Abstract
Although it is widely believed that horizontal patchiness exists in microbial sediment communities, determining the extent of variability or the particular members of the bacterial community which account for the observed differences among sites at various scales has not been routinely demonstrated. In this study, horizontal heterogeneity was examined in time and space for denitrifying bacteria in continental shelf sediments off Tuckerton, N.J., at the Rutgers University Long-Term Ecosystem Observatory (LEO-15). Characterization of the denitrifying community was done using PCR amplification of the nitrous oxide reductase (nosZ) gene combined with terminal restriction fragment length polymorphism analysis. Spatial scales from centimeters to kilometers were examined, while temporal variation was assayed over the course of 1995 to 1996. Sorenson's indices (pairwise similarity values) were calculated to permit comparison between samples. The similarities of benthic denitrifiers ranged from 0.80 to 0.85 for centimeter scale comparisons, from 0.52 to 0.79 for meter level comparisons, and from 0.23 to 0.53 for kilometer scale comparisons. Sorenson's indices for temporal comparisons varied from 0.12 to 0.74. A cluster analysis of the similarity values indicated that the composition of the denitrifier assemblages varied most significantly at the kilometer scale and between seasons at individual stations. Specific nosZ genes were identified which varied at centimeter, meter, or kilometer scales and may be associated with variability in meio- or macrofaunal abundance (centimeter scale), bottom topography (meter scale), or sediment characteristics (kilometer scale).
Measurements of spatial and temporal heterogeneity in microbial communities are of considerable interest to microbial ecologists. Knowledge of the spatial patchiness of bacteria is important for determining the appropriate sampling scales and for addressing basic ecological questions such as which factors may control microbial communities (e.g., top-down control by predators). For instance, marine microbial abundance in seawater is fairly uniform at ca. 109 cells/liter (2, 7). However, when small volumes of water are analyzed (<1 ml), high-density clusters can be revealed (6, 10) and nanoscale patches of bacteria have been induced by the addition of organic substrates (11). So routine marine sampling, which generally involves sample sizes greater than 1 liter, homogenizes bacterial populations, destroying any spatial patchiness information (6). As another example, several studies have shown the process of soil denitrification exhibits both temporal and spatial heterogeneity (8, 15, 16), with one study reporting that 25 to 85% of denitrification occurs in microsites comprising <1% of the soil mass (15). Given that the denitrification process itself exhibits temporal and spatial patchiness, it seems likely that the microbes responsible for the process are also nonrandomly distributed.
In this study, we monitored the temporal and spatial heterogeneity of denitrifying bacteria in a continental shelf environment using the nitrous oxide reductase (nosZ) gene (18, 19). The study was conducted at a Long-Term Ecosystem Observatory (LEO-15) centered on a sand ridge in 15 m of water offshore from the Rutgers University Marine Field Station (22). In order to rapidly characterize the nosZ target genes from the samples, we used terminal restriction fragment length polymorphism (T-RFLP) analysis (1). T-RFLP analysis is a powerful tool that has been used to compare microbial community structure and diversity in a variety of different laboratory and natural settings (4, 9, 12, 17, 21). Our analysis demonstrated variability in denitrifier community structure on horizontal scales ranging from centimeters to kilometers. Additionally, changes at single sites resulting from seasonal differences were observed. This information is critical for understanding the microbial dynamics of denitrifying bacteria in the marine environment and may help elucidate how the oceanic microbial ecosystem is structured and maintained.
MATERIALS AND METHODS
Environmental site.
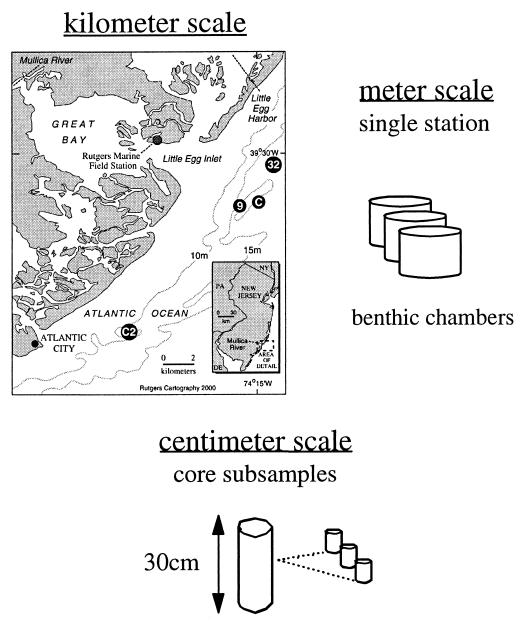
Continental shelf sediment samples were collected on 6 November 1995 (11/6/95), 12/5/95, 5/15/96, 6/25/96, 8/9/96, and 10/13/96 from a long-term ecosystem observatory site (LEO-15) described in detail elsewhere (22). Four stations (stations 9, 32, C, and C-2) were occupied over this time period at the LEO-15 site. Latitude and longitude for the LEO-15 stations are as follows (see Fig. 3): 74°15.73′W, 39°27.68′N (station 9), 74°15.23′W, 39°27.85′N (station C), 74°12.88′W, 39°23.08′N (station C-2), and 74°14.50′W, 39°29.38′N (station 32).
FIG. 3.
Schematic showing the LEO-15 sampling site and the three spatial scales over which denitrifying bacteria were monitored.
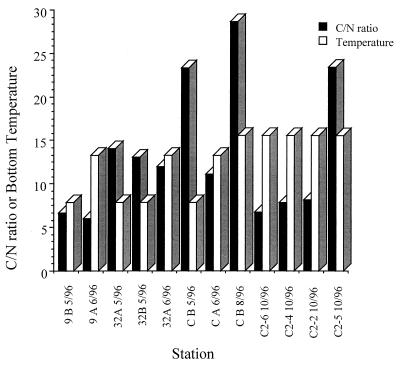
Some general characteristics of the LEO-15 stations are as follows. Station 9 contains medium to coarse sand, with a mean sediment grain size of 1.0 phi unit, and is usually populated by Spisula solidissima (surfclams). [Phi units are calculated as follows: φ = −log2(S); where S is the grain size in millimeters.] Furthermore, station 9 consists of approximately 99.5% sand, with 0.5% silt-clay (20). Station C contains medium to fine sand, with a mean sediment grain size of 1.8 phi units. Sediments at stations 32 and C-2 are predominantly very fine sands, with a mean grain size of 3.7 phi units (5). S. solidissima is rare at these locations; however, polychetes are found in abundance (20). Carbon and nitrogen analyses were performed by the Rutgers University Stable Isotope Facility on a ANCA-GSL elemental analyzer coupled to a Europa Scientific 20/20 isotope mass spectrometer. This analysis indicated that C/N ratios varied from 6 to 28.7 for the different sediments (Fig. 1).
FIG. 1.
Graph of carbon-to-nitrogen ratio and bottom water temperature (in degrees centigrade) during sampling for the various samples used in this study.
Sample collection.
For the 11/6/95, 12/5/95, 5/15/96, and 6/25/96 sampling dates, single sediment cores (size: 30-cm length, 10-cm internal diameter) were hand retrieved by scuba divers. Triplicate subsamples were obtained by collecting the top 2-cm portion of the sediment cores with a sterile, trimmed 3-ml syringe. For the 8/9/96 and 10/13/96 sampling dates, divers retrieved duplicate surficial sediment samples (top 0 to 3 cm) by scraping sterile 50-ml polypropylene centrifuge tubes (Phenix Research Products, Hayward, Calif.) across the sediment surface. The bottom water temperature at the time of collection was recorded for most of the samples (Fig. 1). Despite differences in collection methods, all of the samples used in these studies represent the upper 0 to 3 cm of sediments at the various stations and were stored frozen at −20°C until analysis.
Fingerprinting by T-RFLP analysis.
Total genomic DNA was extracted from approximately 100 mg (wet weight) of sediment, as described previously (18). The Nos661F (5′-CGGCTGGGGGCTGACCAA; labeled at the 5′ end with 6-carboxyfluorescein [6-FAM; Perkin-Elmer]) and Nos1773R (5′-ATRTCGATCARCTGBTCGTT) primers were used to amplify ∼1,100 bp of the nosZ gene. The PCR reactions contained 20 ng of template DNA and 200 pmol of primer, and the amplification conditions were 1 cycle at 94°C for 5 min, followed by 35 cycles of 95°C for 0.5 min, 55°C for 0.5 min, and 72°C for 1.5 min, with a final extension step at 72°C for 10 min (18). After amplification, 15 μl of the PCR products was digested with HinPI restriction enzyme (New England Biolabs) at 37°C for 6 h.
The digested DNA was precipitated with a 0.1 volume of 3 M sodium acetate and 2.0 volumes 95% ethanol, followed by spinning at 16,000 × g in an Eppendorf microcentrifuge for 15 min. The DNA pellet was washed with 70% ethanol, dried, and resuspended in a mixture of 12 μl deionized formamide and 0.5 μl of DNA fragment length internal standard (TAMRA 500; Perkin-Elmer).
Pairwise similarity calculations and cluster analysis.
The fluorescent moiety on the end of the digested PCR product was detected using the PE/ABI GeneScan Software which displays the various T-RFLPs as a series of peaks. The presence or absence of the T-RFLP peaks allows for two samples to be compared using Sorenson's index: Cs = 2Nab/(Na + Nb), where Nab is the number of shared peaks between two samples, and Na and Nb are the number of peaks for samples A and B, respectively (13, 14). Phenograms of denitrifier similarity indices were constructed using unweighted-pair-group mean-average (UPGMA) analysis. All indices and clusters were calculated using the COMbinatorial Polythetic Agglomerative Hierarchical clustering package (COMPAH96; http://www.es.umb.edu/edgwebp.htm).
RESULTS
In this study, T-RFLP technology was used to characterize the target nosZ genes from the samples. In order to accomplish this, it was necessary to select restriction enzyme(s) capable of resolving as many nosZ target genes as possible. We found that HinPI could produce the largest number of diagnostic terminal restriction fragments (Table 1). For example, 15 of the 47 known nosZ sequences could be unambiguously identified, and five TRFLPs contained only 2 nosZ sequences. Only TRFLPs 151, 191, 194, and 248 contained three or more different nosZ genes. Finally, three nosZ genes were found to not cut with this particular restriction enzyme and would remain undetectable.
TABLE 1.
T-RF lengths for various nosZ genes, as generated by the restriction enzyme HinPIa
| Source(s) of nosZ gene | Fragment length (bp) | GenBank accession no(s) |
|---|---|---|
| Paracoccus denitrificans | 28 | X74792 |
| Pseudomonas denitrificans | 28 | AF016059 |
| S32B696I | 36 | AF119918 |
| Ralstonia eutropha | 39 | X65278 |
| Achromobacter cycloclastes | 40 | AF047429 |
| ProR | 77 | AF119937 |
| S32B696E | 79 | AF119949 |
| S32B696G | 107 | AF119948 |
| Sinorhizobium meliloti | 151 | U47133 |
| S32B696B and -Q | 151 | AF119952 and -42 |
| Pseudomonas stutzeri Zobell | 152 | M22628 |
| S321195A and -B | 154 | AF016055 and -56 |
| S321195C | 184 | AF016057 |
| S32B696F and -W | 186 | AF119928 and -25 |
| Bradyrhizobium japonicum | 191 | AJ002531 |
| S32B696C, -J, -O, -S, and -AB | 191 | AF119951, -46, -43, -41, and -39 |
| S32B696D | 192 | AF119950 |
| ProA, -G, -H, -J, -S, and -T | 194 | AF119919, -24, -29, -34, -30, and -31 |
| S32B696K | 199 | AF119945 |
| S32B696L | 202 | AF119927 |
| S32B696R | 248 | AF119926 |
| ProB, -C, -D, and -E | 248 | AF119920, -21, -22, and -23 |
| ProP | 265 | AF119935 |
| ProI and -O | 268 | AF119930 and -33 |
| S32B696M | 283 | AF119944 |
| ProQ | 283 | AF119936 |
| ProV | 383 | AF119938 |
| ProL | 565 | AF119953 |
| S32B696A, -H, and -T | No cut | AF119954, -47, and -40 |
The S32B- and Pro- prefixes are clonal designations that refer to nosZ gene sequences identified from continental shelf sediments. GenBank accession numbers for all sequences are provided.
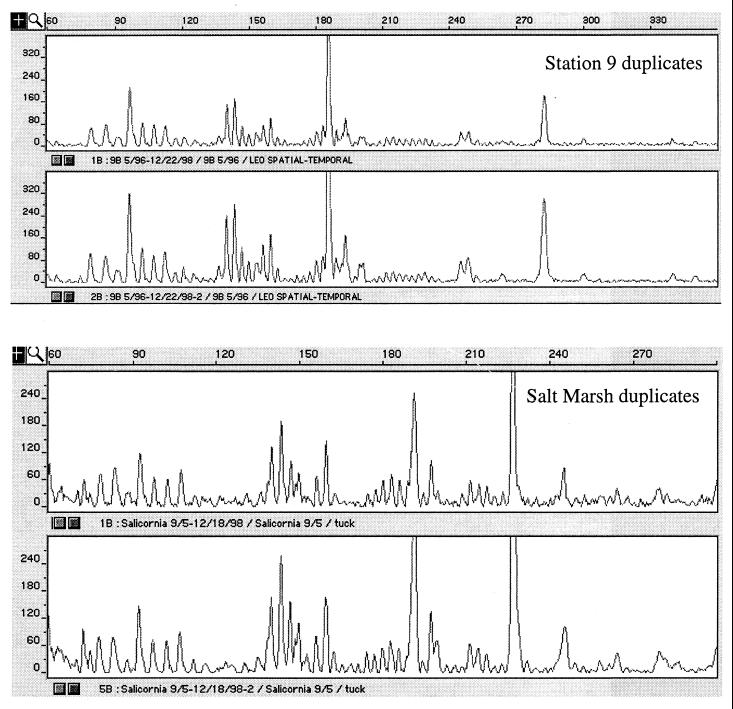
To assess the level of variation inherent in our T-RFLP technique, duplicate samples from station 9 (12/5/95) and a freshwater marsh were fingerprinted. These duplicate samples were extracted, amplified, and digested separately. The fingerprints are found to be nearly identical (Fig. 2). Analysis of the duplicate samples indicated a similarity value (Cs) of 0.99 between duplicate samples from the LEO-15 site and a comparable similarity value of Cs of 0.95 from the freshwater marsh samples. These duplicate fingerprints demonstrating high reproducibility in both banding pattern and peak height suggest that the appearance or disappearance of T-RFLP peaks is not a methodological artifact. However, this analysis did not directly test the extraction efficiency or the potential artifacts from PCR and does not preclude the existence of nosZ genes (denitrifiers) which may remain undetectable using current methodologies.
FIG. 2.
Duplicate T-RFLP fingerprints from marine and freshwater sediment samples.
Heterogeneity of nosZ genes on different spatial scales.
Variability in denitrifier assemblages was measured at LEO-15 over the three horizontal spatial scales indicated in Fig. 3. Centimeter scale variability in the nosZ gene was examined between two subsamples from three cores collected on 11/6/95, 5/15/96, and 6/25/96 at station 32. Meter scale variability was investigated at station C2 on 10/13/96 using four surficial sediment samples (top 0 to 3 cm) taken a few meters apart, and all possible pairwise comparisons were made. The kilometer scale variability of denitrifying bacterial populations was assessed on two occasions, on 5/15/96 and 6/25/96, for stations 9, C, and 32.
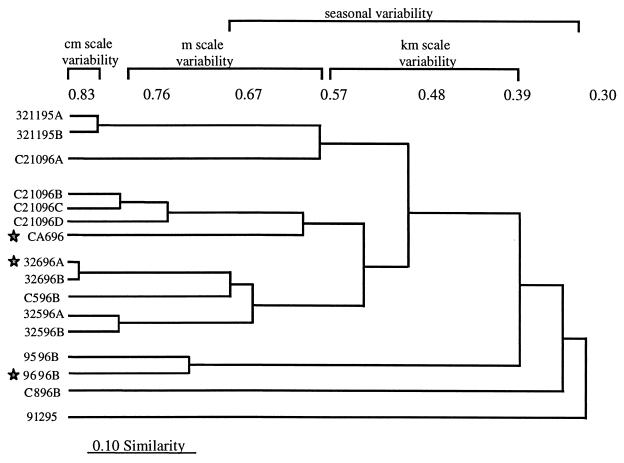
The individual Cs values for the centimeter scale study were 0.82, 0.80, and 0.85 (Fig. 4). The meter scale values ranged between 0.52 and 0.79, exhibiting greater variability than the centimeter scale values. Finally, the kilometer scale comparisons approach the lower end of the range for variability at the meter level, i.e., the similarity values ranged from 0.23 to 0.53. In addition, similarity values were highest for comparisons made between stations 32 and C (Cs = 0.52 to 0.53), while the lowest values were calculated for comparisons between station 9 and either station 32 or station C (e.g., Cs(station 9 versus station 32 [5/96]) = 0.23).
FIG. 4.
Cluster diagram of similarity values for LEO-15 samples. Similarity levels are indicated above the diagram. Similarity values were calculated using Sorenson's index, and clustering was done by the UPGMA method. Stars indicate samples included in the 6/25/96 kilometer level comparison. Phenograms of denitrifier similarity indices were constructed using UPGMA analysis. Samples are indicated using the following code: station identifier (9, 32, C, and C2), sampling date (month and year), and subsample identifier (A, B, etc.). Therefore, 321195A indicates a subsample of the 11/95 station 32 sample.
Temporal variability was examined at all LEO-15 stations for 12 months. The Cs value for all temporal assays ranged between 0.12 and 0.74, which nearly encompasses the range of values seen in the spatial study. In particular, the spread in the temporal values is twice as large as that seen for the meter or kilometer scale comparisons. Station C showed significant variation between sampling dates, but there was no systematic pattern of change between May 1996 (5/96) and 8/96. Station 32 showed a more distinct pattern, with the greatest differences occurring between 11/95 and 5/96 (Cs = 0.49), while the 5/96 and 6/96 samples displayed much greater similarity (Cs = 0.69). The most dramatic seasonal difference occurred at station 9, where the similarity between the 12/95 and 5/96 samples was only 0.12, while between 5/96 and 6/96 the Cs value was 0.74.
Cluster analysis of similarity values.
The COMPAH96 software program was used to perform cluster analysis of the similarity values generated in this study. A phenogram of the output of this analysis was reconstructed using an UPGMA algorithm (Fig. 4). The cluster diagram depicts the very close grouping of the individual centimeter scale comparisons. In addition, the meter level samples are grouped together with samples C2-4, C2-5, and C2-6, forming a tight cluster, and C2-2, splitting off separately. The kilometer scale samples for the 6/25/96 sampling date are indicated by a star. As Fig. 4 shows, these kilometer level samples are grouped at a very low similarity level (∼0.40).
The cluster diagram also illustrates some of the temporal impact on denitrifying bacteria at the various LEO-15 stations. In general, samples tend to cluster according to the time of year. For example, the 11/6/95 station 32 and 10/13/96 station C samples cluster together, as do the station 32 and C samples from 5/15/96 and 6/25/96. The most dramatic temporal changes are seen between the late fall and the 5/15/96 samples at stations 9 and 32.
DISCUSSION
This study demonstrated that the composition of denitrifier assemblages at the various LEO-15 stations change both spatially and temporally, with the greatest dissimilarity occurring over the kilometer and seasonal scales. Our results are consistent with those of other reports. For example, phenotypic variation of 6% in phenanthrene-degrading bacterial populations in intertidal sediments has been noted over spatial scales of centimeters, while temporal effects accounted for 21% of the variation (3). Additionally, a study in the coastal waters of Antarctica on the variability of Archaeal assemblages (14) also yielded a summer-winter comparison (Cs = 0.29), very similar to the fall-late spring results obtained at LEO-15, where Cs(station 9) = 0.12 and Cs(station 32) = 0.49.
Our analysis of T-RFLP number (denitrifier diversity) and the environmental parameters of C/N ratio or bottom water temperature was inconclusive (0.01 ≤ r2 ≤ 0.1). This suggests that it is not a simple matter of a single global regulator controlling all denitrifier populations at this site but is most likely a myriad of regulators affecting individual members of the denitrifying community. Although the variability we describe in denitrifier communities can provide clues as to when and where sampling should occur in the coastal ocean, there is another aspect to the data that is equally important. In particular, the T-RFLP technique can lead directly to testable hypotheses about the ecology of particular denitrifiers in the continental shelf sediment environment.
For example, a testable hypothesis would be that some of the denitrifiers demonstrating variability at the centimeter scale are associated with abiotic or biotic processes which also vary on the centimeter scale at the LEO-15 site. Specifically, the denitrifiers that differed between subsamples of the station 32 samples (e.g., terminal restriction fragments [T-RFs] 112, 154, and 184; station 32, 11/95, A [Table 2]) may be explained by the presence or absence of Ampharetid worm tubes at the site. Figure 4 also illustrates the temporal similarity seen in denitrifiers between the fall and spring samples taken at stations 9 and 32 at LEO-15. Temperature variations between the sampling dates were approximately 5°C and may be responsible for the variation in denitrifiers at these sites and times. For instance, the unique denitrifiers present at station 9 during 12/95 with HinPI T-RFs of 122, 124, 131, or 133 (station 9, 12/95 [Table 3]) may only be found in samples with lower temperatures.
TABLE 2.
T-RFs that differed between centimeter scale subsamples of station 32 sediment coresa
| Station 32 core sampling (mo/yr) | Subsample | T-RF(s) |
|---|---|---|
| 11/95 | A | 112, 154, 184 |
| B | 102, 121, 135, 139, 152, 193, 283 | |
| 5/96 | A | 53, 112, 129, 135, 147, 152, 172, 180, 183, 248, 265, 285 |
| B | 156 | |
| 6/96 | A | 102 |
| B | 156, 160, 183, 200 |
The T-RFs listed in each sample pair were present in only one of the two centimeter scale subsamples; however, some individual T-RFs (e.g., 102) are present more than one time.
TABLE 3.
T-RFs that were unique to specific continental shelf sediment samplesa
| Station | Date (mo/yr) | T-RF(s) |
|---|---|---|
| 9 | 12/95 | 122, 124, 131, 133 |
| 6/96 | 44 | |
| C | 6/96 | 88, 148, 190, 197, 199, 280 |
| 10/96 | 108, 206 |
The T-RFs listed here were found only in the specific samples listed.
The analysis of meter scale variation at station C (10/13/96) provides an interesting example of the possible effects of microsites on the indigenous denitrifying bacteria. Of the four meter level samples examined, three clustered at the 0.75 similarity level or higher. However, C2-2 only grouped with these samples at the 0.52 similarity level. This could indicate that even though the spacing between all four samples was roughly identical, sample C2-2 encompassed some unique feature. The seafloor at station C2 is characterized by sand ridges and troughs, which vary on the scale of meters (5). It is possible that the three similar samples were taken across the crest of a ridge, while the fourth was taken in a trough (or vice versa). The key point is that while station C is a medium to fine sandy environment, troughs adjacent to sand ridges are often characterized by high concentrations of organic “fluff” (5, 20) which could alter the species composition of the denitrifiers present. Therefore, a testable hypothesis to explain meter scale variation in denitrifiers is that denitrifier species vary across a sand ridge, from trough to crest. For example, denitrifiers with T-RFs of 94, 108, or 206 (columns C2-4 and C2-5 in Table 3) might only occur on the crest or troughs of sand ridges.
The kilometer level comparisons also produced results that may reveal the importance of environmental factors on denitrifying bacteria. For the two sampling dates for which kilometer scale comparisons were made (5/15/96 and 6/25/96), comparisons between stations C and 32 displayed much higher similarity values than did comparisons between either stations 9 and 32 or stations 9 and C (Fig. 4). In addition, Table 3 demonstrates that stations 9 and C both contained unique T-RFs for the June samples. Diver observations indicated clear benthic community differences in the three stations for the 5/15/96 sampling date (data not shown). Worm tubes and an organic fluff layer were present at station 32, moon snails and megaripple topography were observed at station 9, and many surfclams were found in the sediments at station C. A testable hypothesis to explain kilometer level variability in denitrifiers is that there are specific suites of denitrifiers associated with the various environments present at LEO-15. For example, denitrifiers with T-RFs of 44 would only be located at station 9.
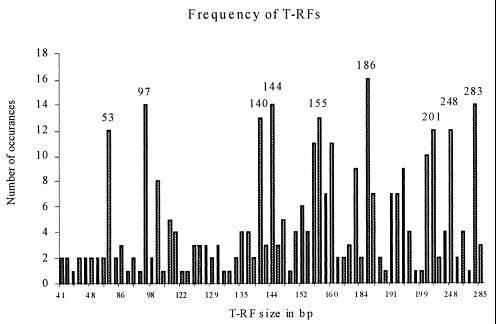
Finally, Fig. 5 shows a frequency distribution of the T-RFs generated in this study. It is apparent from this diagram that one denitrifier was present in all 16 samples examined and that a dozen were present in 10 or more samples. Several of these abundant T-RFs align with known nosZ genes (Table 1), while other common T-RFs (e.g., T-RF of 97 bp) are not currently in the nosZ database. This indicates that there is still much work to be done in order to characterize the denitrifying bacteria at LEO-15. The majority of T-RFs are present in low numbers (i.e., ≤4 samples). This could indicate that there is a core suite of denitrifiers common to the shelf environment, with ancillary species appearing and disappearing over space and time. However, T-RFLP analyses will tend to underestimate the species diversity of a microbial community since species present in low abundance may be below the detection limit or may share a common terminal restriction site, thereby yielding peaks of an identical size.
FIG. 5.
Frequency plot of the individual T-RFs generated in this study. Only the 186-bp T-RF appeared in all 16 samples. Sizes are indicated above those T-RFs appearing in 12 or more samples (75% or higher). Note that the horizontal scale is not linear due to the omission of T-RF sizes with zero occurrences.
In conclusion, denitrifying bacteria exhibit seasonal and spatial variability at the LEO-15 site. Seasonal changes and kilometer scale variations seem to have the greatest effect on denitrifying assemblages, while the smallest differences are observed at the centimeter spatial scale. Although significant temporal variability in denitrifier communities is seen at LEO-15, care must be taken to avoid spatial confounding. It is clear from the data that non-negligible variations occur in as little as a few centimeters, therefore sampling over time must occur at the same location. Otherwise, it may be impossible to fully separate effects of time versus simple variation in sampling location. For example, the variability between the 5/15/96 and 6/25/96 samples does fall within that observed for meter level comparisons, so changes in denitrifying bacteria at these stations cannot unambiguously be ascribed to temporal effects. With care, the T-RFLP methodology has proven to be a capable tool that can be used to address fundamental questions in microbial ecology.
ACKNOWLEDGMENTS
This work was supported in part from NOAA grant NA46RU0149 to Sybil P. Seitzinger and L.J.K., a New York Sea Grant/Hudson River National Estuarine Research Reserve Fellowship to D.J.S., and NSF grant OCE9872024 to L.J.K.
We thank Rose Petrecca, Robert DeKorsey, Andy Laursen, and the divers of the Institute of Marine and Coastal Sciences for their assistance in obtaining sediment samples.
REFERENCES
- 1.Avaniss-Aghajani E, Jones K, Holtzman A, Aronson T, Glover N, Boian M, Froman S, Brunk C F. Molecular technique for rapid identification of mycobacteria. J Clin Microbiol. 1996;34:98–102. doi: 10.1128/jcm.34.1.98-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad T F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 3.Berardesco G, Dyhrman S, Gallagher E, Shiaris M P. Spatial and temporal variation of phenanthrene-degrading bacteria in intertidal sediments. Appl Environ Microbiol. 1998;64:2560–2565. doi: 10.1128/aem.64.7.2560-2565.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruce K D. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR-restriction fragment length polymorphism profiling. Appl Environ Microbiol. 1997;63:4914–4919. doi: 10.1128/aem.63.12.4914-4919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craghan M. Topographic changes and sediment characteristics at a shoreface sand ridge—Beach Haven Ridge, New Jersey. M.S. thesis. Rutgers, N.J: Rutgers University; 1995. [Google Scholar]
- 6.Duarte C M, Vaque D. Scale dependence of bacterioplankton patchiness. Mar Ecol Prog Ser. 1992;84:95–100. [Google Scholar]
- 7.Fenchel T. Suspended marine bacteria as a food source. In: Fasham M J, editor. Flows of energy and materials in marine ecosystems: theory and practice. New York, N.Y: Plenum Press, Inc.; 1984. pp. 301–315. [Google Scholar]
- 8.Groffman P M, Tiedje J M. Denitrification in north temperate forest soils: spatial and temporal patterns at the landscape and seasonal scales. Soil Biol Biochem. 1989;21:613–620. [Google Scholar]
- 9.Knight V, Kerkhof L, Haggblom M. Community analysis of sulfidogenic 2-bromophenol dehalogenating and phenol degrading microbial consortia. FEMS Microbiol Ecol. 1999;29:137–147. [Google Scholar]
- 10.Krembs C, Juhl A R, Long R A, Azam F. Nanoscale patchiness of bacteria in lake water studied with the spatial information preservation method. Limnol Oceanogr. 1998;43:307–314. [Google Scholar]
- 11.Krembs C, Juhl A R, Strickler J R. The spatial information preservation method: sampling the nanoscale spatial distribution of microorganisms. Limnol Oceanogr. 1998;43:298–306. [Google Scholar]
- 12.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magurran A E. Ecological diversity and its measurement. Princeton, N.J: Princeton University Press; 1988. [Google Scholar]
- 14.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and archeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkin T B. Soil microsites as a source of denitrification variability. Soil Sci Soc Am J. 1987;51:1194–1199. [Google Scholar]
- 16.Parkin T B, Starr J L, Meisinger J J. Influence of sample size on measurement of soil denitrification. Soil Sci Soc Am J. 1987;51:1492–1501. [Google Scholar]
- 17.Phelps C, Kerkhof L, Young L. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol Ecol. 1998;27:269–279. [Google Scholar]
- 18.Scala D J, Kerkhof L J. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett. 1998;162:61–68. doi: 10.1111/j.1574-6968.1998.tb12979.x. [DOI] [PubMed] [Google Scholar]
- 19.Scala D J, Kerkhof L J. Diversity of nitrous oxide reductase (nosz) genes in continental shelf sediments. Appl Environ Microbiol. 1999;65:1681–1687. doi: 10.1128/aem.65.4.1681-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snelgrove P V R, Grassle J P, Butman C A. Sediment choice by settling larvae of the bivalve, Spisula solidissima (Dillwyn), in flow and still water. J Exp Mar Biol Ecol. 1998;231:171–190. [Google Scholar]
- 21.van der Maarel M J E C, Artz R R E, Haanstra R, Forney L J. Association of marine Archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Alt C J, Grassle J F. LEO-15: an unmanned long-term environmental observatory. Proc Oceans. 1992;2:849–854. [Google Scholar]