Abstract
A novel amidase involved in bacterial cyclic imide metabolism was purified from Blastobacter sp. strain A17p-4. The enzyme physiologically functions in the second step of cyclic imide degradation, i.e., the hydrolysis of monoamidated dicarboxylates (half-amides) to dicarboxylates and ammonia. Enzyme production was enhanced by cyclic imides such as succinimide and glutarimide but not by amide compounds which are conventional substrates and inducers of known amidases. The purified amidase showed high catalytic efficiency toward half-amides such as succinamic acid (Km = 6.2 mM; kcat = 5.76 s−1) and glutaramic acid (Km = 2.8 mM; kcat = 2.23 s−1). However, the substrates of known amidases such as short-chain (C2 to C4) aliphatic amides, long-chain (above C16) aliphatic amides, amino acid amides, aliphatic diamides, α-keto acid amides, N-carbamoyl amino acids, and aliphatic ureides were not substrates for the enzyme. Based on its high specificity toward half-amides, the enzyme was named half-amidase. This half-amidase exists as a monomer with an Mr of 48,000 and was strongly inhibited by heavy metal ions and sulfhydryl reagents.
A variety of cyclic amide-metabolizing systems occur in nature and play important roles in pyrimidine and purine metabolism, amino acid metabolism (histidine degradation), antibiotic metabolism (β-lactam decomposition), creatinine degradation, etc.
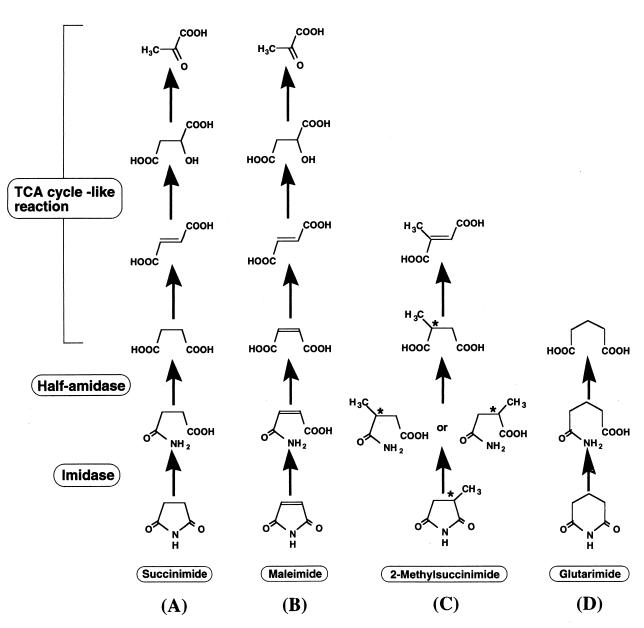
Cyclic imide is a kind of cyclic amide, and the metabolism of cyclic imides has been studied in relation to the detoxification of the antiepileptic agents ethotoin and phensuximide in mammals (3, 36). During the course of a study on cyclic amide transformation for hydantoin from the practical viewpoint of industrial d-amino acid production, we recently found that microorganisms also transform cyclic imides (21, 24–27). Microbial transformation of cyclic imides found in the bacterium Blastobacter sp. strain A17p-4 (22) involves ring opening of cyclic imide to monoamidated dicarboxylate (half-amide) catalyzed by imidase (23), half-amide hydrolysis to dicarboxylate catalyzed by amidase, and subsequent trichloroacetic acid (TCA) cycle-like reactions (Fig. 1). The reactions and enzymes (imidase and amidase) involved in the metabolism have practical potential for production of organic acids from cyclic imides or their metabolites and for fine enzymatic synthesis of useful compounds. For example, pyruvate, an effective precursor in the synthesis of various drugs and agrochemicals, was produced from succinimide or its metabolites (especially fumarate, a cheap material) through cyclic imide-transforming pathway (Fig. 1A and B) (28). Imidase was applied for the regiospecific synthesis of useful half-amide (3-carbamoyl-α-picolinic acid, an intermediate for herbicide) from a cyclic imide (2,3-pyridinedicarboximide) (J. Ogawa, M. Ito, T. Segawa, C.-L. Soong, and S. Shimizu, Abstr. Annu. Meet. '99 Soc. Biosci. Bioeng., abstr. 182, 1999 [in Japanese]). Amidase also has a potential for the chiral resolution of dicarboxylates through the stereoselective hydrolysis of half-amides (Fig. 1C).
FIG. 1.
Proposed pathway for cyclic imide degradation in Blastobacter sp. strain A17p-4.
We report here an amidase catalyzing the second step of cyclic imide transformation. This amidase, named half-amidase, was distinct from known amidases especially in substrate specificity. We also confirmed the physiological role of imidase and half-amidase in cyclic imide transformation by investigating their induction/expression profiles.
MATERIALS AND METHODS
Chemicals.
Glutaramic acid and adipinamic acid were chemically synthesized as described previously (6). α-Ketoglutaramic acid and α-ketosuccinamic acid were prepared by l-amino oxidase from l-glutamine and l-asparagine, respectively (16).
Microorganisms and medium for cultivation.
Blastobacter sp. strain A17p-4 (AKU 990; Faculty of Agriculture, Kyoto University, Kyoto, Japan) was used as the enzyme source and for all experiments. Synthetic minimum liquid medium comprised 1 g of KH2PO4, 1 g of K2HPO4, 0.3 g of MgSO4 · 7H2O, 2 g of NH4Cl, 1 mg of thiamine hydrochloride, 2 mg of riboflavin, 2 mg of nicotinic acid, 2 mg of pantothenic acid, 2 mg of pyridoxine hydrochloride, 0.1 mg of biotin, 1 mg of p-aminobenzoate, 0.1 mg of folic acid, and 10 mg of each metal ion (CaCl2 · 2H2O, MnCl2 · 4H2O, CoCl2, NiCl2 · 6H2O, ZnSO4 · 7H2O, CdCl2 · 2H2O, CuSO4 · 5H2O, FeSO4 · 7H2O, BeSO4 · 4H2O, and RbC1) in 1 liter of deionized water, pH 7.0.
Production of half-amidase.
Minimum liquid medium supplemented with 20 mM sucrose as a carbon source and each test compound at 0.15% (wt/vol) was used for experiment 1; minimum liquid medium supplemented with each test compound at 0.15% (wt/vol) as a sole source of carbon was used for experiment 2. All cultivations were carried out at 28°C for 4 days. Cells from 5 ml of culture were harvested by centrifugation at 10,000 × g for 10 min, then washed twice with 0.85% (wt/vol) NaCl, and suspended in 0.5 ml of 20 mM potassium phosphate buffer (pH 7.0) containing 0.1 mM dithiothreitol. The cell suspension was ultrasonicated at 4°C for 15 min and centrifuged at 14,000 × g for 30 min. The resultant supernatant was used for assay of half-amidase activity as described below.
Expression of half-amidase and imidase.
Four sets of cultivations (A through D) were carried out. All cultures in 2-liter flasks contained minimum liquid medium (200 ml) supplemented with 20 mM sucrose. Cultivations were carried out at 28°C with shaking. At the end of exponential growth (107 h), cultures A and C were supplemented with 20 mM glutarimide and 20 mM sucrose, respectively, while cultures B and D were supplemented with actinomycin D to a final concentration of 1 μg/ml together with addition of 20 mM glutarimide and 20 mM sucrose, respectively. At various intervals of time, a 5-ml aliquot of each culture was sampled and cell growth was measured at an absorbance of 610 nm. Half-amidase and imidase activities were then assayed from the cell extracts of each culture prepared as described above.
Enzyme assays.
The standard half-amidase assay mixture comprised, in 100 μl, 10 μmol of potassium phosphate (pH 7.0), 2 μmol of succinamic acid, and an appropriate amount of enzyme. After incubation at 30°C for 30 to 60 min, the reaction was stopped with 10 μl of 15% (by volume) perchloric acid, followed by neutralization with 90 μl of 500 mM potassium phosphate (pH 7.0). The reaction mixture was centrifuged at 14,000 × g for 10 min, and the supernatant was analyzed for decreases in the concentration of succinamic acid and increases in that of succinic acid. Analysis was performed using a Shimadzu LC-6A high-performance liquid chromatography (HPLC) apparatus at 210 nm on a Cosmosil 5C18 AR-packed column (4.6 by 250 mm; Nacalai Tesque, Kyoto, Japan) at a flow rate of 1.0 ml/min, with 250 mM KH2PO4 (pH 4.4) as the eluent.
Imidase activity was assayed as described previously (23).
One unit of half-amidase and imidase was defined as the amount of enzyme that catalyzed consumption of the substrate or formation of the product at a rate of 1 μmol/min under the assay conditions described above.
Enzyme purification.
All steps were carried out at 0 to 5°C; the buffer used was 20 mM potassium phosphate (pH 7.0) containing 0.1 mM dithiothreitol throughout the purification process unless otherwise stated.
(i) Step 1.
Blastobacter sp. was cultured in minimum liquid medium supplemented with 1% succinimide at 28°C with shaking. Cells (20 g [wet weight] from a 10-liter culture) were harvested by centrifugation (10,000 × g) and suspended in 20 ml of buffer. The cell suspension was disrupted with glass beads (0.25 by 0.50 mm; Dyno-Mill; W. A. Bachofen, Basel, Switzerland) for 30 min. The disrupted cell suspension was centrifuged at 14,000 × g for 60 min, and the resultant supernatant was used as the cell extract (230 ml).
(ii) Step 2.
The cell extract was dialyzed against buffer and then applied to a DEAE-Sephacel column (2.5 by 10 cm) preequilibrated with buffer. After the column was washed with 250 ml of buffer, the enzyme was eluted with a linear gradient of 0 to 0.5 M NaCl in 500 ml of buffer. The active fractions were combined (90 ml) and concentrated by ultrafiltration with a 10,000-kDa-cutoff membrane.
(iii) Step 3.
The enzyme solution was mixed with solid NaCl to obtain a concentration of 4 M and then applied to a phenyl-Sepharose CL-4B column (1.5 by 10 cm) preequilibrated with buffer containing 4 M NaCl. The column was washed with the same buffer (200 ml), followed by a decreasing salt concentration (from 4 to 0 M NaCl) in 500 ml of buffer. The active fractions were combined (65 ml) and concentrated by ultrafiltration.
(iv) Step 4.
The concentrated enzyme was applied to a Sephacryl S-200 column (1.5 by 80 cm) equilibrated with the buffer containing 0.2 M NaCl and eluted with the same buffer. The active fractions were pooled (40 ml) and dialyzed against buffer.
(v) Step 5.
The dialyzed enzyme was applied to a Mono Q HR 5/5 column equilibrated with buffer and then eluted with an increasing salt concentration (from 0 to 0.5 M NaCl). The active fractions were combined (2 ml), dialyzed against buffer, and further purified on a SMART system (Pharmacia LKB, Uppsala, Sweden).
(vi) Step 6.
The dialyzed active fraction was applied to a Mono Q PC 1.6/5 column equilibrated with buffer and then eluted with an increasing salt concentration of 0 to 0.3 M NaCl. The active fractions (0.2 ml) were used for characterization.
Analytical methods for half-amidase.
The relative molecular mass was determined by HPLC on a GS-520 column (7.6 by 500 mm; Asahi Kasei, Tokyo, Japan) and a G-3000 SW column (7.5 by 600 mm; Tosoh, Tokyo, Japan) at 0.3 ml/min with an elution buffer of 20 mM potassium phosphate (pH 7.0) containing 0.2 M NaCl and 0.1 mM dithiothreitol. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12.5% polyacrylamide gel and protein concentration determination were performed as described previously (31). NH2-terminal amino acid sequence analysis was performed as described previously (30).
RESULTS
Production of half-amidase.
The production of half-amidase was significantly enhanced by the addition of cyclic imides such as succinimide and glutarimide (Table 1, experiment 1). The intermediates of cyclic imide metabolism of succinic acid also showed stimulatory effects. Blastobacter sp. was able to grow in minimum medium with succinimide or glutarimide as the sole source of carbon (Table 1, experiment 2). With these cyclic imides as sole sources of carbon, the half-amidase activity increased approximately three to five times compared to that found with sucrose grown cells, and succinimide supported good growth.
TABLE 1.
Effects of various compounds on the production of half-amidasea
| Compound | Mean ± SD
|
Relative activity (%) | |
|---|---|---|---|
| Optical density at 610 nm | Sp act (μmol/min/mg of protein) | ||
| Expt 1 | |||
| Additives in sucrose-containing medium | |||
| None | 0.335 ± 0.012 | 0.176 ± 0.018 | 100 |
| Succinimide | 0.250 ± 0.012 | 0.325 ± 0.010 | 185 |
| Glutarimide | 0.392 ± 0.011 | 0.476 ± 0.008 | 270 |
| Succinamic acid | 0.314 ± 0.011 | 0.163 ± 0.016 | 93 |
| Adipinamic acid | 0.323 ± 0.014 | 0.150 ± 0.008 | 85 |
| Succinic acid | 0.414 ± 0.005 | 0.300 ± 0.015 | 170 |
| Adipic acid | 0.355 ± 0.018 | 0.188 ± 0.008 | 107 |
| Acetamide | 0.223 ± 0.011 | 0.176 ± 0.015 | 100 |
| n-Valeramide | 0.267 ± 0.006 | 0.213 ± 0.012 | 121 |
| Lactamide | 0.222 ± 0.013 | 0.178 ± 0.005 | 99 |
| Benzamide | 0.140 ± 0.010 | Traceb | NDc |
| Dihydrouracil | 0.330 ± 0.019 | 0.213 ± 0.010 | 121 |
| Dihydrothymine | 0.202 ± 0.004 | 0.159 ± 0.013 | 90 |
| Ammonium chloride | 0.236 ± 0.010 | 0.201 ± 0.005 | 114 |
| l-Glutamine | 1.100 ± 0.016 | 0.240 ± 0.004 | 135 |
| Expt 2 | |||
| Additives as sole carbon sources | |||
| Succinimide | 0.233 ± 0.010 | 0.582 ± 0.013 | 331 |
| Glutarimide | 0.101 ± 0.006 | 0.960 ± 0.016 | 545 |
The standard deviations were obtained from four separate determinations. The methods for experiments 1 and 2 are described in Materials and Methods.
Less than the specific activity of 10−3 μmol/min/mg of protein.
ND, not determined.
Amide compounds which act as substrates and inducers of known amidase (acetamide and lactamide for the amidase of Pseudomonas aeruginosa [11, 13]; benzamide for that of Aspergillus nidulans [9]; cyclic ureides for dihydropyrimidinase; ammonium chloride and l-glutamine for ω-amidase) were less effective or somewhat repressive on the production of half-amidase (Table 1).
Expression of half-amidase and imidase in the presence of cyclic imide.
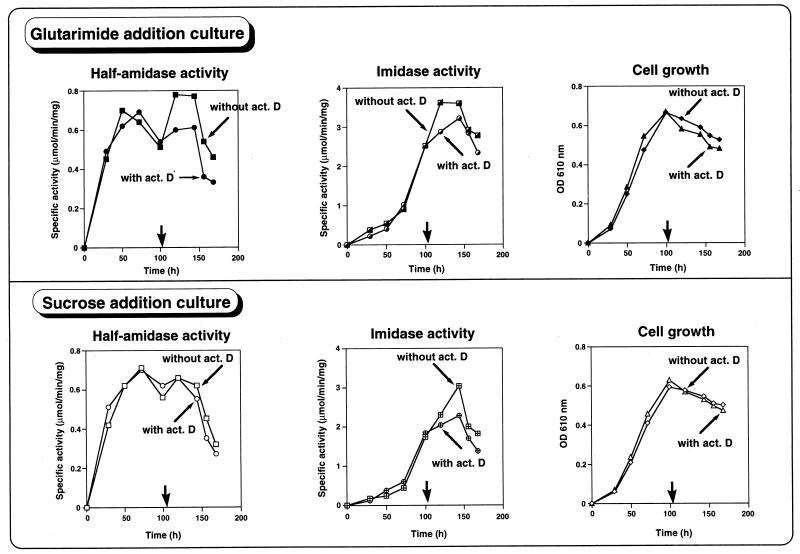
Half-amidase activity increased with cell growth and decreased at the mid-logarithmic phase of growth, but upon the addition of cyclic imide (glutarimide), the production of half-amidase increased rapidly (Fig. 2, glutarimide addition culture, without actinomycin D). However, when cyclic imide and actinomycin D were added together, no enhancement of half-amidase production was observed due to the inhibition of RNA synthesis by actinomycin D (Fig. 2, glutarimide addition culture, with actinomycin D). Similar results were observed with imidase (Fig. 2, glutarimide addition culture). In control experiments, the addition of sucrose did not enhance half-amidase production but increased slightly imidase production (Fig. 2, sucrose addition culture). These results suggested that cyclic imides induce the expression of imidase and specifically half-amidase at gene level.
FIG. 2.
Effect of cyclic imide and the RNA synthesis inhibitor actinomycin (act.) D on the expression of half-amidase and imidase. The experimental conditions (sets A through D) are described in Materials and Methods. Symbols: set A (glutarimide addition without actinomycin D), ■ (half-amidase activity), ┌ (imidase activity), and ⧫ (cell growth); set B (glutarimide addition with actinomycin D), ● (half-amidase activity),  (imidase activity), and ▴ (cell growth); set C (sucrose addition without actinomycin D), □ (half-amidase activity), ⊞ (imidase activity), and ◊ (cell growth); set D (sucrose addition with actinomycin D), ○ (half-amidase activity), ⊕ (imidase activity), and ▵ (cell growth). Addition of glutarimide or sucrose without or with actinomycin D is indicated by a thick arrow. The values of half-amidase activity, imidase activity, and optical density (OD) are the averages of three separate determinations that were reproducible within ±5%.
(imidase activity), and ▴ (cell growth); set C (sucrose addition without actinomycin D), □ (half-amidase activity), ⊞ (imidase activity), and ◊ (cell growth); set D (sucrose addition with actinomycin D), ○ (half-amidase activity), ⊕ (imidase activity), and ▵ (cell growth). Addition of glutarimide or sucrose without or with actinomycin D is indicated by a thick arrow. The values of half-amidase activity, imidase activity, and optical density (OD) are the averages of three separate determinations that were reproducible within ±5%.
Purification of half-amidase and criteria for purity.
Typical results of purification are presented in Table 2. Half-amidase was purified approximately 700-fold from the soluble cell extract of Blastobacter sp. The active fractions from the Mono Q PC 1.6/5 column were ascertained to be pure from the appearance of a single protein band on SDS-PAGE.
TABLE 2.
Purification of half-amidase from Blastobacter sp. strain A17p-4
| Step | Vol (ml) | Total protein (mg) | Total activity (μmol/min) | Sp act (μmol/min/mg) | Recovery (%) | Fold |
|---|---|---|---|---|---|---|
| Cell extract | 230 | 1,890 | 12.4 | 0.00656 | 100 | 1.00 |
| DEAE-Sephacel | 90 | 114 | 11.0 | 0.0965 | 88.7 | 14.7 |
| Phenyl-Sepharose | 65 | 52.7 | 3.10 | 0.0588 | 25.0 | 8.96 |
| Sephacryl S-200 | 40 | 7.00 | 1.01 | 0.144 | 8.15 | 22.0 |
| Mono Q HR 5/5 | 2.0 | 0.103 | 0.209 | 2.03 | 1.69 | 309 |
| Mono Q PC 1.6/5 | 0.2 | 0.0221 | 0.111 | 5.02 | 0.895 | 765 |
Substrate specificity and kinetic properties.
Normal hyperbolic kinetics were observed with all compounds hydrolyzed by the enzyme. The Km, kcat, and kcat/Km values for preferred substrates, calculated from double-reciprocal Lineweaver-Burk plots, are shown in Table 3. The enzyme showed the highest catalytic efficiency (kcat/Km) toward half-amides such as succinamic acid and glutaramic acid but not hydrolyzed diamides such as succinamide, fumaramide, and adipamide. The enzyme also showed activity toward lactamide, the middle-chain amides (n-valeramide, n-caproamide, and lauramide), an unsaturated aliphatic amide (crotoamide), and aromatic amides (benzamide and 2-phenylpropioamide), but with catalytic efficiency very low compared to half-amides.
TABLE 3.
Substrate specificity of half-amidase from Blastobacter sp. strain A17p-4a
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| Succinamic acid | 6.2 ± 0.4 | 5.76 ± 0.30 | 0.93 ± 0.02 |
| Glutaramic acid | 2.8 ± 0.1 | 2.23 ± 0.10 | 1.62 ± 0.06 |
| Adipiamic acid | 8.0 ± 1.4 | 0.36 ± 0.05 | 0.046 ± 0.002 |
| Lactamide | 3.7 ± 0.3 | 0.11 ± 0.01 | 0.033 ± 0.002 |
| n-Valeramide | 5.1 ± 0.2 | 0.18 ± 0.01 | 0.037 ± 0.006 |
| n-Caproamide | 17.0 ± 2.7 | 0.18 ± 0.04 | 0.011 ± 0.001 |
| Crotoamide | 8.8 ± 0.4 | 0.10 ± 0.01 | 0.012 ± 0.001 |
| Benzamide | 6.5 ± 0.1 | 0.13 ± 0.01 | 0.020 ± 0.001 |
| 2-Phenylpropioamide | 3.1 ± 0.1 | 0.078 ± 0.002 | 0.026 ± 0.001 |
Reactions were carried out under standard assay conditions described in Materials and Methods with the variation of substrates, and the amount of ammonia produced was colorimetrically estimated by indophenol method. As for the ester substrates, the reduction of ester or the production of acid was determined by HPLC as described in Materials and Methods. The kinetic constant values are averages ± standard deviations of three separate determinations. The following compounds were judged to be inactive as substrate (less than 0.1% kcat values for succinamic acid; <0.005 s−1): acetamide, fluoroacetamide, chloroacetamide, propioamide, n-butyramide, isobutyramide, acrylamide, oleamide, stearamide, nicotinamide, 2-thiophenecarboxamide, o-aminobenzamide, o-chlorobenzamide, 4,5-imidazoledicarboxamide, o-toluamide, phthalamidic acid, salicylicamide, dl-phenylalanine amide, glycine amide, glycyl-dl-alanine, asparagine, glutamine, fumaramide, succinamide, adipiamide, α-ketoglutaramic acid, α-ketosuccinamic acid, urea, ethylurea, N-methylurea, oxamide, biurea, hydantoic acid, β-ureidopropionate, N-carbamoyl-d- and -l-amino acids, allantoin, pyrazine carboximide, 4-pyridine carboxylate amide, succinate monomethyl ester, succinate monoethyl ester, lactic acid ethyl ester, crotonic acid methyl ester, and isovaleric acid ethyl ester.
The amide compounds judged to be inactive as substrates are some short- and long-chain amides, aromatic amides, amino acid amides, α-keto acid amides, urea, asparagine, glutamine, β-ureidopropionate, and the N-carbamoyl amino acids, which are common substrates of acyl amidase (2, 19, 20), aryl amidase (8, 10), aminopeptidase (1, 5), ω-amidase (4, 17), urease (18), asparaginase (29), glutaminase (29), β-ureidopropionase (21), and N-carbamoyl amino acid amidohydrolases (24, 25, 27), respectively.
Half-amidase did not show amide transferase activity, as determined by acylhydroxamate formation (12) from hydroxylamine and several amides listed in Table 3 as acyl donors. The enzyme did not show esterase and protease activities with methyl or ethyl ester acid and with amino acid amide or dipeptide as substrates, respectively.
Relative molecular mass, subunit structure, and NH2-terminal amino acid sequence.
The half-amidase was determined to be a monomer. Based on the results of HPLC gel filtration and SDS-PAGE analysis, the relative molecular weight of the native enzyme was estimated to be 48,000 and 46,000, respectively. The NH2-terminal amino acid sequence of the enzyme was determined to be Met-Leu-Leu-Ala-Asn-Asp-Pro-Val-His-Ala-Phe-Val-Asp-Tyr-Pro-Ala-Val-Glu-Val-Pro. A BLAST search for similarity with entries in the protein sequence databases (Swissprot, GenBank, and EMBL) found no apparent homology with other reported amidases (12, 15).
Effects of inhibitors.
Enzyme activity, assayed under standard conditions in the presence of various compounds (2 mM), was inhibited by sulfhydryl reagents such as sodium arsenate, 5,5′-dithiobis(2-nitrobenzoate), and N-ethyl maleimide with 98, 79, and 26% inhibition, respectively. Serine protease inhibitors such as phenylmethanylsulfonyl fluoride, tosyl phenylalanyl chloromethyl ketone, and diisopropylphosphofluoridate caused 44, 29, and 27% inhibition, respectively. Metal ion chelators did not show any inhibition. Heavy metal ions such as Hg2+, Cu2+, Zn2+, Ag+, and Cd2+ inactivated enzyme activity by 99, 90, 88, 76, and 79%, respectively. This susceptibility of the half-amidase to inhibitors is similar to findings for other amidases from Rhodococcus sp. (8, 12, 20) and Pseudomonas sp. (2, 10).
Effects of imides, amides, ammonia, and organic acids.
The activity of half-amidase toward succinamic acid was examined in the presence of various substrates of known amidases such as l-glutamine, l-asparagine, and n-butylamide. None of these compounds showed significant inhibitory effects. The half-amidase activity was not inhibited by cyclic imide metabolism intermediates (succinimide, fumarate, l-malate, pyruvate), the products (succinate, ammonium chloride), product-like compounds (glutaric acid, adipic acid), and the TCA cycle intermediates (oxaloacetate, citrate, α-ketoglutarate), while imidase activity was specifically inhibited by succinate (23).
Effects of pH and temperature.
Enzyme activity and stability were assayed in morpholineethanesulfonic acid-NaOH, morpholinepropanesulfonic acid-NaOH, Tris-HCl, and NaOH-Na2CO3 buffer systems at pH 4.5 to 5.1, 6.2 to 8.2, 8.1 to 9.0, and 9.4 to 10.0, respectively. Under the standard assay conditions, the pH optimum was approximately 9.4 to 10.0. More than 80% of the enzyme activity retained at pH 8.0 to 10.0 after incubation at 30°C for 30 min.
The enzyme exhibited maximal activity at 35°C. No enzymatic activity remained after 30 min of incubation above 40°C in 0.2 M Tris-HCl (pH 7.5).
DISCUSSION
The recently found microbial cyclic imide transformation (22, 23, 32, 33) is attracting increasing attention as a novel process for production of useful organic acids and as a new tool for fine enzymatic synthesis of chiral and regioisomeric compounds (34, 35). The half-amidase plays an important role in this transformation, linking cyclic imide metabolism to the TCA cycle. We purified half-amidase from the bacterium Blastobacter sp. strain A17p-4. The properties of the half-amidase were different from those of other known amidases.
The half-amidase is a monomer, and its NH2-terminal amino acid sequence showed no homology with other reported amidases. The half-amidase exhibited narrow substrate specificity toward half-amides, suggesting that the enzyme functions specifically in cyclic imide metabolism and is distinct from known amide-, amino acid amide-, or cyclic amide-transforming enzymes.
The imidase catalyzing the first step of the cyclic imide metabolism showed high activity and affinity toward cyclic imides (23), and the half-amidase showed the highest activity and catalytic efficiency toward the ring-opened products of cyclic imides, i.e., half-amides, suggesting that the half-amidase acts cooperatively with the imidase in the degradation of cyclic imides. The production of both enzymes was enhanced by cyclic imides (Fig. 2; Table 1) (22), indicating that these enzymes were expressed for cyclic imide utilization. In cyclic imide metabolism, half-amidase is positively regulated at the gene level by cyclic imide. Regulation of the imidase gene by cyclic imide is not understood, while it is specifically regulated at the catalytic level by succinate, the product of half-amidase reaction and a key intermediate of cyclic imide metabolism in relation to the TCA cycle (22, 23).
In mammals, cyclic imides were reported to be metabolized by the combined action of dihydropyrimidinase (14) and ω-amidase (7), which are involved in pyrimidine and amino acid metabolism, respectively. Rat liver ω-amidase, which is considered to be a mammalian counterpart of the bacterial half-amidase, has been reported to hydrolyze succinamic acid and glutaramic acid, as well as α-ketoglutaramic acid and α-ketosuccinamic acid, which are not substrates of the half-amidase. Thus, in contrast to the bacterial half-amidase, the rat ω-amidase possessed different substrate specificity and physiological roles, namely, metabolism of glutamine and asparagine (7, 17).
Above all, half-amidase and imidase, which are responsible for bacterial cyclic imide transformation, are unique enzymes with characteristics distinct from those of other cyclic amide-transforming enzymes. The application of these enzymes are now under investigation and will be reported elsewhere.
REFERENCES
- 1.Asano Y, Nakazawa A, Kato Y, Kondo K. Properties of a novel d-stereospecific aminopeptidase from Ochrobactrum anthropi. J Biol Chem. 1989;264:14233–14239. [PubMed] [Google Scholar]
- 2.Ciskanik L M, Wilczek J M, Fallon R D. Purification and characterization of an enantioselective amidase from Pseudomonas chlororaphis B23. Appl Environ Microbiol. 1995;61:998–1003. doi: 10.1128/aem.61.3.998-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudley K H, Roberts S B. Dihydropyrimidinase, stereochemistry of the metabolism of some 5-alkylhydantoins. Drug Metab Dispos. 1978;6:133–139. [PubMed] [Google Scholar]
- 4.Fernald N J, Ramaley R F. Purification and properties of dicarboxylate ω-amidase from Bacillus subtilis 168 and Thermus aquaticus YT-1. Arch Biochem Biophys. 1972;153:95–104. doi: 10.1016/0003-9861(72)90425-0. [DOI] [PubMed] [Google Scholar]
- 5.Hermes H F M, Sonke T, Peters P J H, van Balken J A M, Kamphuis J, Dijkhuizen L, Meijer E M. Purification and characterization of an l-aminopeptidase from Pseudomonas putida ATCC 12633. Appl Environ Microbiol. 1993;59:4330–4334. doi: 10.1128/aem.59.12.4330-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hersh L B. 5-Hydroxy-N-methylpyroglutamate synthetase, purification and mechanism of action. J Biol Chem. 1970;245:3526–3535. [PubMed] [Google Scholar]
- 7.Hersh L B. Rat liver ω-amidase, purification and properties. Biochemistry. 1971;10:2884–2891. doi: 10.1021/bi00791a014. [DOI] [PubMed] [Google Scholar]
- 8.Hirrlinger B, Stolz A, Knackmuss H-J. Purification and properties of an amidase from Rhodococcus erythropolis MP 50 which enantioselectively hydrolyzes 2-arylpropionamides. J Bacteriol. 1996;178:3501–3507. doi: 10.1128/jb.178.12.3501-3507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynes M J, Pateman J A. The use of amides as nitrogen sources by Aspergillus nidulans. J Gen Microbiol. 1970;63:317–324. doi: 10.1099/00221287-63-3-317. [DOI] [PubMed] [Google Scholar]
- 10.Kagayama T, Ohe T. Purification and properties of an aromatic amidase from Pseudomonas sp. GDI 211. Agric Biol Chem. 1990;54:2565–2571. [Google Scholar]
- 11.Kelly M, Clarke P H. An inducible amidase produced by a strain of Pseudomonas aeruginosa. J Gen Microbiol. 1962;27:305–316. doi: 10.1099/00221287-27-2-305. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi M, Komeda H, Nagasawa T, Nishiyama M, Horinouchi S, Beppu T, Yamada H, Shimizu S. Amidase coupled with low-molecular-mass nitrile hydratase from Rhodococcus rhodochrous J1. Eur J Biochem. 1993;217:327–336. doi: 10.1111/j.1432-1033.1993.tb18250.x. [DOI] [PubMed] [Google Scholar]
- 13.Maestracci M, Thiery A, Arnaud A, Galzy P. The amidases from a Brevibacterium strain: study and applications. Adv Biochem Eng Biotechnol. 1988;36:67–115. doi: 10.1007/BFb0047945. [DOI] [PubMed] [Google Scholar]
- 14.Maguire J H, Dudley K H. Dihydropyrimidinase, metabolism of some cyclic imides of different ring size. Drug Metab Dispos. 1978;6:140–145. [PubMed] [Google Scholar]
- 15.Mayaux J-F, Cerbelaud E, Soubrier F, Yeh P, Blanche F, Petre D. Purification, cloning, and primary structure of a new enantiomer-selective amidase from a Rhodococcus strain: structural evidence for a conserved genetic coupling with nitrile hydratase. J Bacteriol. 1991;173:6694–6704. doi: 10.1128/jb.173.21.6694-6704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meister A. Preparation and enzymatic reactions of the keto analogues of asparagine and glutamine. J Biol Chem. 1953;200:571–589. [PubMed] [Google Scholar]
- 17.Meister A, Levintow L, Greenfield R E, Abendschein P A. Hydrolysis and transfer reactions catalyzed by ω-amidase preparations. J Biol Chem. 1955;215:441–460. [PubMed] [Google Scholar]
- 18.Nakano H, Takenishi S, Watanabe Y. Purification and properties of urease from Brevibacterium ammoniagenes. Agric Biol Chem. 1984;48:1495–1502. [Google Scholar]
- 19.Nawas M S, Khan A A, Bhattacharayya D, Siitonen P H, Cerniglia C E. Physical, biochemical, and immunological characterization of a thermostable amidase from Klebsiella pneumoniae NCTR 1. J Bacteriol. 1996;178:2397–2401. doi: 10.1128/jb.178.8.2397-2401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawas M S, Khan A A, Seng J E, Leakey J E, Siitonen P H, Cerniglia C E. Purification and characterization of an amidase from an acrylamide-degrading Rhodococcus sp. Appl Environ Microbiol. 1994;60:3343–3348. doi: 10.1128/aem.60.9.3343-3348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa J, Shimizu S. β-Ureidopropionase with N-carbamoyl-α-l-amino acid amidohydrolase activity from an aerobic bacterium, Pseudomonas putida IFO 12996. Eur J Biochem. 1994;223:625–630. doi: 10.1111/j.1432-1033.1994.tb19034.x. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa J, Soong C-L, Honda M, Shimizu S. Novel metabolic transformation pathway for cyclic imides in Blastobacter sp. strain A17p-4. Appl Environ Microbiol. 1996;62:3814–3817. doi: 10.1128/aem.62.10.3814-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa J, Soong C-L, Honda M, Shimizu S. Imidase, a dihydropyrimidinase-like enzyme involved in the metabolism of cyclic imides. Eur J Biochem. 1997;243:322–327. doi: 10.1111/j.1432-1033.1997.0322a.x. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa J, Miyake H, Shimizu S. Purification and characterization of N-carbamoyl-l-amino acid amidohydrolase with broad substrate specificity from Alcaligenes xylosoxidans. Appl Microbiol Biotechnol. 1995;43:1039–1043. doi: 10.1007/BF00166922. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa J, Chung M C-M, Hida S, Yamada H, Shimizu S. Thermostable N-carbamoyl-d-amino acid amidohydrolase: screening, purification and characterization. J Biotechnol. 1994;38:11–19. doi: 10.1016/0168-1656(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa J, Honda M, Soong C-L, Shimizu S. Diversity of cyclic ureide compounds, dihydropyrimidine-, and hydantoin-hydrolyzing enzymes in Blastobacter sp. A17p-4. Biosci Biotechnol Biochem. 1995;59:1960–1962. [Google Scholar]
- 27.Ogawa J, Shimizu S, Yamada H. N-Carbamoyl-d-amino acid amidohydrolase from Comamonas sp. E222c. Purification and characterization. Eur J Biochem. 1993;212:685–691. doi: 10.1111/j.1432-1033.1993.tb17706.x. [DOI] [PubMed] [Google Scholar]
- 28.Ogawa, J., C.-L. Soong, M. Ito, and S. Shimizu. Enzymatic production of pyruvate from fumarate—an application of microbial cyclic-imide-transforming pathway. J. Mol. Catal. B, in press.
- 29.Ramadan M, El-din A, Asmar F, Greenberg D M. Purification and properties of glutaminase and asparaginase from a Pseudomonad. 1. Purification and physical chemical properties. Arch Biochem Biophys. 1964;108:143–149. doi: 10.1016/0003-9861(64)90365-0. [DOI] [PubMed] [Google Scholar]
- 30.Shimizu S, Ogawa J, Chung M C-M, Yamada H. Purification and characterization of novel enzyme, arylalkyl acylamidase, from Pseudomonas putida Sc2. Eur J Biochem. 1992;209:375–382. doi: 10.1111/j.1432-1033.1992.tb17299.x. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu S, Kataoka M, Chung M C-M, Yamada H. Ketopantoic acid reductase of Pseudomonas maltophilia 845: purification and characterization, role in panthothenate biosynthesis. J Biol Chem. 1988;263:12077–12084. [PubMed] [Google Scholar]
- 32.Soong C-L, Ogawa J, Sukiman H, Prana T, Prana M S, Shimizu S. Distribution of cyclic imide-transforming activity in microorganisms. FEMS Microbiol Lett. 1998;158:51–55. doi: 10.1111/j.1574-6968.1998.tb12799.x. [DOI] [PubMed] [Google Scholar]
- 33.Soong C-L, Ogawa J, Honda M, Shimizu S. Cyclic-imide-hydrolyzing activity of d-hydantoinase from Blastobacter sp. strain A17p-4. Appl Environ Microbiol. 1999;65:1459–1462. doi: 10.1128/aem.65.4.1459-1462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soong, C.-L., J. Ogawa, and S. Shimizu. Cyclic ureide and imide metabolism in microorganisms producing a d-hydantoinase useful for d-amino acid production. J. Mol. Catal. B, in press.
- 35.Syldatk C, May O, Altenbuchner J, Mattes R, Siemann M. Microbial hydantoinases—industrial enzymes from the origin of life? Appl Microbiol Biotechnol. 1999;51:293–309. doi: 10.1007/s002530051395. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y-S, Ramasamy S, Jakoby W B. Rat liver imidase. J Biol Chem. 1993;268:10870–10875. [PubMed] [Google Scholar]