ITS1 and ITS4 for 17 biocontrol isolates of Trichoderma spp.
DNA sequencing.
The ITS1 region of five isolates (isolates 11, 24, 260, 2925, and 2931), representing the two T. harzianum major groups, had been previously sequenced (12). ITS2 sequence was completed for these strains, as well as the further biocontrol strains included in this study. The size of PCR products amplified with the primer pair ITS1-ITS4 ranged from 563 to 602 bp and contained ITS1, ITS2, and the 5.8S rDNA gene and also 11 bp of the 3′ end of the 18S rDNA and 36 bp of the 5′ end of the 28S rDNA.
ITS1 and ITS2 lengths of the 17 biocontrol isolates were split into three groups. The first group included 6 strains, a second group included 10 strains, and a third group included one strain only. The ITS lengths were in agreement with those shown in an earlier study of T. longibrachiatum strains (20). The different ITS lengths are given in Table 2. The length of the 5.8S gene was 158 bp for the 17 strains investigated, and no sequence variation was detected among strains.
TABLE 2.
Infra- and interspecific variation (V) within ITS1 and ITS2 sequences of the BCAs investigated
| Species | No. of sequences | ITS1
|
ITS2
|
||
|---|---|---|---|---|---|
| Length (bp) | V (%) | Length (bp) | V (%) | ||
| Infraspecific | |||||
| T. atroviride | 3 | 184 | 0 | 179 | 0 |
| T. asperellum | 3 | 181–183 | 0.5 | 176–178 | 1.1 |
| T. harzianum s.l. | 10 | 199–203 | 3.0 | 174–175 | 2.8 |
| T. longibrachiatum | 1 | 222 | 171 | ||
| Interspecific | |||||
| T. atroviride versus T. asperellum | 6 | 3.8–3.9 | 5.7–6.8 | ||
| T. atroviride versus T. harzianum s.l. | 13 | 17.6–18.0 | 10.3–10.9 | ||
| T. atroviride versus T. longibrachiatum | 4 | 15.3 | 4.7 | ||
| T. asperellum versus T. harzianum s.l. | 13 | 15.6–16.5 | 9.4–11.8 | ||
| T. asperellum versus T. longibrachiatum | 4 | 14.3–14.4 | 4.7 | ||
| T. harzianum s.l. versus T. longibrachiatum | 11 | 9.5–10.4 | 4.1–6.0 | ||
Four principal ITS sequence types can be recognized, as defined by common sequence motifs and characteristic indels. Type 1 sequences included isolates 24, 2924, 2925, 2926, 2927, 2928, 2929, 2930, 2931, and 2933, and their sequences were similar to those described for strains of T. harzianum s.l. The ITS1 sequence was more variable, although isolates 2927 and 2928 showed identical sequences in this region but differed in one nucleotide in the ITS2. Type 2, observed in strains 11, 260, and 2923, had identical ITS sequences. Their ITS1 sequences were identical to that of T. atroviride DAOM 165779 and had a one-base difference from two T. koningii strains (CBS 457.96 and GJS 91-7), while their ITS2 sequences were different by one base from T. atroviride DAOM 165779 and by three to four bases from T. koningii. Type 3 is represented by strains 3, 25, and ThVA, and their ITS sequences are similar to those of T. asperellum. These three strains showed higher divergences in the ITS2 (1.1%) than in the ITS1 (0.5%) sequences. A fourth type appeared in strain 2932. This strain showed a percentage of similarity of 98.2 to 98.5% with strains of T. longibrachiatum (accession nos. X93932, X93930, X93939, and Z31019) and a significant divergence in its ITS sequences from T. atroviride (ITS1, 15.3%; ITS2, 4.7%), T. asperellum (ITS1, 14.3 to 14.4%; ITS2, 4.7%), and T. harzianum s.l. (ITS1, 9.5 to 10.4%; ITS2, 4.1 to 6.0%). The infraspecies and interspecies variations derived from the nucleotide sequence of ITS1 and ITS2 are shown in Table 2.
Phylogenetic analysis.
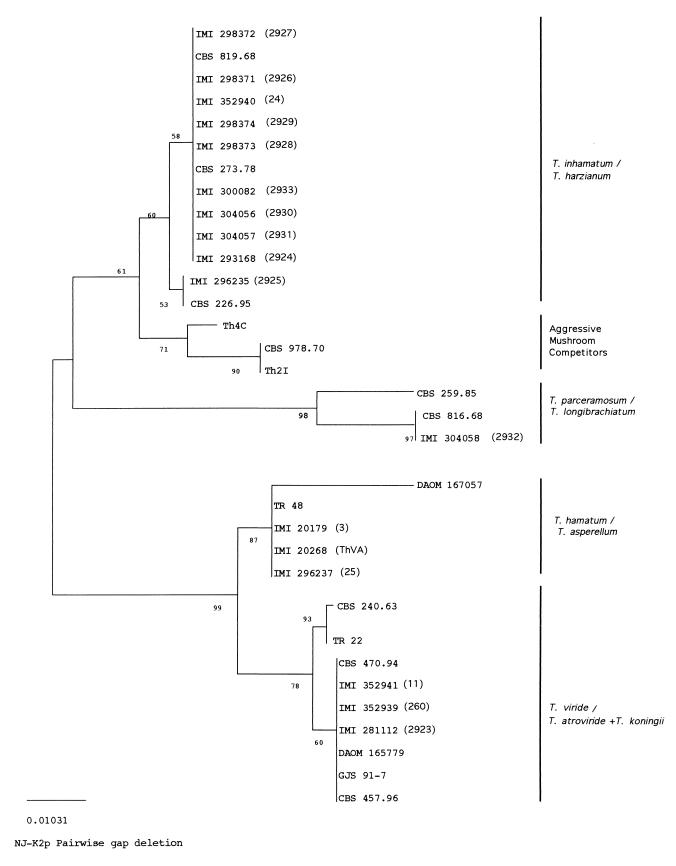
The phylogenetic tree obtained by sequence analysis of ITS1 of 17 biocontrol strains and the sequences of 17 other Trichoderma spp. obtained from sequence databanks is represented in Fig. 1. The ITS1 sequence was chosen for this analysis because it has been showed to be more informative with various sections of the genus Trichoderma (19, 20, 28). Strains of Fusarium sambucinum and Verticillium albo-atrum were used as an outgroup to demonstrate the situation of the root. The outgroup is not shown because the distance between the outgroup and the ingroup is very high and because ingroup branches became very short and thus their representation was difficult. Bootstrap analysis with 1,000 bootstrap replications demonstrated three major branches. On the basis of the bootstrap values, the 34 Trichoderma spp. isolates could be divided into five groups. Group 1, the T. harzianum-T. inhamatum complex, includes 10 of our biocontrol strains with T. harzianum CBS 819.68, T. inhamatum CBS 273.78 (ex-type strain), and T. harzianum CBS 226.95 (ex-neotype strain). This group is supported with a bootstrap value of 60% and contains two subgroups supported by bootstrap values higher than 50%. One of them puts nine of our strains together with the ex-type strain of T. inhamatum. The other subgroup includes the Colombian strain 2925 with the ex-neotype strain of T. harzianum. Group 2 includes three strains representing biotypes Th2 and Th4 of T. harzianum associated with mushroom green mold and is supported by a bootstrap value of 71%. Two distinct subgroups corresponding to both T. harzianum biotypes could be also observed. Group 3 comprises strain 2932 and the ex-type strain T. longibrachiatum CBS 816.68, grouped with a bootstrap stability of 97%, and the ex-type strain T. parceramosum CBS 259.85. This grouping is well supported with a bootstrap value of 98%. Group 4 includes the biocontrol strains 3, 25, and ThVA, which are grouped with the representative strain TR 48 of the new species T. asperellum and the ex-neotype strain DAOM 167057 of T. hamatum. This cluster is supported by a bootstrap value of 87%. Group 5 includes the complex T. viride-T. atroviride and is divided in two subgroups supported by bootstrap values of 93 and 60%. One of these subgroups includes two representative strains of T. viride. The other subgroup contains the biocontrol strains 11, 260, and 2923; two strains of T. atroviride; and two strains of T. koningii. Biocontrol strains included in this study are split into four of the five groups obtained in the tree and are not grouped with strains causing green mold in mushrooms.
FIG. 1.
Phylogenetic relationships of 34 strains of Trichoderma spp. inferred by analysis of ITS1 sequences. The tree was obtained from analysis by the NJ method using the Kimura two-parameter technique of the PHYLIP package (8).
Our results also suggest that 16 of 17 BCAs received as T. harzianum and T. viride were misidentified. Strains 24, 2924, 2926, 2927, 2928, 2929, 2930, 2931, and 2933 were received as T. harzianum, but their position in the T. harzianum-T. inhamatum complex suggests that they should be identified as T. inhamatum. Strain 2932 was received as T. harzianum and is allocated in the tree with the type strain of T. longibrachiatum. Isolates 3, 25, and ThVA are grouped with the typical T. asperellum. Strains 11, 260, and 2923 cannot be classified as T. harzianum since their ITS1 sequences are more similar to those of T. atroviride and T. koningii. However, these strains are more similar to T. atroviride since their ITS2 sequences differ by zero to one base from those of T. atroviride DAOM 165779 and CBS 470.94 and by three to four bases from those of T. koningii GJS 91-7 and CBS 457.96.
Polymorphisms in mtDNA.
BamHI-digested mtDNA from isolate 2924 hybridized with total DNA digested with EcoRI or EcoRV and gave polymorphisms for all isolates studied. In both cases, these hybridizations split the 17 Trichoderma isolates in two major groups, with the exception of ThVA.
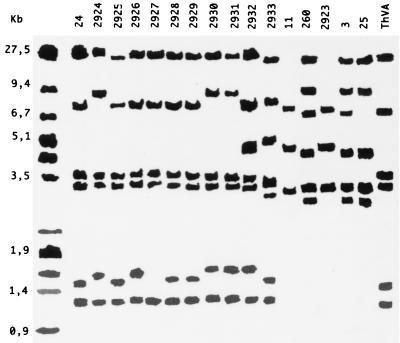
Hybridization of total DNA digested with EcoRI with the probe of mtDNA gave 10 different patterns (Fig. 2). Strains 24, 2925, and ThVA showed a similar pattern, as well as strains 2924, 2930, and 2931, and the strain pairs 2928-2929, 11-2923, and 260-3. Strains 2926, 2927, 2932, 2933, and 25 gave individual specific band patterns.
FIG. 2.
Southern blots of EcoRI-digested genomic DNA of Trichoderma isolates hybridized with mtDNA from T. harzianum 2924 digested with BamHI as a probe. Isolate numbers are listed at the top of the figure. On the left, the molecular size standards of HindIII/EcoRI-HindIII-digested λ DNA are indicated in kilobases.
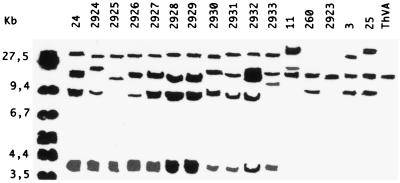
Hybridization of total DNA digested with EcoRV with the probe mentioned above clearly showed two defined patterns which grouped the isolates into 11 and 6 strains (Fig. 3). In the first set of strains four different patterns can be distinguished; those corresponding to (i) strains 24, 2926, 2927, 2928, 2929, 2931, and 2932; (ii) strains 2924 and 2930; (iii) strain 2925; and (iv) strain 2933. The second set showed five distinct patterns corresponding to strains 11, 2923 and ThVA, 260, 3, and 25.
FIG. 3.
Southern blots of EcoRV-digested genomic DNA of Trichoderma isolates hybridized with mtDNA from T. harzianum 2924 digested with BamHI as a probe. Isolate numbers are listed at the top of the figure. On the left, the molecular size standards of HindIII/EcoRI-HindIII-digested λ DNA are indicated in kilobases.
Morphological characteristics in dual cultures.
The parameters with reference to sporulation on the plate, sporulation on the pathogen colony, and production of yellow pigmentation in the culture media obtained after preparation of dual cultures from three strains of T. atroviride, three strains of T. asperellum, nine strains of T. inhamatum, one strain of T. harzianum, and one strain of T. longibrachiatum versus the plant pathogenic fungi P. betae, R. necatrix, B. cinerea, and F. oxysporum f. sp. dianthi on PDA, CMA, and MA were registered. Mean values and standard deviations (SD) for the five species separated in the phylogenetic analysis are shown in Table 3.
TABLE 3.
Dual-culture assay of T. atroviride, T. asperellum, T. inhamatum, T. harzianum, and T. longibrachiatum after 7 days at 25°C with the plant pathogenic fungi P. betae, R. necatrix, B. cinerea, and F. oxysporum f. sp. dianthi on PDA, CMA, and MA
| Parameter and species | Scale results ± SDa on:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDA with:
|
CMA with:
|
MA with:
|
||||||||||
| P. betae | R. necatrix | B. cinerea | F. oxysporum f. sp. dianthi | P. betae | R. necatrix | B. cinerea | F. oxysporum f. sp. dianthi | P. betae | R. necatrix | B. cinerea | F. oxysporum f. sp. dianthi | |
| Sporulation on plate | ||||||||||||
| T. atroviride | 0.9 ± 0.1A | 0.4 ± 0.2 | 0.7 ± 0A | 0.7 ± 0.5A | 0.3 ± 0 | 0.3 ± 0A | 0.3 ± 0A | 0.3 ± 0 | 0.9 ± 0.1A | 0.7 ± 0A | 0.9 ± 0.1A | 0.9 ± 0.1A |
| T. asperellum | 0.7 ± 0AB | 0.2 ± 0.1 | 0.8 ± 0.1A | 0.7 ± 0A | 0.3 ± 0 | 0.3 ± 0A | 0.3 ± 0A | 0.3 ± 0 | 0.7 ± 0B | 0.3 ± 0.3B | 0.8 ± 0.1A | 0.9 ± 0.1A |
| T. inhamatum | 0.5 ± 0.3C | 0.2 ± 0.3 | 0.3 ± 0.3B | 0.4 ± 0.3AB | 0.2 ± 0.1 | 0.1 ± 0.1B | 0.3 ± 0.1A | 0.3 ± 0.1 | 0.3 ± 0.1C | 0.1 ± 0.1C | 0.3 ± 0.2B | 0.3 ± 0.2B |
| T. harzianum | 0.3 ± 0BC | 0 | 0C | 0.3 ± 0AB | 0.3 ± 0 | 0.3 ± 0AB | 0.3 ± 0A | 0.3 ± 0 | 0.3 ± 0C | 0C | 0C | 0C |
| T. longibrachiatum | 0.3 ± 0BC | 0 | 0B | 0B | 0.3 ± 0 | 0.3 ± 0AB | 0B | 0.3 ± 0 | 0.3 ± 0C | 0C | 0.3 ± 0BC | 0.3 ± 0BC |
| Sporulation on pathogen | ||||||||||||
| T. atroviride | 0.9 ± 0.1A | 0 | 0.7 ± 0.5 | 0.7 ± 0.5 | 0A | 0 | 0.3 ± 0A | 0A | 0.9 ± 0.1A | 0 | 1 ± 0A | 0.5 ± 0.3AB |
| T. asperellum | 0.7 ± 0AB | 0 | 0.9 ± 0.1 | 0.7 ± 0 | 0A | 0 | 0.3 ± 0A | 0A | 0.4 ± 0.2B | 0 | 0.8 ± 0.3A | 0.2 ± 0.1A |
| T. inhamatum | 0.4 ± 0.3C | 0 | 0.5 ± 0.4 | 0.4 ± 0.4 | 0.03 ± 0.1A | 0 | 0.2 ± 0.1B | 0A | 0.1 ± 0.1C | 0 | 0.4 ± 0.4B | 0.6 ± 0.4B |
| T. harzianum | 0.3 ± 0BC | 0 | 1 ± 0 | 0.3 ± 0 | 0A | 0 | 0.3 ± 0AB | 0A | 0.3 ± 0BC | 0 | 0B | 0.3 ± 0AB |
| T. longibrachiatum | 0.7 ± 0BC | 0 | 1 ± 0 | 0.3 ± 0 | 0.3 ± 0B | 0 | 1 ± 0C | 0.3 ± 0B | 1 ± 0A | 0 | 1 ± 0A | 0.3 ± 0AB |
| Production of yellow pigment | ||||||||||||
| T. atroviride | 0A | 0.3 ± 0A | 0A | 0A | 0 | 0 | 0 | 0 | 0A | 0.6 ± 0.2A | 0A | 0A |
| T. asperellum | 0A | 0.3 ± 0A | 0A | 0A | 0 | 0 | 0 | 0 | 0A | 0.7 ± 0AB | 0.1 ± 0.1AB | 0A |
| T. inhamatum | 0.7 ± 0.2B | 0.7 ± 0.3B | 0.6 ± 0.3B | 0.7 ± 0.3B | 0 | 0 | 0 | 0 | 1 ± 0B | 0.8 ± 0.3AB | 0.8 ± 0.1C | 0.9 ± 0.1B |
| T. harzianum | 0A | 0.3 ± 0A | 0A | 0.3 ± 0A | 0 | 0 | 0 | 0 | 0.7 ± 0C | 0.7 ± 0AB | 0.3 ± 0B | 0.3 ± 0C |
| T. longibrachiatum | 1 ± 0B | 1 ± 0B | 1 ± 0C | 1 ± 0B | 0 | 0 | 0 | 0 | 1 ± 0B | 1 ± 0B | 1 ± 0C | 1 ± 0B |
Values correspond to the following growth parameters: sporulation on the plate, sporulation on the pathogen colony, and production of a yellow pigment. Results were registered in a scale and are coded as follows: 0, absence; 1, weak; 2, heavy; and 3, very heavy. The mean values from two repetitions were transformed into a scale in which 1 was the maximum rate. The means in each column followed by the same superscript letter are not significantly different (P ≤0.05).
The sporulation on the plate was heavier on PDA and MA than on CMA for T. atroviride, T. asperellum, and T. inhamatum; T. atroviride showed the maximum sporulation, and T. harzianum 2925 and T. longibrachiatum 2932 showed lower sporulation on these media. The different degrees of sporulation were statistically significant (P ≤ 0.05) in the presence of P. betae, B. cinerea, and F. oxysporum f. sp. dianthi on PDA, in the presence of R. necatrix and B. cinerea on CMA, and also in the dual cultures with the four pathogens on MA.
The sporulation of all Trichoderma strains on the pathogen colony was also heavier on PDA and MA. T. longibrachiatum 2932 was the most aggressive strain. With the exception of T. inhamatum against F. oxysporum f. sp. dianthi on MA and T. harzianum against B. cinerea on PDA, these two species showed the lowest degree of sporulation on the four pathogens. B. cinerea was the most sensitive pathogen to the assault of the BCAs, and R. necatrix was the most resistant since all of the Trichoderma strains tested were unable to colonize this pathogen and stopped their growth at the border of its colonies. The different degree of sporulation on the pathogen was statistically significant (P ≤ 0.05) in the presence of P. betae on the three media and in the presence of B. cinerea and F. oxysporum f. sp. dianthi on CMA and MA.
The production of a diffuse yellow pigment in the medium was not observed on CMA, but this feature allowed us to detect significant differences between the productive strains, T. inhamatum, T. harzianum, and T. longibrachiatum and the nonproductive strains T. atroviride and T. asperellum on PDA and MA. This occurs against all the pathogens tested except for the dual cultures with P. betae and B. cinerea on PDA, where T. harzianum was unable to produce yellow pigmentation. As an exception, T. atroviride and T. asperellum produced a yellow pigment in the proximity of the colonies of R. necatrix on PDA and MA. The production of this yellow pigment was significantly different (P ≤ 0.05) for the four pathogens in the dual cultures on PDA and MA.
DISCUSSION
Trichoderma species can act as biocontrol agents through different synergistic mechanisms. However, it is difficult to predict the degree of synergism and the behavior of a BCA in a natural pathosystem. Considering that environmental conditions are important, the right selection of BCAs, which begins with a safe characterization of biocontrol strains in the new taxonomic schemes of Trichoderma, is equally important since the exact identification of strains to the species level is the first step in utilizing the full potential of fungi in specific applications (22).
According to their ITS lengths, the BCAs studied split into three groups (Table 2). T. harzianum s.l. (T. inhamatum and T. harzianum) showed the typical sizes expected for the section Pachybasium. T. atroviride and T. asperellum had a number of nucleotides characteristic of section Trichoderma, and strain 2932 had the species-specific ITS size corresponding to section Longibrachiatum (20). T.. atroviride and T. asperellum, with a similar ITS size and a similar sequence variation with T. harzianum s.l., showed between them nucleotide divergences of 3.8 to 3.9% in ITS1 and 5.7 to 6.8% in ITS2. These percentages fall into the values reported in Trichoderma spp. (infraspecific divergences of between 0 and 2.5% and interspecific divergences of between 2.0 and 20.3% [12, 20]). The use of divergence sequence data for defining species requires careful interpretation, especially because length polymorphisms and inversions can make comparison on a simple basis difficult (4, 39). rDNA sequence divergence can be used as a valid tool for the assignation of rank to taxonomic groups within genera such as Colletotrichum (infraspecific divergences of between 0 and 5% and interspecific divergences of between 7 and 23% for the ITS1 region [42]), but Trichoderma appears to be a more complicated genus since the phylogenetic relationships of many of its members are still unclear (10, 27).
The sequence alignment revealed the presence of hot spots (27) within both ITS regions, which supports the distribution of strains 3, 11, 25, 260, 2923, and ThVA into two separated groups. In general terms, ITS1 showed the highest number of nucleotide substitutions, and it was used for the phylogenetic study. Although studies involving biocontrol and green mold isolates of T. harzianum revealed that the 5.8S rRNA gene is as variable as or even more variable than ITS2 (27), the sequence alignment of our biocontrol isolates did not show variation into this gene.
The results obtained from phylogenetic analysis of ITS1 sequences showed that the 17 Trichoderma BCAs used here can be separated into five different species, and all of them are clearly distinct from T. harzianum Th2 and Th4 (Fig. 1), which is associated with mushroom green mold. These molecular data support the idea that biocontrol Trichoderma strains are not pathogenic to mushrooms. However, rigorous infectivity studies of BCAs in mushrooms are needed since the high value of this information in ecotoxicological studies led to the commercial registration of Trichoderma strains.
Considering the phylogenetic analysis based on ITS1 sequences, T. inhamatum and T. harzianum are grouped as separate clusters. The tree shows that T. harzianum s.l. is divided into these two taxons, which appear in the proximity of the aggressive mushroom competitors. This result is in agreement with previous investigations (10, 28). T. inhamatum makes up the main body of Trichoderma biocontrol strains since nine of them are placed with the ex-type strain of T. inhamatum for one only strain located with the ex-neotype strain of T. harzianum. The separation of these species was proposed by Veerkamp and Gams (45) based on morphological criteria. Other authors have argued that there are not enough reasons to consider T. inhamatum as a different entity to T. harzianum since the ITS sequence homology (20), the RFLP patterns of mitochondrial DNA (25), and Bissett's morphological descriptions (2) suggest that T. harzianum and T. inhamatum are the same species. However, a recent study has defended the separation of both species based on phylogenetic arguments (10). These discrepancies demand a redefinition of the T. harzianum complex (21), but we have wanted to name the two subgroups of T. harzianum s.l. depending on the location of the ex-type species considered in the phylogenetic tree prepared in the present study, referring to the major subgroup, which clusters most of the BCAs, as “T. inhamatum,” and to the other subgroup, with one BCA only, as “T. harzianum.” According to these criteria, the mushroom pathogenic strains should be considered as an independent species.
As an incidental case, T. inhamatum was initially described as separate Colombian isolates from European strains identified as T. harzianum (45), but T. harzianum 2925, the only Colombian strain of this complex, has now been clustered within T. harzianum.
Strain 2932, misidentified as T. harzianum, is located with the ex-type strain T. longibrachiatum CBS 816.68 with a high bootstrap value (97%). It will be interesting to explore new BCAs within the monophyletic group T. longibrachiatum since this result is supported by the fact that the ex-type strain T. parceramosum CBS 259.85, patented as an antiviral antibiotic producer (see references 1 and 20), constitutes a stable branch (see also reference 21) of T. longibrachiatum, with a bootstrap value of 98%.
The two separated groups, T. atroviride and T. asperellum, suggested after ITS sequence variability comparisons, were confirmed in the phylogenetic analysis. This grouping is in agreement with the separation of T. viride from the new species T. asperellum using an integrated molecular approach (22). Three of our isolates, strains 3, 25, and ThVA, are grouped, with a high bootstrap value of 87%, with the biocontrol isolate TR 48 of T. asperellum (22), and strain 3 has an ITS1 sequence identical to that of biocontrol strain T-203 (see reference 22), confirming that this new species can be a good source of BCAs. The ex-neotype strain of T. hamatum DAOM 167057 was also grouped within this clade, in agreement with the close position of T. asperellum to this species (22). We have detected, in our collection of BCAs, new antagonists of Fusarium oxysporum, isolated from compost, which have been characterized as members of T. asperellum.
The fifth group of the tree, T. viride-T. atroviride complex, contains species assigned to section Trichoderma: T. viride, T. atroviride, and T. koningii. Low variation among the ITS sequences has been described among the species of this section, and the maximum variation between morphologically characterized groups within the section was 7 bp (20). However, our phylogenetic analysis separates these species into two subgroups, which are well supported by bootstrap values. One of them includes the two true T. viride strains, and the other subgroup contains three of our BCAs in addition to the reclassified T. atroviride CBS 470.94 1 (10) (formerly T. harzianum Okuda's type 1 [9]), T. atroviride DAOM 165779, and the two T. koningii strains used in the phylogenetic study. The separation into two groups is in agreement with previously observed differences on the basis of conidial morphology and ornamentation (22, 23). Molecular data of ITS1 sequences did not resolve subgroups among T. atroviride and T. koningii. However, both species can be separated by using the divergences in ITS2 sequences (5.7 to 6.8%) and morphological criteria; T. atroviride has smooth and subglobose conidia, and T. koningii has oblong and at most slightly ornamented ones (22). Strains 11, 260, and 2923 produced subglobose conidia rather than oblong to subcylindrical conidia (12). Before the description of T. asperellum as a new species and the redefinition of the T. viride-T. atroviride complex, two subgroups which could correspond with the present concept of T. asperellum and T. atroviride were observed after physiological and biochemical characterization of BCAs (12). Strains of T. atroviride could be grown at 4°C, since they are adapted to low-temperature environments, and T. asperellum was able to grow at 4°C but showed abundant conidiation and colony growth at 28 to 30°C (12). These observations are in agreement with the salient phenotypic features described for these species (22).
Hybridization of genomic DNA with the mtDNA probe showed a grouping of 10 strains in one independent group, all of which showed very similar hybridization profiles, that was related to T. inhamatum and T. harzianum. Strain T. longibrachiatum 2932, excluded from this group, had banding patterns similar to those of strains 2930 and 2931 and was grouped with T. inhamatum 2924, 2930, and 2931 in UPGMA dendrograms derived from morphological, physiological, and biochemical data and from isoenzymatic analyses (12). All of these strains had the same geographic origin, and T. longibrachiatum 2932 is a clear example of the uncertain use of a single gene to characterize a complete organism. No correlation between mtDNA hybridizations and ITS sequences was detected for strains included in the T. atroviride and T. asperellum groups. The different hybridization patterns shown by strains 11, 260, and 2923, with identical ITS sequences, can be considered a discriminatory feature among these BCAs.
The behavior of all Trichoderma strains tested in dual cultures can serve to demonstrate that sporulation and aggressiveness of the BCAs depend on the kind of fungal target and composition of the culture medium. The degree of sporulation of T. asperellum in the presence of P. betae and R. necatrix are notable examples of this relation. In general terms, there is not a particular species of Trichoderma which could be considered the source for the best BCAs. In the case of B. cinerea, T. longibrachiatum was the BCA which showed the heaviest sporulation on the three media. In contrast to this result, F. oxysporum f. sp. dianthi was more sensitive to the attack of T. asperellum and T. atroviride on PDA, of T. longibrachiatum on CMA, and of T. inhamatum on MA.
It is always difficult to extrapolate the biocontrol activity of a given strain from the laboratory to natural environments. However, the significant differences (P ≤ 0.05) observed between T. atroviride and T. inhamatum on P. betae on PDA and MA and between T. atroviride and T. asperellum against the same pathogen on MA are in agreement with the results obtained in the biocontrol at the field level of sugar beet damping-off caused by P. betae (30). In this study strains formerly classified as T. harzianum Rifai were tested, with the present T. atroviride 260 being found to be a better BCA than T. inhamatum 24 and T. asperellum 3 after a 12-week treatment under the experimental conditions of that in planta trial (30).
The very closely related species T. inhamatum and T. harzianum showed differences that were statistically significant (P ≤ 0.05) in the degree of sporulation that occurred in the presence of B. cinerea on PDA and MA and of F. oxysporum f. sp. dianthi on MA. These two species also differed in the production of yellow pigmentation with the four pathogens on PDA and with P. betae, B. cinerea, and F. oxysporum f. sp. dianthi on MA. The behavior of T. atroviride and T. asperellum was very similar under all conditions tested, with only the exception of sporulation in the presence of P. betae and R. necatrix on MA. These statistically significant differences in the growing parameters studied are in agreement with the position of the species in the phylogenetic tree. However, we have not used the biological activities tested here as a tool to define taxonomical categories. As pointed out above, we have detected a certain degree of variability in the behavior of different biocontrol strains, which had previously been grouped phylogenetically. Obviously, any taxonomical study with these species should include a higher number of strains, but we have attempted to distinguish strains of Trichoderma before the exploration of their potential capacity for biocontrol in natural environments without the aim of proposing new diagnostic characteristics for these species.
ACKNOWLEDGMENTS
This research was supported by the Spanish Comisión Interministerial de Ciencia y Tecnología (Project Petri 95-0125-OP) and the Commission of the European Communities (Project FAIR6-CT98-4140).
We thank Gary E. Harman, Antonio Llobell, and Paul Bridge for critical reviewing of the first version of the manuscript. We are also grateful to Eladio Barrio and his Bioinformatic Services at the University of Valencia (Valencia, Spain) for the phylogenetic analysis.
REFERENCES
- 1.Arisan-Atac I, Heidenreich E, Kubicek C P. Randomly amplified polymorphic DNA fingerprinting identifies subgroups of Trichoderma viride and other Trichoderma sp. capable of chestnut blight biocontrol. FEMS Microbiol Lett. 1995;126:249–256. doi: 10.1111/j.1574-6968.1995.tb07426.x. [DOI] [PubMed] [Google Scholar]
- 2.Bissett J. A revision of the genus Trichoderma. II. Intrageneric classification. Can J Bot. 1991;69:2357–2372. [Google Scholar]
- 3.Bissett J. A revision of the genus Trichoderma. III. Section Pachybasium. Can J Bot. 1991;69:2373–2417. [Google Scholar]
- 4.Cannon, P. F., P. D. Bridge, and E. Monte. Linking the past, present and future of Colletotrichum systematics. In D. Prusky, S. Freeman, and M. Dickman (ed.), Host specifity, pathology and host pathogen interaction of Colletotrichum. APS Press, St. Paul, Minn., in press.
- 5.De Meyer G, Bigirimana J, Elad Y, Hofte M. Induced systemic resistance in Trichoderma harzianum T39 biocontrol of Botrytis cinerea. Eur J Plant Pathol. 1998;104:279–286. [Google Scholar]
- 6.Domsch K H, Gams W, Anderson T-H. Compendium of soil fungi. Vol. 1. London, United Kingdom: Academic Press; 1980. [Google Scholar]
- 7.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP: Phylogenetic Inference Package, version 3.5c. Seattle: Department of Genetics. University of Washington; 1993. [Google Scholar]
- 9.Fujimori F, Okuda T. Application of the random amplified polymorphic DNA using the polymerase chain reaction for efficient elimination of duplicate strains in microbial screening. I. Fungi. J Antibiot. 1994;47:173–182. doi: 10.7164/antibiotics.47.173. [DOI] [PubMed] [Google Scholar]
- 10.Gams W, Meyer W. What exactly is Trichoderma harzianum? Mycologia. 1998;90:904–915. [Google Scholar]
- 11.Ghisalberti E L, Rowland G Y. Antifungal metabolites from Trichoderma harzianum. J Nat Prod. 1993;56:1799–1804. doi: 10.1021/np50100a020. [DOI] [PubMed] [Google Scholar]
- 12.Grondona I, Hermosa M R, Tejada M, Gomis M D, Mateos P F, Bridge P, Monte E, García-Acha I. Physiological and biochemical characterization of Trichoderma harzianum, a biological control agent against soilborne fungal plant pathogens. Appl Environ Microbiol. 1997;63:3189–3198. doi: 10.1128/aem.63.8.3189-3198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haran S, Schickler H, Chet I. Molecular mechanisms of lytic enzymes involved in the biocontrol activity of Trichoderma harzianum. Microbiology. 1996;142:2321–2331. [Google Scholar]
- 14.Hjeljord L, Tronsmo A. Trichoderma and Gliocladium in biocontrol: an overview. In: Kubicek C P, Harman G E, editors. Trichoderma and Gliocladium. London, United Kingdom: Taylor & Francis, Ltd.; 1998. pp. 135–151. [Google Scholar]
- 15.Inbar J, Abramshy D, Cohen D, Chet I. Plant growth enhancement and disease control by Trichoderma harzianum in vegetable seedlings grown under commercial conditions. Eur J Plant Pathol. 1994;100:337–346. [Google Scholar]
- 16.Jeffries P, Young T W K. Interfungal parasitic relationships. Wallingford, United Kingdom: CAB International; 1994. p. 296. [Google Scholar]
- 17.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:11–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 18.Kindermann J, El-Ayouti Y, Samuels G J, Kubicek C P. Phylogeny of the genus Trichoderma based on sequence analysis of the internal transcribed spacer region 1 of the rDNA cluster. Fungal Genet Biol. 1998;24:298–309. doi: 10.1006/fgbi.1998.1049. [DOI] [PubMed] [Google Scholar]
- 19.Kuhls K, Lieckfeldt E, Samuels G J, Kovacs W, Meyer W, Petrini O, Gams W, Börner T, Kubicek C P. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivate of the ascomycete Hypocrea jecorina. Proc Natl Acad Sci USA. 1996;93:7755–7760. doi: 10.1073/pnas.93.15.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhls K, Lieckfeldt E, Samuels G J, Meyer W, Kubicek C P, Börner T. Revision of Trichoderma sec. Longibrachiatum including related teleomorphs based on analysis of ribosomal DNA internal transcribed spacer regions. Mycologia. 1997;89:442–460. [Google Scholar]
- 21.Lieckfeldt E, Kuhls K, Muthumeenakshi S. Molecular taxonomy of Trichoderma and Gliocladium and their teleomorphs. In: Kubicek C P, Harman G E, editors. Trichoderma and Gliocladium. London, United Kingdom: Taylor & Francis, Ltd.; 1998. pp. 35–56. [Google Scholar]
- 22.Lieckfeldt E, Samuels G J, Helgard H I, Petrini O. A morphological and molecular perspective of Trichoderma viride: is it one or two species? Appl Environ Microbiol. 1999;65:2418–2428. doi: 10.1128/aem.65.6.2418-2428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer R, Plaskowitz J S. Scanning electron microscopy of conidia and conidial matrix of Trichoderma. Mycologia. 1989;81:312–317. [Google Scholar]
- 24.Miller J H, Giddens J E, Foster A A. A survey of the fungi of forest and cultivated soils of Georgia. Mycologia. 1975;49:779–808. [Google Scholar]
- 25.Muthumeenakshi S. Molecular taxonomy of the genus Trichoderma. Ph.D. thesis. Belfast, United Kingdom: The Queen's University of Belfast; 1996. [Google Scholar]
- 26.Muthumeenakshi S, Mills P R, Brown A E, Seaby D A. Intraspecific molecular variation among Trichoderma harzianum isolates colonizing mushroom compost in the British Isles. Microbiology. 1994;140:769–777. doi: 10.1099/00221287-140-4-769. [DOI] [PubMed] [Google Scholar]
- 27.Ospina-Giraldo M D, Royse D J, Chen X, Romaine C P. Molecular phylogenetic analyses of biological control strains of Trichoderma harzianum and other biotypes of Trichoderma spp. associated with mushroom green mold. Phytopathology. 1999;89:308–313. doi: 10.1094/PHYTO.1999.89.4.308. [DOI] [PubMed] [Google Scholar]
- 28.Ospina-Giraldo M D, Royse D J, Thon M R, Chen X, Romaine C P. Phylogenetic relationships of Trichoderma harzianum causing mushroom green mold in Europe and North America to other species of Trichoderma from world-wide sources. Mycologia. 1998;90:76–81. [Google Scholar]
- 29.Papavizas G C. Trichoderma and Gliocladium: biology and potential for biological control. Annu Rev Phytopathol. 1985;23:23–54. [Google Scholar]
- 30.Pérez de Algaba A, Grondona I, Monte E, García-Acha I. Trichoderma as a biocontrol agent in sugar beet crops. In: Hockenhull J, Jensen D F, Fokkema N J, editors. New approaches in biological control of soil-borne diseases. Copenhagen, Denmark: Organization for Biological Control; 1992. pp. 36–37. [Google Scholar]
- 31.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 32.Rehner S A, Samuels G J. Molecular systematics of the Hypocreales: a teleomorph gene phylogeny and the status of their anamorphs. Can J Bot. 1995;73:S816–S823. [Google Scholar]
- 33.Rifai M A. A revision of the genus Trichoderma. Mycol Papers. 1969;116:1–56. [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Samuels G J. Trichoderma: a review of biology and systematics of the genus. Mycol Res. 1996;100:923–935. [Google Scholar]
- 37.Samuels G J, Lieckfeldt E, Nirenberg H I. Description of T. asperellum sp. nov. and comparison to T. viride. Sydowia. 1999;51:71–88. [Google Scholar]
- 38.Seaby D A. Further observations on Trichoderma. Mushroom J. 1987;197:147–151. [Google Scholar]
- 39.Seifert K A, Wingfield B D, Wingfield M J. A critique of DNA sequence analysis in the taxonomy of filamentous ascomycetes and ascomycetes anamorphs. Can J Bot. 1995;73(Suppl. 1):S760–S767. [Google Scholar]
- 40.Simon A, Sivasithamparan K. Pathogen suppression: a case study of Gaeumannomyces graminis var. tritici in soil. Soil Biol Biochem. 1989;21:331–337. [Google Scholar]
- 41.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 42.Sreenivasaprasad S, Brown A E, Mills P R. DNA sequence variation and interrelationships among Colletotrichum species causing strawberry anthracnose. Physiol Mol Plant Pathol. 1992;41:265–281. [Google Scholar]
- 43.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.United Nations Environment Programme. 1994 Report of Methyl Bromide Technical Options Committee. Montreal protocol on substances that deplete ozone layer. Nairobi, Kenya: United Nations Ozone Secretariat; 1995. [Google Scholar]
- 45.Veerkamp J, Gams W. Los hongos de Colombia-VIII. Some new species of soil fungi from Colombian. Caldasia. 1983;13:709–717. [Google Scholar]
- 46.Von Arx J A. Plant pathogenic fungi. Beihefte zur Nova Hedwigia. 1987;87:288. [Google Scholar]
- 47.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 48.Zimand G, Elad Y, Chet I. Effect of Trichoderma harzianum on Botrytis cinerea pathogenicity. Phytopathology. 1996;86:1225–1260. [Google Scholar]