Abstract
The spatial distribution and diversity of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria (hereinafter referred to as ammonia oxidizers) in the Arctic Ocean were determined. The presence of ammonia oxidizers was detected by PCR amplification of 16S rRNA genes using a primer set specific for this group of organisms (nitA and nitB, which amplifies a 1.1-kb fragment between positions 137 and 1234, corresponding to Escherichia coli 16S rDNA numbering). We analyzed 246 samples collected from the upper water column (5 to 235 m) during March and April 1995, September and October 1996, and September 1997. Ammonia oxidizers were detected in 25% of the samples from 5 m, 80% of the samples from 55 m, 88% of the samples from 133 m, and 50% of the samples from 235 m. Analysis of nitA-nitB PCR product by nested PCR-denaturing gradient gel electrophoresis (DGGE) showed that all positive samples contained the same major band (band A), indicating the presence of a dominant, ubiquitous ammonia oxidizer in the Arctic Ocean basin. Twenty-two percent of the samples contained additional major bands. These samples were restricted to the Chukchi Sea shelf break, the Chukchi cap, and the Canada basin; areas likely influenced by Pacific inflow. The nucleotide sequence of the 1.1-kb nitA-nitB PCR product from a sample that contained only band A grouped with sequences designated group 1 marine Nitrosospira-like sequences. PCR-DGGE analysis of 122 clones from four libraries revealed that 67 to 71% of the inserts contained sequences with the same mobility as band A. Nucleotide sequences (1.1 kb) of another distinct group of clones, found only in 1995 samples (25%), fell into the group 5 marine Nitrosomonas-like sequences. Our results suggest that the Arctic Ocean β-proteobacterial ammonia oxidizers have low diversity and are dominated by marine Nitrosospira-like organisms. Diversity appears to be higher in Western Arctic Ocean regions influenced by inflow from the Pacific Ocean through the Bering and Chukchi seas.
Nitrification, the conversion of ammonia to nitrite and/or nitrate, completes the redox cycle of nitrogen, linking the most-reduced form to the most-oxidized form. Nitrification also supplies nitrate, the substrate for denitrification, potentially resulting in the loss of fixed nitrogen to the atmosphere under oxygen-limiting conditions (30, 44).
The first step in nitrification, the oxidation of ammonia to nitrite, is carried out by chemolithotrophic ammonia-oxidizing bacteria in the β and γ subdivisions of the class Proteobacteria (30, 52, 53). The γ-Proteobacteria contain strains of Nitrosococcus oceanus and Nitrosococcus halophilus, while the β-Proteobacteria contain members of the genera Nitrosomonas (including Nitrosococcus mobilis) and Nitrosospira (including Nitrosolobus and Nitrosovibrio) (11, 38, 39).
It has been difficult to study ammonia-oxidizing bacteria in pure cultures because of their slow growth and contamination of cultures by heterotrophic bacteria. Development of molecular biological approaches using specific oligonucleotides for selective amplification of ribosomal genes has helped overcome this difficulty. Analysis of 16S rRNA gene (rDNA) sequences by techniques such as restriction fragment length polymorphism, denaturing gradient gel electrophoresis (DGGE), cloning and sequencing, or probing have been of particular significance in this regard. Several recent studies have used these techniques to analyze the composition and diversity of ammonia-oxidizing bacterial communities in a range of environments (9, 10, 12, 14, 18, 19, 21, 22, 29, 36, 37, 40, 45, 51). Comparison of ammonia oxidizers in the natural environment with enrichment cultures has shown that enrichment cultures do not represent in situ diversity of ammonia oxidizers. These studies found that Nitrosospira spp. are more abundant in natural environments, whereas Nitrosomonas spp. are more abundant in the enrichment cultures (12, 18, 36, 37).
Stephen et al. (36) recovered ammonia oxidizer rDNA sequences from marine sediment and soil using primers specific for ammonia oxidizers. They grouped these sequences into seven phylogenetic clusters. Marine Nitrosospira-like clones were found in a cluster (group 1) that was phylogenetically distinct from the cultured representatives of Nitrosospira spp. Phillips et al. (29) also used selective primers to recover sequences from suspended particulate material and planktonic samples from the northwest Mediterranean Sea. This study demonstrated that sequences associated with particles were predominantly related to Nitrosomonas eutropha whereas sequences from the free-living bacterial assemblage in the same sample were predominantly related to the marine Nitrosospira group 1 of Stephen et al. (36). These studies have tended to focus on ammonia oxidizers of the β-Proteobacteria, though γ-proteobacterial ammonia oxidizers (e.g., Nitrosococcus oceanus) are also present in the marine environments. Ward and Carlucci (43) and Voytek et al. (41) detected Nitrosococcus oceanus in seawater from the Southern California Bight and in several permanently ice-covered Antarctic saline lakes.
Most studies of ammonia oxidizers have focused on temperate coastal, terrestrial, or freshwater environments or biofilms and bioreactors. Little is known about the biodiversity of ammonia oxidizers in the open ocean in general or cold oceans in particular. Temperature is one of the major environmental factors affecting the growth and metabolic activities of marine microorganisms (34, 35); thus, it might be difficult for inherently slow growing ammonia oxidizers to survive in polar seas. However, Horrigan (13) indicated that nitrifiers are active in cold waters. Jones and Morita (15) and Jones et al. (16) isolated Nitrosomonas cryotolerans from Alaskan coastal waters and showed that this organism was capable of growth at −5°C. Voytek and Ward (40) also detected β-proteobacterial ammonia oxidizers in permanently ice-covered Antarctic lakes.
The goal of the research presented here was to investigate the distribution and diversity of DNA sequences with affinity to ammonia-oxidizing bacteria of the β-Proteobacteria in the Arctic Ocean basin. This perennially ice-covered ocean is surrounded by continents and receives organic matter and nutrients from a number of sources, including the surrounding land masses, phytoplankton production, ice-algal production, and inflow from the Pacific and Atlantic oceans. The organic matter is oxidized in the Arctic Ocean basin (which appears to be net heterotrophic) (49, 50), releasing ammonium that is subsequently oxidized. Our long-range objective is to compare the guild of ammonia oxidizers in the Arctic Ocean with the ammonia oxidizers in other oceans, particularly the Southern Ocean, to improve our understanding of the ecophysiology of this important group of organisms.
MATERIALS AND METHODS
Sample collection.
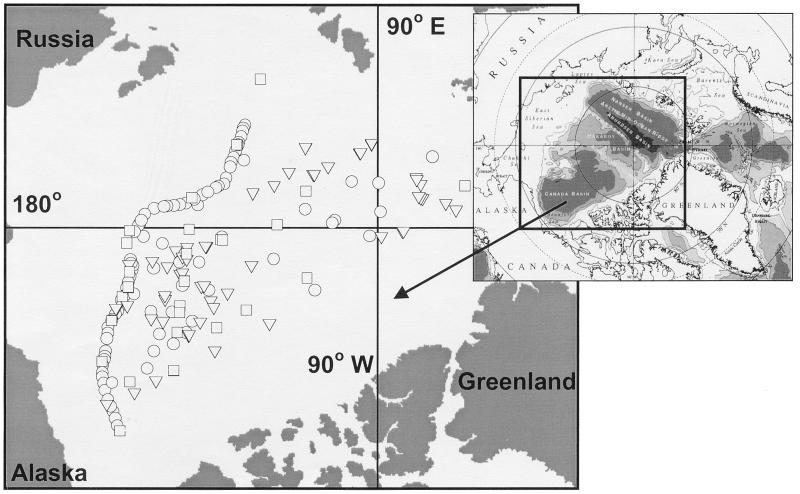
Water samples were collected from depths of 5, 55, 133, and 235 m in the Central Arctic Ocean during the SCICEX 95 (26 March to 8 May 1995), SCICEX 96 (13 September to 28 October 1996), and SCICEX 97 (21 August to 15 October 1997) cruises aboard U.S. Navy submarines Cavalla, Pogy, and Archerfish. Station locations are shown in Fig. 1. Water was collected from a through-hull fitting while submerged or with Niskin bottles when surfaced. Water (6 to 12 liters) was pressure filtered (at 0.4 Pa) through a 0.22-μm-pore-size Sterivex GV filter cartridge (Millipore). Excess water was expelled, and then 1.8 ml of lysis buffer (0.75 M sucrose, 40 mM EDTA, 50 mM Tris [pH 8.3]) was added to each cartridge. The cartridges were frozen and stored at −20°C until processed.
FIG. 1.
Location of Arctic Ocean stations where DNA samples were collected during SCICEX 95 (circle), 96 (triangle), and 97 (square).
DNA extraction.
Total community DNA was extracted from the water samples as described by Ferrari and Hollibaugh (8). Briefly, 40 μl of lysozyme (50 mg ml−1) was added to each cartridge; the cartridges were incubated for 60 min at 37°C; 50 μl of proteinase K (20 mg ml−1) and 100 μl of a 20% (wt/vol) sodium dodecyl sulfate solution were added; and then the cartridges were incubated at 55°C for 2 h. Lysate was removed from the cartridges and placed in clean 15-ml tubes, and then the cartridges were rinsed with 1 ml of lysis buffer. The buffer was withdrawn and combined with the lysate. DNA was purified from 800 μl of the combined lysate by sequential extraction with 800 μl of phenol-chloroform-isoamyl alcohol (25:24:1), chloroform-isoamyl alcohol (24:1), and finally n-butanol. The aqueous phase was removed, placed in a Centricon-100 concentrator (Amicon), mixed with 500 μl of TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]), and centrifuged at 1,000 × g for 10 min; then, 500 μl of TE was added to the Centricon and centrifuged for another 10 min. Blanks were prepared with each set of samples using Sterivex cartridges through which no water had been filtered. The molecular weight and concentration of DNA in extracts were determined by electrophoresing portions of extracts on 1.5% agarose gels with a 1-kb ladder (Promega) as a size marker and using three known concentrations of calf thymus DNA (Pharmacia Biotech) to generate a standard curve. Gels were stained with ethidium bromide (EtBr) and scanned on an FMBIO II (Hitachi) gel scanner. EtBr fluorescence of bands was converted to DNA concentration using the standard curve and FMBIO software.
PCR amplification of 16S rDNA.
All primers used in PCR amplifications were synthesized either by Operon Technologies (Oakland, Calif.) or the University of Georgia Molecular Genetics Instrument Facility (MGIF). Two hundred forty-six samples (8 from 5 m, 153 from 55 m, 57 from 133 m, and 28 from 235 m) were tested for the presence of ammonia-oxidizing bacteria by amplifying 16S rDNA with the nitA and nitB primer set (40) (Table 1), hereinafter referred to as nitAB. These primers amplify a 1.1-kb fragment between position 137 and 1234 of the Escherichia coli 16S rDNA and are specific for ammonia-oxidizing bacteria of the β-subclass of the class Proteobacteria. Reaction mixtures were prepared in a total volume of 100 μl and contained 1× PCR buffer (50 mM Tris-HCl, 100 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 50% glycerol, and 1% Triton X-100 [pH 8.0]), 2.5 mM MgCl2, a 200 μM concentration of each deoxyribonucleotide triphosphate (dATP, dCTP, dGTP, and dTTP), a 0.5 μM concentration of each primer, and 20 to 100 ng of template DNA. All PCRs were performed in a DNA Engine thermocycler (MJ Research) using the following conditions: initial denaturation of the template DNA at 95°C for 10 min with a pause at 82°C to add Taq DNA polymerase (2.5 U; Promega); then, 35 cycles consisting of denaturation (30 s at 94°C), annealing (1 min at 57°C), extension (1.25 min at 72°C), and a final extension at 72°C for 10 min. Reactions containing template DNA from Nitrosomonas cryotolerans (positive control; culture) and E. coli (negative control; Sigma) were included in all sets of amplifications. The success of PCRs was determined by electrophoresis of 6 μl of the reaction mixture in 1.5% (wt/vol) agarose gel in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA [pH 8.3]) with a 1-kb DNA ladder (Promega) as a size marker. Gels were stained with EtBr and examined using a transilluminator (UVP). Not all samples produced detectable PCR products under these conditions, presumably because of the low concentration of compatible template DNA in the sample (all samples yielded PCR product with Bacterial primers used for PCR-DGGE).
TABLE 1.
Sequences of primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| nitA | CTTAAGTGGGGAATAACGCATCG |
| nitB | TTACGTGTGAAGCCCTACCCA |
| AM1 | CCTACGGGAGGCAGCAG |
| AM2 | FaATTACCGCGGCTGCTGG |
| GC-Clamp | CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCC |
F, fluorescein label.
We randomly selected a subset of samples from those that amplified successfully (189 of 246) for analysis of the diversity of ammonia oxidizer sequences by PCR-DGGE. These samples were distributed as follows: 32 samples out of 79 from SCICEX 95 (30 from 55 m and 2 from 133 m), 27 samples out of 69 from SCICEX 96 (2 from 5 m, 16 from 55 m, 6 from 131 m, and 3 from 235 m), and 18 samples out of 41 from SCICEX 97 (14 from 55 m, 3 from 133 m, and 1 from 235 m). The nitAB PCR product from the selected samples was precipitated with 2 volumes of ice-cold ethanol and 0.1 volume of 3 M sodium acetate (pH 5.2) overnight at −20°C and then was centrifuged at 12,000 × g for 30 min at 4°C. The DNA pellet was dried in a DNA SpeedVac (Savant) for 10 min and resuspended in 15 μl of TE (pH 8.0). A portion (4 μl) of this DNA was used as template for reamplification with primers AM1 (Bacterial) and fluorescein-labeled AM2 (universal) (Table 1). These primers amplify positions 341 to 534 in E. coli (23, 24), encompassing hypervariable region 3, which contains a disproportionately large portion of the total variability in the 16S rDNA (4). A 40-bp GC clamp (25) was added to the 5′ end of the AM1 primer. PCR conditions were similar to those used by Ferrari and Hollibaugh (8). The concentration of the resulting PCR product was estimated by the Hoechst dye assay (28) and resolved by DGGE.
DGGE.
DGGE was performed using a CBS Scientific (Del Mar, Calif.) system, essentially following the method of Ferrari and Hollibaugh (8). For each sample, 300 ng of PCR product was loaded on a 6.5% polyacrylamide gel with a 52-to-60% gradient. Gels were run for 15 h at a constant voltage of 75 V in 1× TAE buffer (40 mM Tris, 20 mM sodium acetate, 1 mM EDTA [pH adjusted to 7.4 with acetic acid]) at a constant temperature of 60°C. Gels were scanned using an FMBIO II (Hitachi) gel scanner set to measure fluorescein fluorescence. Bands of interest were excised from the gel, and the DNA was eluted from them into 100 μl of water by incubation at 60°C for 2 h. The eluted DNA was purified using Wizard PCR preps (Promega) and sequenced on an automated sequencer (MGIF) with either AM1 or AM2 or both primers.
Cloning.
Two samples from the SCICEX 95 cruise (95A: 72°16′N, 154°24′W, 55 m; 95B: 72°34′N, 155°47′W, 55 m) and two samples from the SCICEX 96 cruise (96A: 80°28′N, 156°53′W, 132 m; 96B: 83°38′N, 131°16′E, 55 m) were selected to generate clone libraries. The 16S rDNA of each sample was amplified in triplicate using the nitAB primer pair as described above. The triplicate PCRs were combined and purified using Wizard PCR preps (Promega). The PCR product (50 ng) was ligated into PGEM-T Easy Vector (Promega) and transformed into competent E. coli JM 109 cells. The transformed cells were plated on Luria-Bertani (LB) plates containing ampicillin (100 μg ml−1), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (80 μg ml−1), and 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) as recommended by the manufacturer and incubated overnight at 37°C.
A total of 122 white colonies from the four clone libraries (63 from 95A, 21 from 95B, 37 from 96A, and 1 from 96B) were chosen at random, plated on ampicillin-supplemented LB plates, and then incubated overnight. PCR-DGGE was used to identify unique clones for sequencing. DNA was extracted from a few colonies of each clone by boiling in 100 μl of water for 2 min. This DNA was used as template for PCR amplification with AM1-GC and AM2 primers. The PCR product was analyzed by DGGE as described above. Clones were grouped according to their DGGE mobility. Seven clones from library 95A, six clones from library 95B, five clones from library 96A, and one clone from library 96B were chosen for sequencing as representatives of different DGGE bands. These clones were grown in LB medium supplemented with ampicillin, and then plasmid DNA was extracted and purified using a QIA miniprep kit (Qiagen Inc.). Plasmid DNAs were checked for inserts of the correct size by digestion with the enzyme EcoRI (Promega) according to the manufacturer's recommendations.
Phylogenetic analysis.
All sequences were obtained from an automatic sequencer (MGIF). The 1.1-kb nitAB PCR product from a sample that gave one band by DGGE and which was used to construct clone library 96B was sequenced directly using nitAB primers. The 1.1-kb inserts from clones were sequenced using Sp6 and T7 plasmid primers or nitAB PCR primers. Sequences were checked for chimeras using the Ribosomal Database Project's CHIMERA-CHECK program and then were compared to known sequences using BLAST (3). Phylogenetic analyses were conducted by aligning the 16S rDNA sequences with the sequences from the database with the highest BLAST similarity values, using the Genetics Computer Group package (Madison, Wis.). Phylogenetic trees were constructed using Jukes-Cantor distances and the neighbor-joining method (PHYLIP package [7]). Tree robustness was tested by bootstrap analysis (100 replicates).
Nucleotide sequence accession numbers.
The sequences have been deposited in GenBank under accession no. AF142411, AF216675, AF216676, AF230659, AF230660 and AF203511 to AF203523 (Table 2).
TABLE 2.
Accession numbers of sequences used to construct the phylogenetic tree in Fig. 6
| Organism or clone | Accession no. |
|---|---|
| Organisms | |
| Nitrosospira sp. isolate B6 | X84657 |
| Nitrosospira sp. isolate D11. | X84660 |
| Nitrosospira sp. strain Np22-21. | NS16S22 |
| Nitrosospira sp. strain NpAV. | NS16SAV |
| Nitrosospira sp. isolate AF. | X84658 |
| Nitrosospira sp. isolate Ka4 | AJ012106 |
| Nitrosospira briensis strain C-128 | L35505 |
| Nitrosovibrio tenuis strain C-141 | M96397 |
| Nitrosovibrio tenuis strain Nv12 | M96405 |
| Nitrosolobus multiformis (ATCC 25196) | L35509 |
| Nitrosococcus mobilis strain M93 | AF037105 |
| Nitrosomonas sp. isolate:AL212 | AB000699 |
| Nitrosomonas cryotolerans strain Nm 55 | Z46984 |
| Nitrosomonas eutropha strain C-91 | M96402 |
| Nitrosomonas europaea strain C-31 | M96399 |
| Nitrosomonas sp. isolate Koll-21 | AJ224941 |
| Nitrosomonas marina strain C-56 | M96400 |
| Nitrosomonas ureae strain Nm10 | Z46993 |
| Clones | |
| 400 AGG D3 | AF063633 |
| 400 FREE Z14 | AF063636 |
| DT-1.3 | UBU62867 |
| DT-1.5 | UBU62868 |
| DT-1.7 | UBU62885 |
| DT-1.9 | UBU62884 |
| DT-1.12 | UBU62869 |
| EnvA1-21. | Z69091 |
| EnvA2-4 | Z69094 |
| EnvA2-13 | Z69097 |
| EnvB1-17 | Z69104 |
| EnvC2-23 | Z69125 |
| pH7B/D3 | Z69177 |
| pH4.2A/23 | Z69151 |
| pH4.2A/3E | Z69155 |
| pH4.2A/G2 | Z69164 |
| pH4.2A/H2 | Z69165 |
| Enrich ZD5 | Z69146 |
| 96Ba | AF142411 |
| 95A-2 | AF203512 |
| 95A-4 | AF203513 |
| 95A-21 | AF216675 |
| 95A-40 | AF216676 |
| 95A-44 | AF203511 |
| 96A-4 | AF203521 |
| 96A-8 | AF203523 |
| 96A-11 | AF203520 |
| 96A-17 | AF230659 |
| 96A-19 | AF203522 |
| 95B-3 | AF203517 |
| 95B-4 | AF203516 |
| 95B-7 | AF203518 |
| 95B-10 | AF203515 |
| 95B-17 | AF203519 |
| 95B-22 | AF203514 |
| 96B-3 | AF230660 |
Direct sequence of PCR product.
RESULTS
Distribution of ammonia-oxidizing bacteria.
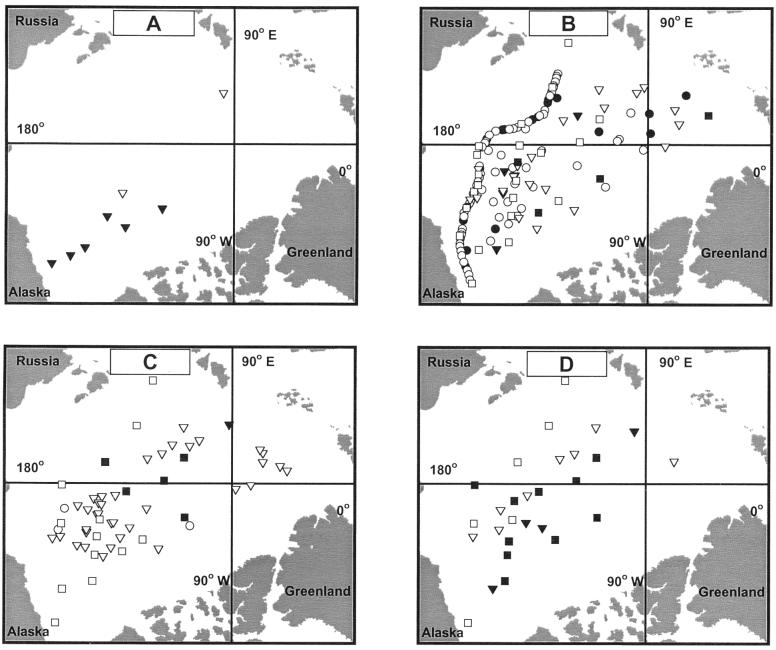
A total of 246 samples were tested for the presence of ammonia-oxidizing bacteria. PCR amplification was successful in 25% of the samples from 5 m (Fig. 2A), 80% of the samples from 55 m (Fig. 2B), 88% of the samples from 133 m (Fig. 2C), and 50% of the samples from 235 m (Fig. 2D). Positive samples yielded a single band of the expected size (1.1 kb), consistent with the presence of ammonia-oxidizing bacteria in these samples.
FIG. 2.
Distribution of ammonia-oxidizing bacteria in the Arctic Ocean at depths of 5 m (A), 55 m (B), 133 m (C), and 235 m (D). Symbols are defined in the legend to Fig. 1; open and filled symbols indicate the presence and lack of detection of ammonia oxidizers, respectively.
Diversity of ammonia-oxidizing bacteria.
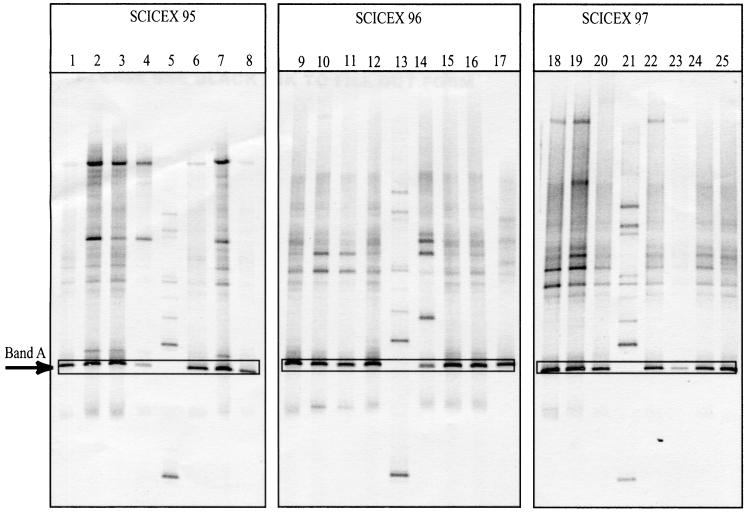
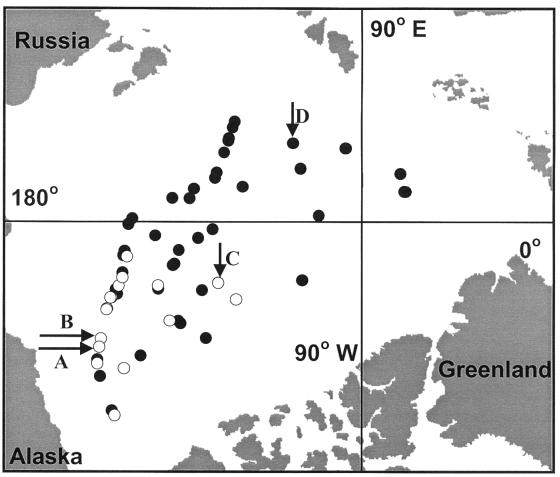
Seventy seven samples were analyzed in 11 different DGGE gels (three gels are shown in Fig. 3) to compare the diversity of ammonia-oxidizing bacteria among different stations and depths. All of the samples contained a major band (band A [Fig. 3]) with the same DGGE mobility, suggesting the presence of the same ammonia oxidizer in all samples. However, 22% of the samples, most of them from the Chukchi Cap and Chukchi Sea shelf break regions (Fig. 4), also contained additional major bands (Fig. 3, lanes 2, 3, 4, 7, 10, 11, 14, 18, and 19), demonstrating a higher diversity of ammonia oxidizers in these samples. In making this assessment, we ignored faint bands that appeared in most samples (Fig. 3, lanes 1, 6, 8, 9, 12, 15, 16, 17, 20, 22, 24, and 25). The banding patterns differed from cruise to cruise (SCICEX 95, 96, and 97), suggesting seasonal variation in the diversity and composition of the ammonia-oxidizing assemblages. The nucleotide sequences of the 150-bp band A fragments excised from the DGGE gels of four samples (two from SCICEX 95 and two from SCICEX 96) were identical, confirming that band A represents the same sequence in different samples.
FIG. 3.
Image of representative DGGE gels containing samples from the Arctic Ocean taken on the SCICEX 95, SCICEX 96, and SCICEX 97 cruises. Lane 1: 55 m, 71°49′N, 152°23′W; lane 2: 55 m, 72°16′N, 154°24′W; lane 3: 55 m, 72°34′N, 155°47′W; lane 4: 55 m, 73°36′N, 161°80′W; lane 6: 55 m, 74°27′N, 163°30′W; lane 7: 55 m, 73°32′N, 160°56′W; lane 8: 55 m, 74°30′N, 164°32′W; lane 9: 55 m, 76°60′N, 162°36′W; lane 10: 132 m, 74°20′N, 163°60′W; lane 11: 55 m, 74°20′N, 163°60′W; lane 12: 55 m, 77°21′N, 150°41′W; lane 14: 132 m, 80°28′N, 156°53′W; lane 15: 55 m, 80°29′N, 156°54′W; lane 16: 55 m, 76°57′N, 161°44′W; lane 17: 55 m, 83°38′N, 131°16′E; lane 18: 131 m, 70°53′N, 141°50′W; lane 19: 235 m, 70°53′N, 141°51′W; lane 20: 55 m, 72°08′N, 154°16′W; lane 22: 55 m, 75°17′N, 170°11′W; lane 23: 55 m, 75°31′N, 179°35′W; lane 24: 55 m, 79°00′N, 189°37′W; lane 25: 55 m, 78°19′N, 164°55′W; lanes 5, 13, and 21 are standards containing a mixture of Clostridium perfringens and Bacillus thuringiensis genomic DNA (Sigma). The nitAB PCR products shown in lanes 2, 3, 14, and 17 were used to generate the clone libraries. The nitAB PCR product shown in lane 17 was also sequenced directly. Band A is the common band present in all samples.
FIG. 4.
Distribution of samples from SCICEX 95, 96, and 97 cruises according to the number of bands in the DGGE gel. Filled circles represent samples that have one major band, and open circles represent samples that have more than one major band. Arrows indicate samples which were used to generate clone libraries 95A (A), 95B (B), 96A (C), and 96B (D).
Phylogenetic analysis.
Direct analysis of the 1.1-kb nitAB PCR product from the sample used to construct the 96B clone library and which contained only DGGE band A (Fig. 3, lane 17) gave a clean, unambiguous sequence. Phylogenetic analysis revealed that the sequence fell into the β-proteobacterial ammonia oxidizer (Nitrosospira-like group 1) clade. BLAST indicated that the sequence was 99.71% similar (3 bp difference) to a clone (400 FREE Z14) in the clone library from a plankton sample collected at a depth of 400 m in the northwest Mediterranean Sea (29). One clone (96B-3) from the clone library of this sample was sequenced. The insert differed at two positions from the sequence obtained directly from the nitAB product of this sample.
Analysis of 121 clones from the other three clone libraries by PCR-DGGE indicated that inserts of 42 clones (67%) from library 95A, 15 clones (71%) from library 95B, and 25 clones (68%) from library 96A were the same as band A in the original samples (Fig. 5). Sequences from three of these clones (95A-44, 95B-10, and 96A-8) differed by 1 to 2 bp and 95B-4 differed by 4 bp from the direct sequence.
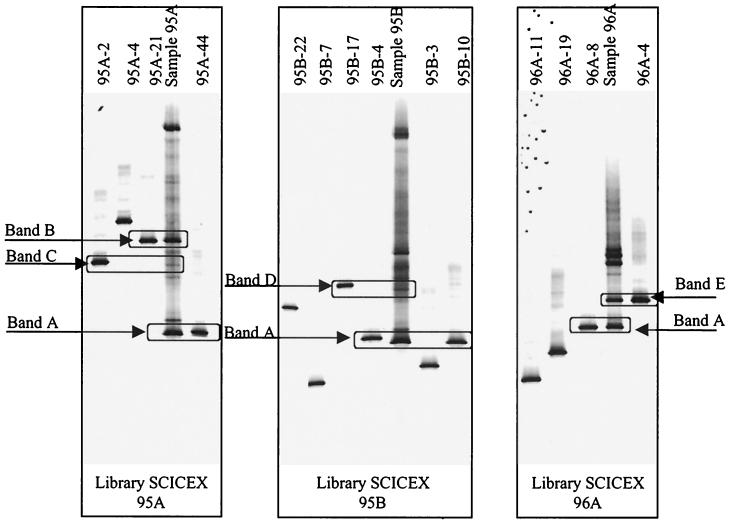
FIG. 5.
Images of DGGE gels comparing bands from clones (which were sequenced) with the band pattern of the PCR product used to generate the clone library. Bands A to E represent the clones with their respective bands in the original samples. Bands of clones 95A-4, 95B-22, 95B-7, 95B-3, 96A-11, and 96A-19 were not detected in the original samples. Clone 95A-21 represents three other clones (95A-13, 95A-14, and 95A-40) that were sequenced, and 96A-4 also represents 96A-17.
Sixteen clones (25%) from library 95A contained inserts that gave DGGE bands with the same mobility as band B (Fig. 5). This was a major band that was present in all 1995 samples that contained more than one band. Four clones (95A-13, 95A-14, 95A-21, and 95A-40) from this library were sequenced. Clones 95A-13 and 95A-14 were found to contain chimeric sequences. Sequences of the other two clones differed from each other by 5 bp and clustered with Nitrosomonas group 5 (Fig. 6).
FIG. 6.
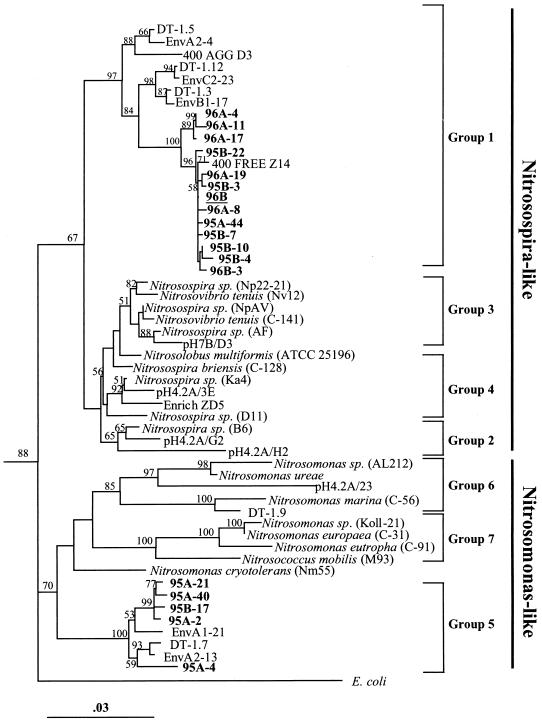
Neighbor-joining tree showing phylogenetic relationship of Arctic Ocean sequences to closely related β-Proteobacteria ammonia oxidizer sequences. The tree was generated by using a 1,040-bp region of the 16S rDNA. Clones from this study are indicated in boldface type, and the direct sequence is underlined. Clusters are numbered according to the method of Stephens et al. (36). Bootstrap values higher than 50% are shown. The tree is unrooted, and E. coli is used as an out group. The bar indicate a Jukes-Cantor distance of 0.03. The accession numbers of the sequences used to make the phylogenetic tree are given in Table 2.
Two clones (3%) from library 95A and one clone (5%) from library 95B gave DGGE bands with the same mobility as bands C and D, respectively (Fig. 5). The sequences of clones 95A-2 (band C) and 95B-17 (band D) both grouped with Nitrosomonas group 5 (Fig. 6). Nitrosomonas-like sequences were only encountered in 1995 samples.
Ten clones (27%) from library 96A (Fig. 5) contained inserts that gave DGGE bands with the same mobility as band E, which was a major band in this sample. The sequences of two representative clones (96A-4 and 96A-17) differed from each other by 1 bp and differed by 8 bp (99.2%) from the direct sequence of band A. Although this band is 99.2% similar to band A, it seems to represent a distinct organism, because 8 bp is a relatively large difference and the clone sequences have the same mobility as a major band E in the sample.
PCR-DGGE screening of the clone libraries revealed that 3 to 5% of the clones were not detected in the original samples (clones 95B-3, 95B-7, 95B-22, 96A-19, 96A-11) (Fig. 5). The sequences of these clones differed by 1 to 3 bp from the direct sequence which gave band A by PCR-DGGE and clustered with Nitrosospira group 1 (Fig. 6). These substitutions occurred within the region amplified by the PCR-DGGE primer set, affecting DGGE mobility of the amplified fragment. While these differences may indicate the presence of rare species in the original sample, it is more likely that they are artifacts of cloning (27). Similarly, we did not detect a DGGE band corresponding to clone 95A-4 in the original sample, but this clone clustered with Nitrosomonas group 5.
DISCUSSION
Amplification of 16S rDNA with specific primers and DGGE have been used successfully in several studies to investigate the diversity of ammonia oxidizers (18, 21, 37). We used a similar approach, employing the nitAB primer set to detect ammonia oxidizers in Arctic Ocean samples and investigating their diversity by DGGE analysis of the nitAB product. The specificity of these primers has been tested by Voytek and Ward (40). They demonstrated that these primers amplify the 16S rDNA of all nine known species of ammonia oxidizers in the β-Proteobacteria. The primers did not amplify the sequences of any closely related, non-ammonia-oxidizing bacteria except Spirillum volutans. These primers were used successfully to amplify DNA collected from a permanently ice-covered Antarctic lake where nitrification was evident but from which no ammonia-oxidizing bacteria have been isolated (40). Ward et al. (45) also used these primers to detect ammonia oxidizers in several lakes in northern Germany. Phylogenetic analysis of the 1.1-kb nucleotide sequences we obtained indicated that they were closely related to ammonia-oxidizing β-Proteobacteria. None of the sequences we obtained had affinity for non-ammonia-oxidizing bacteria, supporting the specificity of the nitAB primer set.
PCR amplification under the conditions we used is at best semiquantitative. It is possible (likely) that some samples which did not yield a PCR product with nitAB contain low concentrations of β-proteobacterial ammonia oxidizer DNA. We did not use the nested PCR approach described by Voytek and Ward (40) in which nonspecific Bacterial primers are used to increase the relative abundance of Bacterial 16S rDNA in samples prior to a secondary amplification with ammonia oxidizer-specific PCR primers. Ward et al. (45) showed that nested PCR yielded several more positive reactions from samples that presumably contain low concentration of ammonium oxidizers.
Although we cannot test the significance of this observation statistically, our results suggest that the ammonia-oxidizing population was vertically stratified, with a maximum in the pycnocline. Direct PCR amplification yielded product most frequently (84%) with samples from 55 to 133 m, suggesting that ammonia oxidizers are widely distributed within this depth range. Ammonia oxidizers were detected less frequently in surface (5 m) and deep (235 m) samples. A strong permanent pycnocline occurs in the Arctic Ocean over this same depth range (30 to 200 m) (32). Organic matter is likely to accumulate at the pycnocline and decompose, releasing ammonium, which may explain the high abundance of ammonia oxidizers in the pycnocline.
The small number of surface samples prevent us from drawing firm conclusions about the apparently low abundance of ammonia oxidizers at the surface. A lower abundance in surface waters might be explained by light inhibition, since light exposure is known to damage ammonia oxidizer cytochromes (26, 42, 44). Alternatively, ammonia oxidizers might be limited by substrate, since ammonia concentrations are lower in the upper mixed layer as a result of uptake by ice algae and phytoplankton. We found fewer ammonia oxidizers at 235 m in the Arctic Ocean. The abundance of ammonia oxidizers may decrease at such a depth because less organic matter is decomposed at these depths, reducing the supply of ammonia available to ammonia oxidizers (50).
PCR coupled with DGGE is a useful tool to obtain a global view of the diversity of the bacteria in a large set of samples (8), particularly when coupled with group-specific primers (18, 22, 46, 47) or sequencing (J. T. Hollibaugh, P. S. Wong, N. Bano, S. K. Pak, and C. Orrego, unpublished data). Our PCR-DGGE analysis showed that all samples positive for ammonia oxidizers contained the same major band, suggesting a broad distribution of this organism throughout the Arctic Ocean. Analysis of this band in four different samples confirmed that it contained the same sequence. However, because this sequence is only 150 bp long, we may not have distinguished closely related bacterial species (20) because discriminating sequence differences could lie outside the PCR-DGGE target region. Sequences of one representative clone from each clone library displaying this band upon PCR-DGGE differed by 1 to 4 bp. Those differences may be due to random errors associated with sequencing or artifacts of cloning (27), since each of these variations was encountered only once, or they may indicate the presence of a group of closely related strains or species. Phylogenetic studies of 16S rDNA sequences revealed that ammonia-oxidizing bacteria in the β-subclass of the proteobacteria are very closely related (11, 38, 39). Recently, Aakra et al. (1) showed that despite the close phylogenetic relationship among the ammonia-oxidizing bacteria, the relative location of the rDNA in the genome appears to vary considerably.
The 1,156-bp sequence obtained directly from the nitAB-amplified PCR product was 99.71% similar to clone 400 FREE (Z14) found in a library generated from free-living bacteria in a sample obtained at a depth of 400 m from the northwest Mediterranean Sea (29). This location is substantially different from the Arctic Ocean, with a higher temperature (13 to 14°C) (5) and lower ammonium concentrations. We found band A corresponding to this sequence in all of the samples we analyzed by PCR-DGGE, suggesting that this organism is ubiquitous and the dominant β-proteobacterial ammonia oxidizer in the Arctic Ocean. Sixty-nine percent of the clones in our libraries contained sequences that were similar (fewer than 4 bp of differences) to this sequence, further supporting the conclusion that it is both ubiquitous and dominant. All of these sequences clustered with marine Nitrosospira-like group 1, which was first detected by Stephen et al. (36) in sediment from the west coast of Scotland. The presence in three different marine environments of this recently detected group suggests that it is widely distributed throughout the ocean; however, it has not been possible to isolate bacteria containing this sequence.
Three to twenty-five percent of our Arctic Ocean clones (bands B, C, and D) from libraries 95A and 95B clustered with the group 5 marine Nitrosomonas-like sequences of Stephen et al. (36). This group also contains only novel Nitrosomonas-like sequences. This group contains sequences that were cloned from a sample of sediment receiving high organic matter loading from a salmon rearing pen (22, 36). Nitrosomonas-like sequences were found in samples from the region of the Arctic Ocean influenced by inflow from the Pacific Ocean. This flow crosses the shallow and productive Bering and Chukchi seas before entering the Arctic Ocean basin and carries a distinctive signal of high nutrients, including regenerated ammonia (33; T. E. Whitledge, personal communication). The Nitrosomonas-like organism may require higher ammonia concentrations than the Nitrosospira-like organism, it may be associated with particles which are more abundant on the shelf, or it may be a sediment organism that is present as a result of resuspension.
The hypothesis that its occurrence is a consequence of higher substrate concentrations is supported by studies indicating that higher ammonia concentrations, such as those found in enrichment cultures, favor the growth of Nitrosomonas sp. while low ammonia concentrations favor the growth of Nitrosospira sp. (12, 30). Phillips et al. (29) found that particle-associated assemblages were dominated by Nitrosomonas-like sequences while Nitrosospira-like sequences were found in the free-living bacterial fraction. Alldredge and Silver (2), Kaltenbock and Herndl (17), and others have demonstrated elevated nutrient concentrations associated with particles of marine snow, and there is abundant evidence that these particles harbor distinct microbial assemblages adapted to conditions (low oxygen, high substrate concentration, the presence of surfaces, etc.) found in particle microenvironments (6, 31, 48). Distinguishing between these hypotheses will require further studies.
ACKNOWLEDGMENTS
We express our appreciation to the officers and crew of the U.S. Navy submarines Cavalla, Pogy, and Archerfish; to Arctic Submarine Laboratory personnel; and to the scientists who collected samples for us on the SCICEX 95, 96, and 97 cruises. These studies would not have been possible without their willing collaboration. Mary Ann Moran, Mandy Joye, and Francoise Lucas, and two anonymous reviewers provided useful comments and other support during the preparation of the manuscript. Feng Chen gave us the Nitrosomonas cryotolerans isolate that was used as a positive control for the PCR amplification. Kitty Williams, Briana Ransom, and Ryan Hollibaugh helped with sample analysis.
This work was supported by NSF awards OPP 95-00237, OPP 96-25131 (reissued as OPP 97-96261), and OPP 98-09971 to J.T.H.
REFERENCES
- 1.Aakra Å, Utåker J B, Nes I F. RFLP of rRNA genes and sequencing of the 16S-23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int J Syst Bacteriol. 1999;47:661–669. doi: 10.1099/00207713-49-1-123. [DOI] [PubMed] [Google Scholar]
- 2.Alldredge A L, Silver M W. Characteristics, dynamics and significance of marine snow. Prog Oceanogr. 1988;20:41–82. [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Barry T, Powell R, Gannon R. A general method to generate DNA probes for microorganisms. Bio/Technology. 1990;8:233–236. doi: 10.1038/nbt0390-233. [DOI] [PubMed] [Google Scholar]
- 5.Conan P, Milton C. Variability of the northern current of Marseilles, western Mediterranean Sea from February to June 1992. Oceanolog Acta. 1995;18:193–205. [Google Scholar]
- 6.Delong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 7.Felsenstein J. PHYLIP: Phylogeny Inference Package (version 3.5). Seattle: University of Washington; 1993. [Google Scholar]
- 8.Ferrari V C, Hollibaugh J T. Distribution of microbial assemblages in the Central Arctic Ocean basin studied by PCR/DGGE: analysis of a large data set. Hydrobiologia. 1999;401:55–68. [Google Scholar]
- 9.Hastings R C, Ceccherini M T, Nerino M, Saunders J R, Bazzicalupo M, McCarthy A J. Direct and biological analysis of ammonia oxidizing bacteria populations in cultivated soil plots treated with swine manure. FEMS Microbiol Ecol. 1997;23:45–54. [Google Scholar]
- 10.Hastings R C, Saunders J R, Hall G H, Pickup R W, McCarthy A J. Application of molecular biological techniques to a seasonal study of ammonia oxidation in a eutrophic freshwater lake. Appl Environ Microbiol. 1998;64:3674–3682. doi: 10.1128/aem.64.10.3674-3682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 12.Hiorns W D, Hastings R C, Head I M, McCarthy A J, Saunders J R, Pickup R W, Hall G H. Amplification of 16S ribosomal RNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of Nitrosospiras in the environment. Microbiology. 1995;141:2793–2800. doi: 10.1099/13500872-141-11-2793. [DOI] [PubMed] [Google Scholar]
- 13.Horrigan S G. Primary production under the Ross ice shelf, Antarctica. Limnol Oceanogr. 1981;26:378–382. [Google Scholar]
- 14.Hovanec T A, Delong E F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones R D, Morita R Y. Low-temperature growth and whole-cell kinetics of a marine ammonium oxidizer. Mar Ecol Prog Ser. 1985;21:239–243. [Google Scholar]
- 16.Jones R D, Morita R Y, Koops H-P, Watson S W. A new marine ammonium-oxidizing bacterium, Nitrosomonas cryotolerans sp. Can J Microbiol. 1988;34:1122–1128. [Google Scholar]
- 17.Kaltenbock E, Herndl G. Ecology of amorphous aggregates (marine snow) in the Northern Adriatic Sea. IV. Dissolved nutrients and the autotrophic community associated with marine snow. Mar Ecol Prog Ser. 1992;87:147–159. [Google Scholar]
- 18.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalchuk G A, Naoumenko Z S, Derikx P J L, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig W, Strunk O, Klugbauer S, Klubauer N, Weizenegger M, Neumaler J, Bachleitner M, Schleifer K H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 21.McCaig A E, Embley T M, Prosser J I. Molecular analysis of enrichment cultures of marine ammonia-oxidizers. FEMS Microbiol Lett. 1994;120:363–368. doi: 10.1111/j.1574-6968.1994.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 22.McCaig A E, Phillips C J, Stephen J R, Kowalchuk G A, Harvey S M, Herbert R A, Embley T M, Prosser J I. Nitrogen cycling and community structure of proteobacterial β subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl Environ Microbiol. 1999;65:213–220. doi: 10.1128/aem.65.1.213-220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers R M, Fischer S G, Lerman L S, Maniatis T. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1985;13:3131–3145. doi: 10.1093/nar/13.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson R J. Differential photoinhibition of marine nitrifying bacteria: a possible mechanism for the formation of the primary nitrate maximum. J Mar Res. 1981;39:227–238. [Google Scholar]
- 27.Pääbo S, Wilson A C. Polymerase chain reaction revealing cloning artifacts. Nature. 1988;334:387–388. doi: 10.1038/334387b0. [DOI] [PubMed] [Google Scholar]
- 28.Paul J H, Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl Environ Microbiol. 1982;43:1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips C J, Smith Z, Embley T M, Prosser J I. Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl Environ Microbiol. 1999;65:779–786. doi: 10.1128/aem.65.2.779-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prosser J I. Autotrophic nitrification in bacteria. Adv Microbiol Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 31.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine-snow associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 32.Rudels B, Jones E P, Anderson L G, Kattner G. On the intermediate depth waters of the Arctic Ocean. In: Johannessen O M, Muench R D, Overland J E, editors. The polar oceans and their role in shaping the global environment, Geophysical Union monograph 85. Washington, D.C.: American Geophysical Union; 1994. pp. 33–46. [Google Scholar]
- 33.Salmon D K, McRoy C P. Nutrient-based tracers in the western Arctic: a new lower holocline water defined. In: Johannessen O M, Muench R D, Overland J E, editors. The polar oceans and their role in shaping the global environment, Geophysical Union monograph 85. Washington, D.C.: American Geophysical Union; 1994. pp. 47–61. [Google Scholar]
- 34.Shiah F K, Ducklow H W. Temperature regulation of heterotrophic bacterioplankton abundance, production, and specific growth-rates in Chesapeake bay. Limnol Oceanogr. 1994;39:1243–1258. [Google Scholar]
- 35.Shiah F K, Ducklow H W. Bacterioplankton growth responses to temperature and chlorophyll variations in estuaries measured by thymidine:leucine incorporation ratio. Aquat Microb Ecol. 1997;13:151–159. [Google Scholar]
- 36.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephen J R, Kowalchuk G A, Bruns M-A V, McCaig A E, Phillips C J, Embley T M, Prosser J I. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol. 1998;64:2958–2965. doi: 10.1128/aem.64.8.2958-2965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teske A, Alm E, Regan J M, Toze S, Rittmann B E, Stahl D A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Utåker J B, Bakken L, Jiang Q Q, Nes I F. Phylogenetic analysis of seven new isolates of ammonia-oxidizing bacteria based on 16S rRNA sequences. Syst Appl Microbiol. 1995;18:549–559. [Google Scholar]
- 40.Voytek M A, Ward B B. Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol. 1995;61:1444–1450. doi: 10.1128/aem.61.4.1444-1450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voytek M A, Priscu J C, Ward B B. The distribution and relative abundance of ammonia-oxidizing bacteria in lakes of the McMurdo Dry Valley, Antarctica. Hydrobiologia. 1999;401:113–130. [Google Scholar]
- 42.Ward B B, Olson R J, Perry M J. Microbial transformation rates in the primary nitrite maximum off Southern California. Deep-Sea Res. 1982;29:247–255. [Google Scholar]
- 43.Ward B B, Carlucci A F. Marine ammonia-and nitrite-oxidizing bacteria: Serological diversity determined by immunofluorescence in culture and in the environment. Appl Environ Microbiol. 1985;50:194–201. doi: 10.1128/aem.50.2.194-201.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward B B. Nitrification and denitrification: probing the nitrogen cycle in aquatic environments. Microb Ecol. 1996;32:247–261. doi: 10.1007/BF00183061. [DOI] [PubMed] [Google Scholar]
- 45.Ward B B, Voytek M A, Witzel K-P. Phylogenetic diversity of natural populations of ammonia oxidizers investigated by specific PCR amplification. Microb Ecol. 1997;33:87–96. doi: 10.1007/s002489900011. [DOI] [PubMed] [Google Scholar]
- 46.Wawer C, Muyzer G. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gradient gel electrophoresis of [NiFe] hydrogenase gene fragments. Appl Environ Microbiol. 1995;61:2203–2210. doi: 10.1128/aem.61.6.2203-2210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wawer C, Jetten S M, Muyzer G. Genetic diversity and expression of the [NiFe] hydrogenase large-subunit gene of Desulfovibrio spp. in environmental samples. Appl Environ Microbiol. 1997;63:4360–4369. doi: 10.1128/aem.63.11.4360-4369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss P, Schweitzer B, Amann R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheeler P A, Gosselin M, Sherr E, Thibault D, Kirchman D L, Benner R, Whitledge T E. Active cycling of organic carbon in the central Arctic Ocean. Nature. 1996;380:697–699. [Google Scholar]
- 50.Wheeler P A, Watkins J M, Hansing R L. Nutrients, organic carbon and organic nitrogen in the upper water column of the Arctic Ocean: implications for the sources of dissolved organic carbon. Deep-Sea Res II. 1997;44:1571–1592. [Google Scholar]
- 51.Whitby C B, Saunders J R, Rodriguez J, Pickup R W, McCarthy A. Phylogenetic differentiation of two closely related Nitrosomonas spp. that inhabit different sediment environments in an oligotrophic freshwater lake. Appl Environ Microbiol. 1999;65:4855–4862. doi: 10.1128/aem.65.11.4855-4862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woese C R, Weisburg W G, Paster B J, Hahn C M, Tanner R S, Kreig N R, Koops H-P, Harms H, Stackebrandt E. The phylogeny of purple bacteria: the beta subdivision. Syst Appl Microbiol. 1984;5:327–336. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- 53.Woese C R, Weisburg W G, Hahn C M, Paster B J, Zablen L B, Lewis B J, Macke T J, Ludwig W, Stackebrandt E. The phylogeny of purple bacteria: the gamma subdivision. Syst Appl Microbiol. 1985;6:25–33. [Google Scholar]