Abstract
Pyrene is a regulated pollutant at sites contaminated with polycyclic aromatic hydrocarbons (PAH). It is mineralized by some bacteria but is also transformed to nonmineral products by a variety of other PAH-degrading bacteria. We examined the formation of such products by four bacterial strains and identified and further characterized the most apparently significant of these metabolites. Pseudomonas stutzeri strain P16 and Bacillus cereus strain P21 transformed pyrene primarily to cis-4,5-dihydro-4,5-dihydroxypyrene (PYRdHD), the first intermediate in the known pathway for aerobic bacterial mineralization of pyrene. Sphingomonas yanoikuyae strain R1 transformed pyrene to PYRdHD and pyrene-4,5-dione (PYRQ). Both strain R1 and Pseudomonas saccharophila strain P15 transform PYRdHD to PYRQ nearly stoichiometrically, suggesting that PYRQ is formed by oxidation of PYRdHD to 4,5-dihydroxypyrene and subsequent autoxidation of this metabolite. A pyrene-mineralizing organism, Mycobacterium strain PYR-1, also transforms PYRdHD to PYRQ at high initial concentrations of PYRdHD. However, strain PYR-1 is able to use both PYRdHD and PYRQ as growth substrates. PYRdHD strongly inhibited phenanthrene degradation by strains P15 and R1 but had only a minor effect on strains P16 and P21. At their aqueous saturation concentrations, both PYRdHD and PYRQ severely inhibited benzo[a]pyrene mineralization by strains P15 and R1. Collectively, these findings suggest that products derived from pyrene transformation have the potential to accumulate in PAH-contaminated systems and that such products can significantly influence the removal of other PAH. However, these products may be susceptible to subsequent degradation by organisms able to metabolize pyrene more extensively if such organisms are present in the system.
Polycyclic aromatic hydrocarbons (PAH) are known to be degradable by a variety of soil bacteria (40). Consequently, the bioremediation of PAH contamination with naturally occurring microorganisms has been attempted at a number of sites (43, 45). Most of the interest in the biodegradation of PAH in the field has been in the removal of the parent compounds, while most research on pure cultures of PAH-degrading bacteria has focused on their ability to grow on or mineralize specific PAH substrates. Relatively little attention has been paid to the potential formation of products from the partial transformation of PAH.
Most of the information that does exist on PAH metabolites has been obtained in the context of identifying transient metabolites formed by isolates during growth on the parent compound (13, 33, 35, 40) or metabolites formed by mutants of wild-type degraders (4, 40). Some bacteria, however, are capable of transforming one or more PAH despite an inability to grow on or mineralize the PAH in question (1, 20, 31, 47). It is important to evaluate the products of such incomplete PAH metabolism because of their potential effects on PAH-degrading microorganisms or on potentially exposed human populations (44). Identification of common products of incomplete metabolism can also be important in assessing the extent to which natural attenuation may be occurring at contaminated sites (44).
In a recent study (1), we described the broad PAH substrate ranges of 11 bacteria isolated from PAH-contaminated soils by enrichment on phenanthrene as a sole carbon source. None of these organisms is capable of mineralizing pyrene, yet all could remove pyrene from solution, and all that were examined further transformed pyrene to unidentified metabolites (1). The objectives of this study were to isolate and identify the major products from the incomplete metabolism of pyrene by four of these organisms, evaluate the potential degradability of these metabolites by an organism known to grow on pyrene as a sole carbon source, and determine the effects of these metabolites on the degradation of phenanthrene, a representative growth substrate for many PAH degraders, and benzo[a]pyrene, a representative carcinogenic PAH that is not known to serve as a growth substrate for any organism.
MATERIALS AND METHODS
Chemicals and media.
Pyrene (99%), osmium tetroxide (>98%), ruthenium dioxide (99.9%), sodium periodate (99.8%), and acetone-d6 (99.9%) were obtained from Aldrich (Milwaukee, Wis.). Phenanthrene (>96%), [7-14C]benzo[a]pyrene (7-14C-labeled BaP) (>98%; specific activity, 26.6 mCi mmol−1), and [4,5,9,10-14C]pyrene (>98%; specific activity, 61 mCi mmol−1) were obtained from Sigma (St. Louis, Mo.). All solvents used were high-pressure liquid chromatography (HPLC) grade or the equivalent. Mineral salts buffer (MSB) was as described in Stucki et al. (39).
cis-4,5-Dihydro-4,5-dihydroxypyrene (PYRdHD) was synthesized by oxidation of pyrene with osmium tetroxide (10, 23). The crude product was extracted with 200 ml of hexane to remove unreacted pyrene, recrystallized from acetone, and dried at 50°C for 24 h. The purity of the synthesized compound was established by the presence of a single peak in chromatograms from HPLC, and the authenticity was established by 1H nuclear magnetic resonance (NMR) spectroscopy. PYRdHD exhibits UV absorbance maxima at 219 and 261 nm. The 1H NMR shifts recorded for PYRdHD were (in parts per million) 7.90 (d, 2), 7.84 (s, 2), 7.82 (d, 2), 7.64 (tr, 2), and 5.15 (s, 2).
Pyrene-4,5-dione (PYRQ) was synthesized by the oxidation of pyrene with ruthenium dioxide and sodium periodate (17). The crude product was dissolved in 50 ml of methylene chloride and passed through a column of silica gel, which was eluted successively with hexane (to remove unreacted pyrene) and methylene chloride. Methylene chloride eluted PYRQ first and then eluted pyrene-1,6-dione and pyrene-1,8-dione. The purity and authenticity of synthesized PYRQ were established by HPLC and 1H NMR spectroscopy, respectively. PYRQ exhibits UV-visible light absorbance maxima at 238, 295, and 432 nm. The 1H NMR shifts for PYRQ were (in parts per million) 8.43 (d, 2), 8.38 (d, 2), 8.05 (s, 2), and 7.88 (tr, 2).
Aqueous solubilities of PYRdHD and PYRQ at room temperature were determined by adding an excess of either compound to 10 ml of MSB in triplicate vials. After 24 h, the samples were filtered through a 0.02-μm-pore-size alumina filter (Whatman, Maidstone, England) and analyzed by HPLC. Concentrations in the filtrate were quantified against external standards of the synthesized compounds.
Organisms.
Pseudomonas saccharophila P15, Pseudomonas stutzeri P16, Bacillus cereus P21, and strain R1 were isolated as described elsewhere (1, 38). Isolate R1 was identified as a strain of Sphingomonas yanoikuyae by MIDI, Inc. (Newark, Del.) on the basis of 16S rRNA gene sequence similarity (500-bp analysis; 1.49% difference from type strain). Mycobacterium PYR-1 was obtained from C. Cerniglia (National Center for Toxicological Research, Jefferson, Ark.).
Strains P15, P16, P21, and R1 cannot mineralize pyrene but they can all transform it to non-mineral products (1). They were selected for this study based on differences in PAH metabolism, as manifested in part by a limited (P16 and P21) or pronounced (P15 and R1) ability to mineralize benz[a]anthracene, BaP, and chrysene (1). Strain PYR-1 was selected for its ability to grow on pyrene as a sole carbon source (26).
Isolation of metabolites.
Bacteria were grown in 500 ml of MSB containing 50 mg of phenanthrene, 5 mg of pyrene, and 20 mg (each) of succinate, peptone, and yeast extract. The cultures were stirred rapidly with a Teflon stir bar for 2 weeks, after which no phenanthrene crystals were visible. The samples were centrifuged, filtered through a 1.0-μm-pore-size filter, acidified to pH 2.5 with 20% H3PO4, and vacuum extracted through 1 g of octadecyl silica (Supelco, Bellefonte, Pa.). Retained products were eluted with 20 ml of methanol, which was evaporated under a gentle stream of N2. The residue was dissolved in 1 ml of acetone and separated by thin-layer chromatography (TLC) (1000-μm-diameter silica gel, 20 by 20 cm; F254; Whatman) with a 50/50 mixture of hexane and benzene, followed by a 90/10 mixture of benzene and acetone and finally a 85/10/5 mixture of benzene, acetone, and acetic acid. Bands that fluoresced strongly under UV light (254 nm) were scraped off the plates, redissolved in acetone, and filtered through a 0.02-μm-pore-size alumina filter (Whatman). The filtrates were evaporated under N2, and then the residue was dissolved in 300 μl of acetonitrile and analyzed by HPLC. Injections (150 μl each) were made on a Supelcosil C18 semipreparative column (25 cm by 10 mm; particle size, 5 μm; Supelco). Fractions were collected manually, and those from each injection were combined. Then, each fraction was evaporated under N2 and dissolved in 0.5 ml of acetone-d6 for NMR analysis.
Experiments with metabolites.
Each organism was grown in 500 ml of MSB containing 50 mg of phenanthrene (for strains P15, P16, P21, and R1) or 50 mg of pyrene (for strain PYR-1) for approximately 1 week, centrifuged, and resuspended in MSB at an optical density at 420 nm (OD420) of 0.1 unless stated otherwise. The PAH-degrading activity of washed-cell suspensions of strains P15, P16, P21, and R1 was verified by measuring the initial rate of phenanthrene degradation (38) prior to each experiment. Subsequent incubations were conducted in the dark or in amber vessels on a rotary shaker at approximately 150 rpm. When added in a particular assay, synthesized PYRdHD or PYRQ was provided in a solution of acetone or acetonitrile, which was evaporated prior to inoculation with cell suspensions.
Metabolite formation from resting cells pregrown on phenanthrene was evaluated by incubating a washed-cell suspension (100 μl) in an HPLC vial insert with approximately 15,000 dpm of [14C]pyrene (1.1 μM) dissolved in 5 μl of dimethyl sulfoxide (DMSO). Reactions were terminated after 24 h by adding 100 μl of acetonitrile. Samples were analyzed by HPLC, and fractions were collected every minute with a Waters (Milford, Mass.) fraction collector. Each fraction was supplemented with 3 ml of scintillation cocktail (ScintiSafe; Fisher Scientific, Pittsburgh, Pa.) and analyzed with a Tri-Carb 1900TR liquid scintillation analyzer (Packard, Meriden, Conn.).
An isotope-trapping experiment was conducted with strain P15, in which washed cells pregrown on phenanthrene were incubated with 50 μM PYRdHD and [14C]pyrene. A similar experiment was conducted with strains P15 and R1, in which each strain was incubated with 1.6 μM PYRQ and 20,000 dpm of 14C-labeled BaP. Samples from these experiments were analyzed by radiochromatography as described above.
The transformation of PYRdHD to PYRQ was evaluated for each strain by incubating triplicate washed-cell suspensions (1 ml each) in MSB containing PYRdHD at concentrations identified below. Reactions were terminated after 3 days by adding 1 ml of acetonitrile, and the supernatants were analyzed by HPLC.
Effects of pyrene metabolites on the growth of strains P15, P16, P21, and R1 were determined by incubating washed-cell suspensions pregrown on phenanthrene (OD420 = 0.01) in vials containing 560 μM phenanthrene (added as a concentrated DMSO solution) alone or with PYRdHD (155 μM) or PYRQ (16 μM). After 3 days, Pierce (Rockford, Ill.) bicinchoninic acid protein assay reagent was added to each vial, and the samples were shaken for another 24 h. The absorbance of the samples at 562 nm was then determined on a Hitachi (Danbury, Conn.) U-3300 spectrophotometer. Protein concentrations were quantified by comparison to bovine serum albumin standards dissolved in MSB and incubated with the samples for 24 h. No growth occurred in controls containing DMSO alone.
Growth of strain PYR-1 on PYRdHD or PYRQ as a sole carbon source was determined with cultures pregrown on pyrene (OD420 = 0.01). PYRdHD or PYRQ was added to vials at concentrations equivalent to 500 μM, and cultures were incubated for 4 weeks. Protein concentrations were determined as described above.
The effects of pyrene metabolites on phenanthrene degradation rates were determined with washed cells pregrown on phenanthrene and incubated in MSB for 24 h to remove any residual phenanthrene. Replicate aliquots (1 ml each) were then added to empty vials or vials containing PYRdHD or PYRQ at concentrations equivalent to 155 or 1.6 μM, respectively. Vials were incubated for 30 min and then supplemented with phenanthrene to a concentration of 5.6 μM. Reactions were terminated by adding 1 ml of acetonitrile at selected intervals up to 20 min, and the remaining concentration of phenanthrene was determined by HPLC. A first-order (in phenanthrene concentration) degradation rate equation was fit to the concentration versus time data with Pro-Stat software (Poly Software International, Sandy, Utah).
The potential reactivity of PYRQ with biomass or buffer constituents was determined with triplicate washed-cell suspensions (5 ml each) of strain R1 or P15 in vials containing 10 μM PYRQ. Reactions were terminated after 3 days by adding 1 ml of acetonitrile, and the supernatants were analyzed for residual PYRQ by HPLC.
The effects of PYRdHD and PYRQ on BaP mineralization by strains P15 and R1 were determined with triplicate washed-cell suspensions (5 ml) in 20-ml scintillation vials containing PYRdHD or PYRQ at concentrations identified below. Each vial contained a test tube in which a fluted strip of filter paper soaked with 150 μl of 2 N KOH served as a CO2 trap (1). Approximately 20,000 dpm of 14C-labeled BaP (equivalent to 0.064 μM) was added to each vial, which was then capped with a Teflon-lined septum. Positive controls did not contain pyrene metabolites, and uninoculated controls contained 5 ml of MSB supplemented with 14C-labeled BaP. Incubations were terminated by injecting 0.5 ml of 20% H3PO4 through the septum. The vials were shaken for an additional 24 h, and then the filter paper strips were placed in 5 ml of scintillation cocktail and another 5 ml of scintillation cocktail was added directly to the sample vials. Samples were analyzed by scintillation counting.
Analyses.
HPLC analyses were conducted on a Waters 600E system with a Supelcosil C18 column (25 cm by 4.6 mm; particle size, 5 μm; Supelco). Pyrene, PYRdHD, and PYRQ were analyzed with a photodiode array detector and quantified at 254 nm, and other putative metabolites were analyzed with fluorescence detection (excitation at 259 nm and emission at 370 nm) or photodiode array detection. The mobile phase for these analyses was initially a 50/50 ratio of acetonitrile to water containing 0.01% trifluoroacetic acid, which was increased to 100% acetonitrile over 20 min; these conditions improved the separation of metabolites compared to analyses performed in earlier work (1). Phenanthrene was analyzed by detection at 254 nm, with a mobile phase of a 70/30 ratio of acetonitrile to water containing 0.01% trifluoroacetic acid that was increased linearly to 100% acetonitrile over 5 min. The 1H NMR spectra were recorded on a Varian Inova 500 spectrometer at 500 MHz, with shifts reported in parts per million relative to the proton resonance of tetramethylsilane.
RESULTS
Isolation of pyrene metabolites.
Growth of strains P15, P16, P21, and R1 in a medium containing phenanthrene as a major carbon source and in the presence of pyrene in excess of its aqueous solubility resulted in the extracellular accumulation of a number of pyrene metabolites. The relative quantities of these metabolites were assessed by the size and strength of the fluorescent bands on TLC plates. The most significant bands that were isolated by TLC were purified further by semipreparative HPLC, and the purified compounds were analyzed by 1H NMR spectroscopy.
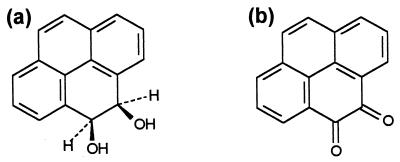
The most significant metabolite from isolates P16 and P21 was identified as PYRdHD (Fig. 1a), which is the first intermediate in the aerobic bacterial pathway for metabolism of pyrene (13, 24, 33, 35, 49). PYRdHD was one of two significant products from strain R1, which also transformed pyrene to PYRQ (Fig. 1b). No other isolated TLC bands were identified, including all those from strain P15, either because there were insufficient amounts for NMR analysis or the NMR spectra could not be characterized. Optical spectra obtained for the unidentified bands did not match published spectra of known intermediates in the bacterial degradation of pyrene (33).
FIG. 1.
Isolated metabolites from pyrene transformation. (a) PYRdHD; (b) PYRQ.
Identification of PYRdHD and PYRQ was confirmed by comparing the NMR spectra of the bacterial metabolites to those of the synthesized compounds as well as published spectra (9, 24, 48). The cis configuration of PYRdHD was established by comparison to published 1H NMR spectra of cis and trans PYRdHD (24). The solubility of PYRdHD in MSB at room temperature is 155 ± 13 μM (36.7 ± 3.1 mg/l) and that of PYRQ is 1.6 ± 0.1 μM (0.38 ± 0.01 mg/l).
Transformation of [14C]pyrene by resting cells.
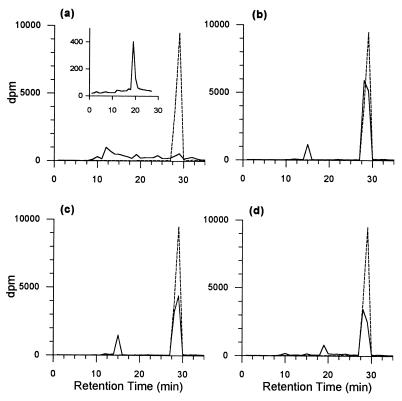
The radiochromatograms of culture fluids from resting cells (Fig. 2) largely confirmed the results obtained from the transformation of pyrene during active growth of the organisms on phenanthrene. On the basis of retention time, resting cells of strains P16 and P21 transformed pyrene primarily to PYRdHD, and strain R1 transformed pyrene to PYRQ and a number of other minor products. Strain P15 removed almost the entire amount of [14C]pyrene in the 24-h incubation period but did not appear to form significant amounts of PYRdHD or PYRQ. The elevated baselines in the radiochromatogram for strain P15 after approximately 8 min (Fig. 2a) indicate the formation of numerous other products, which continued to elute even after the elution of pyrene. Significant amounts of radiolabel also remained in the empty reaction vials for both strain P15 (approximately 15%) and strain R1 (approximately 30%); in contrast, the recovery of radiolabel in MSB amended with acetonitrile in uninoculated controls was approximately 95%. When incubated with [14C]pyrene in the presence of 50 μM PYRdHD, strain P15 accumulated small amounts of a single product that eluted at the same retention time as PYRQ (Fig. 2a). Pyrene removal was considerably slower in the presence of 50 μM PYRdHD.
FIG. 2.
Radiochromatograms of culture fluids from 24-hr incubations of [14C]pyrene with P. saccharophila P15 (a), P. stutzeri P16 (b), B. cereus P21 (c), and S. yanoikuyae R1 (d). The inset to panel a is the chromatogram from incubation of strain P15 with [14C]pyrene in the presence of 50 μM PYRdHD for 24 h, at which time most of the PYRdHD was depleted; the chromatogram is truncated before the much larger peak for unreacted pyrene. Solid lines are sample chromatograms, and dashed lines are chromatograms from uninoculated controls. PYRdHD, PYRQ, and pyrene eluted at 14, 19, and 29 min, respectively, under the HPLC conditions used to generate these chromatograms.
Metabolism of PYRdHD and PYRQ.
Resting cells of strains R1 and P15 both converted PYRdHD nearly stoichiometrically to PYRQ, whereas neither strain P16 nor P21 was able to transform PYRdHD (Table 1). Mycobacterium PYR-1, an organism known to mineralize pyrene, also transformed PYRdHD to PYRQ to various extents depending on the initial concentration of PYRdHD (Table 2). Strain PYR-1 converted all of the PYRdHD to PYRQ at an initial PYRdHD concentration of 200 μM, whereas at an initial concentration of 50 μM only 4% of the diol was converted to the quinone. This organism was able to grow on either PYRdHD or PYRQ as a sole carbon source in MSB medium, as determined by significant increases in optical density and protein concentration relative to inoculated vessels with no carbon source (results not shown). The protein yield from PYRQ was the same as that from 500 μM pyrene under the same conditions, while that from PYRdHD was about one-third the yield from pyrene or PYRQ.
TABLE 1.
Conversion of PYRdHD to PYRQ by resting cells after 3 days
| Strain | PYRdHD removed (μM)a | PYRQ detected (μM) |
|---|---|---|
| P15 | 21.5 ± 0.4 | 20.6 ± 2.1 |
| R1 | 4.1 ± 0.5 | 3.9 ± 0.0 |
| P21 | Noneb | <0.5 |
| P16 | Noneb | <0.5 |
Relative to uninoculated controls, which contained 22.9 ± 0.3 μM PYRdHD. Data are means and standard deviations of triplicate measurements.
Residual PYRdHD was not significantly different from amount obtained from controls.
TABLE 2.
Concentration dependence of PYRQ formation from PYRdHD by Mycobacterium PYR-1 after 3 daysa
| PYRdHD added (μM) | PYRQ detected (μM) |
|---|---|
| 50 | 2.0 ± 1.1 |
| 100 | 18 ± 13 |
| 200 | 205 ± 37 |
All of the added PYRdHD (98.7% ± 6.8%) was recovered in uninoculated vials. No PYRdHD was recovered in any of the sample vials. Data are means and standard deviations of triplicate measurements.
Effects of pyrene metabolites on phenanthrene degradation.
Apparent first-order rates of phenanthrene degradation were measured in the presence and absence of PYRdHD and PYRQ at an initial phenanthrene concentration of 5.6 μM, which is less than its aqueous solubility. PYRdHD strongly inhibited phenanthrene degradation by strains P15 and R1, modestly inhibited phenanthrene degradation by strain P21, and had a negligible effect on strain P16 (Table 3). PYRQ strongly inhibited phenanthrene degradation by strain R1 and had a negligible-to-minor effect on the remaining strains.
TABLE 3.
Effects of pyrene metabolites on apparent rates of phenanthrene degradation
| Strain | Apparent phenanthrene degradation rate in the presence of the indicated metabolite (min−1)a
|
||
|---|---|---|---|
| None | PYRdHD | PYRQ | |
| P15 | 0.10 ± 0.032 | 0.005 ± 0.0005 | 0.10 ± 0.04 |
| P16 | 0.14 ± 0.04 | 0.12 ± 0.03 | 0.13 ± 0.04 |
| P21 | 0.23 ± 0.03 | 0.15 ± 0.03 | 0.16 ± 0.03 |
| R1 | 0.14 ± 0.07 | 0.02 ± 0.02 | 0.01 ± 0.01 |
Best-fit first-order degradation coefficients and the associated 95% confidence interval.
In contrast to the observed inhibitory effects of pyrene metabolites on rates of dissolved phenanthrene degradation by some of the strains, the yield of protein after growth on 560 μM phenanthrene for 3 days in the presence of PYRQ or PYRdHD was equivalent to the yield obtained after growth on phenanthrene alone for all four strains (not shown). There was no growth on PYRQ or PYRdHD by any of these isolates.
Effects of pyrene metabolites on mineralization of BaP.
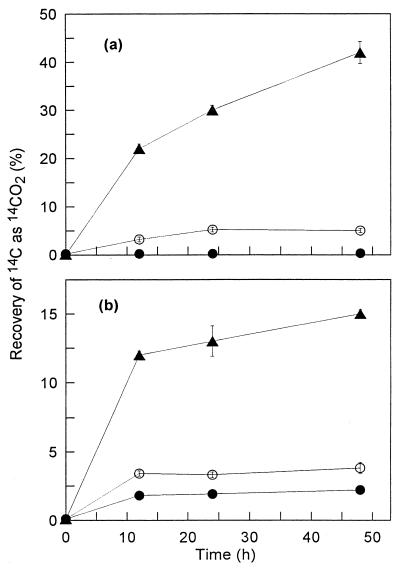
Both PYRdHD and PYRQ at concentrations near their aqueous saturation concentrations strongly inhibited the mineralization of BaP by strains P15 and R1 throughout a 48-h incubation period (Fig. 3). In addition, PYRQ inhibited BaP mineralization by both strains at concentrations lower than its aqueous saturation concentration over a 48-h incubation (Table 4). Radiochromatograms of culture fluids from incubation of either organism with 1.6 μM PYRQ indicated that no metabolites accumulated and less BaP was removed than in positive controls incubated without PYRQ (not shown).
FIG. 3.
Mineralization of BaP by P. saccharophila P15 (a) and S. yanoikuyae R1 (b). Each strain was incubated alone (▴) or in the presence of 155 μM PYRdHD (●) or 1.6 μM PYRQ (○). Data are means and standard deviations of triplicate measurements at each time point. Unobservable error bars are within the size of the symbol. Total recoveries of 14C ranged from 79 to 99% for strain P15 and from 95 to 99% for strain R1.
TABLE 4.
Effect of PYRQ concentration on the mineralization of BaP by strains P15 and R1a
| PYRQ added (μM) | % of added dpm as CO2 after 48 h in strain
|
|
|---|---|---|
| P15 | R1 | |
| None | 52 ± 2.1 | 25 ± 1.8 |
| 0.16 | 45 ± 1.4 | 3.7 ± 0.1 |
| 0.8 | 19 ± 2.6 | 1.2 ± 0.1 |
| 1.6 | 8.0 ± 1.7 | 1.0 ± 0.1 |
| Uninoculated control (1.6 μM) | 0.4 ± 0.2 | 0.1 ± 0.1 |
Data are means and standard deviations of triplicate measurements. Total recoveries of 14C ranged from 83 to 90% for strain P15 and from 88 to 98% for strain R1. Total recoveries in the uninoculated controls were 93 and 98%, respectively.
DISCUSSION
The ability of PAH-degrading organisms to transform PAH that they are incapable of mineralizing has been noted previously (1, 20, 31, 47), but the products of such incomplete metabolism have not been characterized. Partial transformation of pyrene may be particularly relevant because of an apparently limited diversity in the ability of PAH-degrading bacteria to mineralize pyrene. The ability to grow on or mineralize pyrene is primarily associated with actinomycetes (25, 26), whereas a diverse group of bacteria isolated from PAH-contaminated soils by enrichment with phenanthrene can remove pyrene without mineralizing it (1).
Pyrene transformation by P. stutzeri P16 and B. cereus P21 during active growth on phenanthrene leads to the formation of significant amounts of PYRdHD as an apparently terminal, or “dead-end,” metabolite. PYRdHD has been identified as an intermediate in the degradation of pyrene by a number of strains that can grow on or mineralize pyrene (13, 24, 33, 35, 49). It also has been found in pyrene-contaminated marine sediments (30) and has accumulated when pyrene was added to enrichment cultures derived from the sediment (29). The formation of a cis-dihydrodiol from pyrene is consistent with its oxidation via a PAH dioxygenase, an enzyme class for which examples from several bacteria are known to have broad substrate ranges (14, 15, 18, 27, 28, 34, 37, 46).
When grown on phenanthrene in the presence of pyrene, S. yanoikuyae R1 accumulated significant amounts of PYRdHD and PYRQ in the medium, although only PYRQ was detected when [14C]pyrene was incubated with resting cells (Fig. 2d). Formation of PYRQ probably proceeds by subsequent metabolism of PYRdHD, as both strains R1 and P15 converted PYRdHD essentially stoichiometrically to PYRQ (Table 1). PYRQ has not been observed previously as an intermediate in the degradation of pyrene, but o-quinones such as PYRQ can arise from the autoxidation of an o-dihydroxy intermediate (7, 12, 32). We were not able to isolate 4,5-dihydroxypyrene, nor has it been isolated in previous work with pyrene-degrading organisms. Since 4,5-dihydroxypyrene is rapidly oxidized to PYRQ in air at ambient conditions (41), PYRQ presumably results from the autoxidation of 4,5-dihydroxypyrene.
P. saccharophila P15 can convert PYRdHD to PYRQ, but neither metabolite was identified in either growing- or resting-cell incubations of this organism with pyrene. The radiochromatograms from resting cell incubations of strains P15 and R1 (Fig. 2) indicate that other nonmineral products were formed by these organisms. We do not believe that these other products are metabolites further down the known pyrene degradation pathway, such as phenanthrene-dicarboxylic acid or phenanthrene-carboxylic acid. When the HPLC conditions used to analyze culture fluids were optimized for the fluorescence detection of the phenanthrene aromatic skeleton, no such peaks were detected in culture fluids from any of the strains. In addition, degradation beyond phenanthrene-dicarboxylic acid should result in the formation of 14CO2, since the [14C]pyrene used in these experiments is labeled at the 4,5 position.
When isotope trapping with PYRdHD was used to examine the transformation of pyrene by strain P15, a single product accumulated that elutes at the same retention time as PYRQ (Fig. 2a). This result, and the relatively facile oxidation of PYRdHD to PYRQ by this organism, suggests that pyrene metabolism by strain P15 probably does proceed via oxidation at the 4,5 bond. When incubated with a large excess of PYRdHD, intracellular concentrations of 4,5-dihydroxypyrene are likely to be far greater than during the metabolism of pyrene alone. Under these conditions, the autoxidation of 4,5-dihydroxypyrene could be favored over other reactions in which this intermediate may be involved.
The unidentified products from pyrene transformation by strain P15 (and possibly strain R1) most likely represent adducts from the reaction of one or more pyrene metabolites with cellular constituents. The formation of such adducts is consistent with the presence of an insoluble residue when both strains were incubated with radiolabeled pyrene. Based on the accumulation of PYRQ in the isotope-trapping experiment, the reactive metabolite may be pyrene-4,5-dihydriodiol or a reactive intermediate derived from its intracellular oxidation. We rule out covalent reaction of PYRQ with cellular or buffer constituents, even though o-quinones derived from naphthalene and BaP can form conjugates in phosphate and glycine buffers and can also form adducts with nucleophiles such as cysteine and glutathione (32). We recovered 100% of the added PYRQ incubated with strains P15 or R1 for 3 days (not shown), and PYRQ also accumulated stoichiometrically with the removal of PYRdHD by these organisms.
Mycobacterium PYR-1 surprisingly formed significant amounts of PYRQ when incubated with a high concentration of PYRdHD, although it was not formed when strain PYR-1 was grown on pyrene. Similar to the isotope-trapping experiment with strain P15, it is likely that the rate of formation of 4,5-dihydroxypyrene at high concentrations of PYRdHD exceeds the subsequent rate of metabolism, resulting in the accumulation of 4,5-dihydroxypyrene and its consequent autoxidation to PYRQ. A similar phenomenon has been noted before in the bacterial degradation of naphthalene, for which the accumulation of naphthoquinone has been suggested to increase as the rate of naphthalene availability increased (2, 19). Strain PYR-1 is able to consume PYRQ as a growth substrate, which suggests that it would only accumulate transiently if formed by this organism or by other bacteria that might also be transforming pyrene in a complex system. It would be of interest to know if other organisms capable of growing on or mineralizing pyrene are able to degrade PYRQ, as our work suggests that PYRQ has the potential to accumulate in PAH-contaminated systems that contain organisms incapable of more extensive pyrene metabolism.
Effects of pyrene metabolites on the degradation of other PAH.
The present study clearly demonstrated that extracellular metabolites from the partial transformation of one PAH substrate can adversely affect the metabolism of other PAH. Both PYRdHD and PYRQ led to decreased rates of phenanthrene degradation by at least one of the organisms tested (Table 3). These effects did not appear to result from irreversible reactions of the metabolites or reactive species derived from them, since neither metabolite inhibited the growth of any strain when phenanthrene was present as a sole carbon source in excess of its aqueous solubility.
The ability of PYRdHD and PYRQ to virtually block BaP mineralization by both strains P15 and R1 (Fig. 3) is important, as BaP appears to be particularly recalcitrant in contaminated soil systems (5, 11, 21). Cornelissen et al. (11) have suggested that the recalcitrance of BaP and other high-molecular-weight PAH in contaminated soils is due to unexplained biological factors rather than to limitations in the bioavailability of these compounds. Combined with the possibility that BaP degradation can be much slower than that of other PAH (8), the accumulation of inhibitory metabolites could lead to long-term persistence in PAH-contaminated systems.
The mechanisms of inhibition by pyrene metabolites observed in this study are not apparent. It is possible that PYRdHD could competitively inhibit a dihydrodiol dehydrogenase utilized in the degradation of BaP or phenanthrene by strains P15 and R1, as it clearly is a substrate for these organisms. Alternatively, PYRdHD could inhibit BaP mineralization via the formation of PYRQ, which also inhibited BaP mineralization by strains P15 and R1. As discussed above, PYRQ does not appear to react covalently with cellular constituents, although it is possible that only a small concentration of PYRQ is required to react with a critical enzyme or other constituent. However, in this case we would expect inhibition to be independent of PYRQ concentration, since PYRQ would be present in large excess at all concentrations. This was clearly not the case in the mineralization of BaP (Table 4). We also rule out competitive effects of PYRQ as an explanation for the inhibition of BaP oxidation; we would have expected such competition to lead to the accumulation of BaP metabolites, but this was not observed with radiochromatography. A possible explanation for the inhibitory effects of PYRQ is its potential to mediate futile redox reactions that could alter the balance of vital redox cofactors, as has been observed for other PAH o-quinones (32).
Other considerations.
The potential fates of extracellular metabolites such as PYRdHD and PYRQ in a contaminated soil or sediment system are important to consider. Microbial consortia can lead to more extensive degradation of PAH than can be achieved with pure cultures (6, 42), and in some cases such consortia can degrade metabolites that otherwise would accumulate (6, 29). If pyrene-mineralizing organisms are not present in systems in which pyrene metabolites accumulate to inhibitory levels, it may be possible to enhance overall PAH degradation by inoculating the system with organisms capable of degrading the metabolites.
Abiotic reactions of pyrene metabolites with soil constituents, particularly natural organic matter (NOM), may also be important. Radiolabel originating from [14C]pyrene has been shown to interact with NOM at long incubation times in soil (22), and quinones are known to undergo oxidation-reduction reactions with NOM (36). Furthermore, the aqueous solubility of PYRQ (0.37 mg/l) is 2 orders of magnitude lower than that of PYRdHD; thus, PYRQ has the potential to precipitate in some systems, which might influence its subsequent degradation. The potential accumulation of o-quinones derived from PAH in contaminated systems also needs to be evaluated from a human risk assessment standpoint, since a variety of such o-quinones have been shown to be cytotoxic and mutagenic and to form DNA adducts in mammalian cells (32). Transient increases in mutagenicity and toxicity have been observed in PAH-contaminated soils undergoing active bioremediation (3, 16), which might be explained by the presence of PAH metabolites such as those described here.
ACKNOWLEDGMENTS
We thank Ramiah Sangaiah for synthesizing PYRdHD and for performing 1H NMR analyses.
This research was funded by the National Institute of Environmental Health Sciences (grant P42ES05948) under the Superfund Basic Research Program.
REFERENCES
- 1.Aitken M D, Stringfellow W T, Nagel R D, Kazunga C, Chen S-H. Characteristics of phenanthrene-degrading bacteria isolated from soils contaminated with polycyclic aromatic hydrocarbons. Can J Microbiol. 1998;44:743–752. [PubMed] [Google Scholar]
- 2.Auger R L, Jacobson A M, Domach M M. Effect of nonionic surfactant addition on bacterial metabolism of naphthalene: assessment of toxicity and overflow metabolism potential. J Hazard Mater. 1995;43:263–272. [Google Scholar]
- 3.Belkin S, Steiber M, Tiehm A, Frimmel F, Abeliovich A, Werner P, Ulitzur S. Toxicity and genotoxicity enhancement during polycyclic aromatic hydrocarbons' degradation. Environ Toxicol Water Qual. 1994;9:303–309. [Google Scholar]
- 4.Boyd D R, Sharma N D, Agarwal R, Resnick S M, Schocken M J, Gibson D T, Sayer J M, Yagi H, Jerina D M. Bacterial dioxygenase-catalysed dihydroxylation and chemical resolution routes to enantiopure cis-dihydrodiols of chrysene. J Chem Soc Perkins Trans I. 1997;1:1715–1723. [Google Scholar]
- 5.Carmichael L M, Pfaender F K. Polynuclear aromatic hydrocarbon metabolism in soils: relationship to soil characteristics and preexposure. Environ Toxicol Chem. 1997;16:666–675. [Google Scholar]
- 6.Casellas M, Grifoll M, Sabaté J, Solanas A M. Isolation and characterization of a 9-fluorenone-degrading bacterial strain and its role in synergistic degradation of fluorene by a consortium. Can J Microbiol. 1998;44:734–742. [Google Scholar]
- 7.Cavalotti C, Orsini F, Sello G. 1,2-Dihydro-1,2-dihydroxynaphthalene dehydrogenase containing recombinant strains: preparation, isolation and characterization of 1,2-dihydroxynaphthalenes and 1,2-naphthoquinones. Tetrahedron. 1999;55:4467–4480. [Google Scholar]
- 8.Chen S-H, Aitken M D. Salicylate stimulates the degradation of high-molecular weight polycyclic aromatic hydrocarbons by Pseudomonas saccharophila P15. Environ Sci Technol. 1999;33:435–439. [Google Scholar]
- 9.Cho H, Harvey R G. Synthesis of “K-region” quinones and arene oxides of polycyclic aromatic hydrocarbons. Tetrahedron Lett. 1974;16:1491–1494. [Google Scholar]
- 10.Cook J W, Schoental R. Oxidation of carcinogenic hydrocarbons by osmium tetroxide. J Chem Soc. 1948;1948:170–173. doi: 10.1039/jr9480000170. [DOI] [PubMed] [Google Scholar]
- 11.Cornelissen G, Rigterink H, Ferdinandy M M A, Van Noort P C M. Rapidly desorbing fractions of PAHs in contaminated sediments as a predictor of the extent of bioremediation. Environ Sci Technol. 1998;32:966–970. [Google Scholar]
- 12.Davies J I, Evans W C. Oxidative metabolism of naphthalene by soil pseudomonads. J Biochem. 1964;91:251. doi: 10.1042/bj0910251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean-Ross D, Cerniglia C E. Degradation of pyrene by Mycobacterium flavescens. Appl Microbiol Biotechnol. 1996;46:307–312. doi: 10.1007/s002530050822. [DOI] [PubMed] [Google Scholar]
- 14.Denome S A, Stanley D C, Olson E S, Young K D. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J Bacteriol. 1993;175:6890–6901. doi: 10.1128/jb.175.21.6890-6901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Gennaro P, Sello G, Bianchi D, D'Amico P. Specificity of substrate recognition by Pseudomonas fluorescens N3 dioxygenase. J Biol Chem. 1997;272:30254–30260. doi: 10.1074/jbc.272.48.30254. [DOI] [PubMed] [Google Scholar]
- 16.Donnelly K C, Anderson C S, Thomas J C, Brown K W, Manek D J, Safe S H. Bacterial mutagenicity of soil extracts from a bioremediation facility treating wood-preserving waste. J Hazard Mater. 1992;30:71–81. [Google Scholar]
- 17.Fatiadi A J. New applications of periodic acid and periodates in organic and bio-organic chemistry. Synthesis. 1974;4:229–257. [Google Scholar]
- 18.Foght J M, Westlake D W S. Transposon and spontaneous deletion mutants of plasmid-borne genes encoding polycyclic aromatic hydrocarbon degradation by a strain of Pseudomonas fluorescens. Biodegradation. 1996;7:353–366. doi: 10.1007/BF00115749. [DOI] [PubMed] [Google Scholar]
- 19.Ghoshal S, Luthy R G. Biodegradation kinetics of naphthalene in nonaqueous phase liquid-water mixed batch systems: comparison of model predictions and experimental results. Biotechnol Bioeng. 1998;57:356–366. [PubMed] [Google Scholar]
- 20.Grifoll M, Selifonov S A, Gatlin C V, Chapman P J. Actions of a versatile fluorene-degrading bacterial isolate on polycyclic aromatic compounds. Appl Environ Microbiol. 1995;61:3711–3723. doi: 10.1128/aem.61.10.3711-3723.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grosser R J, Warshawsky D, Vestal J R. Mineralization of polycyclic and N-heterocyclic aromatic compounds in hydrocarbon-contaminated soils. Environ Toxicol Chem. 1995;14:375–382. [Google Scholar]
- 22.Guthrie E A, Pfaender F K. Reduced pyrene bioavailability in microbially active soils. Environ Sci Technol. 1998;32:501–508. [Google Scholar]
- 23.Harvey R G, Goh S W, Cortez C. “K-region” oxides and related oxidized metabolites of carcinogenic aromatic hydrocarbons. J Am Chem Soc. 1975;97:3468–3479. doi: 10.1021/ja00845a032. [DOI] [PubMed] [Google Scholar]
- 24.Heitkamp M A, Freeman J P, Miller D W, Cerniglia C E. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl Environ Microbiol. 1988;54:2556–2565. doi: 10.1128/aem.54.10.2556-2565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kästner M, Breuer-Jammali M, Mahro B. Enumeration and characterization of the soil microflora from hydrocarbon-contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH) Appl Microbiol Biotechnol. 1994;41:267–273. [Google Scholar]
- 26.Kelley I, Cerniglia C E. Degradation of a mixture of high-molecular-weight polycyclic aromatic hydrocarbons by a Mycobacterium strain PYR-1. J Soil Contam. 1995;4:77–91. [Google Scholar]
- 27.Kiyohara H, Torigoe S, Kaida N, Asaki T, Iida T, Hayashi H, Takizawa N. Cloning and characterization of a chromosomal gene cluster, pah, that encodes the upper pathway for phenanthrene and naphthalene utilization by Pseudomonas putida OUS82. J Bacteriol. 1994;176:2439–2443. doi: 10.1128/jb.176.8.2439-2443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurie A D, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181:531–540. doi: 10.1128/jb.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X-F, Cullen W R, Reimer K J, Le X-C. Microbial degradation of pyrene and characterization of a metabolite. Sci Total Environ. 1996;177:17–29. [Google Scholar]
- 30.Li X-F, Le X-C, Simpson C D, Cullen W R, Reimer K J. Bacterial transformation of pyrene in a marine environment. Environ Sci Technol. 1996;30:1115–1119. [Google Scholar]
- 31.Mahaffey W R, Gibson D T, Cerniglia C E. Bacterial oxidation of chemical carcinogens: formation of polycyclic aromatic acids from benz[a]anthracene. Appl Environ Microbiol. 1988;54:2415–2423. doi: 10.1128/aem.54.10.2415-2423.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penning T M, Burczynski M E, Hung C-F, McCoull K D, Palackal N T, Tsuruda L S. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox active o-quinones. Chem Res Toxicol. 1999;12:1–18. doi: 10.1021/tx980143n. [DOI] [PubMed] [Google Scholar]
- 33.Rehmann K, Noll H P, Steinberg C E W, Kettrup A A. Pyrene degradation by Mycobacterium sp. strain KR2. Chemosphere. 1998;36:2977–2992. doi: 10.1016/s0045-6535(97)10240-5. [DOI] [PubMed] [Google Scholar]
- 34.Resnick S M, Lee K, Gibson D T. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Ind Microbiol. 1996;17:438–457. [Google Scholar]
- 35.Schneider J, Grosser R, Jayasimhulu K, Xue W, Warshawsky D. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl Environ Microbiol. 1996;62:13–19. doi: 10.1128/aem.62.1.13-19.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott D T, McKnight D M, Blunt-Harris E L, Kolesar S E, Lovley D R. Quinone moieties act as electron acceptors in the reduction of humic substances by humics-reducing microorganisms. Environ Sci Technol. 1998;32:2984–2989. [Google Scholar]
- 37.Selifonov S A, Grifoll M, Eaton R W, Chapman P J. Oxidation of naphthenoaromatic and methyl-substituted aromatic compounds by naphthalene 1,2-dioxygenase. Appl Environ Microbiol. 1996;62:507–514. doi: 10.1128/aem.62.2.507-514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stringfellow W T, Aitken M D. Comparative physiology of phenanthrene degradation by two dissimilar pseudomonads isolated from a creosote-contaminated soil. Can J Microbiol. 1994;40:432–438. doi: 10.1139/m94-071. [DOI] [PubMed] [Google Scholar]
- 39.Stucki G, Gälli R, Ebersold H-R, Leisinger T. Dehalogenation of dichloromethane by cell extracts of Hyphomicrobium DM2. Arch Microbiol. 1981;130:366–371. [Google Scholar]
- 40.Sutherland J B, Rafii F, Khan A A, Cerniglia C E. Mechanisms of polycyclic aromatic hydrocarbon degradation. In: Young L Y, Cerniglia C E, editors. Microbial transformation and degradation of toxic organic chemicals. New York, N.Y: Wiley-Liss; 1995. pp. 269–306. [Google Scholar]
- 41.Tintel C, Terheijden J, Lugtenburg J, Cornelisse J. Photoreduction and photoaddition reactions of pyrenediones. Tetrahedron Lett. 1987;28:2057–2060. [Google Scholar]
- 42.Trzesicka-Mlynarz D, Ward O P. Degradation of polycyclic aromatic hydrocarbons (PAHs) by a mixed culture and its component pure cultures, obtained from PAH-contaminated soil. Can J Microbiol. 1995;41:470–476. doi: 10.1139/m95-063. [DOI] [PubMed] [Google Scholar]
- 43.U.S. Environmental Protection Agency. Innovative treatment technologies: annual status report. 8th ed. Washington, D.C.: EPA-542-R-96-010. U.S. Environmental Protection Agency; 1996. [Google Scholar]
- 44.U.S. Environmental Protection Agency. Use of monitored natural attenuation at superfund, RCRA corrective action and underground storage tank sites. OSWER Directive no. 9200.4-17P. U.S. Washington, D.C.: Environmental Protection Agency; 1999. [Google Scholar]
- 45.Wilson S C, Jones K C. Bioremediation of soil contaminated with polynuclear aromatic hydrocarbons (PAHs): a review. Environ Pollut. 1993;81:229–249. doi: 10.1016/0269-7491(93)90206-4. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Chen R F, Shiaris M P. Metabolism of naphthalene, fluorene, and phenanthrene: preliminary characterization of a cloned gene cluster from Pseudomonas putida NCIB 9816. J Bacteriol. 1994;176:2158–2164. doi: 10.1128/jb.176.8.2158-2164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye D, Siddiqi M A, Maccubbin A E, Kumar S, Sikka H C. Degradation of polynuclear aromatic hydrocarbons by Sphingomonas paucimobilis. Environ Sci Technol. 1996;30:136–142. [Google Scholar]
- 48.Young E R R, Funk R L. A practical synthesis of pyrene-4,5-dione. J Org Chem. 1998;63:9995–9996. [Google Scholar]
- 49.Zylstra G J, Wang X P, Kim E, Didolkar V A. Cloning and analysis of the genes for polycyclic aromatic hydrocarbon degradation. Ann N Y Acad Sci. 1994;721:386–398. doi: 10.1111/j.1749-6632.1994.tb47410.x. [DOI] [PubMed] [Google Scholar]