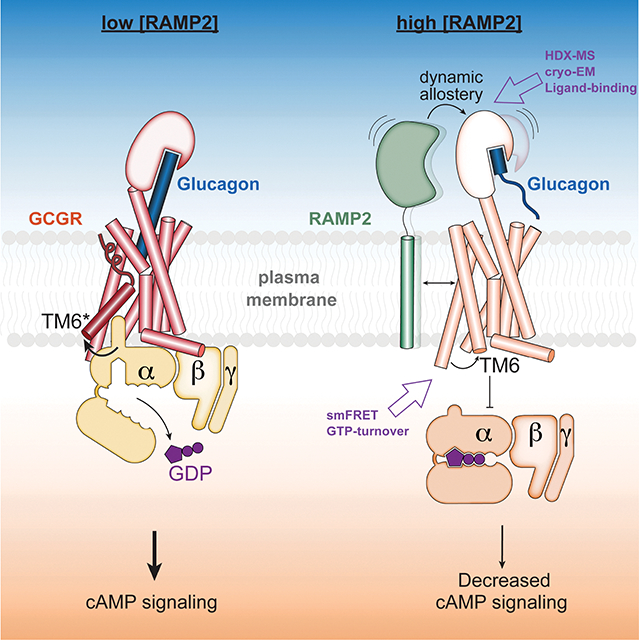
Summary:
Receptor activity-modifying proteins (RAMPs) modulate the activity of many Family B GPCRs. We show that RAMP2 directly interacts with the glucagon receptor (GCGR), a Family B GPCR responsible for blood sugar homeostasis, and broadly inhibits receptor-induced downstream signaling. HDX-MS experiments demonstrate that RAMP2 enhances local flexibility in select locations in and near the receptor extracellular domain (ECD) and in the 6th transmembrane helix, while smFRET experiments show that this ECD disorder results in inhibition of active and intermediate states of the intracellular surface. We determined the cryoEM structure of the GCGR-Gs complex at 2.9 Å resolution in the presence of RAMP2. RAMP2 apparently does not interact with GCGR in an ordered manner, yet the receptor ECD is indeed largely disordered along with rearrangements of several intracellular hallmarks of activation. Our studies suggest that RAMP2 acts as a negative allosteric modulator of GCGR by enhancing conformational sampling of the ECD.
Graphical Abstract
In Brief
Receptor activity-modifying protein 2 functions as a negative allosteric modulator of GCGR by enhancing extracellular receptor dynamics that results in inactive state of the intracellular surface.
Introduction
The glucagon receptor (GCGR) is a Family B heterotrimeric guanine nucleotide-binding protein (G protein)-coupled receptor (GPCR) responsible for maintaining proper blood glucose concentrations1. Glucagon binding to GCGR activates the receptor, resulting in cyclic adenosine monophosphate (cAMP) production predominantly via GCGR interaction with the adenylyl cyclase stimulatory G protein, Gs2. We previously demonstrated that Family B GPCRs, like GCGR, are not as effective at activating Gs as their Family A counterparts, likely due to the kinetically-limiting necessity of a large helical break that occurs in TM6 upon Family B GPCR activation3. We were therefore interested in characterizing modulatory factors that could stimulate this activity. Receptor activity-modifying proteins (RAMPs) are single-pass TM proteins with an N-terminal extracellular domain that bind Family B GPCRs and modify their ligand binding and intracellular signaling4,5. These effects are exemplified by the structural and dynamic changes induced by binding of one of the three RAMPs to either the calcitonin receptor (to form the amylin receptors)6 or the calcitonin receptor-like receptor (to form the calcitonin gene-related peptide receptor or adrenomedullin receptors)7,8. However, the effects of RAMP interaction with other classes of Family B receptors are less well understood. RAMP2 is known to interact with GCGR9 resulting in a disparate array of ligand binding and intracellular consequences, including increased glucagon potency10, decreased potency11 decreased cAMP production over a prolonged period12 or cAMP accumulation10. To address these disparate findings, we used purified RAMP2 and purified GCGR to perform a variety of biochemical, biophysical and structural studies to more directly characterize the effect of non-constitutive RAMP2 binding on GCGR activity.
Results
RAMP2 interacts with and selectively diminishes signaling by GCGR
Our previous study showed that GCGR (along with other Family B GPCRs) is significantly slower at facilitating nucleotide exchange in Gs compared to Family A GPCRs3. We initially speculated that RAMP2 may serve as a cofactor (positive allosteric modulator) to increase GCGR induced Gs turnover. To understand how RAMP2 binding impacts GCGR signaling, we purified monomeric RAMP2 (Fig. S1A) and conducted a time-dependent in vitro GTPase assay (Fig. S1B)13. While agonist (ZP37803)-bound GCGR demonstrates guanine nucleotide exchange factor (GEF) activity that is very weakly sensitive to the initial concentration of GDP ([GDP]i) (Fig. 1A, Fig. S1B), pre-incubation with RAMP2 (2:1 RAMP2:GCGR) results in potent inhibition of GEF activity in a [GDP]i dependent manner. This inhibitory effect is present at all time points over the course of the nucleotide depletion assay (Fig. S1B). RAMP2 has no effect on GCGR-mediated Gs turnover in the absence of GDP, and no impact on basal Gs turnover (data not shown) or turnover stimulated by agonist-bound β2-adrenergic receptor (β2AR, a Family A GPCR) (Fig. S1C), suggesting that the mechanism of inhibition is through specific interaction with GCGR. Because of the [GDP]i-dependent nature of RAMP2 inhibition, we hypothesized that RAMP2 acts by inhibiting receptor-catalyzed nucleotide release from Gs. Consistent with this mechanism of inhibition, pre-incubation of GCGR with RAMP2 decreases the observed Gαs 3H-GDP dissociation rate (koff) from 0.0033 s−1 to ~0.0001 s−1 (Fig. 1B). Thus, contrary to our initial speculation, RAMP2 appears to potently inhibit GCGR signaling through Gs by inhibiting the initial GDP release step; this manifests in the GTP turnover assay as a GDP-titratable inhibitory effect. Because the GTP-depletion rate of GCGR in the presence of RAMP2 is even slower than intrinsic Gs activity alone (Fig. 1A, S1B), we speculated that GCGR/RAMP2 heterodimers may act as a sink for GDP-bound Gs. Indeed, while addition of agonist-bound GCGR to β2AR has little impact on turnover by the β2AR, co-incubation with RAMP2/GCGR/agonist with β2AR results in significant inhibition of β2AR-mediated turnover, suggesting that GCGR/RAMP2 acts to actively sequester Gs away from other Gs-coupled receptors (Fig. S1D).
Figure 1. Biochemical and in-cellulo consequences of RAMP2 interaction with GCGR.
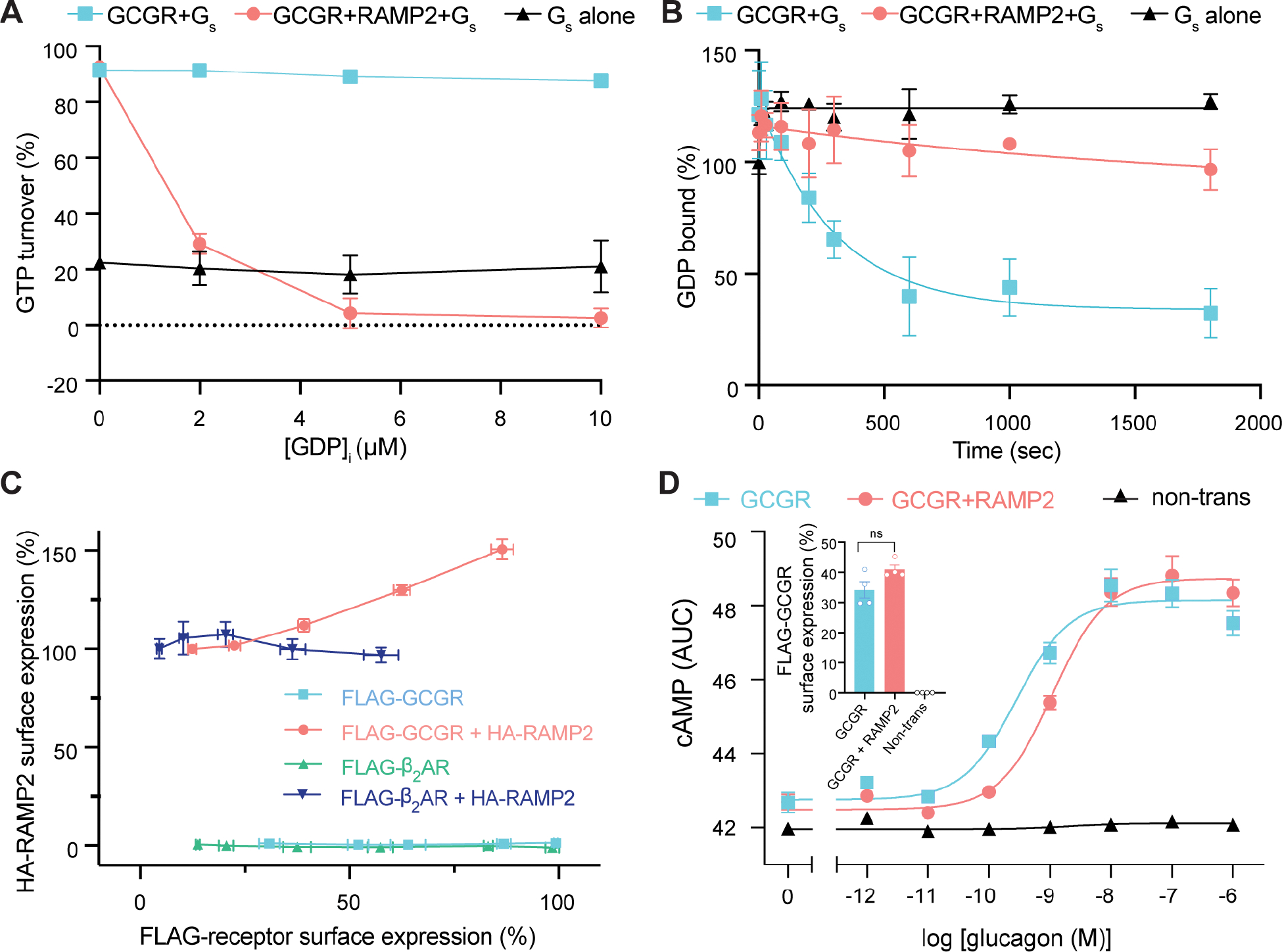
(A) The GTP turnover assay shows that GCGR activation of Gs is largely independent of GDP concentration, but upon pre-coupling of GCGR with RAMP2, Gs activation is potently inhibited in a [GDP] dependent manner and appears to inhibit even intrinsic Gs activity. (B) The rate of 3H-GDP release from Gs is significantly reduced when GCGR is pre-incubated with RAMP2. (C) Increasing the cell-surface expression of the glucagon receptor, but not β2AR, results in an increase in surface expression of RAMP2. (D) Cells transfected with GCGR display increases in cAMP levels upon stimulation with glucagon with an observed EC50 of 0.28 nM (pEC50=9.6 [95% confidence limits 9.8–9.3]); cells expressing the same levels of GCGR in the presence of excess transfected RAMP2 display an EC50 of 1.10 nM (pEC50=9.0 [95% confidence limits 9.1–8.8]), a 4-fold right-shift in potency.
Previous studies interrogating the effect of RAMP2 on GCGR-Gs signaling resulting in discrepant results have been performed with different cell types and/or potentially different RAMP2-receptor ratios10–12. Since overexpression of GCGR (but not β2AR) leads to a concurrent increase in RAMP2 surface expression (Fig. 1C), we carefully optimized the surface-expression levels of GCGR and RAMP2 for cellular signaling assays in HEK293 cells. After selecting transfection conditions for similar surface expression levels of GCGR without and with co-expression of RAMP2, we performed a non-IBMX perturbed cyclic adenosine monophosphate (cAMP) assay as a downstream measure of GCGR-Gs signaling, albeit with potential confounding effects from receptor internalization and intracellular signaling. At similar levels of GCGR surface expression, the presence of RAMP2 results in a significant ~4-fold decrease in glucagon potency (EC50= 0.28 nM vs 1.10 nM) (Fig. 1D), consistent with the Gs inhibition observed with biochemical experiments. This right-shift in observed glucagon potency may be due to RAMP2-induced changes in agonist affinity or efficacy. To address this question, we performed BRET-based TAMRA-labeled glucagon analogue (Fig. 2A) binding assays with N-terminally fused Nanoluc on GCGR and showed that the presence of RAMP2 resulted in a ~2–3-fold decrease in agonist and only a ~1.5-fold decrease in peptide antagonist affinity (Fig. 2B, Fig. S1E). The presence of RAMP2 also results in a change in the total observed BRET change observed for peptide binding (Fig. S1E), perhaps due to RAMP2-induced changes in orientation of the attached luciferase with respect to the fluorophore-labeled peptide, or a decreased capacity to bind peptide. Our biochemical and in cellulo studies suggest that RAMP2 binding to GCGR results in both a diminished glucagon affinity (Fig. 2B, Fig. S1E) and efficacy (Fig. 1A, Fig. S1B).
Figure 2. Design and characterization of a GCGR antagonist and widespread RAMP2-induced GCGR conformational changes.
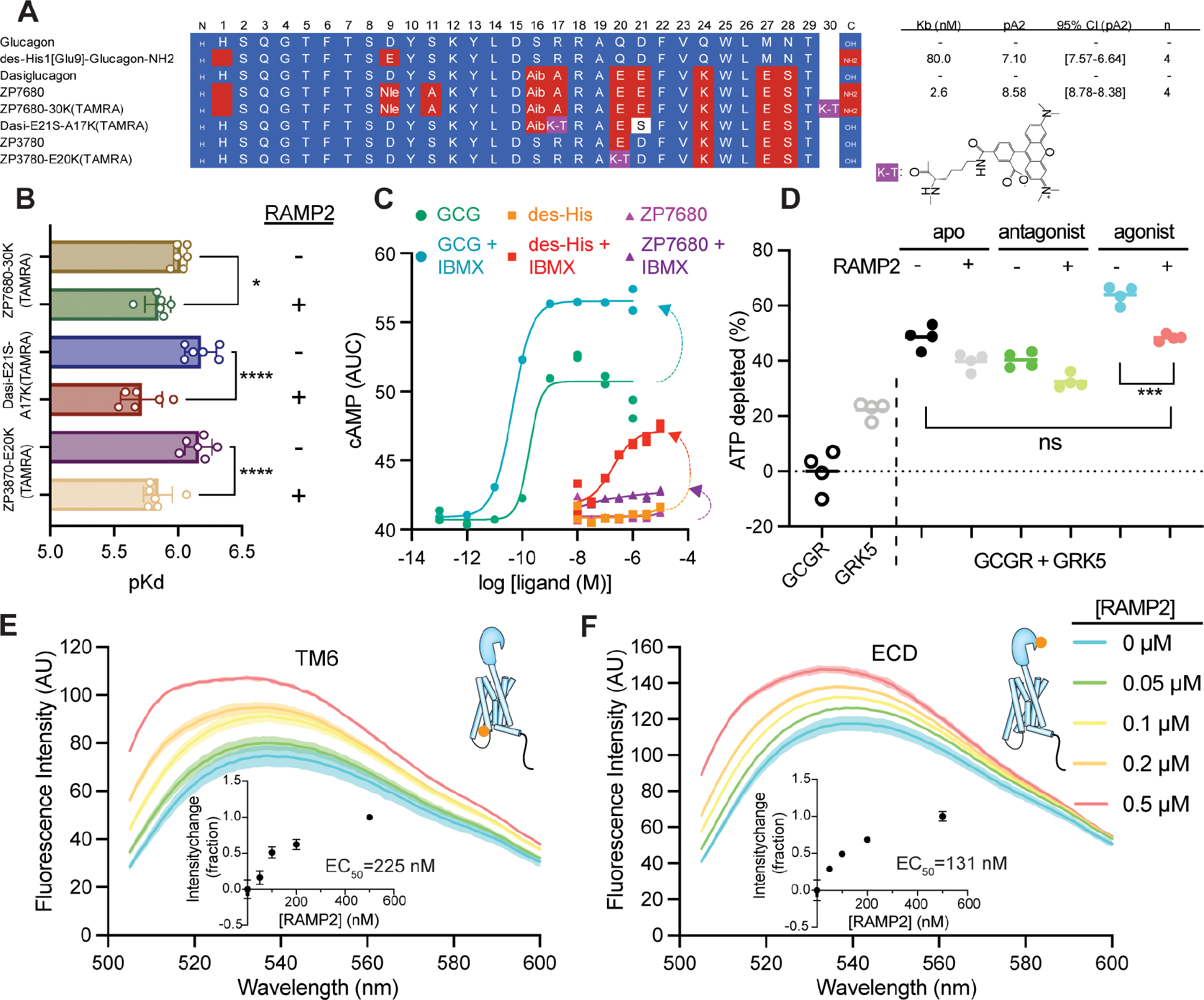
(A) Sequences and estimated affinities of previously described GCGR antagonist des-His1[Glu9]-Glucagon-amide (KB =80 nM) based on the glucagon scaffold, and the high affinity GCGR antagonist ZP7680 (KB =2.6 nM) derived from the dasiglucagon scaffold. See Methods and Fig. S2 for details on the design and characterization of ZP7680 and TAMRA-labeled peptides. Nle; norleucine, Aib=2-aminoisobutyric acid. K-T; TAMRA-labeled lysine. (B) Observed affinities of various TAMRA-labeled peptides in the presence or absence of co-expressed RAMP2. GCGR antagonist (ZP7680-30K(TAMRA)) shows a slight (~1.5-fold, P<0.05) decrease in affinity in the presence of RAMP2, while both agonist peptides (Dasi-E21S-A17K(TAMRA) and ZP3780-E20K(TAMRA)) display 2.9 (P<0.0001) and 2.1-fold (P=0.0001) decreases in affinity, respectively (one way ANOVA, Šidák multiple comparisons test). (C) The antagonist ZP7680 has minimal intrinsic efficacy compared to des-His1[Glu9]-Glucagon-amide. Under conditions allowing for cAMP accumulation by addition of the PDE inhibitor IBMX, des-His1[Glu9]-Glucagon-amide have some agonistic activity whereas ZP7680 is largely silent. (D) In an ATP depletion assay, the presence of RAMP2 inhibits receptor phosphorylation regardless of what is bound in the orthosteric site, though the inhibitory effect is most significant for agonist-induced phosphorylation (P<0.001), where receptor phosphorylation returns to levels seen in the apo-state (P is not significant). Using agonist-bound GCGR site-specifically labeled at the intracellular end of TM6 (E) or on the N-terminal helix of the ECD (F) with the environmentally sensitive fluorophore NBD, addition of increasing amounts of RAMP2 results in titratable environmental changes in both regions of the receptor. P values in B and D are denoted as follows: ns (P>0.05), * (P≤0.05), ** (P≤0.01), *** (P≤0.001), and **** (P≤0.0001).
We developed an engineered, soluble antagonist peptide (Fig. 2A) devoid of apparent intrinsic efficacy (Fig. 2C) to distinguish between potential differences in the behavior of apo receptor and inactive, peptide-bound receptor. The antagonist peptide, ZP7680, retains the solubility of our previously designed agonist peptide3 while enhancing the observed affinity for the receptor relative to a known antagonist peptide, des-His1[Glu9]-Glucagon-NH2 (des-His) (Fig. 2A, Fig. S2A, Fig. S2B) and nearly eliminating the partial activity present in the des-His peptide (Fig. 2C). In addition to G proteins, ligand-activated Family B GPCRs have been shown to signal through ß-arrestins upon being phosphorylated by GPCR kinases (GRKs)12,14–16. Multiple studies have shown that RAMP2 decreases ß-arrestin recruitment to GCGR11,12. To understand if this decrease in ß-arrestin coupling is due to the role of RAMP2 in modulating GRK phosphorylation of GCGR, we measured GRK-mediated GCGR phosphorylation in the presence and absence of RAMP2 with ligands of different efficacy using a luciferase-based phosphorylation assay (Fig. S2D). Compared to unliganded (apo)-receptor, phosphorylation by GRK5 is reduced in antagonist-bound GCGR and increased by our previously developed soluble agonist peptide, ZP3780 (Fig. 2D)3. RAMP2 broadly diminishes GRK5-phosphorylation irrespective of the ligand state of the receptor, aside from the negative allosteric modulator L-168,049 (Fig. 2D; Fig. S2D). This effect is largest for the agonist-bound form of GCGR, where RAMP2 results in GRK5 phosphorylation levels similar to the apo-form of the receptor (Fig. 2D). In-gel staining for phosphorylated protein further confirmed that RAMP2 dramatically inhibits GRK5-mediated phosphorylation of GCGR (Fig. S2E). However, GRK2 phosphorylation of GCGR is not impaired by the presence of RAMP2 (Fig. S2F) suggesting that RAMP2 specifically inhibits phosphorylation of GCGR by certain GRK isoforms.
To probe the impact of RAMP2 binding on the conformation(s) of GCGR, we employed fluorescence spectroscopy, wherein we site-specifically labeled the receptor on an introduced cysteine at the cytoplasmic end of TM6 (F349C) or the ECD (K31C) with the environmentally sensitive fluorophore 4-chloro-7-nitrobenz-2-oxa-1,3-diazole (NBD) in a minimal cysteine GCGR (mC-GCGR) background3. Incremental addition of RAMP2 to agonist-bound mC-GCGR-349C-NBD results in a titratable (EC50~225 nM) increase in fluorescence (Fig. 2E), consistent with increased occupancy of the inactive conformation of TM63. This may provide an explanation for the decrease in activity observed in the GTP turnover assay. In addition to perturbing the intracellular conformational distribution of GCGR, RAMP2 also induces significant changes in fluorescence of the ECD sensor (Fig. 2F, EC50~131 nM). Taken together, these experiments demonstrate that RAMP2 interaction with GCGR induces widespread conformational changes in the receptor from the ECD to the intracellular face of the receptor.
RAMP2 binding enhances local conformational dynamics of GCGR
Next, we performed hydrogen-deuterium exchange monitored by mass spectrometry (HDX-MS) to quantify changes in local conformational flexibility across the receptor upon binding of RAMP2 (Fig. 3A, Fig. S3A,B). For HDX-MS we used antagonist-bound GCGR as our pull-down experiment showed RAMP2 interacts most stably with antagonist-bound GCGR (Fig. S3C). HDX-MS was performed on GCGR in the absence or presence of excess RAMP2. We obtained high-quality peptide coverage for 66% of the GCGR sequence (average of two replicates), with most peptides reporting on the ECD, TM1, TM2, TM6, TM7, and the C-terminus (Fig. S3A,B). We also performed HDX-MS on RAMP2 alone. We obtained high-quality coverage of 39.5% of the RAMP2 sequence (average of two replicates), with peptides primarily derived from the single-pass transmembrane segment (Fig. S3A,B). Peptides within the RAMP2 transmembrane segment, spanning residues 147–163, exhibited reduced deuterium uptake in the presence of GCGR (Fig. 3A,B). While not a quantitative measure of complex formation (due to the excess RAMP2 in the complex study), reduced exchange in this transmembrane segment is consistent with an AlphaFold2-predicted model17 of the GCGR–RAMP2 complex, in which the RAMP2 transmembrane segment forms an extended binding interface with TM3, TM4 and TM5 of GCGR (Fig. 3A).
Figure 3. RAMP2 binding broadly increases the conformational heterogeneity in GCGR.
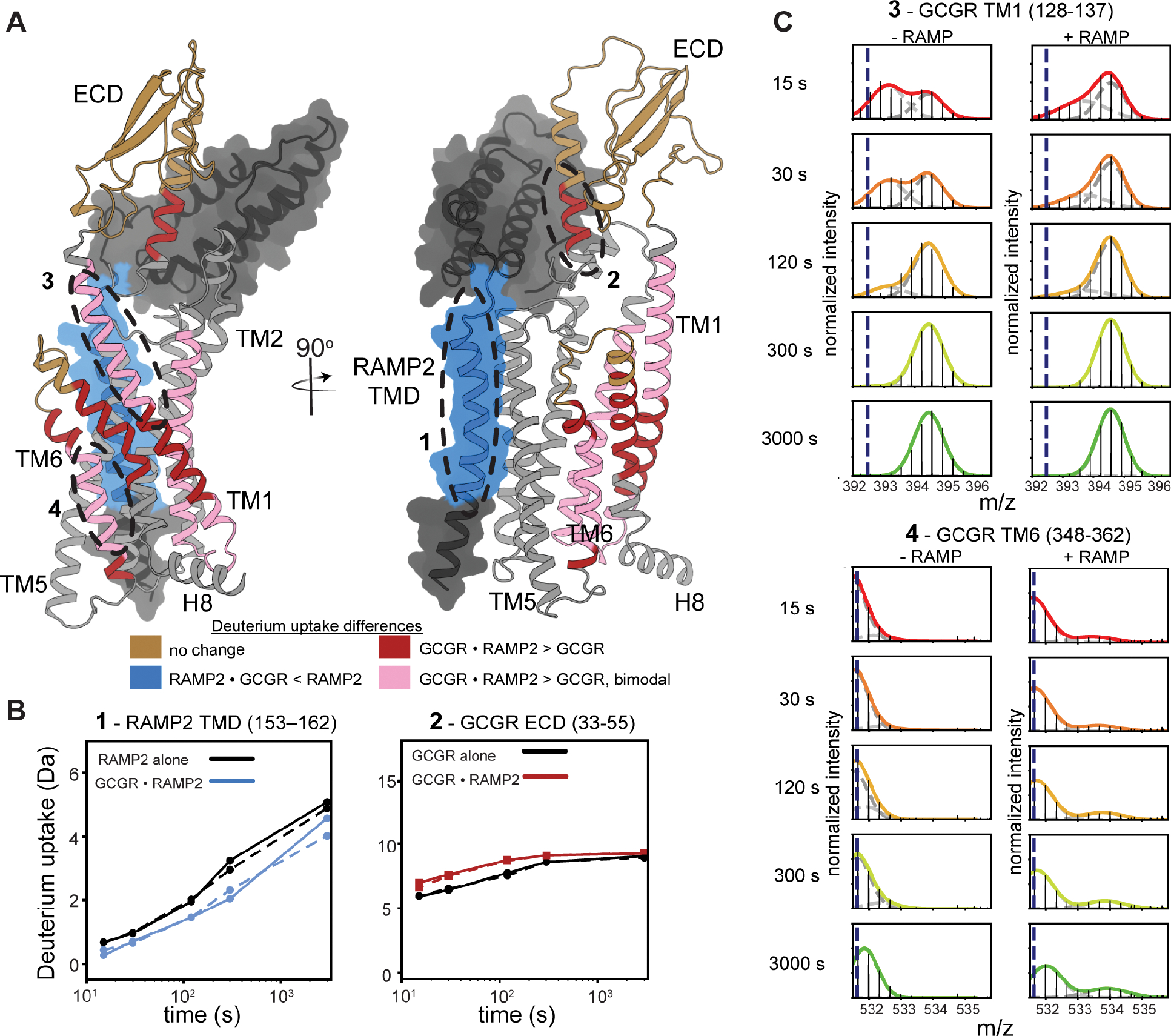
(A) Alphafold model of GCGR-RAMP2 complex onto which changes in HDX upon complex formation are plotted. (B) Example HDX-MS exchange curves in the presence and absence of excess RAMP2 (independent replicates in solid and dashed lines) show that several key regions of the RAMP2 (153–162) and receptor (33–55) are impacted by heterodimer complex formation. (C) HDX-MS plots showing bimodal distribution in GCGR TM1 (residues 128–137), an indication of conformational heterogeneity. This heterogeneity is increased in the presence of RAMP2. Though not present in the absence of RAMP2, a bimodal distribution is induced in TM6 (348–362) upon addition of RAMP2.
For GCGR in the presence of RAMP2, we found a surprising enhancement of deuterium uptake in the N-terminal region of the ECD, including a peptide corresponding to residues 33–55 (Fig. 3B). The region of this peptide responsible for enhanced exchange (32–38) is adjacent to the NBD-labeling position in the ECD, and the changes responsible for the increase in HDX in this region likely contribute to the NBD fluorescence change (Fig. 2F). Structural elements within the remainder of the ECD exhibited no difference in deuterium uptake upon RAMP2 binding (Fig. S3A), indicating that RAMP2 likely perturbs the ECD through a rigid-body motion and not by altering its local structure. Multiple studies have shown the importance of ECD dynamics in Family B receptor ligand binding kinetics, activation and signaling18–21.
In addition to the ECD, RAMP2 binding also resulted in complex changes in hydrogen exchange behavior elsewhere in the receptor, indicating RAMP2-enhanced conformational heterogeneity. This effect is most easily seen in peptides that display bimodal behavior, both in terms of an increased number of peptides with bimodal behavior and by a change in the ratio of the two peaks for those that show bimodal behavior in the isolated receptor. Bimodal mass-isotope distributions are seen in peptides that include the N-terminal portion of TM1 in the presence and absence of RAMP2, as well as in portions of TM2 and TM6 in the presence of RAMP2 (Fig. 3C, Fig. S3A,B). Such bimodal behavior can arise when the rate of labeling is faster than the rate of interconversion between a closed, exchange-incompetent conformation and an open, exchange-competent conformation (EX1 or EXX kinetics)22. For example, RAMP2 binding increases the population of the heavier (right-hand) peak for peptides covering the beginning of TM1 (residues 128–137) (Fig. 3C), suggesting that RAMP2 increases the rate of conversion between two conformations that TM1 populates within the native conformational ensemble of GCGR in the absence of RAMP2. While in CLR-RAMP structures (which are obligate dimers) TM1 is not in direct contact with RAMP2, in GCGR the extracellular end of TM1 might unravel to accommodate ECD conformational changes upon RAMP binding. Mutations in this region have been shown to decrease agonist peptide binding23, suggesting a possible functional link to RAMP2-induced changes in conformational flexibility. Additionally, within TM6, peptides containing the PXXG motif display bimodal behavior only in the presence of RAMP2 (Fig. 3C). The PXXG motif of TM6 has been shown to be pivotal for receptor activation24. Increased hydrogen exchange upon RAMP2 binding is not universal: other locations in GCGR, including in the C-terminus, do not exhibit changes in deuterium uptake upon RAMP2 binding (Fig. S3A), demonstrating that the increases in observed dynamics are not a result of broad-scale receptor destabilization. Taken together, our HDX-MS results show that RAMP2 selectively modulates the intrinsic conformational heterogeneity of several key structural regions within GCGR, including the ECD and TM6.
Effects of RAMP2 on the activation dynamics of GCGR observed by single-molecule FRET
We next used single-molecule fluorescence resonance energy transfer (smFRET) to study the effect of RAMP2 on the conformational dynamics of the intracellular face of GCGR. Using our mC-GCGR construct, we site-specifically labeled a previously characterized pair of introduced cysteines, one in TM4 as a reference site (265C) and one in the activation-sensitive TM6 (345C)3, with cysteine-reactive versions of LD555 (donor) and LD655 (acceptor) fluorophores to probe TM6 outward movement upon ligand and RAMP2 binding. We used nitroxide spin labels observed by cwEPR to characterize and minimize background labeling (see Fig. S4A for more details). The fluorescently-labeled mC-GCGR-265C/345C protein was immobilized at low density on a PEG-passivated coverslip with a biotinylated anti-FLAG antibody, and imaged using total internal reflection (TIRF) microscopy25,26. In the apo-state, GCGR exhibits a broad distribution of FRET efficiencies with a major peak centered at ~0.83 (Fig. 4A, black). This high-FRET state likely corresponds to the inactive, inward conformation of TM6, which brings the intracellular end of TM6 into close proximity to the intracellular end of TM4, as seen previously3,13. Representative individual traces of apo-GCGR largely occupy the high-FRET state with occasional transitions to a mid-FRET state with a FRET efficiency ~0.63 (Fig. 4C). Antagonist peptide binding with or without RAMP2 addition have a minimal effect on the FRET distribution relative to apo receptor (Fig. S4B), consistent with all three conditions representing a similar ensemble of inactive states. In the presence of full agonist peptide, heterogeneity in the distribution of FRET values remains, but the mid-FRET peak ~0.63 becomes dominant at the expense of the inactive, high-FRET state along with an increase in a low-FRET peak at FRET efficiency ~0.32 (Fig. 4A, dark blue). This mid-FRET peak appears to be an agonist-specific intermediate state of the receptor rather than an average of inactive/active states because we are able to resolve transitions to and from this state when imaging with a 100 ms frame rate27 (Fig. 4C; Fig. S4C). This is in apparent contradiction with previous DEER results suggesting that GCGR has no significant change in distance between TM4 and TM6 upon binding to glucagon, though some subtle but significant changes in the bimane fluorescence spectra upon agonist binding were observed3. We speculate that subtle changes in conformation at the intracellular end of TM6 that are not observable by the pure distance changes measured by DEER are amplified by the large, flexible dyes used for smFRET experiments, as well as the inherently complex nature of observed FRET efficiencies28.
Figure 4. Enhanced conformational heterogeneity stabilizes an inactive intracellular conformation of TM6.
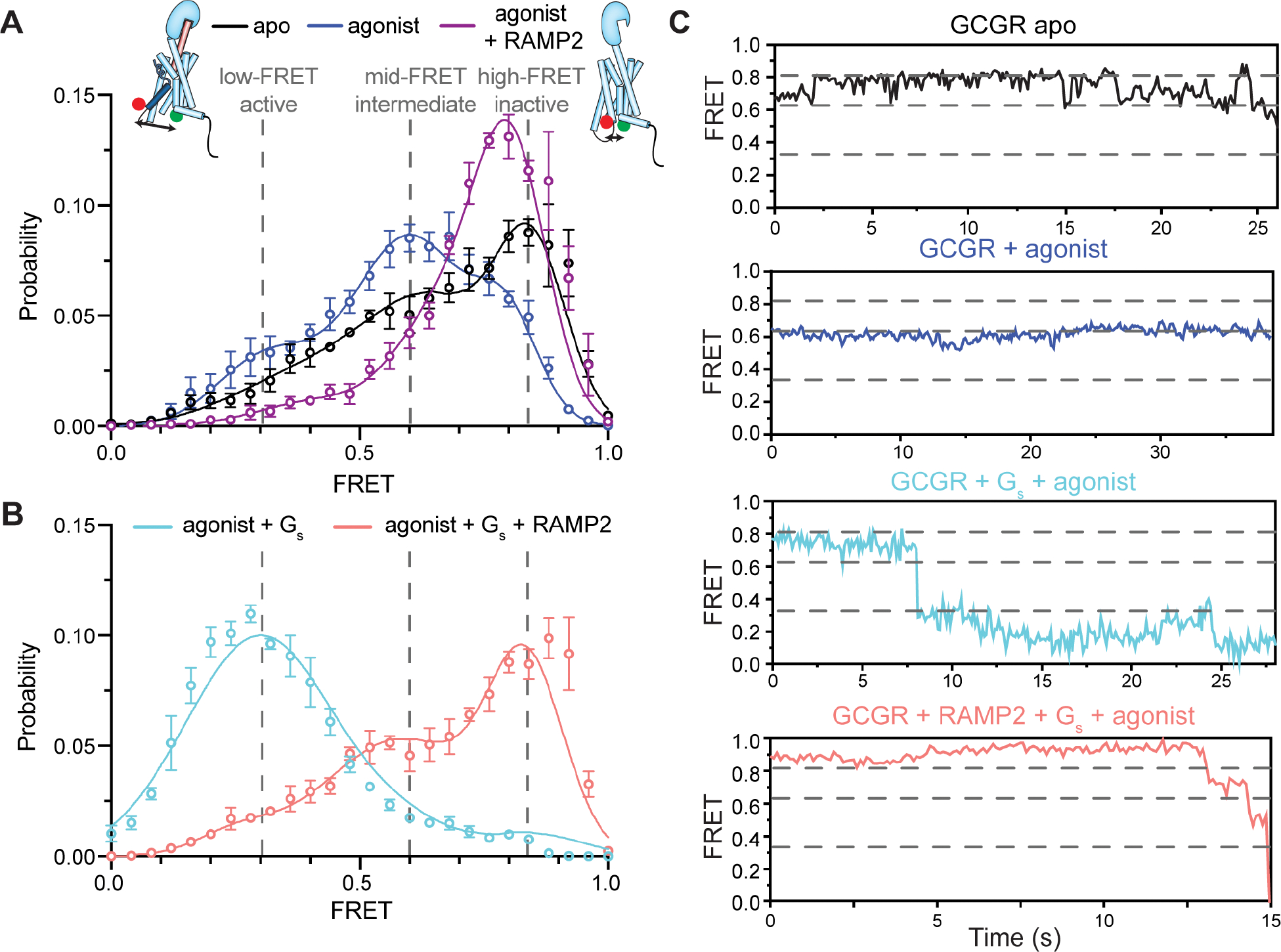
(A) smFRET experiments show that a dominant high-FRET state (~0.83) of GCGR labeled with donor and acceptor fluorophores at the intracellular ends of TM4 and TM6 is present in the absence of orthosteric agonist (A, black, N=162) with brief excursions to mid- (~0.63) FRET (~0.32) states (C, black), and the binding of agonist peptide results in a dominant mid-FRET state at the expense of the high-FRET state (A, blue, N=150; C). The mid-FRET intermediate and low-FRET active conformations of TM6 induced by agonist peptide are largely abrogated by the presence of RAMP2 (A, purple, N=117) as the distribution shifts towards and inactive-like conformation. Gs coupling to agonist-bound GCGR increases the population of the low-FRET, suggesting a full outward movement of TM6 at the expense of the mid-FRET agonist-specific intermediate state and high-FRET inactive state (B, cyan, N=158; C). However, pre-coupling of agonist-bound GCGR with RAMP2 potently inhibits a Gs-induced increase in the population of the fully outward TM6 conformation as well as the agonist-associated intermediate state (B, salmon, N=179; C). Histograms are shown with a 3-Gaussian fit to the data.
Formation of a nucleotide-free GCGR-Gs complex results in a substantial increase in occupancy of the low-FRET state (~0.32) (Fig. 4B, 4C, cyan), suggesting that this state is the fully outward, active state of TM6 observed in the cryo-EM structure of GCGR/Gs complex3. Addition of RAMP2 to agonist-bound receptor results in a near complete elimination of fully-active and agonist-associated intermediate states of TM6 in favor of an inactive-like conformation (Fig. 4A, purple). Further, the addition of RAMP2 to agonist-bound GCGR in the presence of Gs shifts occupancy back to the high-FRET inactive state that is comparable to, or even above, that of the apo-state (Fig. 4B, 4C, salmon) and interestingly, not to the agonist-bound intermediate state. Hence, by inhibiting the formation of the agonist-associated intermediate and fully active conformations of GCGR, RAMP2 binding decreases the probability of productive agonist-induced GCGR-Gs interaction, conceivably explaining at least part of the decreased GTP turnover (Fig. 1A) and GDP release rates (Fig. 1B).
Structure of GCGR/Gs in the presence of RAMP2
The observation that GCGR/RAMP2 co-complexes can suppress basal GTP turnover of Gs (Fig. 1A) suggests that RAMP2 does not fully prevent GCGR association with Gs but leads to unproductive coupling (Fig. S1D), which has been observed for other GPCRs29. To understand how RAMP2 induces this unproductive coupling with Gs, we obtained a structure of the GCGR/RAMP2/Gs complex by cryoEM. In the smFRET experiments with RAMP2, performed at low concentrations of receptor and Gs for a short time (~30 minutes), RAMP2 binding resulted in nearly full inhibition of active state(s), though Gs is still able to populate the fully outward conformation to some extent (Fig. 4B, salmon) relative to the distribution observed for agonist-bound receptor in the presence of RAMP2 (Fig. S4B, purple). However, we can force GCGR/RAMP2/Gs complex formation by incubating at high concentrations for longer times with excess stabilizing Nb35 (Fig. S5A–C). We ensured any GCGR/Gs present in the sample was complexed with RAMP2 by pulling down directly on FLAG-tagged RAMP2. Co-complex formation was confirmed by size-exclusion chromatography (SEC) and SDS-PAGE (Fig. S5B–C). This GCGR/RAMP2/Gs/Nb35 complex was subjected to cryoEM imaging to yield a final density map at a global nominal resolution of 2.90 Å (Fig. 5A, Fig. S5D, Table 1). We observed clear density for the agonist peptide and Gs but no density was obtained for RAMP2 (Fig. S5D). Additionally, though most parts of the receptor can be modeled unambiguously (Fig. S5E), some regions of GCGR cannot be built due to lack of density. These regions include the ECD, ICL3 and H8. While the GCGR ECD in the absence of RAMP2 exhibits well resolved density (Fig. 5C), in the presence of RAMP2 the GCGR ECD is not well-defined (Fig. 5B), consistent with experiments presented above showing RAMP2 enhances dynamics of this domain (Fig. 5B). The ECD of GCGR plays an important role in determining peptide potency20; the observed flexibility induced by RAMP2 is consistent with the decreased glucagon potency observed in cells (Fig. 1D).
Figure 5. Structure of GCGR-Gs in the presence of RAMP2.
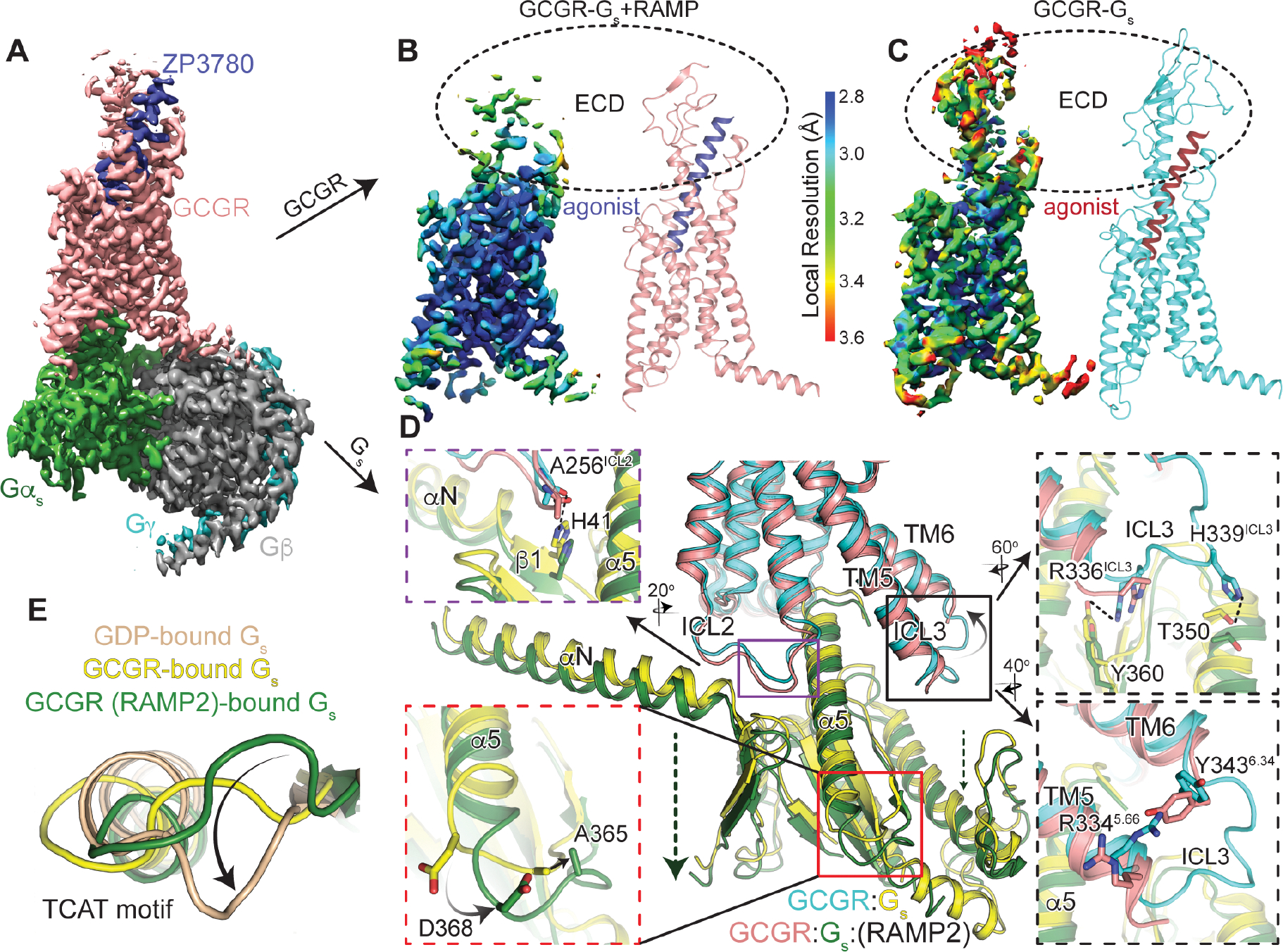
(A) Cryo-EM density map of the ZP3780 and Gs heterotrimer bound GCGR in the presence of RAMP2 colored by subunit. Salmon, GCGR; blue, ZP3780; Gαs, green; Gβ, grey; Gγ, cyan. Cryo-EM density for ZP3780-bound GCGR in the presence (B) and absence (C) of RAMP2 colored by local resolution demonstrates a significant disordering of the receptor ECD in the presence of RAMP2. The intracellular interface of GCGR with Gαs is perturbed by RAMP2 in several significant ways (D), including inducing disorder in ICL3 and the resulting loss of contacts with Gαs (black, top), a downward movement of ICL2 (purple), and a lack of stabilizing contacts across TM5/TM6 (black, bottom). Further, there are rearrangements in the backbone and side chains in the TCAT motif at the base of the α5 helix of Gαs. Comparing this motif to GDP-bound Gs (E) it is clear that this important loop in the G protein occupies a distinct conformation.
Table 1:
CryoEM data collection, model refinement and validation
| Data Collection | |
|
| |
| Voltage (kV) | 300 |
| Magnification | 96,000× |
| Total electron dose (e−/Å2) | 56.6 |
| Defocus range (μm) | −0.7 to −2.0 |
| Calibrated pixel size (Å) | 0.852 |
| Micrographs collected | 4343 |
|
| |
| Data Processing | |
|
| |
| Extracted particles | 2,154,997 |
| Particles used for final reconstruction | 212,196 |
| Final map resolution (Å, 0.143 FSC) | 2.90 |
| Map resolution range (Å) | 2.5 – 3.2 |
| Map sharpening B factor (Å2) | 103.2 |
|
| |
| Model Content | |
|
| |
| Initial models used (PDB code) | 6WPW (GCGR/Gs/Nb35) |
| Total number of atoms | 8,834 |
| No. of protein residues | 1,112 |
| No. of ligands | 0 |
|
| |
| Model Validation | |
|
| |
| CC map vs. model (%) | 77 |
| RMSD | |
| Bond lengths (Å) / Bond angles (°) | 0.002 / 0.598 |
| Ramachandran plot statistics | |
| Favored (%) | 97.4 |
| Allowed (%) | 2.6 |
| Outliers (%) | 0.0 |
| Rotamer outliers (%) | 0 |
| C-beta deviations | 0 |
| Clash score | 3.83 |
Several specific interactions stabilizing the GCGR ECD are missing or modulated in the presence of RAMP2. A key disulfide bond between the N-terminal helix of the ECD (C43ECD) and a loop (C67ECD) is no longer observed and might be broken (Fig. S6A). This disulfide is highly conserved among Family B GPCRs and might be important to preserve the ECD fold30. Even regions that remain structured in the extracellular portion of the receptor are perturbed; a hydrogen bond between K37ECD of the N-terminal ECD helix and S213ECL1 in ECL1 of the receptor is broken, along with a cation-π interaction stabilizing the N-terminal ECD helix (K35ECD/F31ECD) (Fig. S6B). The breaking of ECD interactions and specific structural elements affects peptide agonist/receptor interactions. E20 of the peptide agonist is no longer within hydrogen-bonding distance of Q1311.29 in TM1 (Fig. S6C), and deeper in the core of the receptor D1952.68 in TM2 forms an intrahelical salt bridge with R1992.72 at the expense of interaction with agonist, (Fig. S6D). The cryoEM density as well as the solution HDX-MS studies, which demonstrates increased ECD conformational sampling, suggest a linkage between perturbations in ECD conformation and intracellular TM6 conformation as observed by smFRET measurements; breakage and rearrangement of these interactions in the receptor core may provide a route for this negative allosteric communication.
ICL2 and ICL3 form an important part of the receptor-G protein binding interface (Fig. 5D)3,31. In the absence of RAMP2, ICL2 points into a hydrophobic pocket made up of residues in the αN/β1 hinge of Gαs e.g. the carbonyl oxygen of A256ICL2 hydrogen bonds with H41 of Gαs (Fig. 5D, purple box). However, in the presence of RAMP2, A256ICL2 is not within bonding distance of H41 of Gαs (Fig. 5D, purple box), due to the entire Gs being ~ 3 Å further removed from the core of the receptor (Fig. 5D). Additionally, in the presence of RAMP2, no density is seen for ICL3 presumably due to its increased flexibility, which might be a result of losing stabilizing interactions across TM5-6 like the loss of a cation-p interaction between R3345.66 and Y3436.34 in the presence of RAMP2 (Fig. 5D, bottom black box). This ICL3 disorder results in the loss of a number of interactions with Gαs such as H339ICL3 with T350 (Gαs) and R336 ICL3 with Y360 (Gαs) (Fig. 5D, top black box). The lack of interaction with ICL3 could contribute to the observed G protein shift away from the receptor core.
Binding of Gs begins with the translation of α5 to engage the receptor core. This leads to the rearrangement of the β6-α5 loop that contains the TCAT motif, which directly contacts the guanosine base of GDP32. The TCAT motif in Gs bound to GCGR in the presence of RAMP2 has moved ~5 Å (Ca of A365) away from the nucleotide-binding site compared to in the absence of RAMP2 (Fig. 5D, red box). This conformation of the TCAT might be more consistent with a conformation that cannot coordinate nucleotide. Gs bound to RAMP2-saturated GCGR displays a distinct TCAT motif relative to both GDP-bound Gs alone as well as GCGR-bound Gs (Fig. 5E). It is tempting to speculate that, in the presence of RAMP2, Gs adopts a conformation that cannot bind GTP and cause productive turnover, consistent with the Gs-sequestering effect observed previously (Fig. S1D).
GCGR extracellular and intracellular conformational dynamics by 3D variability analysis
In order to better interpret the lack of local density in portions of the cryoEM structure of GCGR/Gs in the presence of RAMP2 we mapped the conformational heterogeneity in our final set of particles onto 3 principal components using 3D variability analysis (Fig. S6E)33. Within the principal components we see the typical “normal” motional modes observed for large biomolecular complexes (Fig. S6E; PC1, PC2). However, within the major principal component (PC0) there is evidence of concerted structural changes between extracellular and intracellular regions of the receptor. At one end of the continuum of structural snapshots (cyan), the density throughout the receptor and peptide is very comparable to the structure of GCGR/Gs in the absence of RAMP2 (Fig. 6A), including the ECD conformation “capping” the peptide agonist (red) and ICL3/H8 conformations consistent with fully Gs-engaged, nucleotide-free complex3. At the alternate end of the PC0 continuum (salmon), the receptor ECD is in a completely distinct conformation having shifted away from the canonical peptide-bound conformation along with the top half of the agonist (blue). The TM region of the receptor does not display such conformational heterogeneity, consistent with the high resolution in the TM region (Fig. 5B). However, concurrent with the ECD movement away from the peptide binding region of the TM core, we observe H8 movement away from Gs upon loss of stable contacts (Fig. 4A). Such H8 contacts are important and common for Family B GPCR activity34. ICL3 has been shown to be important for receptor efficacy and G-protein selectivity31,34; it also occupies a distinct orientation in the alternate conformation observed from the principal component analysis (Fig. 6A). Thus, the likely explanation for the lack of structured density in the receptor ECD, ICL3, and H8 in our RAMP2-engaged complex is the presence of an ensemble of conformations in the final sample, ranging from those similar to the fully-active, canonical Gs-bound state to a distinct, RAMP2-induced conformation lacking many contacts necessary for Family B GPCR signaling through G-proteins.
Figure 6. RAMP2-induced conformational dynamics in the GCGR ECD inhibits intracellular activation.
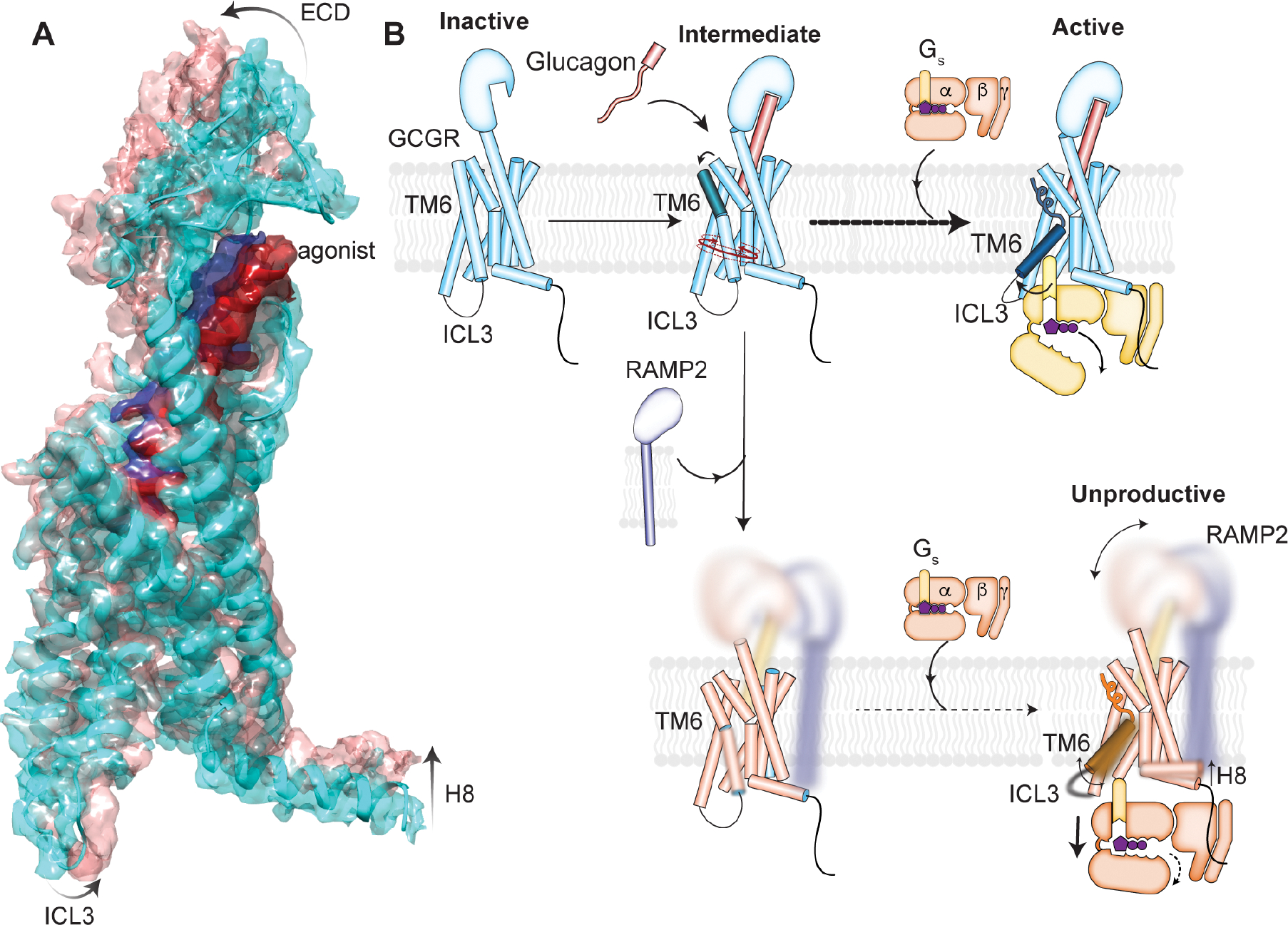
(A) The major principal component of the 3D variability analysis directly demonstrates that changes in the GCGR ECD and agonist conformation are linked to changes in the intracellular conformation including ICL3 and H8. (B) Proposed model for RAMP2-induced inhibition of GCGR activation. Glucagon binding to GCGR promotes population of a temporally distinct intermediate conformation of TM6, followed by full outward movement upon binding to Gs. RAMP2 binding to GCGR causes enhanced ECD conformational sampling as observed in both HDX-MS solution experiments as well as cryo-EM structural analysis. This increase in dynamics in the ECD inhibits formation of both intermediate and active states of TM6 observed by smFRET experiments, and any engagement with Gs results in a largely unproductive complex.
Discussion
In this work we present data on signaling modulation of a Family B GPCR, GCGR, by RAMP2. Our study investigates the effect of non-obligate heterodimerization of RAMP2 and GCGR, unlike previous studies that have focused on obligate receptor/RAMP heterodimers (e.g. CLR). RAMPs were first discovered as chaperones of the calcitonin-like receptor (CLR), with which they form obligate dimers and enhance the cell surface expression. Recently, however it has become clear that RAMPs alter trafficking, signaling and pharmacology of some Family B receptors with which they form non-obligate dimers. For example, RAMP2 binding to PTH1 receptor modulates signaling by causing a specific and selective increase in Gs activation35. On the other hand, RAMP2 has no effect on Gs activation by the type 1 corticotrophin releasing factor receptor (CRF1)36 and VPAC14. We demonstrate using in-cell, biochemical and biophysical assays that RAMP2 interacts with GCGR and this interaction has profound implications for GCGR signaling. In agreement with previous studies in cells, we found that GCGR overexpression enhances cell surface levels of RAMP237 and the presence of RAMP2 decreases glucagon potency at GCGR11. In a GTP-turnover assay, RAMP2 decreases GCGR-induced Gs activation in a GDP-dependent manner by slowing GDP release from the G protein. In addition to inhibiting Gs, RAMP2 has been shown to decrease ß-arrestin recruitment to GCGR12. We provide here biochemical data to demonstrate that this change in trafficking behavior of GCGR when engaged with RAMP2 is likely due to its inhibition of GRK5 phosphorylation. We show that RAMP2 broadly increases GCGR dynamics, including in the ECD, ICL3 and H8, and specifically stabilizes TM6 in an inactive state. Taken together our data suggests a model in which RAMP2 binding to GCGR increases ECD dynamics (Fig. 6B) which translates to decreased ligand potency. Several studies have shown single point mutations in the ECD, even if they are not in the direct ligand interacting residues, change ligand efficacy20. The role of Family B GPCR ECD conformation(s) in relation to intracellular activation remains underexplored, though there are likely differences between evolutionarily distinct subclasses of receptors. These differences in ECD conformations found in distinct subclasses of Family B GPCRs might alter their mode of interaction with RAMPs. For example, all three RAMPs interact with the calcitonin subfamily of receptors by “clamping” the receptor ECD in a manner where the extreme N-terminal helix is near parallel to the bilayer (Fig. S7A–C). To explore the conformation of GCGR ECD in the presence of RAMP2, we prepared covalently crosslinked GCGR/RAMP2/Gs complex and were able to observe low-resolution density for GCGR/RAMP2 containing micelles. These 2D classes showed a similar orientation to that predicted from AlphaFold (Fig. S6F) with the RAMP and GCGR ECDs appearing upright next to each other. Therefore, in contrast to calcitonin receptors, the ECD of peptide-bound glucagon receptor subfamily members is in an upright orientation with an N-terminal helix that is perpendicular to the bilayer (Fig. S7D). The GCGR ECD is probably prevented from occupying a “clamped” calcitonin-like conformation due to the differences in the helical nature of their peptide ligands, wherein glucagon is completely helical and calcitonin peptides are not.
RAMP2 binding appears to increase the disorder in the ECD of the receptor resulting in an enhanced population of the inward, inactive state of TM6 (Fig. 6B), though this does not seem to preclude G protein binding. The presence of RAMP2 slows GCCR-induced GDP release from Gs and inhibits GEF activity to levels lower than intrinsic basal nucleotide exchange; both suggest that interaction of RAMP2 with GCGR results in unproductive coupling to Gs. Hence, RAMP2 is a negative allosteric modulator of GCGR whose allosteric effects appears to be driven by increasing flexibility in parts of GCGR ECD that translates to unproductive Gs coupling and thereby, signaling. This kind of “dynamic allostery” has been proposed for other RAMP complexes5,8 and such flexibility driven allosteric effects play an underappreciated role in protein–protein interactions in GPCRs. The unproductive GCGR-Gs coupling in the presence of RAMP2 seems to sequester Gs away from other GPCRs. This type of sequestration has been observed for other receptors especially viral GPCRs. The human cytomegalovirus GPCRs, US27 and US28 couple to host G proteins and function as a G protein ‘sink’ so as to blunt the host immune response mounted thorough the chemokine receptors-G protein signaling29,38. Though the exact effect(s) of this Gs sequestration is unclear, a potentially interesting consequence may be found in glucagon signaling from the liver versus the kidney. We show here that RAMP2:GCGR ratios are important for achieving the observed changes in glucagon potency (Fig. 1D); the RAMP2:GCGR expression ratio is very low in liver (0.08:1) suggesting minimal inhibition and sequestration occurs, though in kidney the ratio is much higher (1.71:1)39 where the function of the glucagon receptor is not to regulate blood sugar but to modulate excretion of certain ions40. Differential expression of interacting proteins can significantly diversify biological functions41. Thus, the expression profiles of GCGR and RAMP2 may be uniquely titrated as a strategy to finely calibrate the way in which individual tissues, even individual cells, respond to the same extracellular stimulus. Understanding the effect of these GPCR-effector tissue specific interactions may allow for exploiting such variation as a source of targeted GPCR signaling output selectivity in drug development.
Limitations of the study
While this study employed a variety of biochemical, biophysical and structural experiments to probe the impact of RAMP2 binding on GCGR activation, these experiments all relied on biochemically purified versions of these proteins solubilized in detergent micelles. The effect of detergent rather than lipid bilayer incorporation of membrane proteins remains highly variable3,42 but we have previously shown comparable activity of GCGR in the two environments3. Further, the biochemical inhibition of GCGR signaling observed by RAMP2 is recapitulated in vitro (Fig. 1D) although these experiments rely on heterologously expressed GCGR and RAMP2. Our biochemical results suggest that binding of RAMP2 to GCGR substantially changes its activation behavior, by largely (but not universally) inhibiting downstream activation pathways; RAMP2 inhibition of GCGR phosphorylation by GRK5 is not inconsistent with previous studies suggesting a RAMP2-induced modulation of GCGR trafficking behavior12. Further experiments in RAMP2 knock out mice in response to glucagon stimulation would be highly useful to probe the role of this interaction in various cell types and organs in vivo.
STAR Methods text
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brian Kobilka (kobilka@stanford.edu).
Materials availability
Plasmids generated in this study will be distributed upon request.
Data and code availability
The cryo-EM density map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-29453 and model coordinates have been deposited in the Protein Data Bank (PDB) under accession number 8FU6.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains
E. coli BL21 (DE3) cells (Sigma-Aldrich) were used to express Nb35. E. coli Rosetta 2 (DE3) cells were used to express Gas.
Cell lines
Spodoptera frugiperda Sf9 cells (Expression Systems) were used for baculovirus generation and expression of wild type GCGR, RAMP2, GRK2, and GRK5. Trichuplusia ni cells (Expression Systems) were used for expression of heterotrimeric Gs and Gβ1γ2. Both insect cell systems were maintained at 27°C with shaking.
Inducible Expi293 cells (Thermo Fisher) were used to express mutants of GCGR. HEK293 cells (ATCC) were used for BRET binding assays, and the same cells with a stably expressing FRET cAMP biosensor43 were used to measure cAMP levels. Both HEK cell derivatives were maintained at 37°C in a humidity and CO2 controlled incubator and Expi293 cells were maintained with shaking.
METHOD DETAILS
In vitro assay cell lines and constructs
HEK293 cells stably expressing an Epac1-based fluorescence resonance energy transfer (FRET) cAMP biosensor capable of reporting intracellular cAMP levels (cAMP biosensor cells) described previously43 were transfected with expression vector DNA encoding a FLAG-tagged human GCGR alone or together with HA-tagged RAMP2. The FLAG-tagged GCGR (FLAG-GCGR) consisted of a HA-signal peptide (MKTIIALSYIFCLVFA) followed by a FLAG tag (DYKDDDD), a small linker comprising a NotI restriction site (AAA) and the mature human GCGR (UNIPROT ID: P47871, residues 26–477). Likewise, a FLAG-tagged β2-adrenergic receptor was generated. The HA-tagged RAMP2 (HA-RAMP2) consisted of the HA-signal peptide followed by the HA-tag (YPYDVPDYA), a small linker comprising a NotI restriction site (AAA) and the mature human RAMP2 (UNIPROT ID: O60895, residues 36–175;). cAMP biosensor cells were maintained in growth medium (D-MEM, Gibco; supplemented with 10% fetal bovine serum, Gibco; 1% sodium pyruvate, Gibco; 1% MEM non-essential amino acids, Gibco; and 1% penicillin–streptomycin Solution, Gibco) and 50 μg/ml zeocin for selection of cell expressing the FRET-based cAMP biosensor construct.
For characterization of the antagonistic properties of ZP7680, the FLAG-tagged human GCGR was stably expressed in the HEK293 Epac1 biosensor cell line. A clone with an expression level corresponding to the transiently expressed FLAG-tagged human GCGR described above was used. The stable expressing cell line was maintained in growth medium with 50 μg/ml zeocin and 0.5 mg/ml G418, latter for selection of cells expressing the FLAG-GCGR construct.
cAMP assay
To assess the impact of RAMP2 on GCGR signaling, the cAMP biosensor cells were transiently transfected in suspension as described previously44 with different amounts of vector DNA encoding the FLAG-GCGR without or in presence of a fixed concentration of vector DNA encoding the HA-RAMP2. In brief, cAMP biosensor cells were brought into suspension by trypsination and resuspended in growth medium to a density of 200,000 cells/ml. For 1 ml of cell suspension transfected, 3 μL Fugene6 (Promega) in 57 μl OptiMEM (Gibco) was mixed with a total of 1 μg DNA in 25 μl OptiMEM. To vary surface expression levels (monitored by cell surface ELISA below) a range of 3–50 ng vector DNA encoding FLAG-GCGR supplemented with vector DNA to a total of 1 μg or with 800 ng vector DNA encoding HA-RAMP2 and vector DNA to a total of 1 μg was used. The Fugene6/DNA mixture was incubated for 20 min at room temperature and then added to 1 ml of cell suspension. Transfected cells were then seeded in black poly-D-lysine coated 96-well plates at 100 μl/well. Amounts were scaled up depending on the number of wells to be assayed. The cAMP assay was performed 48 hours after transfection. Prior to the assay, cells were washed once in 100 μl assay buffer (HBSS; Gibco, 20 mM HEPES pH 7.5, 1 mM CaCl2, 1 mM MgCl2, 0.1% BSA), and replaced with 100 μl of fresh assay buffer. After 15 min preincubation, cells were stimulated with glucagon diluted to different concentrations in 50 μl assay buffer and cAMP levels monitored continuously for 60 min by recording the change in FRET of the cAMP biosensor using an Envision plate reader (Perkin Elmer) equipped with filters for measuring the fluorescence of mCerulean fluorescent protein (474 nm) and mCitrine fluorescent protein (524 nm) following excitation of mCerulean (434 nm). To determine the EC50 and Emax of glucagon, the area under the curve for the individual cAMP traces was calculated and plotted against the concentration of glucagon and fitted by nonlinear regression to a 4-parameter logistic curve using GraphPad Prism.
Cell surface ELISA assay
To assess the expression level of GCGR and the impact of GCGR and RAMP2 on surface expression levels, the FLAG-tagged GCGR and HA-tagged RAMP2 constructs described above were used in a direct cell ELISA assay as described previously45. In brief, cells transfected for the cAMP assay above were seeded in white poly-D-lysine coated 96-well plates at 100 μl/well. At the same time as the functional assay, cells were fixated with 4% paraformaldehyde in DPBS, blocked in 3% dry milk in DPBS, incubated for 1 hour with HRP-conjugated anti-Flag antibody (Sigma Aldrich), or HRP-conjugated anti-HA antibody (R&D Systems), both diluted 1:2000 in 3% dry milk, followed by washing 4 times first in 3% dry milk in DPBS and then in DPBS. Surface expressed proteins were then quantified by addition of 60 μl DPBS buffer and 20 μl HRP substrate (Bio-Rad) per well, incubation for 10 min and detection of luminescence using an EnVision plate reader (Perkin Elmer). FLAG-GCGR luminescence was normalized to that of 50 ng vector DNA transfected and HA-RAMP2 luminescence normalized to that of 800 ng HA-RAMP2 vector DNA alone and plotted against each other.
In vitro characterization of ZP7680
The glucagon antagonist ZP7680 was designed by removing His1 and replacing Asp9 with Nle and Ser11 with Ala46 in the dasiglucagon backbone47 to obtain a soluble and potent glucagon antagonist with minimal intrinsic efficacy. ZP7680 was characterized in vitro using the HEK293 Epac1 biosensor cell line stably transfected with the FLAG-tagged GCGR. Measurement of cAMP levels was determined as described above. Concentration response curves were generated in absence (as above) or presence of 500 μM IBMX to assess the intrinsic efficacy of ZP7680 and the previously described glucagon antagonist des-His1[Glu9]-Glucagon-NH248. For generation of Schild plots to calculate antagonist KB values, the antagonists were preincubated at different concentrations for 15 min prior to adding glucagon at increasing concentrations. No IBMX was added. The GraphPad Prism Gaddum/Schild EC50 shift function was used to determine antagonist KB-values.
Nanoluc-GCGR binding assay
The Nanoluc-GCGR fusion protein (Nluc-GCGR) used to assess binding affinities of TAMRA-labeled peptides in absence or presence of RAMP2 consisted of the IL6-signal peptide, the Nanoluc protein, a flexible linker sequence (SGGGGSSGRPQGA) followed by the mature human GCGR (amino acids 26–477; Uniprot no. P47871). The fusion protein was generated by subcloning cDNA encoding the linker and the mature human GCGR into the pNLF1-secN [CMV/Hygro] Vector (Promega).
The antagonist peptide ZP7680 was modified by addition of a Lys-TAMRA moiety following the T29 residue [ZP7680-30K(TAMRA]. The agonist peptide ZP3780 was modified by substituting the E20 with a Lys-TAMRA moiety [ZP3780-E20K(TAMRA)], and dasiglucagon was modified by substituting A17 with a Lys-TAMRA moiety, and E21S [Dasi-E21S-A17K(TAMRA)] (Fig. 2A).
To perform the BRET binding assay, HEK293 cells were transiently transfected with Nluc-GCGR in presence or absence of RAMP2 using the protocol described above except that 6 ng Nluc-GCGR or 12 ng Nluc-GCGR with 800 ng RAMP2 was used to achieve similar expression levels of the NLuc-GCGR in absence or presence of RAMP2 measured by Nanoluc-luminescence counts. 48 hours after transfection, cells were detached in DPBS without calcium and magnesium (Gibco) and 2 mM EDTA (Sigma), counted, and resuspended to a density of 1.2e6 cells per milliliter in ice-cold binding buffer (50 mM HEPES pH 7.4, 5 mM EGTA [Sigma], 5 mM MgCl2, 0.05% casein solution [Sigma]) and placed on ice. TAMRA-labeled peptides were prepared and diluted in binding buffer to indicated concentrations. Non-specific binding was determined for same TAMRA-labeled peptide concentrations in presence of 100 μM ZP3780. TAMRA-peptide dilutions were added to wells of a 384-well Opti-Plate (Perkin Elmer), with or without 100 μM ZP3780 to a final volume of 12.5 μL and placed on ice to prevent receptor internalization upon addition of cells. To initiate the binding assay, 12.5 μL cell suspension was added to each well, and incubated with shaking at 4°C for 6 hr. To determine the TAMRA-labeled peptide binding, 25 μL of 1:500 diluted NanoBRET™ Nano-Glo® Substrate (Promega) in binding buffer was added to each well, incubated for 5 min at 4°C while shaking and then transferred to an Envision plate reader equipped with 470 nm and 595 nm emission filters to quantify Nanoluc luciferase luminescence and TAMRA fluorescence, respectively. The ratio of TAMRA fluorescence over Nanoluc luciferase luminescence was calculated to determine total and non-specific (in presence of 100 μM ZP3780) binding signals and subtracted to obtain the specific binding. The specific binding signal was plotted against the concentration of the TAMRA-labeled peptide and fitted to a one-site binding isotherm by non-linear regression using GraphPad prism.
Purification of wild type GCGR
Human glucagon receptor (Q27-F477) with an N-terminal FLAG and C-terminal octahistidine tag with 3C sites to remove tags was expressed as previously described3 using the baculovirus method in Spodoptera frugiperda (Sf9) cells. L-168,049 was added to 10 μM final upon infection and cells were collected 48 hours later and stored at −80°C until purification. GCGR was extracted from cell membranes with 1% lauryl maltose neopentyl glycol (L-MNG; Anatrace) and 0.1% cholesterol hemisuccinate (CHS) in 20 mM HEPES pH 7.5, 150 mM sodium chloride (NaCl), 20% glycerol, 5 mM imidazole, 30 μM NNC0640 ligand, and the protease inhibitors benzamidine and leupeptin, along with benzonase (Sigma-Aldrich). After douncing and stirring at 4 °C 1.5 hours, the solubilization mixture was centrifuged to pellet cell debris, and the supernatant was applied to nickel-chelating sepharose resin. After binding to nickel resin for 2 hours, the resin was washed with 20 mM HEPES pH 7.5, 150 mM NaCl, 0.1% L-MNG, 0.01% CHS, 20% glycerol, 5 mM imidazole, and leupeptin/benzamidine, followed by elution in the same buffer supplemented with 200 mM imidazole. 2 mM calcium chloride was added to the elution from the nickel resin and subsequently applied to M1 anti-FLAG immunoaffinity resin. The protein was washed in 20 mM HEPES pH 7.5, 100 mM NaCl, 0.05% L-MNG, 0.005% CHS with 2 mM calcium chloride, and eluted in the same buffer (without calcium chloride) with 5 mM ethylenediaminetetraacetic acid (EDTA) and FLAG peptide. Finally, monomeric GCGR was separated from multimers and aggregates via size exclusion chromatography on an S200 10/300 Increase gel filtration column (GE Healthcare) in 20 mM HEPES pH 7.5, 100 mM NaCl, 0.02% L-MNG and 0.002% CHS. Pure, monomeric GCGR was then spin concentrated to ~300 μM and flash frozen at −80 °C until further use.
Purification of minimal cysteine GCGR
The minimal cysteine version of GCGR (C171T, C240A, C287A, C401V) and related constructs used for bulk fluorescence and single-molecule FRET experiments (F31C, F245C introductions) were expressed as previously described3 in Expi293F cells. Constructs were transfected into Expi293F (Thermo Fisher) cells expressing the tetracycline repressor with the Expifectamine transfection kit (Thermo Fisher) following manufacturer’s directions with induction 2 days post-transfection with doxycycline (4 μg/mL and 5 mM sodium butyrate) with 1 μM L-168,049 (Tocris Bioscience). Pellets were frozen and stored at −80°C for later purification. Cells were dounced and solubilized with 20 mM HEPES pH 7.5, 100 mM NaCl, 20% glycerol, 1% L-MNG, 0.1% CHS, protease inhibitors and benzonase, followed by purification on anti-FLAG immunoaffinity chromatography as above. Following FLAG purification, minimal cysteine constructs were labeled with fluorophores as described below followed by size exclusion chromatography on Superdex 200 Increase 10/300 gel filtration column (GE Healthcare) in 20 mM HEPES pH 7.5, 100 mM NaCl, 0.02% L-MNG and 0.002% CHS. Aliquots of labeled protein were flash frozen and used as below.
Purification of RAMP2
The sequence for human RAMP2 (UNIPROT ID: O60895, residues 43–175)(cDNA.org) was cloned with with an N-terminal FLAG tag, C-terminal eGFP followed by a hexahistidine tag along with a 3C protease site between RAMP2 and eGFP was cloned into the pFastBac vector. Protein was expressed using the baculovirus method in Sf9 cells as for GCGR. Pellets were frozen at −80°C until purification. Cells were solubilized with 20 mM HEPES pH 7.5, 750 mM sodium chloride, 20% glycerol, 1% n-dodecyl-ß-D-malopyranoside (DDM; Anatrace), protease inhibitors and benzonase and dounced on ice. After stirring at 4°C 1.5 hours, solubilized RAMP-eGFP was separated from cell debris by centrifugation. 2 mM Ca2+ was added to the supernatant before purification on M1 anti-FLAG immunoaffinity resin. Following FLAG purification, 3C protease was added 1:10 w/w with RAMP2-eGFP to cleave off the eGFP overnight at 4°C. Cleaved, monomeric RAMP2 was separated from eGFP and excess 3C via reverse nickel chromatography, followed by purification of monomeric RAMP2 with size exclusion chromatography on Superdex 200 Increase 10/300 gel filtration columns (GE Healthcare) in 20 mM HEPES pH 7.5, 200 mM NaCl, and 0.05% DDM. Aliquots of monomeric RAMP2 were flash frozen for later experiments. Typically, a single SEC run is sufficient to remove nearly all multimeric RAMP2; the SEC-pure, concentrated, flash frozen RAMP2 used for biochemical and structural experiments herein was subjected to further analytical SEC on the same Superdex 200 Increase 10/300 gel filtration column (GE Healthcare) to confirm its monodisperse nature (Fig. S1A).
Purification of GRKs
Full length wild type human GRK5 and bovine GRK2 were cloned into pVL1392 vector with C-terminal hexa-histidine tags for baculovirus production. GRKs were expressed and purified as previously described49,50. In brief, Sf9 cells were infected with BestBac baculovirus at a density of 4.0 × 106 cells per mL and collected 48 hours post-infection. Cells were pelleted and frozen for later purification. Cells were dounced and solubilized with 20 mM HEPES pH 7.5, 300 mM NaCl, 1 mM dithiothreitol (DTT; Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich), 1 mM EDTA, 0.02% Triton X-100 (Sigma), protease inhibitors and benzonase. Following 1.5 hour incubation at 4 °C, cell debris was pelleted by centrifugation. The supernatant was incubated with nickel-chelating sepharose resin for 2 hours at 4 °C after adding 20 mM imidazole and 5 mM magnesium chloride. Following washing nickel resin with 20 mM HEPES pH 7.5, 300 mM NaCl, 1 mM DTT, 1 mM PMSF, protease inhibitors and 0.05% DDM with 40 mM imidazole, the protein was eluted with the same buffer supplemented with 200 mM imidazole. The eluate was applied to a MonoS 10/100 column (GE Healthcare) for cation-exchange chromatography purification and eluted with a linear sodium chloride gradient. Monomeric kinase was purified by size exclusion chromatography on Superdex 200 Increase 10/300 gel filtration columns in 20 mM HEPES pH 7.5, 200 mM NaCl, 100 μM tris(2-carboxyethyl)phosphine) (TCEP; Sigma), and 0.05% DDM. Aliquots of purified human GRK5 and bovine GRK2 were flash frozen for later experiments.
Expression & purification of heterotrimeric G-proteins
Heterotrimeric Gs were expressed in Trichoplusia ni (T. ni) using the BestBac method (Expression Systems) and purified as previously described3,13. Briefly, two baculoviruses were used, one encoding the respective Gα subunit and the other encoding both Gβ1 and Gγ2, along with a histidine tag and HRV 3C protease site at the amino terminus of the β-subunit. T. ni cells were infected with the two baculoviruses for 48 hours and were subsequently harvested by centrifugation and lysed in 10 mM Tris, pH 7.5, 100 μM MgCl2, 5 mM β-mercaptoethanol (βME), 20 μM GDP and protease inhibitors. The resulting membranes were harvested by centrifugation and solubilized in a buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl, 1% sodium cholate, 0.05% DDM, 5 mM MgCl2, 5 mM βME, 5 mM imidazole, 20 μM GDP and protease inhibitors. The solubilization reaction was homogenized with a Dounce homogenizer and incubated with stirring at 4 °C for 1.5 hours followed by centrifugation. The soluble fraction was loaded to Ni-chelated Sepharose and gradually detergent-exchanged into 0.1% DDM. The protein was eluted in the above buffer supplemented with 200 mM imidazole and dialyzed overnight into 20 mM HEPES pH 7.5, 100 mM NaCl, 0.1% DDM, 1 mM MgCl2, 5 mM βME and 20 μM GDP along with HRV 3C protease.
The 3C-cleaved heterotrimer was further purified away from 3C and uncleaved protein with reverse Ni chromatography and the resulting G protein was dephosphorylated with lambda protein phosphatase (NEB), calf intestinal phosphatase (NEB), and Antarctic phosphatase (NEB) in the presence of 1 mM manganese chloride (MnCl2). After dephosphorylation, the protein was bound to a MonoQ 10/100 GL column (GE Healthcare) in 20 mM HEPES pH 7.5, 50 mM NaCl, 1 mM MgCl2, 0.05% DDM, 100 μM TCEP, and 20 μM GDP and washed in the same buffer, followed by elution with a linear gradient from the loading buffer to the same buffer with 500 mM NaCl. The main peak with G protein heterotrimer was collected and dialyzed into 20 mM HEPES pH 7.5, 100 mM NaCl, 0.02% DDM, 100 μM TCEP and 20 μM GDP overnight, followed by spin-concentration to <250 μM, addition of 20% glycerol, and flash-freezing in liquid nitrogen and storage at −80°C until further use.
Purification of Gαs and Gβ1γ2
Gαs and Gβ1γ2 were expressed and purified for the GDP release assay as previously described3. Gβ1γ2 was expressed and purified in an identical manner to the heterotrimer purification above (without co-infection with Gαs virus) aside from the linear elution gradient from MonoQ 10/100 GL column (GE Healthcare), which for Gβ1γ2 is from the loading concentration of 50 mM NaCl to a final 250 mM NaCl concentration over 7.5 column volumes. Human Gαs subunit with an amino-terminal hexahistidine tag with a HRV 3C protease site to cleave off the histidine tag was expressed in Rosetta 2 (DE3) cells (EMD Millipore) in a pET28a vector. Cells transformed with the above vector were grown in Terrific Broth to an OD600 of ~0.6 followed by induction of protein production with 0.5 mM isopropyl B-D-1-thiogalactopyranoside (IPTG; Sigma-Aldrich). After growth overnight at room temperature, cells were harvested and resuspended in lysis buffer composed of 50 mM HEPES pH 7.5, 100 mM NaCl, 1 mM MgCl2, 50 μM GDP, 5 mM βME, 5 mM imidazole and protease inhibitors. Cells in lysis buffer with subjected to sonication with a 50% duty cycle, 70% power for 4 cycles of 45 seconds. Intact cells and cell debris was removed with centrifugation, and the resulting supernatant was bound to Ni-chelated Sepharose resin and washed several times with lysis buffer in batch, followed by several column volumes washing on-column and elution with lysis buffer supplemented with 200 mM imidazole. Ni-purified protein was dialyzed overnight at 4°C against 20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM MgCl2, 20 μM GDP, 5 mM βME and 5 mM imidazole along with HRV 3C protease to cleave off the amino-terminal histidine tag. Cleaved Gα subunit was passed through Ni-chelated sepharose resin to remove uncleaved protein, 3C protease, and amino-terminal histidine tags. The protein was then concentrated and run on a Superdex 200 10/300 Increase column in 20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM MgCl2, 20 μM GDP, and 100 μM TCEP.
Purification of Nb35
Nb35 was expressed & purified as previously described51. Briefly, plasmid was transformed into Escherichia coli BL21 cells and grown to an optical density 0.7–1.0 and expression was induced with 1 mM IPTG overnight at room temperature. Cells were harvested and lysed followed by purification via nickel affinity chromatography and size exclusion chromatography on a Superdex S200 10/300 Increase gel filtration column (GE Healthcare) in 20 mM HEPES pH 7.5 with 150 mM sodium chloride. Purified Nb35 was flash frozen and stored at −80°C for later use.
Formation & purification of GCGR-RAMP2-Gs-Nb35 complex
GCGR-RAMP2-Gs-Nb35 complex was formed as previously described for GCGR-Gs-Nb353 complex aside from the addition of separately purified RAMP2 (described above). 1% L-MNG/0.1% CHS was added to purified Gαsß1γ2 for 1 hour on ice in order to exchange detergents from the DDM used for initial purification. ZP3780 was dissolved to 5 mM in dH2O and added to purified apo-GCGR to 500 μM final and incubated at room temperature for 1 hour. A 1.2-fold molar excess of agonist-bound GCGR was added to purified RAMP2 and incubated for an additional hour at room temperature. Detergent-exchanged Gαsß1γ2 was then added to the GCGR-ZP3780-RAMP2 complex at a 1.5-fold molar excess with receptor and incubated at room temperature for 2 hours before addition of a 2-fold molar excess of Nb35 (with respect to Gαsß1γ2) and further incubation on ice for 1.5 hours. Finally, apyrase (1 unit, NEB) and 3C protease (1:10 w/w ratio with GCGR) were added to stabilize the nucleotide-free state of Gαsß1γ2 and cleave off the M1 FLAG tag of the receptor. The final molar ratios of protein components was 1:1.2:1.8:3.6 (RAMP2:GCGR:Gs:Nb35). The complex was purified with M1 anti-FLAG affinity chromatography as previously described3 but using the M1 FLAG tag on the RAMP2 protein rather than on GCGR to ensure that any GCGR/Gs complex present was bound to RAMP2. Briefly, a 4-fold volume of 20 mM HEPES pH 7.5, 100 mM sodium chloride, 0.8% L-MNG/0.08% CHS, 0.27% glyco-diosgenin (GDN; Anatrace)/0.027% CHS, 1 mM magnesium chloride, 10 μM ZP3780, and 2 mM calcium chloride was added to the complexing reaction, followed by purification with M1 anti-FLAG chromatography. The complex was loaded to M1 anti-FLAG resin equilibrated with 20 mM HEPES pH 7.5, 100 mM sodium chloride, 0.3% L-MNG/0.03% CHS, 0.1% GDN/0.01% CHS, 5 μM ZP3780, and 2 mM calcium chloride and washed with 2 column volumes of the same. The complex was further washed with 4 additional column volumes of the same buffer with progressively lower detergent concentrations, followed by elution with 20 mM HEPES pH 7.5, 100 mM sodium chloride, 0.00075% L-MNG/0.000075% CHS, 0.00025% GDN/0.000025% CHS, 10 μM ZP3780, 5 mM EDTA, and FLAG peptide. The eluted complex was supplemented with 100 μM TCEP, analyzed by gel and size exclusion chromatography and concentrated to ~9 mg ml−1 for electron microscopy.
Crosslinking of the GCGR/RAMP2/Gs complex
The complex above containing GCGR, agonist, RAMP2, Gs, and Nb35 was covalently crosslinked by adding disuccinimidyl dibutyric urea (DSBU, Thermo Scientific) to a final concentration of 3 mM for 1 hour on ice. Covalent crosslinking was confirmed by SDS-PAGE gel analysis as observed by a loss of monomeric complex components, and the resulting crosslinked complex was analyzed by size exclusion chromatography to confirm a lack of multimeric or aggregated species (data not shown).
Cryo-EM data acquisition & processing
An aliquot of purified GCGR-RAMP2-Gs-Nb35 complex was applied to glow-discharged 200 mesh grids (Quantifoil R1.2/1.3) at a concentration of ~9 mg ml−1 and vitrified using a Vitrobot Mark IV (Thermo Fisher Scientific) at 100% humidity at 4 °C after blotting for 3 s with a blot force of 3. CryoEM images were collected on a Titan Krios operated at 300 kV at a nominal magnification of 96,000x using a Gatan K3 Summit direct electron camera in counting mode corresponding to a pixel size of 0.85 Å. A total of 4343 image stacks were obtained with a dose rate of 16.4 e−/pixel/s and total exposure time of 2.5 s with 0.05 s per frame, resulting in a total dose of 56.6 electrons per Å2. The defocus range was set to −0.7 to −2.0 μm.
All subsequent processing was performed with CryoSPARC52. Dose-fractionated image stacks were imported to CryoSPARC and subjected to patch-based beam-induced motion correction and contrast transfer function (CTF) estimation. A total number of 2,612,672 particles were picked with a template-based auto-picking protocol, followed by removal of particles from micrographs with a CTF resolution estimation of greater than 3.5 Å. A subset of 434,551 particles were selected after 2 rounds of 2-dimensional (2D) classification. These particles were further subjected to 3-dimensional (3D) classification with heterogeneous classification into 4 classes, resulting in a class containing 212,196 final particles, which were then reconstructed to 2.90 Å nominal resolution at FSC of 0.143 with non-uniform refinement53. Local resolution was estimated within CryoSPARC followed by 3D-variability analysis33 of the final particle set into 3 motional modes filtered to 5 Å.
Model building and refinement
The initial template for GCGR, Gs, agonist peptide, and scFv16 were derived from the previously determined structure in the absence of RAMP2 (PDB 6WPW)3. Models were docked into the EM density map using UCSF Chimera54, followed by manual building in Coot55. The final model was subjected to global refinement and minimization in real space using phenix.real_space_refine in Phenix56. Residues with weak side chain density were stubbed to their Cβ position while preserving sequence information. Molprobity57 was used to evaluate geometry. The final model parameters are shown in Table 1.
Phosphorylation ATP depletion assay
GCGR was diluted to 40 μM in phosphorylation buffer composed of 100 mM HEPES pH 7.5, 35 mM sodium chloride, 5 mM magnesium chloride, 0.01% L-MNG/0.001% CHS, 20 μM diC8-PtdIns(4,5)P2, 100 μM TCEP and 80 μM ATP. Various ligands were added to 150 μM and bound to receptor at room temperature for 1 hour. Aliquots were split in half and RAMP2 added 0.75:1 with receptor and incubated for a further 1 hour at room temperature. GRK5 was diluted to 4 μM in the phosphorylation buffer above without ATP and incubated on ice until the start of the reaction, when equal volumes of receptor solution with different ligands and RAMP2 and GRK5 solution were mixed. Total ATP was measured by mixing receptor with phosphorylation buffer in the absence of GRK5, and intrinsic GRK5 activity was measured by mixing GRK5 solution with phosphorylation buffer without receptor. The reaction was allowed to proceed for 90 minutes at room temperature before quenching with equal volume 20 mM HEPES pH 7.5, 400 mM NaCl, and 10 mM EDTA on ice. Residual ATP was measured by adding equal volume of GTPase-GLO™ luciferase detection reagent for 10 minutes, and luminescence detected on SpectraMax Paradigm plate reader. The method is described graphically in Fig. S2C. Raw luminescence values were converted to percent depleted values by normalizing to receptor alone (0% depletion) and zero signal (100% depletion). Statistical differences between conditions were calculated in GraphPad Prism 9 and the P values displayed are described in the figure panel.
Phosphorylation gel assay
GCGR (2.0 μM final) was equilibrated in phosphorylation buffer (100 mM HEPES pH 7.5, 35 mM sodium chloride, 5 mM magnesium chloride, 0.01% L-MNG/0.001% CHS, 20 μM diC8-PtdIns(4,5)P2, 100 μM TCEP and 1 mM ATP) and agonist ZP3780 was added to 10 μM final. For samples with GRK2, purified Gß1γ2 was added to 1 μM final. RAMP2 was added to 5 μM final. The reaction was equilibrated for 1 hour at room temperature to allow binding of agonist and RAMP2 to receptor before kinase (GRK2 or GRK5) was added in 200 nM increments (1:10 with GCGR each addition). The reaction(s) were sampled at various time points by quenching with 4x Laemmli Sample Buffer (Bio-Rad) with βME. Samples were analyzed by gel electrophoresis and stained with Pro-Q™ Diamond phosphoprotein gel stain (Thermo Fisher). Total protein was measured by staining with Coommassie blue or SYPRO Ruby (Thermo Fisher) protein gel stain (Thermo Fisher). Pro-Q™- and SYPRO™-stained gels were imaged with a Typhoon laser-scanner (Cytiva) using a 532 nm excitation wavelength and a 560 nm longpass filter.
GTP turnover assay
The GTP turnover assay was done using a modified version of the GTPase-GLO™ assay (Promega) as previously described3,13. Briefly, the final reaction was composed of 20 mM HEPES pH 7.5, 100 mM sodium chloride, 10 mM magnesium chloride, 100 μM TCEP, 0.01% L-MNG/0.001% CHS, variable concentrations of GDP, 10 μM GTP. Prior to initiating the reaction, GCGR was incubated in the presence of excess (20 μM) agonist ZP3780 in buffer with 20 mM HEPES pH 7.5, 100 mM sodium chloride, 0.01% L-MNG/0.001% CHS and 20 μM GTP for 1 hour in order to bind agonist. Concurrently, G protein was exchanged to L-MNG/CHS by adding 1% L-MNG/0.1% CHS for 1 hour on ice followed by dilution to 2 μM into G-protein buffer containing 20 mM HEPES pH 7.5, 100 mM sodium chloride, 20 mM magnesium chloride, 200 μM TCEP, 0.01% L-MNG/0.001% CHS and various concentrations of GDP. The solutions containing Gs and GCGR at 2-fold their final reaction concentrations were mixed to initiate the reaction. After incubation for various times (Fig. S1B) GTPase-Glo reagent supplemented with 10 μM adenosine 5’-diphosphate (ADP) was added and incubated with the sample for 30 minutes at room temperature. Luminescence was measured after addition of detection reagent for 10 minutes at room temperature using a MicroBeta Counter or using a SpectraMax Paradigm plate reader. Tests of statistical significance were performed in GraphPad Prism 9 and are described in the relevant figure legends.
GDP release assay Gαs and Gβ1γ2
The single turnover GDP release assay was performed similar to previously described3 with slight modifications. Briefly, Gαs in 20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 100 μM TCEP and 5 μM GDP was diluted to 0.8 μM in GDP-loading buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM EDTA and 100 μM TCEP) with 2.5 μM [3H]-GDP (Perkin Elmer). Gαs was incubated in GDP-loading buffer for 1 hour at room temperature, after which, purified Gß1γ2 was added to a 1.2-fold molar excess and allowed to form complex with Gαs for 30 minutes at room temperature. GCGR was diluted to 10 μM in 20 mM HEPES pH 7.5, 100 mM sodium chloride, 0.01% MNG/0.001% CHS, and 2 mM GTP and incubated with 100 μM ZP3780 for 1 hour at room temperature. The reaction was initiated by mixing the agonist-bound GCGR with [3H]-GDP loaded Gs heterotrimer at final concentrations of 5 μM and 200 nM, respectively. The reaction was stopped at various time points by dilution of the 20 μL reactions with 500 μL ice-cold wash buffer (20 mM HEPES pH 7.5, 150 mM sodium chloride, 20 mM magnesium chloride) and immediately filtered using a microanalysis filter holder (EMD Millipore) and pre-wet mixed cellulose filters (25 mM, 0.22 um). The filter was subsequently washed three times with 500 μL ice-cold wash buffer. Radioactivity was measured by liquid scintillation spectrometry. [3H]-GDP off-rates were calculated in GraphPad Prism 9.
RAMP2/GCGR pulldown assay
GCGR was diluted to 15 μM in 20 mM HEPES pH 7.5, 100 mM sodium chloride and 0.01% MNG/0.001% CHS, 5 mM magnesium sulfate and 2 mM calcium chloride along with a 1:10 w/w amount of 3C protease to cleave off the FLAG tag on GCGR. RAMP2 was added to ~10 μM final. The sample was split and various ligands (full agonist ZP3780, partial agonists GLP-1 and oxyntomodulin, negative allosteric modulator L-168,049, and antagonist peptide ZP7680) were added to 150 μM (10-fold molar excess with GCGR) final concentration and incubated overnight on ice. The next day, samples were purified on M1 anti-FLAG affinity resin using the residual M1 FLAG tag on the RAMP2. Samples were bound to M1 anti-FLAG resin equilibrated with 20 mM HEPES pH 7.5, 100 mM sodium chloride and 0.01% MNG/0.001% CHS and 2 mM calcium chloride for 20 minutes at room temperature, followed by washing over 15 minutes with ~5 column volumes of the same buffer to remove unbound GCGR. Samples were eluted with 20 mM HEPES pH 7.5, 100 mM sodium chloride and 0.01% MNG/0.001% CHS, 5 mM EDTA and FLAG peptides and analyzed by SDS-page chromatography (Fig. S3C).
Receptor labeling with IANBD-amide
Purified minimal cysteine GCGR with introduced cysteines at either position 345 (F345C, TM6) or position 31 (F31C, ECD) were diluted to 50 μM in 20 mM HEPES pH 7.5, 100 mM sodium chloride and 0.02% MNG/0.002% CHS. N,N’-dimethyl-N-(iodoacetyl)-N’-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD-amide; Thermo Fisher) was solubilized in DMSO to make a 25 mM stock, then added to GCGR at 5-fold molar excess (250 μM) and incubated at room temperature 45 minutes. The reaction was quenched with excess cysteine and the reaction mixture was purified by size-exclusion chromatography on a Superdex 200 Increase 10/300 gel filtration column in 20 mM HEPES pH 7.5, 100 mM sodium chloride and 0.01% MNG/0.001% CHS. The IANBD-amide-labeled minimal cysteine GCGR was concentrated and aliquots were flash frozen for subsequent experiments.
Bulk fluorescence experiments
The aliquots of 31C- or 345C-IANBD-amide labeled minimal cysteine GCGR was diluted to 2 μM in 20 mM HEPES pH 7.5, 100 mM sodium chloride, 0.01% MNG/0.001% CHS and 50 μM ZP3780 and incubated at room temperature for 1 hour in order to fully bind agonist. RAMP2 was added to varying fold molar excess with GCGR (0, 0.25, 0.5, 1, 2.5) and incubated at room temperature for 1 hour. Samples were diluted 10-fold with the same buffer for a final GCGR-minimal cysteine-IANBD concentration of 200 nM and final concentrations of RAMP ranging from 0 to 500 nM. Fluorescence data was collected in a 150 μL cuvette with FluorEssence v3.8 software on a Fluorolog instrument (Horiba) in photon counting mode. NBD fluorescence was measured by excitation at 420 nm with excitation and emission bandwidth passes of 5 nm, and the emission spectra was recorded from 505 to 605 nm with 1 nm increments and a 0.5 s integration time. Raw fluorescence counts were divided by 1000 for plotting. The fluorescence at the peak maximum of 539 nm for all conditions was used for the inset panels Fig. 2E & Fig. 2F as a function of RAMP2 concentration by normalizing the highest [RAMP2] to 1.0 and original fluorescence value to 0.0 in order to more quantitatively interpret binding of RAMP2 to GCGR. The fitted EC50 values were calculated using GraphPad Prism 9 using the one site total binding equation.
Preparation of smFRET samples with minimal cysteine GCGR
GCGR-mincys-265C-345C was diluted to 10 μM and incubated with 200 μM RSSR (Enzo Life Sciences) for 2 hours at room temperature. Next the sample was incubated with 14 mM iodoacetamide at RT for 30 minutes followed by 1.5 hours at 4°C. The sample then underwent size exclusion chromatography on a Superdex 200 Increase 10/300 gel filtration column. Monomeric receptors were collected and concentrated to 10 μM and incubated with 100 μM TCEP for 1 hour at room temperature to remove RSSR spin label. The sample was then incubated with 100 μM maleimide LD555 and 100 μM maleimide LD655 for 1 hour at room temperature followed by incubation with 5 mM L-cysteine for 10 minutes followed by removal of free fluorophore, cysteine, and TCEP with size exclusion chromatography, again on a Superdex 200 Increase 10/300 gel filtration column. Aliquots of specifically-labeled, pure protein were flash frozen for later use.
smFRET data collection
To inhibit nonspecific protein adsorption, flow cells for single-molecule experiments were prepared as previously described25 using mPEG (Laysan Bio) passivated glass coverslips (VWR) and doped with biotin PEG16. Before each experiment, coverslips were incubated with NeutrAvidin (Thermo Fisher), followed by 10 nM biotinylated antibody (mouse anti-FLAG, Genscript). Between each conjugation step, the chambers were flushed to remove free reagents. The antibody dilutions and washes were done in T50 buffer (50 mM NaCl, 10 mM Tris, pH 7.5). To achieve sparse immobilization of labeled receptors on the surface, purified labeled receptor was diluted (ranging from 100-fold to 1000-fold dilution) and applied to coverslips. After achieving optimum surface immobilization (~400 molecules in a 2,000 μm2 imaging area), unbound receptors were washed out of the flow chamber and the flow cells were then washed extensively (up to 50 times the cell volume).
Receptors were imaged for smFRET in imaging buffer consisting of 3 mM Trolox, 100 mM NaCl, 2 mM CaCl2, 20 mM HEPES pH 7.5, 0.01% L-MNG, 0.001% CHS and an oxygen scavenging system (0.8% dextrose, 0.8 mg ml−1 glucose oxidase, and 0.02 mg ml−1 catalase). All buffers were made in UltraPure distilled water (Invitrogen). Samples were imaged with a 1.65 na X60 objective (Olympus) on a total internal reflection fluorescence microscope with 100 ms time resolution unless stated otherwise. Lasers at 532 nm (Cobolt) and 633 nm (Melles Griot) were used for donor and acceptor excitation, respectively. Fluorescence was passed through a Chroma ET550lp filter and split into donor and acceptor signals with a Chroma T635lpxr. FRET efficiency was calculated as (IA−0.1ID)/(ID+IA), in which ID and IA are the donor and acceptor intensity, respectively, after back-ground subtraction. Imaging was with 100 millisecond acquisition time (10 Hz) with a Photometrics Prime 95B cMOS camera.
smFRET data processing
SPARTAN version 3.758 was used to analyze fluorescence movies. Donor and acceptor channels were aligned using the first 10 frames of each movie while excluding particles closer than 3.5 pixels using an integration window of 12 pixels. Single-molecule intensity traces showing single-donor and single-acceptor photobleaching with a stable total intensity for longer than 5 seconds (50 frames), SNRbg > 15 and donor/acceptor correlation coefficients < 0.0 were collected (20–30% of total molecules per imaging area). Individual traces were smoothed using a nonlinear filter59 with following filter parameters: window = 2, M = 2 and P = 15 for histograms. Each experiment was performed ≥4 times to ensure reproducibility. smFRET histograms were compiled from ≥100 molecules per condition (100 millisecond time resolution). Error bars in the histograms represent the standard error from ≥4 independent movies. To ensure that traces of different lengths contribute equally, histograms from individual traces were normalized to one before compiling. Histograms were fit to 3 Gaussians using a custom Python script.
Hydrogen–deuterium exchange labeling reaction
Prior to HDX, purified RAMP2 in 20 mM HEPES pH 7.5, 100 mM sodium chloride and 0.05% DDM/0.005% CHS, and GCGR in 20 mM HEPES pH 7.5, 100 mM sodium chloride and 0.02% MNG/0.002% CHS, were both separately incubated overnight with PNGaseF to facilitate peptide digestion during LC/MS. Subsequently, 10 μM GCGR was incubated in the presence of saturating RAMP2 (at least 1.5:1 RAMP2:GCGR) at 22 °C for at least one hour. Additionally, 10 μM GCGR was incubated with a volume of 0.05% DDM/0.005% CHS equivalent to the volume of RAMP2 added in preparation of the complex. Similarly, 10 μM RAMP was incubated with a volume of 0.02% LMNG/0.002% CHS equivalent to the volume of GCGR added in preparation of the complex.
To prepare deuterated buffer for HDX, NaCl and HEPES were resuspended in D2O (Sigma-Aldrich) to a final concentration of 100 mM NaCl, 20 mM Hepes, pHread=7.3. pHread was adjusted with DCl (Sigma-Aldrich) and NaOD (Sigma-Aldrich). To initiate exchange, samples were diluted 1:10 into D2O buffer and quenched 1:1 (3 M urea, 20 mM TCEP, pH 2.4), for a total sample volume of 70 μL. At selected time points, 0.5 μL of porcine pepsin (10 mg/ml; Sigma Aldrich) and 0.5 μL of aspergillopepsin (10 mg/ml; Sigma Aldrich) were added to each sample, which was then rapidly vortexed, returned to ice for 3 minutes, and flash frozen in liquid N2. Samples were stored at −80 °C prior to LC/MS analysis. Note that proteases were resuspended in 100 mM NaCl, 20 mM HEPES, pH 7.5 to 10 mg/ml and filtered (0.22 μm filter, Corning), aliquoted and stored at −80 °C prior to use.
Liquid chromatography/mass spectrometry analysis
Samples were thawed and injected into a cooled valve system (Trajan LEAP) coupled to an LC (Thermo Ultimate 3000) flowing buffer A (0.1% formic acid) at 200 μL/min. Sample time points were injected in non-consecutive order. The valve chamber, trap column, and analytical column were kept at 2 °C.
Peptides were desalted for 4 minutes on a trap column (1 mM ID × 2 cm, IDEx C-128) manually packed with POROS R2 reversed-phase resin (Thermo Scientific). Peptides were then separated on a C8 analytical column (Thermo Scientific BioBasic-8 5 μm particle size 0.5 mM ID × 50 mM) with buffer B (100% acetonitrile, 0.1% formic acid) flowing at a rate of 40 μL/min, increasing from 5% to 50% over the first 26 minutes and from 50% to 90% B over 90 s. The analytical column was washed using two repeating sawtooth gradients and equilibrated at 5% buffer B prior to the next injection. After every fourth injection, a blank injection was performed to monitor for run-to-run carry over. Peptides were eluted directly into a Q Exactive Orbitrap Mass Spectrometer (ThermoFisher) operating in positive ion mode (MS1 settings: resolution 140000, AGC target 3e6, maximum IT 200 ms, scan range 300–1500 m/z). Separate samples of GCGR and of RAMP2 were subject to tandem mass spectrometry analysis (MS1 settings same as above but with resolution 70000, and MS2 settings as follows: resolution 17500, AGC target 2e5, maximum IT 100 ms, loop count 10, isolation window 2.0 m/z, NCE 28, charge state 1 and >8 excluded, dynamic exclusion 15.0 s).
Peptide identification and analysis
MS2 data were processed using Byonic (Protein Metrics), which resulted in a list of reference peptides for both GCGR and RAMP2. This reference set included both unmodified sites and de-glycosylated sites within the RAMP and GCGR extracellular domains. Deuterium uptake data were analyzed with HD-Examiner (Version 3.1, Sierra Analytics). HD-Examiner was used with default settings; uptake values were adjusted to account for 1:10 dilution of undeuterated sample into deuterated buffer. For unimodal peaks, changes in deuterium uptake were determined by subtracting the mass centroid of the undeuterated peptide from that of the deuterated peptide. For bimodal peaks, peptides were analyzed using scripts adapted from60. Briefly, bimodal peaks were globally fit to sums of Gaussians, such that the widths (and the centers, in the case of bimodals exhibiting EX1 behavior) were constant across time points. Fitting was performed using the lmfit package in Python61. We only analyzed peptides that met the following criteria: less than 40 residues in length, had sequence overlap with other peptides, and were not marked as ‘low-confidence’ by HDExaminer. Deuteration differences for bimodal peptides displayed in the Woods plot (Fig. S3B) were based on the centroids of the far-right (higher mass) peaks, identified using HD-Examiner’s automated bimodal fitting method, when applicable.
Figure Preparation
All graphical panels aside from HDX-MS panels and single-molecule FRET traces were prepared using GraphPad Prism 9. Graphical panels of HDX-MS data were prepared with custom Python software. Single-molecule FRET traces were extracted using Spartan and prepared using custom software. Cryo-EM images, class 2D class averages, and FSC curves were prepared in cryoSPARC. Protein structure figures were prepared using Pymol. Cryo-EM density figures were made using either UCSF Chimera54 or Chimera X62. Figures were constructed in Adobe Illustrator.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification methods and tools used are described in each relevant section of the methods or figure legends. Unless otherwise noted, error bars correspond to standard deviations of the data point. P values for comparisons between data sets follow the following standard definitions: ns (P>0.05), * (P≤0.05), ** (P≤0.01), *** (P≤0.001), and **** (P≤0.0001). P values were calculated using GraphPad Prism using an unpaired t test assuming Gaussian distributions.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HRP-anti-FLAG monoclonal antibody | Sigma Aldrich | Cat# A8592 |
| HRP-anti-HA monoclonal antibody | R&D Systems | Cat# HAM0601 |
| Biotinylated mouse anti-FLAG antibody | Genscript | Cat# A01429 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lauryl maltose neopentyl glycol (L-MNG) | Anatrace | Cat# NG310 |
| n-dodecyl-β-D-maltoside (DDM) | Anatrace | Cat# D310 |
| Cholesteryl Hemisuccinate | Steraloids | Cat# C6823 |
| PNGase F | New England Biolabs | Cat# P0704S |
| Benzamidine hydrochloride hydrate | Sigma Aldrich | Cat# B6506-100G |
| Leupeptin | Sigma Aldrich | Cat# 50-568-49 |
| Lambda Protein Phosphatase | New England Biolabs | Cat# P0753L |
| Calf intestinal phosphatase | New England Biolabs | Cat# M0290S |
| Antarctic phosphatase | New England Biolabs | Cat# M0289S |
| TCEP | Thermo Fisher Scientific | Cat# 77720 |
| FLAG peptide | Stanford Pan Facility | N/A |
| GDP | Sigma Aldrich | Cat# G7127 |
| GDN (glyco-diosgenin): Synthetic Digitonin Substitute | Anatrace | Cat# GDN101 |
| EDTA | Sigma Aldrich | Cat# E5134-500G |
| ESF921 culture medium | Expression Systems | Cat# 96-001 |
| 2-Mercaptoethanol | Sigma Aldrich | Cat# M6250-100ML |
| Apyrase | New England Biolabs | Cat# M0398S |
| Fugene6 | Promega | Cat# E2693 |
| OptiMEM | Gibco | Cat# 31985-070 |
| HBSS | Thermo Fisher | Cat# 14025092 |
| Paraformaldehyde | Sigma Aldrich | Cat# P6148 |
| DPBS | Gibco | Cat# 14040-09 |
| HRP substrate | Bio-Rad | Cat# 170-5060 |
| ZP7680 | This study | N/A |
| ZP3780 | Ref. 3 | N/A |
| IBMX | Sigma Aldrich | Cat# I5879 |
| NanoBRET™ Nano-Glo Substrate | Promega | Cat# N1572 |
| Casein solution | Sigma Aldrich | Cat# C4765 |
| EGTA | Sigma Aldrich | Cat# E3889 |
| EDTA | Sigma Aldrich | Cat# 27285 |
| 3C protease | This study | N/A |
| L-168,049 | Tocris | Cat# 2311 |
| NNC0640 | Novo Nordisk A/S | N/A |
| HEPES | Sigma-Aldrich | Cat# H7006 |
| Sodium chloride | Fisher Scientific | Cat# S64010 |
| Benzonase | Sigma Aldrich | Cat# E1014 |
| Glycerol | Fisher Scientific | Cat# AC332030010 |
| Imidazole | Sigma-Aldrich | Cat# 56750 |
| Chelating Sepharose Fast Flow Resin | GE Healthcare | Cat# 17-0575-02 |
| Calcium chloride (2 M) | Quality Biological | Cat# 351-130-061 |
| Doxycycline hyclate | Sigma-Aldrich | Cat# D9891-5G |
| Sodium butyrate | Sigma-Aldrich | Cat# 303410-100G |
| Expi293 Expression Medium | Thermo Fisher Scientific | Cat# A1435101 |
| ExpiFectamine 293 Transfection Kit | Thermo Fisher Scientific | Cat# A14525 |
| Dithiothreitol (DTT) | Sigma-Aldrich | Cat# D0632-25G |
| Phenylmethylsulfonyl fluoride (PMSF) | Sigma-Aldrich | Cat# 78830-5G |
| Triton X-100 | Sigma | Cat# X100-100ML |
| Manganese chloride (MnCl2) | Sigma-Aldrich | Cat# M3634-100G |
| Isopropyl B-D-1-thiogalactopyranoside (IPTG) | Goldbio | Cat# I2481C50 |
| Disuccinimidyl dibutyric urea (DSBU) | Thermo Scientific | Cat# A35459 |
| diC8-PtdIns(4,5)P2 (C8-PIP2) | Avanti Polar Lipids | Cat# 850185P-500ug |
| GLP-1 | This study | N/A |
| Glucagon | This study | N/A |
| Oxyntomodulin | This study | N/A |
| 4× Laemmli Sample buffer | Bio-Rad | Cat# 161-0747 |
| Pro-Q Diamond phosphoprotein gel stein | Thermo Fisher | Cat# P33301 |
| SYPRO Ruby protein gel stain | Thermo Fisher | Cat# S12000 |
| [3H]-GDP | Perkin Elmer | Cat# NET966250UC |
| N,N’-dimethyl-N-(iodoacetyl)-N’-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD-amide) | Thermo Fisher | Cat# D2004 |
| L-cysteine | Sigma | Cat# C7352-25G |
| RSSR | Enzo Life Sciences | Cat# ALX-430-102-M025 |
| Iodoacetamide | Sigma-Aldrich | Cat# I1149 |
| Maleimide LD555 | Lumidyne Technologies | Cat# 04 |
| Maleimide LD655 | Lumidyne Technologies | Cat# 10 |
| mPEG-SVA, MW 5,000 | Laysan Bio | Cat# MPEG-SVA-5000 |
| BIO-PEG-SVA, MW 5,000 | Laysan Bio | Cat# BIO-PEG-SVA-5000 |
| NeutrAvidin | Thermo Fisher | Cat# 31000 |
| Trolox | Millipore Sigma | Cat# 238813 |
| Dextrose | Millipore Sigma | Cat# DX0145 |
| Glucose oxidase | Millipore Sigma | Cat# G7141 |
| Catalase | Millipore Sigma | Cat# E3289 |
| UltraPure distilled water | Invitrogen | Cat# 10977015 |
| D2O | Sigma Aldrich | Cat# 151882 |
| DCl | Sigma Aldrich | Cat# 543047 |
| NaOD | Sigma Aldrich | Cat# 372072 |
| Urea | IBI Scientific | Cat# IB72060 |
| Porcine pepsin | Sigma Aldrich | Cat# P6887 |
| Aspergillopepsin | Sigma Aldrich | Cat# P2143 |
| Formic acid | Fisher Scientific | Cat# A117-50 |
| Acetonitrile | Thermo Fisher | Cat# 51101 |
| Critical Commercial Assays | ||
| GTPase-Glo™ assay | Promega | Cat# V7682 |
| Deposited Data | ||
| GCGR-Gs (RAMP2) coordinates | This paper | PDB: 8FU6 |
| GCGR-Gs (RAMP2) cryo-EM map | This paper | EMDB: EMD-29453 |
| Experimental Models: Cell Lines | ||
| Spodoptera frugiperda Sf9 cells | Expression Systems | N/A |
| Trichuplusia ni Hi5 cells | Expression Systems | N/A |
| Expi293F inducible cells | Thermo Fisher Scientific | Cat# A39241 |
| HEK-293 cells | ATCC | Cat# CRL-1573 |
| cAMP biosensor cells | Ref. 42 | N/A |
| E. coli Rosetta 2 (DE3) cells | Millipore Sigma | Cat# 70954-3 |
| E. coli BL21 cells | Sigma-Aldrich | Cat# CMC0014 |
| Recombinant DNA | ||
| pFastbac-GCGR | Ref. 3 | N/A |
| pVL1392-Gα | Ref. 3 | N/A |
| pVL1392duet-Gbg | Ref. 3 | N/A |
| Human RAMP2 coding sequence | cDNA.org | Cat# RAMP200000 |
| pFastbac-RAMP2 | This paper | N/A |
| pcDNA3.1(+)neo-HA-FLAG-GCGR | This paper | N/A |
| pcDNA3.1(+)neo-HA-HAtag-RAMP2-eGFP | This paper | N/A |
| pcDNA3.1(+)neo-HA-FLAG-B2AR | This paper | N/A |
| pcDNA-TetO-mincysGCGR | Ref. 3 | N/A |
| pcDNA-TetO-mincysGCGR-F31C | Ref. 3 | N/A |
| pcDNA-TetO-mincysGCGR-F245C | Ref. 3 | N/A |
| pcDNA-TetO-mincysGCGR-265C-345C | Ref. 3 | N/A |
| pVL1392-hisGRK5 | Ref. 48 | N/A |
| pVL1392-hisGRK2 | Ref. 48 | N/A |
| Nb35 | Ref. 50 | N/A |
| pNLF1-secN [GMV/Hygro] | Promega | Cat# N1371 |
| pNLF1-secN-GCGR | This paper | N/A |
| Software and Algorithms | ||
| cryoSPARC (v3.3) | Ref. 51 | https://cryosparc.com/ |
| Phenix | Ref. 55 | https://www.phenix-online.org/ |
| UCSF Chimera | Ref. 53 | https://www.cgl.ucsf.edu/chimera/ |
| PyMol | Schrödinger | https://pymol.org/2/ |
| Chimera X | Ref. 60 | https://www.rbvi.ucsf.edu/chimerax/ |
| COOT | Ref. 54 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Molprobity | Ref. 56 | http://molprobity.biochem.duke.edu/ |
| Prism (v9) | GraphPad | http://www.graphpad.com |
| Byonic | Protein Metrics | https://proteinmetrics.com/byonic/ |
| HD-Examiner (v3.1) | Sierra Analytics | https://www.leaptec.com/products/hdexaminer-from-sierra-analytics |
| Bimodal fitting | This paper | N/A |
| lmfit | Ref. 61 | https://lmfit.github.io/lmfit-py/ |
| SPARTAN | Ref. 57 | https://www.scottcblanchardlab.com/spartan-download |
| MATLAB 2018a | Mathworks | https://www.mathworks.com |
| Micromanager | Micromanager | https://micro-manager.org/ |
| Illustrator (v27) | Adobe | https://www.adobe.com/products/catalog.html |
| Other | ||
| Microfluidic imaging chambers for TIRF-based smFRET | Ref. 26 | N/A |
Highlights.
RAMP2 binds to the glucagon receptor and acts as a negative allosteric modulator
RAMP2 promotes extracellular receptor dynamics resulting in inactive cytosolic face
CryoEM shows unproductive complex formation and confirms dynamic extracellular domain
Dynamic allostery as an endogenous GPCR regulatory mechanism
Acknowledgements
We thank S. Reedtz-Runge (Novo Nordisk A/S) for providing the ligand NNC0640. We thank Betsy White for providing pure G-proteins. We thank Lars-Bo Hansen (Zealand Pharma A/S) for peptide synthesis. We thank S. Shoemaker and N. Dall for assistance with HDX-MS analysis scripts. K.K. was supported by the American Diabetes Association (ADA) Postdoctoral Fellowship. E.S.O. was supported by the American Heart Association (AHA) Postdoctoral Fellowship. N.R.L. was supported by a Miller Postdoctoral Fellowship. The work is supported by NIH grant GM050945 to S.M.; S.M. and B.K.K. are Chan Zuckerberg Biohub investigators. Cryo-EM data were collected at the Stanford cryo-EM center (cEMc).
Footnotes
Declaration of interests:
B.K.K. is a cofounder of and consultant for ConfometRx. J.M.M. and I.T. are employees of Zealand Pharma A/S.
Inclusion and Diversity
We support inclusive, diverse and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Müller TD, Finan B, Clemmensen C, Di Marchi RD, and Tschöp MH (2017). The new biology and pharmacology of glucagon. Physiol. Rev. 97, 721–766. 10.1152/physrev.00025.2016. [DOI] [PubMed] [Google Scholar]
- 2.Mayo KE, Miller LJ, Bataille D, Dalle S, Göke B, Thorens B, and Drucker DJ (2003). International Union of Pharmacology. XXXV. The glucagon receptor family. Pharmacol. Rev. 55, 167–194. 10.1124/pr.55.1.6. [DOI] [PubMed] [Google Scholar]
- 3.Hilger D, Kumar KK, Hu H, Pedersen MF, O’Brien ES, Giehm L, Jennings C, Eskici G, Inoue A, Lerch M, et al. (2020). Structural insights into differences in G protein activation by family A and family B GPCRs. Science 369, eaba3373. 10.1126/science.aba3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christopoulos A, Christopoulos G, Morfis M, Udawela M, Laburthe M, Couvineau A, Kuwasako K, Tilakaratne N, and Sexton PM (2003). Novel receptor partners and function of receptor activity-modifying proteins. J. Biol. Chem. 278, 3293–3297. 10.1074/jbc.C200629200. [DOI] [PubMed] [Google Scholar]
- 5.Pioszak AA, and Hay DL (2020). RAMPs as allosteric modulators of the calcitonin and calcitonin-like class B G protein-coupled receptors. In Advances in Pharmacology (Elsevier Inc.), pp. 115–141. 10.1016/bs.apha.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J, Belousoff MJ, Liang Y-L, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, et al. (2022). A structural basis for amylin receptor phenotype. Science 375, eabm9609. 10.1126/science.abm9609. [DOI] [PubMed] [Google Scholar]
- 7.Liang YL, Khoshouei M, Deganutti G, Glukhova A, Koole C, Peat TS, Radjainia M, Plitzko JM, Baumeister W, Miller LJ, et al. (2018). Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature 561, 492–497. 10.1038/s41586-018-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang Y-L, Belousoff MJ, Fletcher MM, Zhang X, Khoshouei M, Deganutti G, Koole C, Furness SGB, Miller LJ, Hay DL, et al. (2020). Structure and Dynamics of Adrenomedullin Receptors AM 1 and AM 2 Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins. ACS Pharmacol. Transl. Sci. 3, 263–284. 10.1021/acsptsci.9b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzen E, Dodig-Crnković T, Kotliar IB, Pin E, Ceraudo E, Vaughan RD, Uhlèn M, Huber T, Schwenk JM, and Sakmar TP (2019). Multiplexed analysis of the secretin-like GPCR-RAMP interactome. Sci. Adv. 5, eaaw2778. 10.1126/sciadv.aaw2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weston C, Lu J, Li N, Barkan K, Richards GO, Roberts DJ, Skerry TM, Poyner D, Pardamwar M, Reynolds CA, et al. (2015). Modulation of glucagon receptor pharmacology by receptor activity-modifying protein-2 (RAMP2). J. Biol. Chem. 290, 23009–23022. 10.1074/jbc.M114.624601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cegla J, Jones BJ, Gardiner JV, Hodson DJ, Marjot T, McGlone ER, Tan TM, and Bloom SR (2017). RAMP2 influences glucagon receptor pharmacology via trafficking and signaling. Endocrinology 158, 2680–2693. 10.1210/en.2016-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGlone ER, Manchanda Y, Jones B, Pickford P, Inoue A, Carling D, Bloom SR, Tan T, and Tomas A (2021). Receptor Activity-Modifying Protein 2 (RAMP2) alters glucagon receptor trafficking in hepatocytes with functional effects on receptor signalling. Mol. Metab. 53, 101296. 10.1016/j.molmet.2021.101296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregorio GG, Masureel M, Hilger D, Terry DS, Juette M, Zhao H, Zhou Z, Perez-Aguilar JM, Hauge M, Mathiasen S, et al. (2017). Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature 547, 68–73. 10.1038/nature22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krilov L, Nguyen A, Miyazaki T, Unson CG, Williams R, Lee NH, Ceryak S, and Bouscarel B (2011). Dual Mode of glucagon receptor internalization: Role of PKCα, GRKs and β-arrestins. Exp. Cell Res. 317, 2981–2994. 10.1016/j.yexcr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark LJ, Krieger J, White AD, Bondarenko V, Lei S, Fang F, Lee JY, Doruker P, Böttke T, Jean-Alphonse F, et al. (2020). Allosteric interactions in the parathyroid hormone GPCR–arrestin complex formation. Nat. Chem. Biol. 16, 1096–1104. 10.1038/s41589-020-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wootten D, Miller LJ, Koole C, Christopoulos A, and Sexton PM (2017). Allostery and biased agonism at class B G protein-coupled receptors. Chem. Rev. 117, 111–138. 10.1021/acs.chemrev.6b00049. [DOI] [PubMed] [Google Scholar]
- 17.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi K, Kawakami K, Kusakizako T, Miyauchi H, Tomita A, Kobayashi K, Shihoya W, Yamashita K, Nishizawa T, Kato HE, et al. (2022). Endogenous ligand recognition and structural transition of a human PTH receptor Endogenous ligand recognition and structural transition of a human PTH receptor. Mol. Cell 82, 1–16. 10.1016/j.molcel.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y-L, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, et al. (2017). Phase-plate cryo-EM structure of a class B GPCR–G-protein complex. Nature 546, 118–123. 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao LH, Yin Y, Yang D, Liu B, Hou L, Wang X, Pal K, Jiang Y, Feng Y, Cai X, et al. (2016). Differential requirement of the extracellular domain in activation of class B G protein-coupled receptors. J. Biol. Chem. 291, 15119–15130. 10.1074/jbc.M116.726620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koth CM, Murray JM, Mukund S, Madjidi A, Minn A, Clarke HJ, Wong T, Chiang V, Luis E, Estevez A, et al. (2012). Molecular basis for negative regulation of the glucagon receptor. Proc. Natl. Acad. Sci. 109, 14393–14398. 10.1073/pnas.1206734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodge EA, Benhaim MA, and Lee KK (2020). Bridging protein structure, dynamics, and function using hydrogen/deuterium-exchange mass spectrometry. Protein Sci. 29, 843–855. 10.1002/pro.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siu FY, He M, De Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, et al. (2013). Structure of the human glucagon class B G-protein-coupled receptor. Nature 499, 444–449. 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Graaf C, Song G, Cao C, Zhao Q, Wang M, Wu B, and Stevens RC (2017). Extending the Structural View of Class B GPCRs. Trends Biochem. Sci. 42, 946–960. 10.1016/j.tibs.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Vafabakhsh R, Levitz J, and Isacoff EY (2015). Conformational dynamics of a class C G-protein-coupled receptor. Nature 524, 497–501. 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habrian CH, Levitz J, Vyklicky V, Fu Z, Hoagland A, McCort-Tranchepain I, Acher F, and Isacoff EY (2019). Conformational pathway provides unique sensitivity to a synaptic mGluR. Nat. Commun. 10, 1–13. 10.1038/s41467-019-13407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Li M, Liang W, Shi X, Fan J, Kong R, Liu Y, Zhang J, Chen T, and Lu S (2022). Delineating the activation mechanism and conformational landscape of a class B G protein-coupled receptor glucagon receptor. Comput. Struct. Biotechnol. J. 20, 628–639. 10.1016/j.csbj.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loura LMS (2012). Simple estimation of förster resonance energy transfer (FRET) orientation factor distribution in membranes. Int. J. Mol. Sci. 13, 15252–15270. 10.3390/ijms131115252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutsumi N, Maeda S, Qu Q, Vögele M, Jude KM, Suomivuori CM, Panova O, Waghray D, Kato HE, Velasco A, et al. (2022). Atypical structural snapshots of human cytomegalovirus GPCR interactions with host G proteins. Sci. Adv. 8. 10.1126/sciadv.abl5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pioszak AA, and Xu HE (2008). Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc. Natl. Acad. Sci. U. S. A. 105, 5034–5039. 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao A, Han S, Li X, Li Z, Zhao P, Dai A, Chang R, Tai L, Tan Q, Chu X, et al. (2020). Structural basis of Gs and Gi recognition by the human glucagon receptor. Science 367, 1346–1352. [DOI] [PubMed] [Google Scholar]
- 32.Hilger D, Masureel M, and Kobilka BK (2018). Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12. 10.1038/s41594-017-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punjani A, and Fleet DJ (2021). 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702. 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
- 34.Liang YL, Belousoff MJ, Zhao P, Koole C, Fletcher MM, Truong TT, Julita V, Christopoulos G, Xu HE, Zhang Y, et al. (2020). Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation. Mol. Cell 77, 656–668. 10.1016/j.molcel.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Nemec K, Schihada H, Kleinau G, Zabel U, Grushevskyi EO, Scheerer P, Lohse MJ, and Maiellaro I (2022). Functional modulation of PTH1R activation and signaling. Proc Natl Acad Sci U S A 119, 1–12. 10.1073/pnas.2122037119/-/DCSupplemental.Published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wootten D, Lindmark H, Kadmiel M, Willcockson H, Caron KM, Barwell J, Drmota T, and R PD (2013). Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. Br. J. Pharmacol. 168, 822–834. 10.1111/j.1476-5381.2012.02202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris M, Mackie DI, Pawlak JB, Carvalho S, Truong TT, Safitri D, Yan Yeung H, Routledge S, Harper MT, Al-Zaid B, et al. (2021). RAMPs regulate signalling bias and internalisation of the GIPR. bioRxiv, 2021.04.08.436756. [Google Scholar]
- 38.Okashah N, Wright SC, Kawakami K, Mathiasen S, Zhou J, Lu S, Javitch JA, Inoue A, Bouvier M, and Lambert NA (2020). Agonist-induced formation of unproductive receptor-G12 complexes. Proc. Natl. Acad. Sci. U. S. A. 10.1073/pnas.2003787117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Tissue-based map of the human proteome. Science 347. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 40.Bankir L, Bouby N, Blondeau B, and Crambert G (2016). Glucagon actions on the kidney revisited: Possible role in potassium homeostasis. Am. J. Physiol. - Ren. Physiol. 311, F469–F486. 10.1152/ajprenal.00560.2015. [DOI] [PubMed] [Google Scholar]
- 41.Fraser HB, Hirsh AE, Wall DP, and Eisen MB (2004). Coevolution of gene expression among interacting proteins. Proc. Natl. Acad. Sci. U. S. A. 101, 9033–9038. 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey L, Lakomek N-A, Riek R, and Bibow S (2017). Micelles, Bicelles, and Nanodiscs: Comparing the Impact of Membrane Mimetics on Membrane Protein Backbone Dynamics. Angew. Chemie Int. Ed. 56, 380–383. 10.1002/anie.201608246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vedel L, Bräuner-Osborne H, and Mathiesen JM (2015). A cAMP biosensor-based high-throughput screening assay for identification of Gs-coupled GPCR ligands and phosphodiesterase inhibitors. J. Biomol. Screen. 20, 849–857. 10.1177/1087057115580019. [DOI] [PubMed] [Google Scholar]
- 44.Seven AB, Barrow-Alvarez X, de Lapeyriere M, Papasergi-Scott M, Robertson MJ, Zhang C, Nwokonko RM, Gao Y, Meyerowitz JG, Rocher J, et al. (2021). G-protein activation by a metabotropic glutamate receptor. Nature 595, 450–454. 10.1038/s41586-021-03680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papasergi-Scott MM, Robertson MJ, Seven AB, Panova O, Mathiesen JM, and Skiniotis G (2020). Structures of metabotropic GABAB receptor. Nature 584, 310–314. 10.1038/s41586-020-2469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unson CG, and Merrifield RB (1994). Identification of an essential serine residue in glucagon: Implication for an active site triad. Proc. Natl. Acad. Sci. U. S. A. 91, 454–458. 10.1073/pnas.91.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hövelmann U, Bysted BV, Mouritzen U, Macchi F, Lamers D, Kronshage B, Møller DV, and Heise T (2018). Pharmacokinetic and pharmacodynamic characteristics of dasiglucagon, a novel soluble and stable glucagon analog. Diabetes Care 41, 531–537. 10.2337/dc17-1402. [DOI] [PubMed] [Google Scholar]
- 48.Unson CG, Gurzenda EM, and Merrifield RB (1989). Biological activities of des-His1[Glu9]glucagon amide, a glucagon antagonist. Peptides 10, 1171–1177. 10.1016/0196-9781(89)90010-7. [DOI] [PubMed] [Google Scholar]
- 49.Huang W, Masureel M, Qu Q, Janetzko J, Inoue A, Kato HE, Robertson MJ, Nguyen KC, Glenn JS, Skiniotis G, et al. (2020). Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 579, 303–308. 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beyett TS, Fraley AE, Labudde E, Patra D, Coleman RC, Eguchi A, Glukhova A, Chen Q, Williams RM, Koch WJ, et al. (2019). Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe. Proc. Natl. Acad. Sci. U. S. A. 116, 15895–15900. 10.1073/pnas.1818547116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasmussen SGF, Devree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. (2011). Crystal structure of the β 2 adrenergic receptor-Gs protein complex. Nature 477, 549–557. 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 53.Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221. 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, and Cowtan K (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, et al. (2011). The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106. 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Z, Wasserman MR, Zhao H, Altman RB, Juette MF, Blanchard SC, and Terry DS (2016). Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344. 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haran G (2004). Noise reduction in single-molecule fluorescence trajectories of folding proteins. Chem. Phys. 307, 137–145. 10.1016/j.chemphys.2004.05.017. [DOI] [Google Scholar]
- 60.Costello SM, Shoemaker SR, Hobbs HT, Nguyen AW, Hsieh CL, Maynard JA, McLellan JS, Pak JE, and Marqusee S (2022). The SARS-CoV-2 spike reversibly samples an open-trimer conformation exposing novel epitopes. Nat. Struct. Mol. Biol. 29, 229–238. 10.1038/s41594-022-00735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newville M, Stensitzki T, Allen DB, and Ingargiola A (2014). LMFIT: Non-Linear Least-Square Minimization and Curve-Fitting for Python. [Google Scholar]
- 62.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-29453 and model coordinates have been deposited in the Protein Data Bank (PDB) under accession number 8FU6.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


