Abstract
A seminested PCR assay was developed in order to amplify the kinetoplast minicircle of Leishmania species from individual sand flies. The kinetoplast minicircle is an ideal target because it is present in 10,000 copies per cell and its sequence is known for most Leishmania species. The two-step PCR is carried out in a single tube using three primers, which were designed within the conserved area of the minicircle and contain conserved sequence blocks. The assay was able to detect as few as 3 parasites per individual sand fly and to amplify minicircle DNA from at least eight Leishmania species. This technique permits the processing of a large number of samples synchronously, as required for epidemiological studies, in order to study infection rates in sand fly populations and to identify potential insect vectors. Comparison of the sequences obtained from sand flies and mammal hosts will be crucial for developing hypotheses about the transmission cycles of Leishmania spp. in areas of endemicity.
Visceral leishmaniasis (kala-azar) is a serious health problem in the greater Athens area in Greece. From 1962 to 1992 the Greek Ministry of Health recorded 1,005 cases, pointing out that 90% of the infected individuals lived either on or near the slopes of the mountains surrounding the Athens basin, in or near the hills situated in it, or close to quarries (25). Data concerning the epidemiology of leishmaniasis (transmission cycles and geographical distribution) have changed during the past four decades in the Old World, particularly in the Mediterranean Basin, where visceral leishmaniasis was traditionally zoonotic. Leishmaniasis has become an opportunistic disease, since immunocompromised patients, such as human immunodeficiency virus (HIV) patients (6), can serve as human reservoirs.
The leishmaniases are the most diverse and complex of all vector-borne diseases in their ecology and epidemiology because they encompass 21 species of human-infective parasites (11), several reservoirs and vector species, and a wide range of topographically different foci. There are two classical methods for estimation of infection rates in reservoir hosts or vectors, microscopic analysis and isolation in culture; both are laborious and inaccurate, as many species and subspecies of flagellate protozoa are often morphologically indistinguishable. The use of Leishmania-specific monoclonal antibodies (1) in radioimmunoassay and in indirect immunofluorescence has been described and provides one approach to the problem. Leishmania kinetoplastic DNA (kDNA)-specific probes have been used for hybridization to squash-blotted sand flies for the detection and identification of Leishmania parasites (7, 17). These techniques have proved useful for epidemiological field studies because large number of samples can be handled at the same time. However, due to the low sensitivity of these approaches, dissection of sand fly alimentary tracts is still required to avoid the inhibition effects of abdominal compounds.
Diagnostic assays for leishmaniasis have been developed based on the amplification of several DNA targets such as the minicircle of kDNA (1, 24), the rRNA gene (8), the miniexon-derived RNA (9), and repeated genomic sequences (16). The minicircle (0.8 to 1 kb in length) of kDNA is an ideal target, since it is present in 10,000 copies per cell that are distributed among about 10 different sequence classes. In addition, the minicircle sequence is known for most Leishmania species, and it possesses a variable region that offers accurate discrimination between species.
The aim of the present research was to study the infection rate in sand fly populations, as well as to identify potential insect vectors and latent infections present in areas where leishmaniasis is endemic. The limiting factors for the existing techniques are their low sensitivities and the difficulty of processing large number of samples, as is required for epidemiological studies. To overcome these difficulties, a simple method was developed based on the minicircle sequence of Leishmania donovani described by Smyth et al. (24). The procedure consisted of a seminested PCR assay which takes place in a single tube and uses three primers. This method will permit the comparison of Leishmania sequences amplified from sand fly vectors to those from kala-azar patients, which is crucial in order to investigate the patterns of Leishmania transmission.
MATERIALS AND METHODS
Sand fly collection.
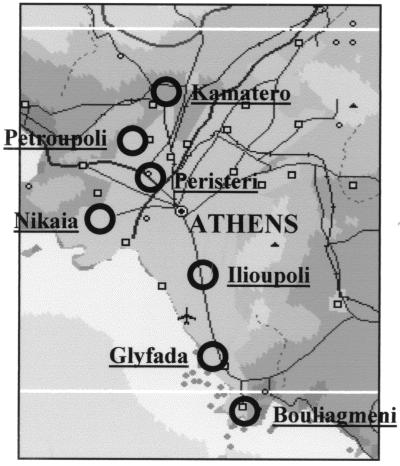
Sand flies were caught in seven areas of the greater Athens area (Fig. 1) where cases of visceral leishmaniasis had been reported: Glyfada (three collection stations), Peristeri (one station), Bouliagmeni (two stations), Ilioupoli (one station), Kamatero (one station), Petroupoli (two stations), and Nikaia (three stations). Insects were captured by means of CDC miniature light traps, which were placed overnight in corrals with chickens, rabbits, or ducks. Specimens were collected from the traps with a manual-capture tube and were stored immediately in liquid nitrogen. Only female sand flies were selected for the present study.
FIG. 1.
Map of the greater Athens area. Black circles show the areas where sand flies were collected for the present study.
Dissection and morphological identification of sand flies.
Sand flies were washed in 1% detergent (washing-up liquid) solution for 5 min and dissected in a drop of 1× PBS (phosphate-buffered saline) using minute pins, under a stereoscope. Heads and last abdominal segments were kept for morphological identification based on the keys described by Leger et al. (12). Individual code names, consisting of a letter(s) taken from the collection area name followed by a number were assigned; for example, “G133” represents the 133rd female sand fly collected in Glyfada.
Leishmania isolates.
All Leishmania species were obtained from the Association pour la Promotion de la Recherche et des Echanges en Parasitologie (Laboratoire de Parasitologie, Institut de Botanique, Montpellier, France) and cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 500 U of penicillin/ml, and 500 μg of streptomycin/ml. The identification numbers of the Leishmania strains are as follows: Leishmania infantum, LEM 235 (MHOM/TN/80/IPT1); L. donovani, LEM 703 (MHOM/IN/80/DD8); L. major, LEM 134 (MHOM/SU/73/5-ASKH); L. aethiopica, LEM 144 (MHOM/ET/72/L100); L. tropica, LEM 419 (MHOM/SU/74/K27); L. amazonensis, LEM 690 (MHOM/BR/73/M226); L. mexicana, LEM 695 (MHOM/BZ/82/BEL21); and L. braziliensis, LEM 2252 (MHOM/BR/84/LTB300).
DNA extraction.
DNA was extracted as described by Aransay et al. (3). Briefly, individual sand fly bodies were homogenized with a sealed Pasteur pipette in 1.5-ml tubes. One hundred fifty microliters of extraction buffer (1% sodium dodecyl sulfate [SDS]–25 mM NaCl–25 mM EDTA) was added, and samples were placed at 65°C for 30 min. Following the addition of 100 μl of 3 M potassium acetate (pH 7.2), the homogenates were incubated on ice for 30 min and then centrifuged for 15 min at 13,000 × g. Supernatants were recovered, and DNA was precipitated with the addition of 600 μl of 100% ethanol. DNA pellets were resuspended in 50 μl of 0.5× Tris-EDTA (TE) (pH 8.0). Five-microliter portions of these DNA extracts were used for PCR amplification.
Leishmania cultures were washed twice in 1× PBS and were then resuspended in extraction buffer and extracted as described above for the insect samples.
Amplification of kinetoplastic minicircle DNA from sand flies.
Primers LINR4, LIN17, and LIN19 were designed within the conserved area of the minicircle and contained conserved sequence blocks (CSB) (4) CSB-3, CSB-2, and CSB-1 respectively. The combination of primers LINR4 (forward) (5′-GGG GTT GGT GTA AAA TAG GG-3′), LIN17 (reverse) (5′-TTT GAA CGG GAT TTC TG-3′), and LIN19 (reverse) (5′-CAG AAC GCC CCT ACC CG-3′) was used in a seminested PCR technique. The first amplification reaction was carried out in a total of 10 μl containing 1× Taq polymerase buffer (GIBCO-BRL), 1.5 mM MgCl2, 0.2 μM deoxynucleoside triphosphates (dNTPs), 1 μM LINR4, 0.2 μM LIN17, 1 U of Taq polymerase (GIBCO-BRL), and 5 μl of DNA extract, overlaid with mineral oil. The mixture was incubated in a Perkin-Elmer thermocycler (GeneAmp PCR System 9600) at 94°C for 5 min followed by 17 cycles, each consisting of 30 s at 94°C, 30 s at 52°C, and 30 s at 72°C. After the last cycle, the extension was continued for a further 10 min. The seminested amplification was carried out, with the addition of a 90-μl solution containing buffer, MgCl2, dNTPs, and Taq polymerase as described above for the first round, and LIN19 to a final concentration of 1 μM, for 33 cycles (94°C for 30 s, 58°C for 30 s, and 72°C for 1 min). Twenty microliters of the amplification reaction product was resolved in a 1.5% agarose gel and visualized under UV transillumination.
Standard PCR with primers LINR4 and LIN17 or LINR4 and LIN19 was carried out in a total of 100 μl containing 1× Taq polymerase buffer (GIBCO-BRL), 1.5 mM MgCl2, 0.2 μM dNTPs, 1 μM LINR4, 1 μM LIN17 or LIN19, and 1.5 U of Taq polymerase (GIBCO-BRL). The reaction mixtures were incubated at 94°C for 5 min, followed by 40 cycles, each consisting of 30 s at 94°C, 30 s at 52°C (for LINR4 and LIN17) or 58°C (for LINR4 and LIN19), and 1 min at 72°C, and a final extension at 72°C for 10 min. Products were resolved as described above.
Sequencing.
PCR products were purified with GFX PCR DNA and a Gel Band purification kit (Pharmacia Biotech).
Sequencing of 200 ng of the PCR product amplified from sand fly samples was carried out by cycle sequencing with a Sequitherm EXCEL II Long-Read DNA Sequencing Kit (EPICENTRE TECHNOLOGIES, BIOZYM), using the fluorescent primers LIN-R4-(700) (5′-GGT TGG TGT AAA ATA GGG-3′) and LIN-19-(800) (5′-GAA CGC CCC TAC CCG-3′). After 1 min of preheating to 95°C, 30 amplification cycles (each cycle consisting of 30 s at 95°C, 30 s at 55°C, and 1 min at 70°C) and an extra elongation step (5 min at 72°C) were performed. Then 1.5 μl of the sequencing reaction was loaded in a 4.6% acrylamide gel. Sequences were resolved in a LI-COR 4200 automated sequencer.
Sequence analyses.
Overlaps of both DNA strand sequences were performed for each sample using DNAMAN for windows, version 2.6 (C. Woffelman, Lynon Biosoft, Institute of Molecular Plant Sciences, Leiden University), and sequence similarity searching for each single sample was performed using the BLAST program (2). All sequences were assembled by SeqPUP (D. G. Gilbert, Biology Department, Indiana University, 1995), and multiple sequence alignments were achieved using the CLUSTAL W Multiple Sequence Alignment Program, version 1.7 (D. G. Higgins, J. D. Thomson, and T. J. Gibson), applying the default settings (gap opening penalty, 10.00; gap extension penalty, 0.05; delay divergent sequences, 40%; DNA transitions weight, 0.50).
Identification of sand fly species by PCR and RFLP.
Molecular identification of individual sand flies was carried out by restriction fragment length polymorphism (RFLP) of amplified small-subunit (SSU) rRNA genes as previously described (3).
RESULTS
Morphological identification of sand flies.
A total of 645 female sand flies from seven different collection areas were studied, from which seven phlebotomine species were morphologically identified: Phlebotomus (Phlebotomus) papatasi (n = 327), Phlebotomus (Paraphlebotomus) alexandri (n = 25), Phlebotomus (Paraphlebotomus) sergenti (n = 1), Phlebotomus (Larroussius) neglectus (n = 68), Phlebotomus (Larroussius) tobbi (n = 70), Phlebotomus (Adlerius) simici (n = 123), and Sergentomyia minuta (n = 31) (authorities for all species are given by Seccombe et al. [22]). The distribution of the species for each station is outlined in Table 1.
TABLE 1.
Number of sand flies collected in the greater Athens area in 1992 and number of sandflies found with Leishmania DNA
| Species | Total no. (no. positivea) for the following station (code):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Glyfada (G) | Peristeri (P) | Bouliagmeni (B) | Ilioupoli (I) | Kamatero (K) | Petroupoli (PP) | Nikaia (N) | Total | |
| P. (Phlebotomus) papatasi | 165 (9) | 93 (15) | 0 (0) | 1 (0) | 6 (0) | 18 (2) | 28 (1) | 327 (27) |
| P. (Larroussius) neglectus | 5 (0) | 1 (1) | 16 (0) | 0 (0) | 23 (1) | 0 (0) | 20 (1) | 68 (3) |
| P. (Larroussius) tobbi | 33 (3) | 4 (1) | 19 (1) | 0 (0) | 25 (0) | 1 (0) | 0 (0) | 70 (5) |
| P. (Adlerius) simici | 98 (4) | 1 (0) | 7 (0) | 0 (0) | 14 (1) | 1 (0) | 9 (0) | 123 (5) |
| P. (Paraphlebotomus) alexandri | 22 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (0) | 25 (1) |
| P. (Paraphlebotomus) sergenti | 0 (0) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) |
| S. minuta | 5 (0) | 0 (0) | 0 (0) | 4 (0) | 11 (0) | 5 (0) | 6 (0) | 31 (0) |
| Total | 328 (17) | 99 (17) | 42 (1) | 6 (0) | 79 (2) | 25 (2) | 66 (2) | 645 (41) |
Number of sand flies from which Leishmania DNA was amplified.
Sensitivity and specificity of the seminested PCR assay.
Combinations of several primers designed within the conserved area of the kDNA minicircle were tested for their ability to increase the sensitivity of the standard PCR. The set of LINR4, LIN17, and LIN19 was successfully used in a seminested PCR assay that was carried out in two amplification steps, but in a single tube.
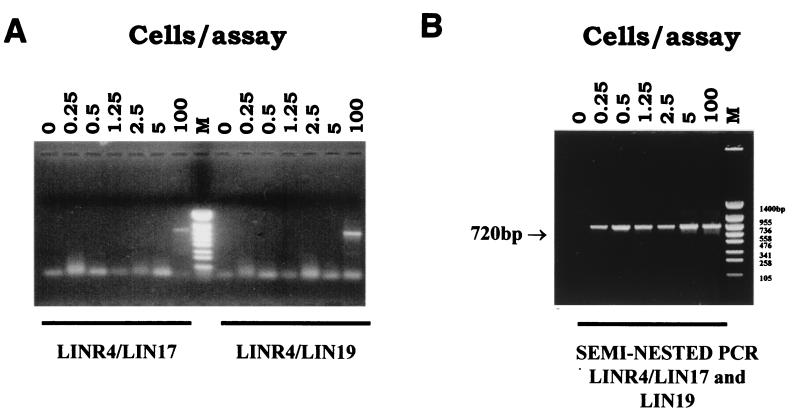
Different concentrations of L. infantum parasites were extracted together with individual sand flies so that 100, 5, 2.5, 1.25, 0.5, 0.25, and 0 cells were used per amplification reaction. With the seminested PCR it was possible to detect as few as 0.25 parasite per reaction (Fig. 2B), while a standard PCR with primers LINR4 and LIN17 or LINR4 and LIN19 failed to detect 5 parasites (Fig. 2A). Occasionally, two faint nonspecific bands (around 1,000 and 600 bp) appeared in addition to the expected 720-bp band in the amplification reactions with the higher concentrations of promastigotes (Fig. 2B).
FIG. 2.
Sensitivity comparison between standard PCR with primers LINR4 and LIN17 or LINR4 and LIN19 (A) and seminested PCR with the set of primers LINR4, LIN17, and LIN19 (B). The DNA size marker (lane M) is PUC19 TaqI/PUC19 Sau3A.
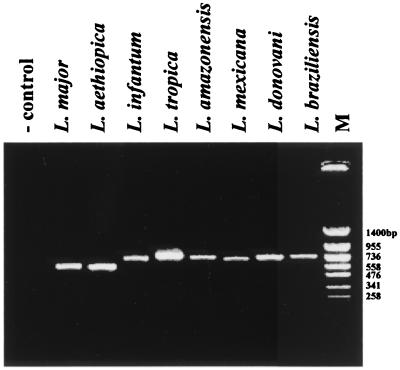
The specificity of the assay was tested by amplification of the kDNA minicircle from eight Leishmania species. All PCR products were about 720 bp long, except for those amplified from L. major and L. aethiopica, which were around 650 bp (Fig. 3).
FIG. 3.
Specificity of the seminested PCR assay with primers LINR4, LIN17, and LIN19. The DNA size marker (lane M) is PUC19 TaqI/PUC19 Sau3A.
Evaluation of the method for sand flies collected in the wild.
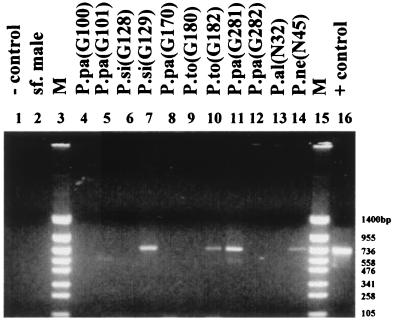
The method was validated on sand flies collected during an epidemiological study in the area of endemicity of greater Athens during the summer of 1992. Minicircle DNA was amplified from 35 out of 522 (6.7%) female sand flies that did not contain a blood meal when collected: 23 P. (Phlebotomus papatasi flies, 3 P. (Larroussius) neglectus flies, 5 P. (Larroussius) tobbi flies, 5 P. (Adlerius) simici flies, and 1 P. (Paraphlebotomus) alexandri fly. Minicircle DNA was also amplified from 6 [all belonging to P. (Phlebotomus) papatasi] out of 123 (4.8%) specimens that contained blood.
The size of all the amplified products was about 720 bp (Fig. 4), as expected from the results obtained from the reference strains. There were five additional samples that resulted in faint amplified products of about 600 bp (Fig. 4, lane 5). Among the positive samples, 12 had two more amplified bands (around 1,000 and 600 bp) in addition to the expected 720-bp band (Fig. 4, lane 11).
FIG. 4.
Seminested PCR with primers LINR4, LIN17, and LIN19 for detection of Leishmania DNA within sand flies collected in parts of the greater Athens area. − control, reaction without DNA; sf. male, male sandfly specimen; M, DNA size marker PUC19 TaqI/PUC19 Sau3A; P.pa, P. (Phlebotomus) papatasi; P.si, P. (Adlerius) simici; P.to, P. (Larroussius) tobbi; P.al, P. (Paraphlebotomus) alexandri; P.ne, P. (Larroussius) neglectus; + control, L. infantum (LEM 235).
Cross-contamination was monitored by negative controls for sample extraction and PCR; all were negative. False-negative results due to inhibition were also controlled by adding Leishmania DNA to duplicates of arbitrary negative samples. No inhibition was detected in any of the samples tested.
Sequencing of PCR products amplified from sand fly samples and sequence analyses.
Direct sequencing of the PCR products was carried for both strands. (GenBank and EMBL accession numbers of the sequences are listed in Table 2.) The sequencing of those samples whose amplification pattern consisted of more than one band resulted in a single clear sequence. All sequences were compared for similarities to the EMBL and GenBank databases. Most of the sequences were related to L. donovani and L. infantum kinetoplast minicircle DNA. The sequences obtained were different from those of positive controls that were used in the laboratory, namely, LEM 235 (L. infantum) (GenBank accession number AF190550; EMBL accession number AJ270144) and LEM703 (L. donovani) (GenBank accession number AF190551; EMBL accession number AJ270145). The smaller products (600 bp) were proven to be nonspecific PCR products unrelated to Leishmania kDNA sequences.
TABLE 2.
List of codes and species of sand flies from which Leishmania DNA was amplified, and the corresponding GenBank and EMBL accession numbers of the amplified sequences
| Specimen code | Species | GenBank (EMBL) accession no. |
|---|---|---|
| G121 | P. (Adlerius) simici | AF190509 (AJ270103) |
| G129 | P. (Adlerius) simici | AF190510 (AJ270104) |
| G138 | P. (Larroussius) tobbi | AF190511 (AJ270105) |
| G142 | P. (Phlebotomus) papatasi | AF190512 (AJ270106) |
| G146 | P. (Phlebotomus) papatasi | AF190513 (AJ270107) |
| G160 | P. (Phlebotomus) papatasi | AF190514 (AJ270108) |
| G166 | P. (Adlerius) simici | AF190515 (AJ270109) |
| G167 | P. (Larroussius) tobbi | AF190516 (AJ270110) |
| G169 | P. (Adlerius) simici | AF190517 (AJ270111) |
| G181 | P. (Phlebotomus) papatasi | AF190518 (AJ270112) |
| G182 | P. (Larroussius) tobbi | AF190519 (AJ270113) |
| G184 | P. (Paraphlebotomus) alexandri | AF190520 (AJ270114) |
| G227a | P. (Phlebotomus) papatasi | AF190521 (AJ270115) |
| G281 | P. (Phlebotomus) papatasi | AF190522 (AJ270116) |
| G297a | P. (Phlebotomus) papatasi | AF190523 (AJ270117) |
| G304 | P. (Phlebotomus) papatasi | AF190524 (AJ270118) |
| G319a | P. (Phlebotomus) papatasi | AF190525 (AJ270119) |
| P12 | P. (Phlebotomus) papatasi | AF190526 (AJ270120) |
| P26 | P. (Larroussius) tobbi | AF190527 (AJ270121) |
| P27a | P. (Phlebotomus) papatasi | AF190528 (AJ270122) |
| P35 | P. (Phlebotomus) papatasi | AF190529 (AJ270123) |
| P38 | P. (Phlebotomus) papatasi | AF190530 (AJ270124) |
| P49 | P. (Phlebotomus) papatasi | AF190531 (AJ270125) |
| P57 | P. (Phlebotomus) papatasi | AF190532 (AJ270126) |
| P67 | P. (Phlebotomus) papatasi | AF190533 (AJ270127) |
| P70 | P. (Phlebotomus) papatasi | AF190534 (AJ270128) |
| P81 | P. (Phlebotomus) papatasi | AF190535 (AJ270129) |
| P84 | P. (Phlebotomus) papatasi | AF190536 (AJ270130) |
| P89 | P. (Phlebotomus) papatasi | AF190537 (AJ270131) |
| P90 | P. (Phlebotomus) papatasi | AF190538 (AJ270132) |
| P92 | P. (Larroussius) neglectus | AF190539 (AJ270133) |
| P95a | P. (Phlebotomus) papatasi | AF190540 (AJ270134) |
| P97a | P. (Phlebotomus) papatasi | AF190541 (AJ270135) |
| P98 | P. (Phlebotomus) papatasi | AF190542 (AJ270136) |
| K35 | P. (Adlerius) simici | AF190543 (AJ270137) |
| K46 | P. (Larroussius) neglectus | AF190544 (AJ270138) |
| PP19 | P. (Phlebotomus) papatasi | AF190545 (AJ270139) |
| PP23 | P. (Phlebotomus) papatasi | AF190546 (AJ270140) |
| N15 | P. (Phlebotomus) papatasi | AF190547 (AJ270141) |
| N45 | P. (Larroussius) neglectus | AF190548 (AJ270142) |
| B8 | P. (Larroussius) tobbi | AF190549 (AJ270143) |
This sand fly contained a blood meal at the time of collection.
Multiple alignments of the sequences obtained were carried out in order to recognize any correlation between different sequences and areas or times of collection or species of host sand fly. Eight classes of sequences were recognized after comparison of the 41 sequences (three groups of sequences and five single sequences), but no obvious relationship between the assembled sequences and their origins was found. The first class of sequences was found in specimens G121, G169, G319, P12, P49, and P95, with slight differences in base composition. The second class was found in samples G138, G142, G160, G166, G167, G181, G182, G184, G297, G304, P26, P27, P35, P38, P57, P67, P70, P81, P84, P89, P90, P92, P97, P98, K35, PP19, PP23, and N45. The third was found in specimens G146 and N15, and the other five belonged to the amplified products from samples G129, G227, G281, K46, and B8.
Sample P81 was amplified on three different occasions and direct sequencing of the amplified products was carried out to check whether there was any difference in the amplification of the minicircle sequence. The sequences of the three products were identical.
Sand fly species identification by PCR-RFLP of SSU ribosomal DNAs (rDNAs).
In order to confirm the species of the sand flies from which Leishmania DNA was amplified, SSU rRNA genes were amplified from all positive samples. The RFLP profiles obtained agreed with the species-specific patterns described by Aransay et al. (3) and with morphological identifications (data not shown).
DISCUSSION
Control of leishmaniasis in areas of endemicity requires a thorough knowledge of Leishmania ecology and epidemiology. There is a major problem for epidemiologists both in the identification of reservoir hosts and in the detection of vectors. Therefore, finding naturally infected sand flies is essential in identifying a species as a vector of Leishmania and in studying infection rates in areas of endemicity.
The applicability of kDNA for the detection and identification of Leishmania within sand flies by DNA hybridization have been shown previously (17, 19, 20). However, nonradiolabeled and more-sensitive methods are required for epidemiological studies. The highly sensitive technique of PCR has been used formerly to detect Leishmania DNA in sand fly species from New World (5) and India (14). A highly sensitive method, for both screening for the presence of Leishmania within sand flies collected in the wild and identification of Leishmania species, was developed. The high sensitivity of the assay developed is needed because detection in crude biological samples is hampered by the inhibitory effects of other substances present and by impaired accessibility of the kinetoplast (18). The ability to amplify DNA from at least eight Leishmania species is important, making this technique useful in areas of endemicity where these species are present. It is also worth mentioning that the proposed technique takes place in a single tube, and therefore, there is no extra manipulation of the first-round amplification product, as is required for the nested-PCR technique (23). This feature reduces the risk of false-positive results due to cross-contamination.
Sand fly samples which contained a blood meal did not require any extra manipulations such as are needed for squash blots (13), since the DNA extraction procedure used (without phenol or chloroform extractions) eliminates any inhibitor that could affect the Taq polymerase enzyme activity. Furthermore, the same DNA extract from single sand flies was successfully used for accurate identification of sand fly species by PCR-RFLP as described previously (3).
Direct sequencing of the amplified minicircle kDNA products offers a viable alternative to the PCR-RFLP approach described by Noyes et al. (15), since the high variability of minicircle sequences does not impose any limitations on the sequence analysis. The fact that clear and reproducible sequences were obtained could suggest either that there is amplification of a predominant minicircle class present in Leishmania parasites or that primer sequences are not conserved in all the minicircle classes.
The high infection rate of sand flies observed in this study (5.4% of the tested specimens without blood in their guts) could be attributed to the high sensitivity of the assay. In addition, many of the infected samples were collected from the same corral, which means that they probably fed on the same infected source.
Leishmania DNA was found in some P. (Larroussius) neglectus samples, the only proven vector of L. infantum in Greece, as well as in P. (Larroussius) tobbi, P. (Adlerius) simici, P. (Paraphlebotomus) alexandri, and P. (Phlebotomus) papatasi. P. (Larroussius) tobbi, which also belongs to the Larroussius subgenus, had been previously investigated for its vectorial capacity (21), and it is considered a suspected vector. The other three species [P. (Adlerius) simici, P. (Paraphlebotomus) alexandri, and P. (Phlebotomus) papatasi) have never been implicated in the transmission of Leishmania in Greece. However, P. (Adlerius) simici and P. (Paraphlebotomus) alexandri are proven vectors of L. infantum and L. donovani, respectively, in different regions of China (10). Detection of Leishmania DNA does not imply that a sand fly species is a vector, since the assay cannot distinguish between the presence of Leishmania amastigotes (the stage in macrophages) from an infected blood meal and promastigotes (the Leishmania stage which is developed in the gut of a sand fly vector). Therefore, the technique described is useful for investigating the presence of Leishmania in populations, but it cannot be used as the only tool for identifying a certain sand fly species as a vector. Experimentally infected sand flies can then be used in transmission assays in order to demonstrate that the suspected species are vectors and to investigate possible transmission cycles (10).
The combination of the newly developed method with the molecular tool for sand fly species identification (3) will make the laborious dissection of sand flies unnecessary. As a result, highly skilled microscopists will not be required for accurate diagnosis of parasite and vector species. In addition, large numbers of samples can be screened synchronously, as is required for epidemiological studies.
ACKNOWLEDGMENTS
A.M.A. was a recipient of a predoctoral fellowship from the Basque Government.
We are grateful to B. Papadopoulos and G. Gozalo for assistance in sampling.
REFERENCES
- 1.Adini I, Jacobson R L, Kasap M, Schlein Y, Jaffe C L. Species-specific detection of Leishmania in sandflies using an enzyme-linked immunosorbent assay. Trans R Soc Trop Med Hyg. 1998;92:35–37. doi: 10.1016/s0035-9203(98)90946-4. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Aransay A M, Scoulica E, Chaniotis B, Tselentis Y. Typing of sandflies from Greece and Cyprus by DNA polymorphism of 18S rRNA gene. Insect Mol Biol. 1999;8:179–184. doi: 10.1046/j.1365-2583.1999.820179.x. [DOI] [PubMed] [Google Scholar]
- 4.Brewster S, Aslett M, Barker D C. Kinetoplast DNA minicircle database. Parasitol Today. 1998;14:437–438. doi: 10.1016/s0169-4758(98)01330-1. [DOI] [PubMed] [Google Scholar]
- 5.de Bruijn M H, Barker D C. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop. 1992;52:45–58. doi: 10.1016/0001-706x(92)90006-j. [DOI] [PubMed] [Google Scholar]
- 6.Dejeux P. Global control and Leishmania HIV co-infection. Clin Dermatol. 1999;17:317–325. doi: 10.1016/s0738-081x(99)00050-4. [DOI] [PubMed] [Google Scholar]
- 7.Esseghir S, Ftaiti A, Ready P D, Khdraoui B, Zaafouri B, Dellagi K, Ben-Ismail R. The squash blot technique and the detection of Leishmania major in Phlebotomus papatasi in Tunisia. Arch Inst Pasteur Tunis. 1993;70:493–496. [PubMed] [Google Scholar]
- 8.Guevara P, Alonso G, da Silveira J F, de Mello M, Scorza J V, Anez N, Ramirez J L. Identification of New World Leishmania using ribosomal gene spacer probes. Mol Biochem Parasitol. 1992;56:15–26. doi: 10.1016/0166-6851(92)90150-i. [DOI] [PubMed] [Google Scholar]
- 9.Hassan M Q, Ghosh A, Ghosh S S, Gupta M, Basu D, Mallik K K, Adhya S. Enzymatic amplification of mini-exon-derived RNA gene spacers of Leishmania donovani: primers and probes for DNA diagnosis. Parasitology. 1993;107:509–517. doi: 10.1017/s0031182000068086. [DOI] [PubMed] [Google Scholar]
- 10.Killick-Kendrick R. The biology and control of phlebotomine sandflies. Clin Dermatol. 1999;17:279–289. doi: 10.1016/s0738-081x(99)00046-2. [DOI] [PubMed] [Google Scholar]
- 11.Lainson R, Shaw J J. Evolution, classification and geographical distribution. In: Peters W, Killick-Kendrick R, editors. The leishmaniases in biology and medicine. 1. Biology and epidemiology. London, United Kingdom: Academic Press; 1987. pp. 1–120. [Google Scholar]
- 12.Leger N, Pesson B, Madulo-Leblond G. Les phlebotomes de Grece. Biol Gallo-Hellenica. 1986;11:165–192. [Google Scholar]
- 13.McNerney R, Frame I A, Vexenat J A, Fonseca de Castro J A, Howard M K, Dillon R, Wilson S, Miles M A. Visceral leishmaniasis in Teresina, N.E. Brazil: towards a DNA probe kit and its adaptation to processing blood-contaminated samples. Arch Inst Pasteur Tunis. 1993;70:405–418. [PubMed] [Google Scholar]
- 14.Mukherjee S, Hassan M Q, Ghosh A, Ghosh K N, Bhattacharya A, Adhya S. Short report: Leishmania DNA in Phlebotomus and Sergentomyia species during a kala-azar epidemic. Am J Trop Med Hyg. 1997;57:423–425. doi: 10.4269/ajtmh.1997.57.423. [DOI] [PubMed] [Google Scholar]
- 15.Noyes H A, Reyburn H, Bailey J W, Smith D. A nested-PCR-based schizodeme method for identifying Leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropica in Pakistan. J Clin Microbiol. 1998;36:2877–2881. doi: 10.1128/jcm.36.10.2877-2881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piarroux R, Azaiez R, Lossi A M, Reynier P, Muscatelli F, Gambarelli F, Fontes M, Dumon H, Quilici M. Isolation and characterization of a repetitive DNA sequence from Leishmania infantum and development of a visceral leishmaniasis polymerase chain reaction. Am J Trop Med Hyg. 1993;49:364–369. doi: 10.4269/ajtmh.1993.49.364. [DOI] [PubMed] [Google Scholar]
- 17.Ready P D, Smith D F, Killick-Kendrick R. DNA hybridizations on squash-blotted sandflies to identify both Phlebotomus papatasi and infecting Leishmania major. Med Vet Entomol. 1988;2:109–116. doi: 10.1111/j.1365-2915.1988.tb00060.x. [DOI] [PubMed] [Google Scholar]
- 18.Rodgers M R, Popper S J, Wirth D F. Amplification of kinetoplast DNA as a tool in the detection and diagnosis of Leishmania. Exp Parasitol. 1990;71:267–275. doi: 10.1016/0014-4894(90)90031-7. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez N, Aguilar C M, Barrios M A, Barker D C. Detection of Leishmania braziliensis in naturally infected individual sandflies by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1999;93:47–49. doi: 10.1016/s0035-9203(99)90176-1. [DOI] [PubMed] [Google Scholar]
- 20.Rogers W O, Burnheim P F, Wirth D F. Detection of Leishmania within sandflies by kinetoplast DNA hybridization. Am J Trop Med Hyg. 1988;39:434–439. doi: 10.4269/ajtmh.1988.39.434. [DOI] [PubMed] [Google Scholar]
- 21.Schlein Y, Polacheck I, Yuval B. Mycoses, bacterial infections and antibacterial activity in sandflies (Psychodidae) and their possible role in the transmission of leishmaniasis. Parasitology. 1985;90:57–66. doi: 10.1017/s0031182000049015. [DOI] [PubMed] [Google Scholar]
- 22.Seccombe A K, Ready P D, Huddleston L M. A catalogue of Old World phlebotomine sandflies (Diptera: Psychodidae, Phlebotominae) Occas Pap Syst Entomol. 1993;8:1–57. [Google Scholar]
- 23.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Brown A J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth A J, Ghosh A, Hassan M Q, Basu D, De Bruijn M H, Adhya S, Mallik K K, Barker D C. Rapid and sensitive detection of Leishmania kinetoplast DNA from spleen and blood samples of kala-azar patients. Parasitology. 1992;105:183–192. doi: 10.1017/s0031182000074096. [DOI] [PubMed] [Google Scholar]
- 25.Tselentis Y, Gikas A, Chaniotis B. Kala-azar in Athens basin. Lancet. 1994;343:1635. doi: 10.1016/s0140-6736(94)93085-6. [DOI] [PubMed] [Google Scholar]