Abstract
Production of 2,4-diacetylphloroglucinol (2,4-DAPG) in the rhizosphere by strains of fluorescent Pseudomonas spp. results in the suppression of root diseases caused by certain fungal plant pathogens. In this study, fluorescent Pseudomonas strains containing phlD, which is directly involved in the biosynthesis of 2,4-DAPG, were isolated from the rhizosphere of wheat grown in soils from wheat-growing regions of the United States and The Netherlands. To assess the genotypic and phenotypic diversity present in this collection, 138 isolates were compared to 4 previously described 2,4-DAPG producers. Thirteen distinct genotypes, one of which represented over 30% of the isolates, were differentiated by whole-cell BOX-PCR. Representatives of this group were isolated from eight different soils taken from four different geographic locations. ERIC-PCR gave similar results overall, differentiating 15 distinct genotypes among all of the isolates. In most cases, a single genotype predominated among isolates obtained from each soil. Thirty isolates, representing all of the distinct genotypes and geographic locations, were further characterized. Restriction analysis of amplified 16S rRNA gene sequences revealed only three distinct phylogenetic groups, one of which accounted for 87% of the isolates. Phenotypic analyses based on carbon source utilization profiles revealed that all of the strains utilized 49 substrates and were unable to grow on 12 others. Individually, strains could utilize about two-thirds of the 95 substrates present in Biolog SF-N plates. Multivariate analyses of utilization profiles revealed phenotypic groupings consistent with those defined by the genotypic analyses.
Strains of fluorescent Pseudomonas spp. have been studied extensively as biological control agents for soilborne plant pathogens (6, 14, 16, 29, 32). Strains that produce the antifungal compound 2,4-diacetylphloroglucinol (2,4-DAPG) play an important role in the suppression of some root diseases when introduced into the rhizosphere via seed or soil treatments (13, 26, 30). The genetic locus responsible for the biosynthesis of 2,4-DAPG contains phlD, a unique bacterial gene with similarity to plant genes encoding chlacone/stilbene synthases (2, 3). The phlD gene has been used as a genetic marker to detect 2,4-DAPG-producing Pseudomonas strains from several soils because the occurrence of the gene is correlated with the production of 2,4-DAPG in vitro (12, 18).
The fungus Gaeumannomyces graminis var. tritici attacks the roots of wheat and barley, causing the disease take-all (1, 8). Continuous cropping of wheat in fields that have take-all results in a gradual decline in the incidence and severity of the disease, a phenomenon called take-all decline (TAD) (1, 10, 27). The production of phloroglucinol derivatives by phlD-containing (phlD+) Pseudomonas strains plays an important role in TAD (18, 19, 24). Populations sizes of phlD+ Pseudomonas strains were significantly larger in the rhizosphere of wheat grown in TAD (i.e., suppressive) soils than in non-TAD (i.e., conducive) soils from Washington State (18). In addition, phlD+ strains were associated with the roots of wheat grown in soils from other regions of the United States and The Netherlands with a history of wheat monoculture (21, 24). Both 2,4-DAPG and phlD+ pseudomonads have been isolated directly from the rhizosphere of wheat grown in a TAD soil (4, 20). Pasteurization of a TAD soil resulted in the concurrent loss of phlD+ Pseudomonas and suppressiveness to take-all, while transfer of relatively small amounts of TAD soil into a conducive soil resulted in the establishment of 2,4-DAPG-producing populations sufficient to suppress the disease (19).
Several distinct groups of 2,4-DAPG-producing Pseudomonas strains have been identified (12, 26). Two groups were differentiated on the basis of antibiotic production, one that produced 2,4-DAPG, hydrogen cyanide, and pyoluteorin (PLT) and the other that did not produce PLT but did produce the first two substances. The former group contained strains that were genotypically quite homogeneous, despite having been isolated from the roots of different crop plants grown in soils from different geographic locations (12). In contrast, eight distinct genotypes were observed among 32 isolates known to produce 2,4-DAPG but not PLT (12). Sharifi-Tehrani et al. reported that the 2,4-DAPG-producing, PLT-negative strains as a group were more effective as biological control agents against Pythium ultimum on cucumbers and Fusarium oxysporum f.sp. radicis-lycopersici on tomatoes, and they noted that the amount of 2,4-DAPG produced by these strains in vitro correlated with the amount of disease suppression in the latter system (26). These results suggest that there is diversity among 2,4-DAPG-producing Pseudomonas spp. in nature and that strains will differ in their effectiveness as biological control agents.
Given the association of phlD-containing Pseudomonas spp. with the phenomenon of TAD, we initiated this study to better define the diversity of these bacteria in different wheat-growing regions. Knowledge of the genotypic and phenotypic characteristics of these strains may implicate specific subsets of 2,4-DAPG producers in TAD and will allow us to distinguish strains that have different abilities to suppress soilborne root pathogens of wheat.
MATERIALS AND METHODS
Soils and plants.
A collection of soils from wheat-growing regions of the United States and The Netherlands was assembled (Table 1). Soils from fields near Lind or Quincy, Wash., that had supported long-term wheat cropping or native vegetation were described previously (18) and were designated 4L and W or QT, QTN, Q, Q8r, QX-87, and QV, respectively. Other United States soils were collected during 1997. Soils designated FTAD1R and FFL1R were from fields on the campus of North Dakota State University (NDSU), Fargo, that had been cropped continuously to wheat since 1882 (NDSU research plot no. 2) or to flax since 1894 (NDSU research plot no. 30). The HT soil was from a field near Hallock, Minn., that had been cropped continuously to wheat for 10 years. The OC soil was from a field on the campus of Cornell University, Ithaca, N.Y., which historically had been cropped primarily to legumes but on which wheat had been planted in 1997. Soils CV and CC were collected near Caldwell, Kans., from a roadside site near a field with a 100-year history of wheat monoculture. Soils JMP and D27B were from fields near Woensdrecht, The Netherlands, that had been cropped continuously to wheat for 27 and 14 years, respectively. Soils were air dried, sieved, and homogenized by hand and then maintained at room temperature prior to potting.
TABLE 1.
Soils and strains used in this study
| Prefix or straina | Isolate no. | rep-PCR genotypesb | Soil source | Historyc | Sampled | Reference |
|---|---|---|---|---|---|---|
| Prefix | ||||||
| CV | 1-1, 2-3, 2-4, 2-8, 3-3, 3-6, 4-1, 4-3, 4-5, 4-7 | H, I | Caldwell, Kans. | Not cropped | 1998, g.c. | This study |
| CC | 1-1, 1-3, 2-1, 2-8, 5-1 | H, I | Caldwell, Kans. | Not cropped | 1998, g.c. | This study |
| 3-1, 3-6, 4-1 | J, J | Caldwell, Kans. | Not cropped | 1998, g.c | This study | |
| FTAD1R | 25, 26, 27, 33, 34, 35, 38 | D, D1 | Fargo, N.D. | LTW | 1997, g.c. | This study |
| 36 | I, I | Fargo, N.D. | LTW | 1997, g.c. | This study | |
| FFL1R | 8, 10, 13, 14, 17, 22 | J, J | Fargo, N.D. | LTF | 1997, g.c. | This study |
| 18, 21, 25 | G, I | Fargo, N.D. | LTF | 1997, g.c. | This study | |
| 9 | D, E | Fargo, N.D. | LTF | 1997, g.c. | This study | |
| HT | 5-1, 5-5, 5-8, 5-10, 5-12, 6-2, 6-4, 6-7 | N, N | Hallock, Min. | LTW | 1998, g.c. | This study |
| OC | 4-1, 4-2 | D, D2 | Ithaca, N.Y. | Mixed, W | 1998, g.c. | This study |
| 4L | 1, 3, 5, 6, 7, 8, 9, 10, 11, 12 | D, D1 | Lind, Wash. | LTW | 1996, g.c. | This study |
| W | 2-6, 2-9 | D, D1 | Lind, Wash. | LTW | 1998, field | This study |
| 4-4 | L, L | Lind, Wash. | LTW | 1998, field | This study | |
| QT | 1-6, 3-3, 3-1, 3-2, 4-2, 6-1 | E, F | Quincy, Wash. | LTW | 1998, g.c. | This study |
| 1-5, 5-1, 5-2 | D, D1 | Quincy, Wash. | LTW | 1998, g.c. | This study | |
| Q | 1-3, 1-4, 2-1, 2-6, 2-10, 2-12 | B, C | Quincy, Wash. | LTW | 1998, field | This study |
| 2-2, 2-19 | E, F | Quincy, Wash. | LTW | 1998, field | This study | |
| 2-5 | D, E | Quincy, Wash. | LTW | 1998, field | This study | |
| QV | 1, 2, 5, 7, 12, 14, 7V16, 7V18, 8V4, 8V8 | D, E | Quincy, Wash. | Not cropped | 1996, g.c. | 19 |
| Q8r | 1, 2, 6, 7, 8, 9, 10, 15, 20 | D, D1 | Quincy, Wash. | LTW | 1996, g.c. | 19, 20 |
| 10 | D, E | Quincy, Wash. | LTW | 1996, g.c. | 19 | |
| QX-87 | 1, 2, 4, 5, 9, 12, 13, 88 | B, C | Quincy, Wash. | LTW | 1987, lab | 9, 12 |
| 37 | E, F | Quincy, Wash. | LTW | 1987, lab | 12 | |
| 128 | D, D1 | Quincy, Wash. | LTW | 1987, lab | 9 | |
| D27BB | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 | M, M | Woensdrecht 1 | LTW | 1998, g.c. | This study |
| JMP | 6, 9, 10, 11, 12, 16, 17, 18, 22 | F, G | Woensdrecht 2 | LTW | 1998, g.c. | This study |
| 7 | F, H | Woensdrecht 2 | LTW | 1998, g.c. | This study | |
| Strain | ||||||
| CHA0 | A, A | Switzerland | 29 | |||
| F113 | K, K | Ireland | 25 | |||
| Pf5 | A, B | Texas | 11, 15 | |||
| PILH1 | M, M | Italy | 12 |
Prefixes apply to soils and strains used in this study. CV and CC refer to two different isolations from soil obtained from the same geographic location.
BOX- and ERIC-defined phlD-positive genotypes are given (see Results).
LTW, long-term wheat cropping; LTF, long-term flax cropping; W, uncertain history, cropped to wheat currently.
The year of isolation is given followed by the location where wheat plants were grown. g.c., growth chamber.
Triticum aestivum cultivars used in this study included cultivar Penawawa for the experiments carried out in United States soils and cultivar Bussard for isolations from the two Dutch soils. Plants were incubated in a growth chamber under controlled conditions (15°C, 12 h light:12 h darkness) or grown in the field during the summer of 1998 (Table 1).
Strain isolation and maintenance.
Strains designations followed those given to the soils from which the strains originated. While most strains were isolated from wheat plants grown under controlled conditions, some were obtained from plants grown in the field during the summer of 1998 at Lind or Quincy, Wash. (Table 1). Soils from Quincy, Wash., were collected at the same site, but these soils were incubated differently prior to isolation of phlD+ Pseudomonas spp. (see references 17 to 19 for details). For example, the Q8 and QV soils were cycled multiple times to wheat in pots at 4-week intervals (19). Reference strains CHA0, Pf5, F113, and PILH1 were isolated in a variety of ways from other crop plants (11, 12, 15, 25, 29).
The large majority of phlD+ Pseudomonas strains were isolated by procedures similar to those described by Raaijmakers et al. (18). Briefly, 1 g of roots was obtained from wheat plants grown in each soil. Root samples were maintained in sterile test tubes at 4°C prior to processing. For processing, sterile distilled water was added to each tube and bacteria were dislodged from the rhizosphere by 1 min of vortexing followed by a 1-min incubation in a sonication bath. Samples were serially diluted and plated onto a modified King's medium B (KMB) agar, KMB+++, which has been shown to be selective for fluorescent Pseudomonas spp. (28) (20 g of proteose peptone [Difco Laboratories, Detroit, Mich.] per liter), 1.2 g of KH2PO4, 1.5 g of MgSO4 · 7H2O, 10 ml of glycerol, and 15 g of agar; supplemented with ampicillin [40 μg/ml], chloramphenicol [13 μg/ml], and cycloheximide [200 μg/ml] [Sigma Chemical Co., St. Louis, Mo.]). In some instances, one-third-strength KMB medium supplemented with the same concentrations of antibiotics was used (1/3 KMB+++). Pseudomonas isolates containing the phlD gene were identified as previously described (18). Briefly, colonies that hybridized with the phlD probe were isolated, and the presence of phlD was confirmed by PCR amplification with the gene-specific primers Phl2A and Phl2B. These strains generally produced a reddish-brown pigment, a phenotype which has been correlated with the presence of 2,4-DAPG (2). In all, 138 phlD-containing Pseudomonas strains were identified. Frozen stock cultures of all strains were stored in Luria-Bertani broth plus 40% glycerol at −80°C.
Genotypic analyses.
Colonies growing on 1/3 KMB+++ or full-strength KMB agar medium were suspended in sterile distilled water in polystyrene 96-well microtiter plates (Costar, Corning, N.Y.) to an optical density at 600 nm (OD600) of 0.15 to 0.45 and then subjected to freeze-thaw lysis. Analyses were replicated by amplifying at least two independent colonies of each strain on separate occasions.
Genomic fingerprints were obtained by amplification with BOX or enterobacterial repetitive intergenic consensus sequence (ERIC) primers (22). Individual reaction mixtures consisted of 5 μl of 5× Gitschier buffer [83 mM (NH4)2SO4, 335 mM Tris-HCl (pH 8.8), 33.5 mM MgCl2, 33.5 μM EDTA, 150 mM β-mercaptoethanol], 0.3 μl of bovine serum albumin solution (14 mg/ml), 1.25 μl of dimethyl sulfoxide, 9.64 μl of sterile distilled water, 1.56 μl of the deoxynucleoside triphosphates (2.0 mM each, mixed 1:1:1:1), 1.0 μl of solutions of each primer (50 pmol/μl), 0.33 μl of Taq DNA polymerase (5 U/μl; Promega, Madison, Wis.), and 5 μl of thawed template (corresponding to 104 to 106 bacteria prior to lysis). Negative-control reaction mixtures, containing no cell lysate, were used for each amplified set. Amplification reactions were processed in PTC200 thermocyclers (MJ Research, Inc., Watertown, Mass.). Cycling conditions for ERIC-PCR were as follows: 95°C for 7 min and then 30 cycles of 94°C for 1 min, 51°C for 1 min, and 65°C for 8 min, followed by a 16-min incubation at 65°C; after cooling to 4°C, the PCR products were stored at −20°C. Cycling conditions for BOX-PCR were the same except that the annealing temperature was 50°C. Amplified products were separated on 1.5% agarose gels in 0.5× Tris-borate-EDTA buffer for 5 to 6 h at 140 V and 10°C. DNA banding patterns were visualized by staining with ethidium bromide and were analyzed with the software GelCompar 4.0 (Applied Maths, Kortrijk, Belgium) by correlation-based clustering (22, 23). Groups were defined by the 95th percentile (near-minimum) similarity coefficient of replicate assays for identical strains.
Amplified ribosomal DNA restriction analysis (ARDRA) was performed as follows. Full-length 16S rRNA gene sequences were obtained from each strain by PCR amplification with the conserved eubacterial primers 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-ACG GCT ACC TTG TTA CGA CTT-3′), based on those defined by Weisburg et al. (31). Reaction master mixes were prepared as described above except that 25 pmol of each primer was used and 0.5 μg of RNase was added for each reaction. The cycling conditions were as follows: 95°C for 5 min and then 30 cycles of 94°C for 1 min, 50°C for 1 min, and 65°C for 4 min, followed by an 8-min incubation at 65°C; after a 4°C soak, the PCR products were stored at −20°C. Restriction digestions consisted of 10 μl of a PCR mixture in a total volume of 30 μl with 10 U of a single restriction enzyme (New England Biolabs Inc., Beverly, Mass.). Reaction mixtures were incubated at either 37°C (HaeIII, HhaI, HinfI, MspI, RsaI, or Sau96I) or 60°C (BstUI or TaqI) for 2 to 4 h and then stored at −20°C. Digestion products were subjected to electrophoresis on 2% agarose gels in 0.5× Tris-borate-EDTA buffer for 2 to 3 h at 140 V. Banding patterns were visualized by ethidium bromide staining and scored by comparison to a 100-bp DNA ladder.
Phenotypic tests.
Substrate utilization profiles were generated by using Biolog SF-N plates (Biolog, Inc., Hayward, Calif.). Colonies (≤10 days old) grown on KMB agar were suspended in 1 ml of sterile distilled water, pelleted by centrifugation, and resuspended in 0.5 ml of sterile distilled water. The concentration of washed cells was adjusted with water to an OD600 of 0.05 to 0.10 by using a microplate spectrophotometer (MR5000; Dynatech Inc., Burlington, Mass.). A 10-μl aliquot of washed cells (approximately 106 cells) was added to each well of a Biolog SF-N plate preloaded with 90 μl of sterile distilled water per well. The plates were incubated at room temperature in the dark. Bacterial growth was assayed spectrophotometrically at 600 nm after 3 and 7 days of incubation. Multivariate statistical analyses of the data obtained were performed with the program MULTIV 1.2.1 (Valerio De Patta Pillar, Department of Ecology, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil). Groups were defined by the 95th percentile (near-minimum) similarity coefficient of replicate assays for identical strains.
RESULTS
Genomic fingerprinting of strains.
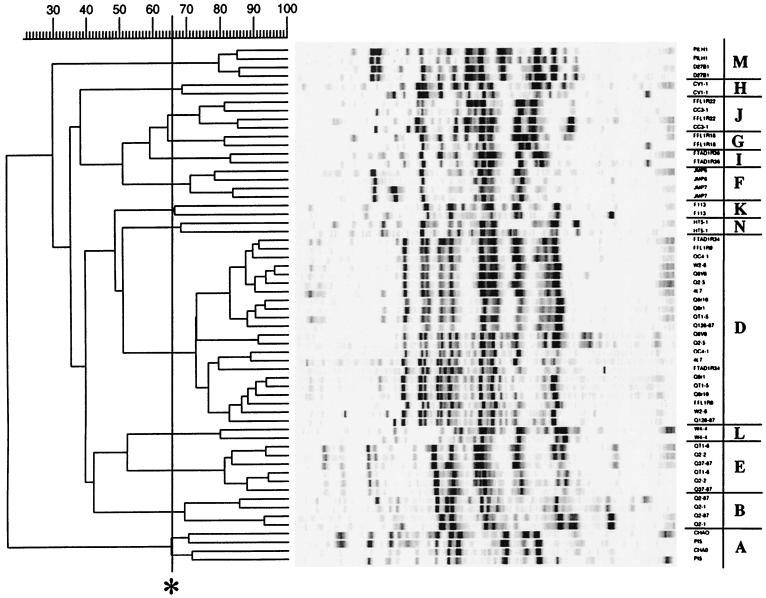
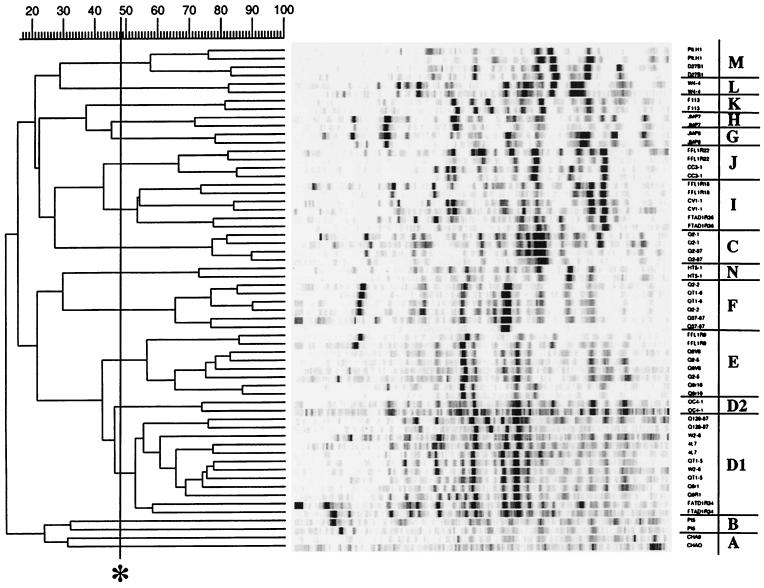
Whole-cell rep-PCR amplifications of the isolates yielded complex genomic fingerprints consisting of 10 to 30 amplification products ranging in size from 100 to 3,000 bp. Among the 142 phlD+ isolates, 13 distinct genomic fingerprints were observed by rep-PCR performed with the primer BOXA1R (Fig. 1). A similar, but not identical, clustering was obtained with the ERIC primer set (Fig. 2). Three differences were observed. First, strains CHA0 and Pf5 were separated by ERIC- but not by BOX-PCR (Table 2). This was likely due to the low signal-to-noise ratio obtained by using the whole-cell amplification protocol for these two strains, because the ERIC patterns for these two strains clustered together when isolated genomic DNA was used as template (data not shown). Second, the large BOX-defined D genotype was split into three distinct clusters, D1, D2, and E, when the ERIC data was used (Table 2). However, visual inspection of the fingerprint patterns of these groups indicated that a majority of amplified bands comigrated, and although the three clusters were distinct by the statistical criterion applied, they were closely related (>42.5% similar, compared to the 95th percentile near-minimum definition of identity of 48.5% similarity) (Fig. 2). A similar split occurred with the JMP isolates; ERIC genotypes G and H corresponded to BOX genotype F (Table 2). The third difference involved the splitting of ERIC genotype I into BOX genotypes G, H, and I and the close association of these groups with genotype J as defined by both BOX and ERIC fingerprint patterns (cf. Fig. 1 and 2; Table 2). Again, the similarity scores calculated for these clusters were close to the values defining identity, except that BOX genotype H appeared very distinct.
FIG. 1.
Cluster analysis of genomic fingerprint patterns of phlD-containing Pseudomonas strains generated by PCR amplification of whole-cell template with the BOXA1R primer. Only patterns of strains representing unique genotypes isolated from individual soils are shown. Two independent amplifications were used for each strain. Using GelCompar 4.0, the UPGMA algorithm was applied to the similarity matrix generated from the tracks of the whole patterns by using Pearson's correlation coefficient. The similarity coefficient used to define distinct groups (see Materials and Methods) is noted (∗). Distinct groups of genotypes, labeled alphabetically A through N, are discussed more fully in the text.
FIG. 2.
Cluster analysis of genomic fingerprint patterns of phlD-containing Pseudomonas strains generated by PCR amplification of whole-cell template with the ERIC primer set. Only patterns of strains representing unique genotypes isolated from individual soils are shown. Two independent amplifications were used for each strain. Using GelCompar 4.0, the UPGMA algorithm was applied to the similarity matrix generated from the tracks of the whole patterns by using Pearson's correlation coefficient. The similarity coefficient used to define distinct groups (see Materials and Methods) is noted (∗). Distinct groups of genotypes, labeled alphabetically A through N, are discussed more fully in the text.
TABLE 2.
Genotypic and phenotypic diversity of phlD+ Pseudomonas spp.
| Type straind | Genotypea
|
Phenotype at dayb:
|
Consensusc
|
||||
|---|---|---|---|---|---|---|---|
| BOX | ERIC | 16S | 3 | 7 | Split | Lump | |
| CHA0 | A | A | A | A | A | A1 | A |
| Pf5 | A | B | A | B | B | B | A |
| Q2-87 | B | C | B | C | D | C | C |
| Q2-1 | B | C | B | C | D | C | C |
| Q8r1 | D | D1 | B | D | D | D1 | D |
| Q8R10 | D | E | B | D | D | D3 | D |
| Q128-87 | D | D1 | B | D | D | D1 | D |
| QT1-5 | D | D1 | B | D | D | D1 | D |
| 4L7 | D | D1 | B | D | D | D1 | D |
| W2-6 | D | D1 | B | D | D | D1 | D |
| OC4-1 | D | D2 | B | D | D | D2 | D |
| FTAD1R34 | D | D1 | B | D | D | D1 | D |
| Q8V8 | D | E | B | D | D | D3 | D |
| Q2-5 | D | E | B | D | D | D3 | D |
| FFL1R9 | D | E | B | D | D | D3 | D |
| Q37-87 | E | F | B | D | D | E | E |
| QT1-6 | E | F | B | D | D | E | E |
| Q2-2 | E | F | B | D | D | E | E |
| JMP6 | F | G | B | E | D | F1 | F |
| JMP7 | F | H | B | E | D | F2 | F |
| FFL1R18 | G | I | B | D | D | G1 | G |
| CV1-1 | H | I | B | D | D | G2 | G |
| FTAD1R36 | I | I | B | D | D | G3 | G |
| FFL1R22 | J | J | B | D | D | H | H |
| CC3-1 | J | J | B | D | D | H | H |
| F113 | K | K | C | D | D | I | I |
| W4-4 | L | L | B | F | F | J | J |
| D27B1 | M | M | B | D | D | K | K |
| PILH1 | M | M | B | D | D | K | K |
| HT5-1 | N | N | C | G | D | L | L |
| Totale | 13 | 15 | 3 | 7 | 4 | 17 | 11 |
Genotypes were defined by rep-PCR (ERIC, BOX) and ARDRA with eight different restriction enzymes.
Phenotypes were defined based on substrate utilization profiles developed on Biolog SF-N plates after 3 and 7 days of growth.
Designations incorporate either data from both sets (split) or the single least-discriminating set (lump) of genotypic information based on rep-PCR.
These type strains represent all unique genotypes isolated from individual soils. Prefix designations are the same as those noted in Table 1.
Total number of distinct groups.
The largest set of genotypically similar isolates, defined as BOX group D or ERIC groups D1, D2, and E, contained over 30% of all isolates. This set included isolates from eight soils (FTAD1R, FFL1R, OC, 4L, W, QT, Q8r, and QV) in four different locations (Fargo, Ithaca, Lind, and Quincy). We refer to this apparently cosmopolitan set of isolates as the Q8r1 supergroup. In contrast, most of the other genotypic groups contained isolates from a single geographic location. Only three other groups, ERIC I, J, and M, contained isolates from more than one location (Table 2).
Isolates from the same soil that displayed the same genomic fingerprint are listed together in Table 1. In general, only one or two phlD+ strains were isolated from the rhizosphere of wheat grown in each soil under a given set of conditions. Multiple genotypes were obtained from Quincy and Fargo soils (Table 3), but these were isolated from wheat plants that had been grown under different light, temperature, and moisture regimes for the Quincy soils and different cropping histories at Fargo (Table 1). Four distinct ERIC genotypes were isolated from the Quincy soils; however, only one genotype predominated under each set of conditions. For example, 8 of 10 isolates from soil QX-87 were ERIC genotype C, while 6 of 9 isolates from soil QT were ERIC genotype F. Similarly, Fargo soils FTAD1R, from a long-term wheat field, and FFL1R, from an adjoining long-term flax field, yielded four distinct genotypes, but a single dominant genotype was isolated from each (ERIC genotypes D1 and J, respectively). Thirty type strains, representing the distinct rep-PCR genotypes present in each soil, were identified for further analyses (Table 2).
TABLE 3.
Number of isolates representing distinct genotypes of phlD-containing Pseudomonas strains isolated from Quincy and Fargo soils
ARDRA.
Patterns of restriction fragment length polymorphisms generated by using eight different restriction enzymes indicated that amplified 16S rRNA gene sequences were very similar for most of the 30 representative phlD+ strains (Table 4). Sequences from CHA0 and Pf5 differed from those of all other phlD+ strains in four of the eight digests. The enzyme TaqI revealed similarities between these two strains, F113 and HT5-1. No restriction fragment length polymorphisms were found among 16S rRNA gene sequences from phlD+ strains upon digestion with BstUI, HinfI, or Sau96I.
TABLE 4.
Amplified 16S rRNA gene restriction analyses of phlD+ strains
| Enzyme | Pattern | Observed fragmentsa (bp) | Members |
|---|---|---|---|
| BstUI | A | 425, 300, 125 | All strains |
| HaeIII | A | 850, 225, 150, 125 | CHA0, Pf5 |
| B | 700, 225, 150, 125 | All others | |
| HhaI | A | 425, 275, 250, 150, 125 | CHA0, Pf5 |
| B | 425, 375, 275, 250, 225 | All others | |
| HinfI | A | 1050, 225, 125 | All strains |
| MspI | A | 575, 150, 125 | CHA0, Pf5 |
| B | 575, 525, 125 | All others | |
| RsaI | A | 900, 375, 150 | CHA0, Pf5 |
| B | 950, 375, 150 | All others | |
| Sau96I | A | 625, 275, 200, 175, 125 | All strains |
| TaqI | A | 950, 375, 200 | CHA0, Pf5, F113, HT5-1 |
| B | 625, 375, 325, 200 | All others |
The sizes of DNA fragments were estimated by using a 100-bp marker to a precision of ±25 bp. Multiple fragments of similar size may occur at the size classes listed. Fragments smaller than 100 bp were not considered in the analyses due to lack of resolution.
Growth on 95 different substrates.
The same phlD+ isolates representing distinct genotypes were examined for their ability to grow in vitro on a variety of carbon sources. After 3 days of incubation, each strain grew on 63 to 68 of the 95 different substrates present in Biolog SF-N plates, except CHA0 and Pf5, which grew on no more than 61 of the substrates. The number of substrates sustaining growth of each strain increased by approximately 10% after 7 days. However, not all of the strains used the same set of substrates. Forty-nine substrates were utilized by all phlD+ strains (Table 5); 17 of these allowed for rapid growth to saturation (OD600 ≥0.375 after 3 days). However, 12 substrates did not support the growth of any strain (Table 5). Strains CHA0 and Pf5 grew on significantly fewer substrates than the other phlD+ strains, and they differed from the other strains in the utilization of seven substrates (Table 5). The carbon source utilization profiles of strains lacking phlD differed greatly both from one another and from those of the phlD+ strains (data not shown).
TABLE 5.
Substrate utilizationa by phlD+ Pseudomonas strains
| Strains | Substrate(s) supporting growth on Biolog SF-N plates |
|---|---|
| All phlD-positive strains | Tween 40, Tween 80, d-fructose, α-d-glucose, m-inositol, d-mannitol, d-mannose, cis-acotinate, citrate, d-gluconate, β-hydroxybutyrate, α-ketoglutarate, dl-lactate, malonate, propionate, quinic acid, succinate, bromosuccinate, alaninamide, d-alanine, l-alanine, l-alanyl-glycine, l-asparagine, l-aspartate, l-glutamate, glycyl-l-glutamate, l-histidine, hydroxy-l-proline, l-leucine, l-proline, l-pyroglutamate, dl-carnitine, γ-aminobutyrate, urocanate, inosine, uridine, putrescine, 2-aminoethanol, glycerol, dl-α-glycerophosphate, dextrin, d-arbitol, maltose, sucrose, trehalose, methylpyruvate, d-glucosaminic acid, succinamic acid, l-serine |
| All except CHA0 and Pf5 | l-Arabinose, d-galactose, d-sorbitol, d-galactonic acid lactone, d-galacturonate, d-glucuronate |
| Only CHA0 and Pf5 | Glucose-1-phosphate |
| No phlD-positive strains | N-Acetyl-d-galactosamine, adonitol, α-d-lactose, lactulose, β-methyl-d-glucoside, xylitol, formate, α-hydroxybutyrate, α-ketobutyrate, d-serine, thymidine, phenylethylamine |
Utilization was defined as growth determined spectrophotometrically if cultures obtained an OD600 of >0.125 after 7 days of incubation. Substrates allowing for growth to an OD600 of >0.375 after 3 days of incubation are highlighted in boldface.
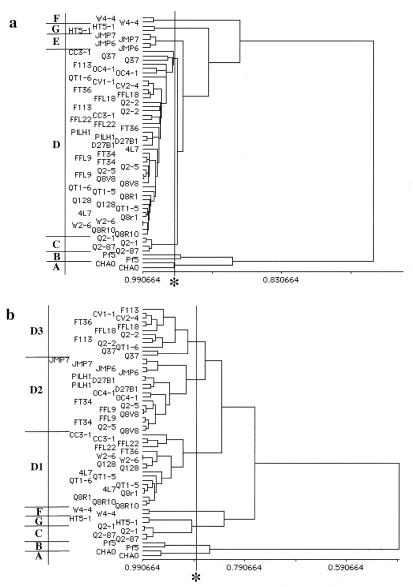
Multivariate analyses of the utilization profiles of the representative strains were performed (Fig. 3). Four to 11 distinct clusters were observed for the 30 phlD+ isolates, depending on the algorithm used for clustering (Fig. 3) and the length of incubation (data not shown). Three groups were conserved in all of the analyses, so they can be considered phenotypically distinct. These groups are represented by strains CHA0, Pf5, and W4-4. Three other groups, represented by HT5-1, JMP6 or JMP7, and Q2-87 or Q2-2, also were distinct in most cases. The remaining 21 strains were consolidated into a single cluster by the simple linkage algorithm but split into three distinct groups by the complete linkage algorithm (Fig. 3). To determine which clustering might better describe these strains, two types of ordination analyses, with different mathematical underpinnings, were used. Both principal coordinate and principal component analyses more strongly supported the model of a single distinct group (data not shown). Data from 3-day and 7-day incubations gave very similar results in terms of the topology of the dendrograms; however, the longer incubations resulted in fewer distinct groups because most strains had grown to their maximal density on a greater percentage of substrates (data not shown).
FIG. 3.
Cluster analysis of carbon source utilization patterns of phlD-containing Pseudomonas strains cultured for 3 days. Only strains representing unique genotypes isolated from individual soils were assayed. Two independent assays were performed on each strain. Using MULTIV 1.2.1, the simple linkage (a) and complete linkage (b) algorithms were applied to the similarity matrix generated by using Pearson's correlation coefficient. The similarity coefficient used to define distinct groups (see Materials and Methods) is noted (∗). Distinct groups of phenotypes, labeled alphabetically A through G, are discussed more fully in the text.
DISCUSSION
While phlD+ Pseudomonas spp. have been found in soils naturally suppressive to take-all (18, 19), it is not clear that isolates from such soils contribute equally to the phenomenon of TAD. As a first step toward characterizing these populations, we studied the genotypic and phenotypic diversity of a large collection of isolates from soils of different wheat-growing regions of the United States and The Netherlands. Using rep-PCR, we identified at most 17 genotypes distributed among three ARDRA groups (Table 2). Previously, Keel et al. characterized a smaller collection of phloroglucinol-producing strains and found seven randomly amplified polymorphic DNA-defined genotypes belonging to three ARDRA groups (12). The genotypic distinctions of the 14 strains included in both studies (CHA0, Pf5, F113, PILH1, and all QX-87 isolates) were entirely consistent with one another. Each of the newly identified genotypes in our study occurred in soils from locations not previously examined. Only one or two genotypes were isolated from each soil under a particular set of conditions. One possible explanation for this observation is that a single genotypically distinct population of phlD+ Pseudomonas strains dominates in the rhizosphere under a given set of environmental conditions. Among the three BOX genotypes isolated from the Quincy TAD soil, there were at least two distinctly different phenotypes based on carbon source utilization (Fig. 3 and Table 2). It may be that the different types of 2,4-DAPG producers had differentially adapted to changing environmental conditions and/or that some sort of microbial succession had taken place. The dominant genotypes obtained from the two Fargo soils, FTAD1R34 and FFL1R22, also differed phenotypically (Table 2). These two soils differed in their cropping history, indicating that wheat and flax roots may enrich for different phlD+ genotypes over time.
The strains isolated in this study may differ in their effectiveness against fungal root pathogens of wheat and barley. Different genotypes of 2,4-DAPG-producing Pseudomonas strains have been reported to differ in their capacity to inhibit the growth of Fusarium on tomatoes and of Pythium on cucumbers (26). Differences in biocontrol capacities may be due to one or more of the following factors. First, different genotypes produce different amounts of 2,4-DAPG and other antifungal metabolites in vitro (12, 26), and such differences also may occur in situ. Second, the degree to which strains can colonize the rhizosphere may impact their ability to suppress invading pathogens. Indeed, evidence that DAPG producers differ substantially in their ability to colonize wheat roots when introduced as a seed or soil treatment is now mounting (20, 21; B. B. McSpadden Gardener and D. M. Weller, unpublished data). Lastly, there may be undiscovered phenotypic characteristics of certain genotypes that positively contribute to disease suppression, even though the production of 2,4-DAPG alone can explain the ability of some strains to suppress fungal root pathogens (7, 13, 30).
The genotypic data presented in this study could aid substantially in the identification and selection of new 2,4-DAPG-producing biocontrol agents. Currently, most biocontrol studies are initiated by collecting large numbers of isolates and then laboriously screening them for the ability to suppress the growth of plant pathogens (5). Because it is reasonable to assume that isolates of the same genotype will perform similarly, fewer isolates may need to be tested in greenhouse and field studies. With rep-PCR, novel genotypes can be easily recognized and selected for more-intensive analyses. Additionally, our screening program can be focused on genotypes that are known to be particularly effective on wheat. In our collection, nearly one-third of the isolates were genotypically similar to strain Q8r1-96. This strain very effectively colonizes wheat roots and can suppress take-all when applied at low doses (19, 21). Preliminary evidence suggests that other isolates of this genotype also have this ability (B. B. McSpadden Gardener and D. M. Weller, unpublished data). The widespread occurrence of this genotype may be indicative of the generalist nature of Q8r1-like pseudomonads and their ability to adapt successfully to the wheat rhizosphere regardless of the prevailing soil conditions. It is possible, too, that subtle phenotypic differences that represent site-specific adaptations may be discovered by comparing isolates of this group.
Of the assays used in this study, rep-PCR was the most discriminating because it revealed the largest number of distinct groups. Previously, it was noted that rep-PCR has a greater capacity to distinguish strains than do 16S rRNA sequence-based approaches (22). However, there has not been a similar comparison of taxonomic abilities of rep-PCR and phenotypic assays based on carbon source utilization. In this instance, rep-PCR proved to be more effective in discriminating between strains than the Biolog SF-N assays. In our study of carbon source utilization, over 75% of the substrates used by any individual strain could be utilized by all of the phlD+ isolates (Table 5). Most differences among strains were not in the number or types of carbon sources used but rather in the degree to which they supported growth. This homogeneity of growth phenotypes is reflected in the relatively high similarity coefficients used to define distinct clusters (Fig. 3). While only at most seven groups were defined by using the Biolog data, these groups were all strongly associated with particular genotypes. Thus, there is hidden genotypic diversity within these phenotypic groups, suggesting that there is also hidden phenotypic diversity which is not detected with the Biolog assay. The carbon sources present in the Biolog assays were selected for their ability to discriminate microbial taxa and included some substrates not likely to be found in the rhizosphere. While a modified system using substrates found in wheat rhizospheres may be more useful in identifying strains with different abilities to colonize this environment, it remains to be seen whether such an in vitro test can detect the same degree of diversity as the genomic fingerprint assays.
REFERENCES
- 1.Asher M J C, Shipton P J. Biology and control of take-all. New York, N.Y: Academic Press; 1981. [Google Scholar]
- 2.Bangera M G, Thomashow L S. Characterization of a genomic locus required for synthesis of the antibiotic 2,4-diacetylphloroglucinol by the biological control agent Pseudomonas fluorescens Q2-87. Mol Plant-Microbe Interact. 1996;9:83–90. doi: 10.1094/mpmi-9-0083. [DOI] [PubMed] [Google Scholar]
- 3.Bangera M G, Thomashow L S. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J Bacteriol. 1999;181:3155–3163. doi: 10.1128/jb.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonsall R F, Weller D M, Thomashow L S. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl Environ Microbiol. 1997;63:951–955. doi: 10.1128/aem.63.3.951-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook R J. Making greater use of introduced microorganisms for biological control of plant pathogens. Annu Rev Phytopathol. 1993;31:53–80. doi: 10.1146/annurev.py.31.090193.000413. [DOI] [PubMed] [Google Scholar]
- 6.Cook R J, Thomashow L S, Weller D M, Fujimoto D, Mazzola M, Bangera G, Kim D-S. Molecular mechanisms of defense by rhizobacteria against root disease. Proc Natl Acad Sci USA. 1995;92:4197–4201. doi: 10.1073/pnas.92.10.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenton A M, Stephens P M, Crowley J, O'Callaghan M, O'Gara F. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol. 1992;58:3873–3878. doi: 10.1128/aem.58.12.3873-3878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutteridge R J, Hornby D, Hollins T W, Prew R D. Take-all in autumn-sown wheat, barley, triticale and rye grown with high and low inputs. Plant Pathol. 1993;42:425–431. [Google Scholar]
- 9.Harrison L A, Letendre L, Kovacevich P, Pierson E, Weller D M. Purification of an antibiotic effective against Gaumannomyces graminis var. tritici produced by a biocontrol agent, Pseudomonas aureofaciens. Soil Biol Biochem. 1993;25:215–221. [Google Scholar]
- 10.Hornby D. Suppressive soils. Annu Rev Phytopathol. 1983;21:65–85. [Google Scholar]
- 11.Howell C R, Stipanovic R D. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology. 1979;69:480–482. [Google Scholar]
- 12.Keel C, Weller D M, Natsch A, Défago G, Cook R J, Thomashow L S. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62:552–562. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger U, Wirthner P, Haas D, Defago G. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant-Microbe Interact. 1992;5:4–13. [Google Scholar]
- 14.Lemanceau P, Alabouvette C. Biological control of Fusarium diseases by fluorescent Pseudomonas and non-pathogenic Fusarium. Crop Protect. 1991;10:279–286. [Google Scholar]
- 15.Nowak-Thompson B, Gould S J, Kraus J, Loper J E. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can J Microbiol. 1994;40:1064–1066. [Google Scholar]
- 16.O'Sullivan D J, O'Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierson E A, Weller D M. Use of mixtures of fluorescent pseudomonads to suppress take-all and improve the growth of wheat. Phytopathology. 1994;84:940–947. [Google Scholar]
- 18.Raaijmakers J, Weller D M, Thomashow L S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl Environ Microbiol. 1997;63:881–887. doi: 10.1128/aem.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raaijmakers J, Weller D M. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant-Microbe Interact. 1997;11:144–152. [Google Scholar]
- 20.Raaijmakers J, Bonsall R F, Weller D M. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology. 1999;89:470–475. doi: 10.1094/PHYTO.1999.89.6.470. [DOI] [PubMed] [Google Scholar]
- 21.Raaijmakers J, Hayes K, Thomashow L S, Weller D M. Diversity and rhizosphere competence of 2,4-diacetylphloroglucinol-producing Pseudomonas strains. Phytopathology. 1999;89:S63. [Google Scholar]
- 22.Rademaker J L W, de Bruijn F J. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis. In: Caeteno-Anolles G, Gresshoff P M, editors. DNA markers: protocols, applications, and overviews. New York, N.Y: John Wiley and Sons; 1997. pp. 151–171. [Google Scholar]
- 23.Rademaker J L W, Louws F J, Rossbach U, de Bruijn F J. Computer assisted pattern analysis of molecular fingerprints and data base construction. In: Akkermans A D L, van Elsas J D, DeBruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 1–33. [Google Scholar]
- 24.Schroeder K L, Raaijmakers J M, Kalloger S E, Mavrodi D V, Thomashow L S, Cook R J, Weller D M. Distribution of 2,4-diacetylphloroglucinol-producing Pseudomonas spp. with extended monoculture. Phytopathology. 1998;88:S80. [Google Scholar]
- 25.Shanahan P, O'Sullivan D J, Simpson P, Glennon J D, O'Gara F. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl Environ Microbiol. 1992;58:353–358. doi: 10.1128/aem.58.1.353-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharifi-Tehrani A, Zala M, Natsch A, Moenne-Loccoz Y, Defago G. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur J Plant Pathol. 1998;104:631–643. [Google Scholar]
- 27.Shipton P J, Cook R J, Sitton J W. Occurrence and transfer of a biological factor in soil that suppresses take-all of wheat in eastern Washington. Phytopathology. 1973;63:511–517. [Google Scholar]
- 28.Simon A, Ridge E H. The use of ampicillin in a simplified selective medium for the isolation of fluorescent pseudomonads. J Appl Bacteriol. 1974;37:459–460. doi: 10.1111/j.1365-2672.1974.tb00464.x. [DOI] [PubMed] [Google Scholar]
- 29.Stutz E, Defago G, Kern H. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology. 1986;76:181–185. [Google Scholar]
- 30.Vincent M N, Harrison L A, Brackin J M, Kovacevich P A, Mukerji P, Weller D M, Pierson E A. Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl Environ Microbiol. 1991;57:2928–2934. doi: 10.1128/aem.57.10.2928-2934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weisburg W G, Barnes S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weller D M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]