Abstract
Reduction of aerobic fermentation on sugars by altering the fermentative/oxidative balance is of significant interest for optimization of industrial production of Saccharomyces cerevisiae. Glucose control of oxidative metabolism in baker's yeast is partly mediated through transcriptional regulation of the Hap4p subunit of the Hap2/3/4/5p transcriptional activator complex. To alleviate glucose repression of oxidative metabolism, we constructed a yeast strain with constitutively elevated levels of Hap4p. Genetic analysis of expression levels of glucose-repressed genes and analysis of respiratory capacity showed that Hap4p overexpression (partly) relieves glucose repression of respiration. Analysis of the physiological properties of the Hap4p overproducer in batch cultures in fermentors (aerobic, glucose excess) has shown that the metabolism of this strain is more oxidative than in the wild-type strain, resulting in a significant reduced ethanol production and improvement of growth rate and a 40% gain in biomass yield. Our results show that modification of one or more transcriptional regulators can be a powerful and a widely applicable tool for redirection of metabolic fluxes in microorganisms.
Modification and control of metabolic fluxes is an important goal in most industrial applications of microorganisms. This is the case for baker's yeast, which has applications in production of heterologous proteins, brewing, and baking. Maximization of biomass yields of baker's yeast growing on sugars is hampered by the cell's strong tendency to produce ethanol, even under aerobic conditions when sugars are present in excess. This alcoholic fermentation might be prevented by manipulation of the carbon flux distribution between fermentation and respiration.
Attempts at redirection of carbon fluxes by interference in expression levels of single enzymes of glycolytic or fermentative pathways have so far not been successful (24, 25). Reduced activities of fermentative enzymes such as pyruvate decarboxylase result in impaired growth on glucose (8, 9) and deprive the cells of their fermentative capacity necessary for raising of dough. Increasing respiratory activity seems to be a better approach, but numerous studies have shown that overproduction of a single enzyme results in either little or no increase of flux through a metabolic pathway (20). This is because flux control is usually not exerted by only a single enzyme (2, 20, 28). A more rational approach would therefore be to manipulate the activity of a regulatory protein involved in control of all key enzymes of one or more specific metabolic pathways. The potential of this approach is illustrated by the partial alleviation of glucose control of sucrose and galactose metabolism as a result of disruption of the glucose repressor Mig1p alone or in combination with Mig2p (16, 17). These modifications do not, however, result in a significant change in respiratory functions, ethanol production, or biomass yield.
Aiming at redirection of fermentative flux toward oxidative carbon flux, we focused on the Hap4p activator subunit of the transcriptional complex involved in carbon source-dependent regulation of respiratory function (3, 6, 11). Transcription of this subunit is glucose repressible (11), suggesting that Hap4p is the key component of the complex in terms of its control of transcriptional activity in response to carbon source. This study shows that raising the expression level of Hap4p on glucose results in a partial alleviation of glucose repression of respiratory genes and function. The resulting significant changes in carbon metabolism, growth rate, and biomass yield demonstrate the pivotal role of the transcriptional regulator Hap4p in control of respiro-fermentative metabolism in S. cerevisiae.
MATERIALS AND METHODS
Overexpression of HAP4 in Saccharomyces cerevisiae strain DL1.
A 1.7-kb fragment containing the coding region of HAP4 was isolated from pSLF406 (11) by digestion with BspHI (which cleaves HAP4 at position −1 relative to the start codon the coding sequence), blunting, and subsequent digestion with PstI. YCplac111::ADH1 (which was derived from Ycplac111 [12] and contains a 723-bp EcoRV ADH1 promoter fragment from pBPH1 [28]) was cleaved with SmaI and PstI and ligated with the HAP4 fragment to generate YCplac111::ADH1-HAP4. Yeast strain DL1 (30) was transformed by the lithium acetate method (14). Transformants were selected on plates containing 2% glucose, 2% agar, and 0.67% yeast nitrogen base (Difco) supplied with the required amino acids but lacking leucine.
Growth of yeast in flask-batch cultures.
For shake flask cultivation, yeast cells were grown in either YPD (1% yeast extract, 1% Bacto Peptone, 3% d-glucose), YPEG (yeast extract, 1% Bacto Peptone, 2% ethanol, 2% glycerol), YPL [lactate medium; 1.5% lactic acid, 2% sodium lactate, 0.1% glucose, 8 mM MgSO4, 45 mM (NH)2HPO4, 0.5% yeast extract], or selective mineral medium (0.67% yeast nitrogen base) containing 3% d-glucose.
Growth of yeast in fermentor-batch cultures.
Transformed yeast cells were grown in selective mineral Evans medium (7) containing 30 g of d-glucose per liter and supplemented with filter-sterilized 40 mg each of uracil and l-histidine per liter. Instead of citrate, nitrilotriacetic acid (2 mM) was used as a chelator, and silicone antifoaming agent (1 ml/20 liters) was added to the medium. After heat sterilization of the medium at 120°C (glucose sterilized separately at 110°C), filter-sterilized vitamins were added to final concentrations per liter as follows: myoinositol, 0.55 mM; nicotinic acid, 0.16 mM; Ca-d-(+)-panthothenate, 0.02 mM; pyridoxine-HCl, 0.013 mM; thiamine-HCl, 0.006 mM; biotin, 0.02 μM. Cultivation was performed at 28°C in New Brunswick Scientific Bioflow fermentors, at a stirrer speed of 900 rpm. The pH was kept constant at 5.0 via the automatic addition of 2 mol of NaOH per liter. Antifoam (BDH) was added automatically at fixed time intervals. An airflow of 0.8 liter min−1 maintained the dissolved oxygen tension above 40% of air saturation. The starting working volume of the cultures was 1.0 or 1.4 liters. Samples of 30 ml were taken every hour for analysis of culture purity, optical cell density (OD; spectrophotometrically at 600 nm), substrate and product concentrations, and dry biomass weight (by centrifugation of 10.0 ml of culture, washing cells with demineralized H2O, and drying the cell pellet overnight at 80°C; parallel samples varied by less than 1%). The dissolved carbon dioxide concentration was continuously monitored by a Servomex IR PA404 gas analyzer and oxygen was measured by a Taylor Servomex OA 272 gas analyzer. The absolute amounts of gas consumption/production during the time course of the experiment were calculated by the mean of the gas concentration, corrected for the decreasing volume of the culture. Specific consumption/production rates of metabolites (q; millimoles consumed or produced per gram of dry yeast biomass per hour) were calculated from the slope of a plot of metabolite concentration versus dry biomass concentration, amplified by the specific growth rate. The correlation of metabolite concentration versus dry biomass concentration was linear for all metabolites during the independent experiments.
Substrate consumption and product formation in liquid medium.
Concentrations of extracellular carbon compounds were determined by high-pressure liquid chromatography analysis using an Aminex HPX87H organic acids column (Bio-Rad) at 65°C. The column was eluted with 5 mM H2SO4. Detection was by means of a 2142 refractive index detector (LKB) and SP4270 integrator (SpectraPhysics).
Analysis of O2 consumption.
For oxygen consumption capacity measurements of flask-batch-grown cells, the cells were harvested, washed three times with ice-cold demineralized H2O, and resuspended in oxygraph buffer [1% yeast extract, 0.1% KH2PO4, 0.12% (NH4)2SO4 (pH 4.5)] at 200 OD units ml−1. Oxygen consumption capacity of the cells was measured with a Clark-type oxygen electrode, with 0.1 mM ethanol as substrate. Rates were determined from the slope of a plot of O2 concentration versus time.
RNA isolation, Northern analysis, and labeling of DNA fragments.
Cells were harvested, and RNA was isolated, separated, on a nondenaturing 1.2% agarose gel, and transferred to a nitrocellulose filter as described previously (5). Prehybridization was performed in hybridization buffer (50% formamide, 25 mM NaPi [pH 6.5], 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt's solution) containing 50 μg of single-stranded salmon sperm DNA per ml. DNA fragments used as probes in this study include an 840-bp HindIII-SalI fragment from pJH1 (5), a 1.6-kb BamHI-KpnI fragment containing the yeast actin gene (19), a 2.5-kb HindIII-SalI fragment from YE23SH containing the QCR2 gene (22), a 1,333-bp NcoI-HindIII fragment from pAZ6 containing the yeast PDA1 gene (32), and a 1.2-kb BamHI-HindIII fragment from YE23R-SOD/SUC containing the SUC2 gene (31). Fragments were 32P labeled by nick translation as described by Maniatis et al. (18). Labeled probes were added to the prehybridization buffer, and hybridization was performed overnight at 42°C. Blots were washed once with 2× SSC–0.1% sodium dodecyl sulfate (SDS), twice with 1× SSC–0.1% SDS, and finally with 0.5× SSC–0.1% SDS. Blots were air dried completely, and autoradiography was performed with Kodak X-Omat 100 film or analyzed by a Storm 840 Phosphorimager (Molecular Dynamics).
RESULTS
Hap4p overexpression on glucose results in derepression of respiratory chain components.
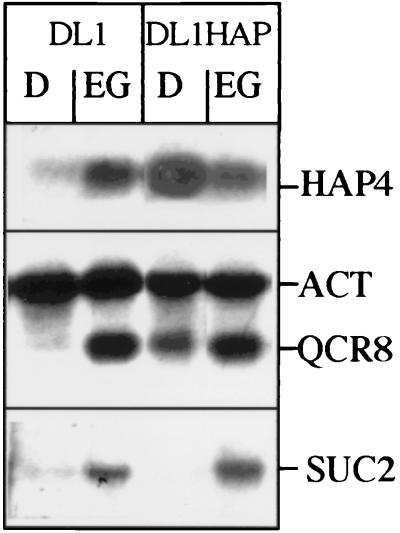
To elevate the repressed endogenous level of Hap4p in S. cerevisiae growing on glucose, we introduced a centromeric plasmid containing the coding region of HAP4 under control of the glucose-inducible ADH1 promoter in a laboratory S. cerevisiae strain. As a reference to this Hap4p overproducer (DL1HAP), a wild-type strain was transformed with the same plasmid but lacking the HAP4 gene (DL1). The expression level of HAP4 mRNA in both strains under fermentative and nonfermentative growth conditions is depicted in Fig. 1. Expression of HAP4 in wild-type cells is strongly repressed by glucose. Introduction of the plasmid with HAP4 under control of the ADH1 promoter leads to an approximately 10-fold increased expression level of HAP4 in DL1HAP which is grown on medium containing 2% glucose. This level is comparable to the expression level of HAP4 in wild-type cells when grown on medium containing ethanol-glycerol, which induces transcription of HAP4 about ninefold (Fig. 1 and reference 4).
FIG. 1.
Expression of several mRNAs in a Hap4p overexpression strain. DL1 and DL1HAP were grown to mid-logarithmic phase in medium containing 2% glucose (D) or 2% ethanol–2% glycerol (EG); 20 μg of total RNA was hybridized with probes specific for HAP4, ACT (actin), QCR8, or SUC2 mRNA.
To study the effect of HAP4 overexpression on transcriptional control of respiratory function, we first studied the mRNA levels of different genes encoding components of the respiratory chain. As shown in Fig. 1, the elevated level of Hap4p in glucose-containing medium leads to derepression of transcription of QCR8, the gene encoding the 11-kDa subunit of the yeast ubiquinol-cytochrome c oxidoreductase (QCR) complex of the respiratory chain. Hap4p overexpression does not result in full induction levels of QCR8 as found on nonfermentable carbon sources such as ethanol-glycerol, which is probably due to the involvement of other factors in transcriptional control of QCR8. Comparable results were obtained for a number of other genes encoding respiratory components, such as QCR2, QCR7, and CYC1 (not shown). The sustained glucose repression of SUC2, without a Hap2/3/4/5p binding box in the promoter region, shows that Hap4p overproduction does not alleviate glucose repression in general.
Respiratory capacity is increased in Hap4p overproducer.
To test whether the increased level of mRNAs of respiratory components results in an higher respiratory capacity of the Hap4p-overproducing strain, we measured oxygen consumption rates of cells grown to mid-logarithmic phase in shake flask cultures. Respiratory capacity of DL1HAP cells collected from media containing glucose was nearly twofold greater than that of wild-type cells (18.1 versus 9.4 nmol of glucose produced/min/mg [dry weight] of yeast biomass). When Hap4p-producing strains were grown in the presence of the nonfermentable carbon source lactate, the respiratory capacity is further increased approximately fivefold, to a level similar to that for the wild-type strain grown on lactate (86.7 versus 88.1 nmol/min/mg of yeast biomass). This indicates that an elevated level of Hap4p only partially relieves repression of respiratory function, which is in agreement with the partially derepressed transcript levels of respiratory genes.
In vivo carbon catabolism is shifted from fermentation toward respiration.
To follow a possible change from fermentative toward a more oxidative catabolism during growth on excess glucose, we further characterized the physiological properties of the Hap4p-overproducing strain. Since cultivation in shake flask cultures is subject to oxygen limitations and variable pH conditions that can influence carbon metabolism, cells were grown under controlled conditions in fermentors (constant pH, aeration, and stirring) in well-defined mineral media. Aerobic growth of the wild-type strain carrying the empty plasmid was compared with growth of DL1HAP cells in a defined mineral salts medium containing glucose (30 g/liter [3%]). As calculated from three independent growth curves (one example shown in Fig. 2), the wild type grew exponentially with a specific growth rate of 0.17 ± 0.01 h−1, whereas the growth rate of DL1HAP was 0.20 ± 0.01 h−1. Overproduction of Hap4p thus results in an increased growth rate of approximately 17%, which was also observed during early logarithmic flask-batch growth (data not shown).
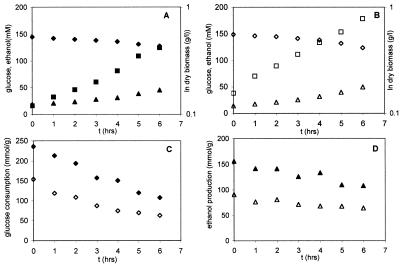
FIG. 2.
Growth characteristics and glucose consumption and ethanol production profile in fermentor batch cultivations. Shown are dry yeast biomass formation (□, ■) and glucose (◊, ⧫) and ethanol (▵, ▴) concentrations present in the culture supernatant of the wild-type strain DL1 (A) and the Hap4p overproducer DL1HAP (B) and glucose consumption (C) and ethanol production (D) relative to the amount of dry yeast biomass formed in wild-type DL1 (closed symbols) and DL1HAP (open symbols) strains.
During a 6-h period of exponential growth, samples were taken at hourly intervals to measure substrate consumption and biomass and product formation. Exponential growth (biomass formation) of both the wild-type (Fig. 2A) and Hap4p-overproducing (Fig. 2B) strain are shown, along with the concentrations of residual glucose and produced ethanol present in the culture supernatant. Although from this figure only the difference in growth between the two strains seems apparent, the difference in carbon catabolism becomes evident when the data are presented relative to the amount of biomass formed (Fig. 2C and D). Calculation of the specific ethanol production and glucose consumption rates (Table 1) showed that Hap4p overexpression results in a significant reduction in the specific glucose consumption rate (qglucose; 15% reduction) and specific ethanol production rate (qethanol; 25% reduction). As a consequence, the biomass yield, i.e., the amount of biomass formed per 100 g of consumed glucose, is 39% higher in the Hap4p overproducer: 9.9 ± 0.4 g for DL1 and 13.7 ± 0.4 g for DL1HAP.
Analysis of production rates of other carbon compounds present in the fermentor effluent (Table 1) showed that the specific glycerol production rate is significantly decreased in the Hap4p overproducer (27% of the wild-type level), whereas the amount of produced acetate is 2.2-fold increased in DL1HAP cells. The change in product pattern of these compounds is directly attributable to an increased ability to reoxidize redox equivalents via respiration. In accordance with this, the carbon flux through the trichloroacetic acid cycle was increased approximately twofold as calculated directly from either the oxygen consumption rate or from the total CO2 production rate corrected for ethanol production. The respiratory coefficient (qCO2/qO2 ratio) of the DL1HAP strain is hence significantly lower than that of the wild-type strain, which is also indicative for a less fermentative catabolism of the Hap4p-overproducing strain. All data are thus consistent with a shift of carbon metabolism from fermentative toward oxidative metabolism due to an increased expression level of HAP4.
DISCUSSION
Interference in molecular regulatory circuits can be used as a powerful tool for redirection of metabolic fluxes in order to optimize industrial use of microorganisms. We have exemplified this by redirection of the fermentative-oxidative carbon balance in baker's yeast in order to reduce aerobic alcoholic fermentation triggered by the presence of glucose. This has been achieved by raising the expression level of Hap4p, the major regulatory protein of the Hap2/3/4/5p complex required for transcriptional induction of respiratory components. Although previously it has been suggested that regulation of the transcript level of HAP4 would be the main determinant for the activity of the Hap complex (3, 11), we now present the first experimental evidence for this hypothesis. Our results also show that the DNA binding factors of the complex, Hap2p, Hap3p, and Hap5p, are constitutively present at sufficient levels and that posttranslational modifications do not play an important role in glucose control of the Hap complex, in contrast to other transcriptional regulators like Cat8p (23) and Mig1p (21). Hence, an artificially elevated expression level of Hap4p is sufficient to increase mRNA levels of respiratory genes on glucose. This resulted in an increased respiratory capacity and in vivo carbon flux through the tricarboxylic acid cycle and respiration. The alleviation of repression of respiration and redirection of respiro-fermentative carbon flux is only partial, which can be explained by the control of other factors and mechanisms on regulation of oxidative function. Nevertheless, this partial effect results in a significant gain in specific growth rate and biomass yield during growth on an excess of glucose due to the large difference in the energetic efficiency between respiration and fermentation.
When grown under anaerobic conditions, Hap4p-overexpressing strains are identical to wild-type cells with respect to growth rate, ethanol production, and biomass yield (data not shown). This implies that overexpression of HAP4 exhibits its effect only during aerobic growth of yeast. Processes depending on anaerobic alcoholic fermentation, like brewing or dough leavening, will be unaffected by HAP4 overexpression. HAP4-modified strains should therefore be suited to optimize biomass yields in the aerobic production phase of industrial yeast strains. Experiments are in progress to test whether HAP4 overexpression has an effect on the rapid triggering of alcoholic fermentation, which occurs during local and transient exposure to an excess of glucose due to imperfect mixing in sugar-limited, aerobic industrial fed-batch cultivations.
The profound effect of Hap4p overproduction on cell physiology during growth on excess of glucose suggest a broad spectrum of effects on different functional groups of genes involved in carbon and energy metabolism. A switch from fermentative to respiratory growth, which normally occurs during the diauxic shift upon depletion of glucose, is correlated with widespread changes in the expression of genes involved in fundamental cellular processes such as carbon metabolism, mitochondrial assembly, stress response, protein synthesis, and carbohydrate storage (4, 15). Although many genes contain Hap2/3/4/5p consensus binding sites in their promoter regions (10), only few have been reported to be regulated by the HAP complex (3, 6). We are currently performing genome-wide transcript profiling studies that will reveal the total spectrum of gene families that are directly or indirectly affected by Hap4p overexpression. Insight into gene families that are affected or unaffected by Hap4p overproduction may provide clues toward modification of other regulatory proteins. The use of transcript-profiling analysis techniques (1, 26) will be invaluable in the exploration of regulators of metabolic pathways that should be modified for biotechnological applications.
We have presented a clear example of how modification of a transcriptional regulator may serve as a powerful tool for manipulation of the cell's physiology and as an attractive alternative to the unsuccessful alteration of expression levels of single enzymes. It should, however, be realized that overexpression of transcriptional activators can have a deleterious effect on growth which is attributable to squelching of general transcription factors, as observed for GAL4 (13) and GCN4 (27), but also in studies with GAL4-HAP4 fusions on 2μm plasmids (J. Stebbins and S. Triezenberg, Abstr. Yeast Genet. Mol. Biol. Meet., abstr. 517, p. 307, 1998). Undesired pleiotropic effects can also occur in case of interference in factors with a central role high in signaling cascades, such as the general repressor complex Tup1/Ssn6 or the Snf1/Snf4 kinase with a central role in the glucose response (reviewed in reference 15). Our study involving a modest change of a family-specific transcriptional regulator is one of the first successful and promising examples of genetic engineering of metabolic fluxes in yeast.
TABLE 1.
Effect of HAP4 overexpression on carbon flux distributiona
| q | Fluxa
|
|
|---|---|---|
| DL1 | DL1HAP | |
| qglucose | −9.84 | −8.08 |
| qethanol | 14.49 | 10.84 |
| qglycerol | 2.31 | 0.62 |
| qacetate | 0.28 | 0.64 |
| qCO2 | 17.4 | 17.4 |
| qO2 | 3.3 | 6.9 |
Determined as described in Materials and Methods; calculated from the linear correlations of metabolite concentration versus dry biomass formed during 6 h of exponential growth in a fermentor-batch cultivation, amplified by the specific growth rate. Data are presented as the average of calculations based on duplicate metabolite and biomass determinations from two independent experiments. Data varied by less than 5%.
ACKNOWLEDGMENTS
We thank Frank Lubbers for excellent technical assistance and Jack Pronk (Technical University of Delft) and André Boorsma for helpful suggestions and comments on the experiments and the manuscript.
This work was supported by the Dutch Association for Biotechnological Research Schools (ABON) and is currently part of a Dutch Economical and Ecological Technologies (EET) project (EET-97.018).
REFERENCES
- 1.Brown P O, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 2.Cornish-Bowden A, Hofmeyr J H. Determination of control coefficients in intact metabolic systems. Biochem J. 1995;298:367–375. doi: 10.1042/bj2980367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang V D, Valens M, Bolotin-Fukuhara M, Daignan-Fornier B. A genetic screen to isolate genes regulated by the yeast CCAAT-box binding protein hap2p. Yeast. 1994;10:1273–1283. doi: 10.1002/yea.320101004. [DOI] [PubMed] [Google Scholar]
- 4.DeRisi J L, Vishwanath R I, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 5.De Winde J H, Grivell L A. Global regulation of mitochondrial biogenesis in yeast: ABF1 and CPF1 play opposite roles in regulating expression of the QCR8 gene, encoding subunit VIII of the mitochondrial ubiquinol cytochrome c oxidoreductase. Mol Cell Biol. 1992;12:2872–2883. doi: 10.1128/mcb.12.6.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Winde J H, Grivell L A. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog Nucleic Acids Res Mol Biol. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]
- 7.Evans C G T, Herbert D, Tempets D W. The continuous cultivation of micro-organisms. II. Construction of a chemostat. Methods Microbiol. 1970;2:277–327. [Google Scholar]
- 8.Flikweert M T, de Swaaf M, van Dijken J P, Pronk J T. Growth requirements of pyruvate-decarboxylase-negative Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;174:73–79. doi: 10.1111/j.1574-6968.1999.tb13551.x. [DOI] [PubMed] [Google Scholar]
- 9.Flikweert M T, Vanderzanden L, Janssen W M T M, Steensma H Y, Vandijken J P, Pronk J T. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C247::AID-YEA911%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Fondrat C, Kalogeropoulos A. Approaching the function of new genes by detection of their potential upstream activation sequences in Saccharomyces cerevisiae: application to chromosome III. Comput Appl Biosci. 1996;12:363–374. doi: 10.1093/bioinformatics/12.5.363. [DOI] [PubMed] [Google Scholar]
- 11.Forsburg S L, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Sugino A. New-yeast Escherichia coli shuttle vectors constructed with in vitro mutagenized genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 13.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 14.Ito H, Fukuda Y, Murata M, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J R, Broach J R, editors; Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. 2. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 16.Klein C J L, Olsson L, Ronnow B, Mikkelsen J D, Nielsen J. Alleviation of glucose repression of maltose metabolism by MIG1 disruption in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62:4441–4449. doi: 10.1128/aem.62.12.4441-4449.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein C J L, Rasmussen J J, Ronnow B, Olsson L, Nielsen J. Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J Biotechnol. 1999;68:197–212. doi: 10.1016/s0168-1656(98)00205-3. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 19.Ng R, Abelson J. Isolation and sequence of the gene for actin in S. cerevisiae. Proc Natl Acad Sci USA. 1980;77:3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederberger P, Prasad R, Miozzari G, Kacser H. A strategy for increasing an in vivo flux by genetic manipulations. Biochem J. 1992;287:473–479. doi: 10.1042/bj2870473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostling J, Ronne H. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 22.Oudshoorn P, van Steeg H, Swinkels B W, Schoppink P, Grivell L A. Subunit II of yeast QH2:cytochrome-c oxidoreductase: nucleotide sequence of the gene and features of the protein. Eur J Biochem. 1987;163:97–103. doi: 10.1111/j.1432-1033.1987.tb10741.x. [DOI] [PubMed] [Google Scholar]
- 23.Randez-Gil F, Bojunga N, Proft M, Entian K D. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol Cell Biol. 1997;17:2502–2510. doi: 10.1128/mcb.17.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozenzweig R F. Regulation of fitness in yeast overexpressing glycolytic enzymes. Genet Res. 1992;59:35–48. doi: 10.1017/s0016672300030159. [DOI] [PubMed] [Google Scholar]
- 25.Schaaff I J, Heinisch J, Zimmermann F K. Overproduction of glycolytic enzymes in yeast. Yeast. 1989;5:285–290. doi: 10.1002/yea.320050408. [DOI] [PubMed] [Google Scholar]
- 26.Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 27.Tavernarakis N, Thireos G. Transcriptional interference caused by GCN4 overexpression reveals multiple interactions mediating transcriptional activation. Mol Gen Genet. 1995;247:571–578. doi: 10.1007/BF00290348. [DOI] [PubMed] [Google Scholar]
- 28.van Heeswijk W C, Bakker B M, Teusink B, Kholodenko B N, Somsen O J, Snoep J L, Westerhoff H V. Live control of the living cell. Biochem Soc Trans. 1999;27:261–264. doi: 10.1042/bst0270261. [DOI] [PubMed] [Google Scholar]
- 29.van Loon A P G M, Brandli A W, Pesold-Hurt B, Blank D, Schatz G. Transport of proteins to the mitochondrial intermembrane space: the ‘matrix-targeting’ and the ‘sorting’ domains in the cytochrome c1 presequence. EMBO J. 1987;6:2433–2439. doi: 10.1002/j.1460-2075.1987.tb02522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Loon A P G M, van Eijk E, Grivell L A. Biosynthesis of the ubiquinol-cytochrome c reductase complex in yeast. Discoordinate synthesis of the 11 kDa subunit in response to increased gene copy number. EMBO J. 1983;2:1765–1770. doi: 10.1002/j.1460-2075.1983.tb01655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Steeg H, Oudshoorn P, van Hell B, Polman J E M, Grivell L A. Targeting efficiency of a mitochondrial pre-sequence is dependent on the passenger protein. EMBO J. 1986;5:3643–3650. doi: 10.1002/j.1460-2075.1986.tb04694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wenzel T J, Teunissen A W, Steensma H Y. PDA1 mRNA: a standard for quantitation of mRNA in Saccharomyces cerevisiae superior to ACT1 mRNA. Nucleic Acids Res. 1995;23:883–884. doi: 10.1093/nar/23.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]