Abstract
The effect of lactic acid on the outer membrane permeability of Escherichia coli O157:H7, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium was studied utilizing a fluorescent-probe uptake assay and sensitization to bacteriolysis. For control purposes, similar assays were performed with EDTA (a permeabilizer acting by chelation) and with hydrochloric acid, the latter at pH values corresponding to those yielded by lactic acid, and also in the presence of KCN. Already 5 mM (pH 4.0) lactic acid caused prominent permeabilization in each species, the effect in the fluorescence assay being stronger than that of EDTA or HCl. Similar results were obtained in the presence of KCN, except for P. aeruginosa, for which an increase in the effect of HCl was observed in the presence of KCN. The permeabilization by lactic and hydrochloric acid was partly abolished by MgCl2. Lactic acid sensitized E. coli and serovar Typhimurium to the lytic action of sodium dodecyl sulfate (SDS) more efficiently than did HCl, whereas both acids sensitized P. aeruginosa to SDS and to Triton X-100. P. aeruginosa was effectively sensitized to lysozyme by lactic acid and by HCl. Considerable proportions of lipopolysaccharide were liberated from serovar Typhimurium by these acids; analysis of liberated material by electrophoresis and by fatty acid analysis showed that lactic acid was more active than EDTA or HCl in liberating lipopolysaccharide from the outer membrane. Thus, lactic acid, in addition to its antimicrobial property due to the lowering of the pH, also functions as a permeabilizer of the gram-negative bacterial outer membrane and may act as a potentiator of the effects of other antimicrobial substances.
Lactic acid, as produced by lactic acid starter culture bacteria or as an additive to foods, functions as a natural antimicrobial having a generally recognized as safe status. As reviewed by Doores (8), lactic acid is able to inhibit the growth of many types of food spoilage bacteria, including gram-negative species of the families Enterobacteriaceae and Pseudomonadaceae. Among other organic acids, lactic acid is recognized as a biopreservative in naturally fermented products (25), and numerous applications for decontamination of meat by lactic acid have been described (7, 10, 22, 29, 32, 33). The antibacterial action of lactic acid is largely, but not totally, assigned to its ability in the undissociated form to penetrate the cytoplasmic membrane, resulting in reduced intracellular pH and disruption of the transmembrane proton motive force (25).
The relative efficacy of lactic acid against gram-negative bacteria is not unexpected considering that as a small water-soluble molecule lactic acid gains access to the periplasm through the water-filled porin proteins of the outer membrane (OM), as reviewed by Nikaido (18). The OM, however, functions as an efficient permeability barrier that is able to exclude macromolecules (such as bacteriocins or enzymes) and hydrophobic substances (i.e., hydrophobic antibiotics). The permeability barrier property of the OM is largely due to the presence of a specific lipopolysaccharide (LPS) layer on the membrane surface. LPS molecules consist of a lipid part, termed lipid A, and a hydrophilic heteropolysaccharide chain protruding outward and providing the cell with a hydrophilic surface (11). Certain external agents that either release LPS and other components from the OM or intercalate in the membrane can abolish the integrity of the OM. In both cases there is a concomitant loss of the permeability barrier function. Such agents are called permeabilizers (31); examples include EDTA, which chelates divalent cations that stabilize molecular interactions in the OM so that LPS is released, and polycations such as polyethyleneimine (12) or polymyxin B nonapeptide, which cause OM damage without LPS release. Permeabilizers as such need not be bactericidal or bacteriostatic to gram-negative cells but, by enabling other compounds to penetrate, an increased susceptibility to hydrophobic antibiotics, detergents, lysozyme, or bacteriocins is achieved. Accordingly, food-grade permeabilizers in combination with other antimicrobials would be ideal as part of the hurdle concept in inhibiting gram-negative spoilage bacteria and pathogens in food materials (13).
Roth and Keenan reported in 1971 (26) that lactic acid is able to cause sublethal injury to Escherichia coli, and similar properties have also been assigned to acetic acid (23); indirect evidence inferred that such injury involved disruption of the LPS layer. A permeabilizer function of lactic acid would not only be utilizable in decontamination procedures and in protective cultures but it would also provide a mechanistic explanation supporting the antimicrobial and health-promoting effects of probiotic lactic acid bacteria (28). We have investigated here the effects of lactic acid on the permeability properties of OM of three gram-negative bacterial species associated with food safety and food spoilage.
(Some of these results were presented at the 99th General Meeting of The American Society for Microbiology, Chicago, Ill., May 30 to June 3, 1999.)
MATERIALS AND METHODS
Chemicals.
Lactic acid (mixture of d and l forms; pro analysis grade), potassium lactate, and Triton X-100 were from BDH (Poole, England); chicken egg white lysozyme (EC 3.2.1.17), HEPES, n-heptadecanoic acid methyl ester, 1-N-phenylnaphthylamine (NPN), and sodium dodecyl sulfate (SDS) were from Sigma-Aldrich (Steinheim, Germany); and EDTA was from Riedel-de-Haen (Seelze, Germany). Proteinase K (EC 3.4.21.64) and KCN were from Merck (Darmstadt, Germany). A stock solution of NPN (0.5 M) was prepared in acetone and diluted to 40 μM into 5 mM HEPES (pH 7.2) for the fluorometric assays. All materials for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were obtained from Novex (San Diego, Calif.), including precast Tris-glycine gels (18% acrylamide).
Bacteria.
E. coli ATCC 35150 (O157:H7), Pseudomonas aeruginosa ATCC 9027, and Salmonella enterica serovar Typhimurium SL696 (34) were cultivated in Luria-Bertani broth as described previously (12).
Permeability assay based on the uptake of NPN.
NPN is a very hydrophobic probe whose quantum yield is greatly enhanced in a glycerophospholipid environment compared to an aqueous environment (30). Uptake of NPN by bacterial membrane(s) is manifested as fluorescence, and it indicates damage in the gram-negative bacterial OM, which normally is able to exclude hydrophobic substances (18). This permeability assay was recently adapted for the automated spectrofluorometer Fluoroskan (Labsystems, Helsinki, Finland), whereby fluorescent readings are made from microtiter plates (15). For these experiments, bacteria were grown to the mid-logarithmic phase of growth (optical density at 630 nm of 0.5 ± 0.02), deposited by centrifugation, and suspended into a half-volume of 5 mM HEPES buffer (pH 7.2). Such suspensions were then supplemented with lactic acid at 0.05 or 0.1% (wt/vol) corresponding to 5 mM and 10 mM, respectively, and causing the pH to drop to 4.0 ± 0.1 and 3.6 ± 0.1. Parallel suspensions were made in which pH was adjusted to the above values with hydrochloric acid. In assays involving KCN to de-energize cells, this salt was included in the buffers. The pH-adjusted suspensions (100 μl) were then pipetted into microtiter plate wells (Cliniplate Black, catalog no. 9502 867; Labsystems), which already contained 50 μl of pH-adjusted cell-free buffer and 50 μl of a 40 μM solution of NPN in buffer, yielding an end concentration of 10 μM NPN. In assays utilizing potassium lactate (10 mM), EDTA (1 mM), or MgCl2 (5 mM), these were included in the cell-free buffer. Immediately after the cells were mixed with the other constituents, the plates were read for fluorescence in the Fluoroskan, using an excitation filter of 355 nm (half bandwidth, 38 ± 3 nm) and an emission filter of 405 nm (half bandwidth, 50 ± 5 nm). The fluorescence values were subtracted with the simultaneously recorded value of cell suspension in HEPES at pH 7.2 in the presence of 10 μM NPN. Four parallel wells of each sample were recorded, and three to seven independent assays were performed; experiments involving KCN were performed twice.
Bacteriolysis.
Sensitization of bacteria to the action of lytic agents by acids or KCN was measured as described recently (12), utilizing turbidometric monitoring of cell lysis with the Multiskan MCC/340 spectrophotometer (Labsystems).
Release of LPS and phospholipid.
The release of LPS from serovar Typhimurium was assayed by SDS-PAGE and by fatty acid analysis (gas chromatography of fatty acid methyl esters) of cell-free supernatants after treatment of the bacterial suspensions with either lactic acid, HCl, or EDTA. The protocols for these experiments were recently described in detail (14). The concentration of lactic acid in the release assay was 5 mM, yielding a pH value of 3.5 in the 10 mM Tris buffer initially adjusted to pH 7.2 by HCl. In parallel, cell suspensions in the above buffer were adjusted to pH 3.5 by HCl alone. Treatment with EDTA was at 1 mM at pH 7.2. From 10-ml suspensions, 8.6 ml of cell-free supernatant was taken for fatty acid analysis, and 0.5 ml was taken for SDS-PAGE. Both aliquots were freeze-dried before processing.
Statistical methods.
For the NPN uptake values and the bacteriolysis values, the two-tailed unpaired Student's t test was used to determine differences; a P value of <0.05 was considered significant.
RESULTS
NPN uptake induced by acids.
Table 1 summarizes the results of NPN uptake experiments with lactic acid, HCl, and EDTA, including the effect of addition of the MgCl2 or the presence of KCN in the assay buffer. For all bacteria, lactic acid brought about a significantly higher NPN uptake than hydrochloric acid. The effect was seen already at 5 mM lactic acid (pH 4.0); only with E. coli was the NPN uptake further enhanced by the higher concentration of lactic acid (10 mM, pH 3.6). The strongest response to lactic acid was observed in serovar Typhimurium, but each test organism reacted more strongly to lactic acid than to the classical permeabilizer EDTA. The addition of an equimolar concentration of MgCl2 together with lactic acid decreased the NPN uptake slightly but significantly for E. coli and P. aeruginosa; in serovar Typhimurium such an effect was insignificant. MgCl2 also diminished the effect of HCl, but only in E. coli; surprisingly, P. aeruginosa reacted with higher uptake values to HCl in the presence of MgCl2 than in its absence. The responses to EDTA differed characteristically among the three bacterial species, P. aeruginosa reacting most prominently and E. coli with the lowest figures; MgCl2 abolished the effect in all cases. The above effects were generally similar in the presence of KCN; except for P. aeruginosa, for which the effect of HCl with KCN was enhanced to a level similar to that obtained by lactic acid. Potassium lactate at concentrations of up to 10 mM (pH 6.8 in the cell suspension) had no NPN uptake-enhancing activity on serovar Typhimurium (data not shown).
TABLE 1.
NPN uptake induced by acids
| Strain (n)a | Relative fluorescence ± SDb at:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| pH 3.6 ± 0.1
|
pH 4.0 ± 0.1
|
pH 7.2 ± 0.1
|
||||||
| Lactic acid (10 mM) | HCl | Lactic acid (5 mM) | Lactic acid + MgCl2 | HCl | HCl + MgCl2 | EDTA (1 mM) | EDTA + MgCl2 | |
| E. coli (7)b | 357 ± 75 | 191 ± 22 | 288 ± 40 | 201 ± 31 | 138 ± 24 | 38 ± 6 | 40 ± 13 | 2 ± 1 |
| E. coli + KCN (2) | ND | ND | 379 ± 2 | 218 ± 35 | 145 ± 41 | 55 ± 10 | 62 ± 8 | 4 ± 1 |
| P. aeruginosa (3) | 373 ± 38 | 262 ± 16 | 377 ± 29 | 246 ± 24 | 280 ± 29 | 378 ± 1 | 273 ± 21 | 10 ± 5 |
| P. aeruginosa + KCN (2) | ND | ND | 397 ± 16 | 270 ± 8 | 346 ± 41 | 374 ± 33 | 343 ± 49 | 2 ± 2 |
| Serovar Typhimurium (4) | 529 ± 38 | 328 ± 100 | 531 ± 56 | 463 ± 41 | 288 ± 126 | 122 ± 64 | 99 ± 39 | 12 ± 3 |
| Serovar Typhimurium + KCN (2) | 496 ± 14 | 229 ± 104 | 480 ± 22 | 426 ± 35 | 147 ± 112 | 66 ± 36 | 67 ± 15 | 15 ± 2 |
n, Number of independent experiments.
The NPN uptake values for the cell controls subtracted from the total fluorescence values to obtain the values shown in the table were 99 ± 21, 70 ± 22 (MgCl2), 116 ± 13 (KCN), and 87 ± 5 (KCN + MgCl2) for E. coli; 124 ± 12, 107 ± 9 (MgCl2), 171 ± 13 (KCN), and 129 ± 26 (KCN + MgCl2) for P. aeruginosa; and 145 ± 22, 98 ± 16 (MgCl2), 141 ± 18 (KCN), and 78 ± 11 (KCN + MgCl2) for serovar Typhimurium.
Effect of acids on bacteriolysis.
To further investigate the permeabilizer effect of lactic acid, its effect on the sensitivity of bacteria toward lysozyme and the detergents SDS and Triton X-100 was measured. In parallel, similar assays with HCl (pH 3.6) and KCN (1 mM) were performed. The results are summarized in Table 2. Lactic acid had a strong sensitizing effect to SDS in each species; similar effects to the nonionic detergent Triton X-100 were also noted, especially in P. aeruginosa. This strain was also strongly sensitized by lactic acid to the lytic action of lysozyme. Hydrochloric acid brought about significantly weaker sensitizing effects than lactic acid to SDS in the enteric bacteria. However, P. aeruginosa was strongly affected; P. aeruginosa was also sensitized to Triton X-100 by HCl, but less so to lysozyme than by lactic acid. Although KCN had a slight effect on the SDS sensitivity of each species and also a minimal effect with Triton X-100 on P. aeruginosa, it was evident that de-energization of the bacterial cells did not have any major impact on their permeability properties.
TABLE 2.
Sensitization of gram-negative bacteria to lytic agents
| Strain and lytic substance | Concn | Relative turbidity (%) at 4 mina
|
|||
|---|---|---|---|---|---|
| Control | Lactic acid (10 mM, pH 3.6) | HCl (pH 3.6) | KCN (1 mM, pH 7.2) | ||
| E. coli O157:H7 | |||||
| Lysozyme | 10 μg/ml | 101 ± 2 | 99 ± 1 | 100 ± 1 | 100 ± 1 |
| Triton X-100 | 0.1% | 104 ± 1 | 102 ± 1 | 104 ± 1 | 103 ± 2 |
| Triton X-100 | 1% | 95 ± 1 | 92 ± 1 | 95 ± 1 | 95 ± 1 |
| SDS | 0.1% | 102 ± 1 | 52 ± 11 | 102 ± 1 | 97 ± 2 |
| SDS | 1% | 97 ± 1 | 47 ± 7 | 95 ± 1 | 83 ± 3 |
| P. aeruginosa | |||||
| Lysozyme | 10 μg/ml | 99 ± 1 | 23 ± 1 | 53 ± 26 | 101 ± 1 |
| Triton X-100 | 0.1% | 102 ± 3 | 80 ± 1 | 55 ± 20 | 104 ± 1 |
| Triton X-100 | 1% | 98 ± 1 | 69 ± 1 | 45 ± 19 | 92 ± 1 |
| SDS | 0.1% | 104 ± 2 | 18 ± 1 | 17 ± 1 | 101 ± 1 |
| SDS | 1% | 101 ± 1 | ND | ND | 94 ± 1 |
| Salmonella serovar Typhimurium | |||||
| Lysozyme | 10 μg/ml | 101 ± 1 | 102 ± 2 | 100 ± 1 | 102 ± 1 |
| Triton X-100 | 0.1% | 104 ± 3 | 101 ± 4 | 103 ± 1 | 104 ± 1 |
| Triton X-100 | 1% | 97 ± 1 | 91 ± 2 | 92 ± 2 | 94 ± 5 |
| SDS | 0.1% | 102 ± 3 | 60 ± 10 | 94 ± 2 | 98 ± 1 |
| SDS | 1% | 97 ± 2 | 53 ± 10 | 85 ± 3 | 77 ± 11 |
ND, not determined.
Acids induce LPS release.
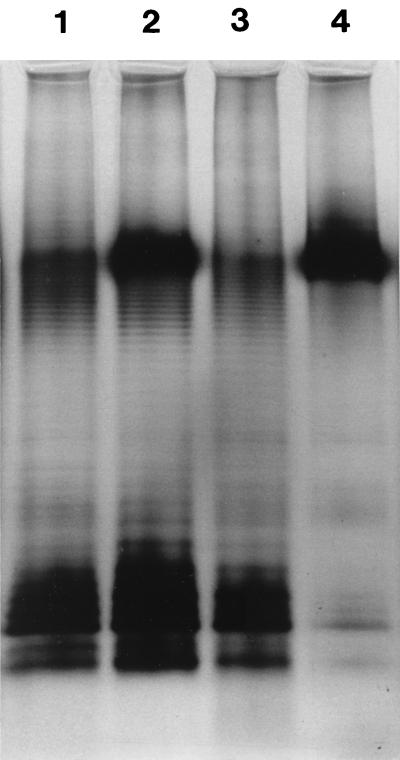
Cell-free supernatants after treatment of serovar Typhimurium with lactic acid, HCl, or EDTA were processed for SDS-PAGE to investigate the possible release of LPS. A silver-stained gel showing the result is pictured in Fig. 1. Whereas very little LPS was present in the supernatant of untreated cells, the supernatants of acid-treated suspensions yielded prominent ladder patterns characteristic of serovar Typhimurium smooth-type LPS. Based on visual estimation of the intensity of staining, the supernatant of lactic acid-treated bacteria contained more LPS than those derived from treatments by HCl or EDTA. The supernatants were also subjected to fatty acid analysis to obtain both qualitative and quantitative data on the released lipid material. Analysis results (Table 3) confirmed that lactic acid had been the most active acid with respect to LPS release, as indicated by the greatest sum of LPS-specific (35) fatty acids C12:0, C14:0, and C3-OH-14:0. In addition to LPS, also other lipid material (glycerophospholipids) was released, represented by the unsaturated fatty acids found in the supernatants. However, LPS-specific fatty acids accounted for a greater proportion in the acid supernatants compared to that of the control, suggesting a preferential release of LPS.
FIG. 1.
Silver-stained SDS-polyacrylamide gel (18% acrylamide) of proteinase K-treated cell-free supernatants of serovar Typhimurium SL696 exposed to HCl (pH 3.6) (lane 1), lactic acid (5 mM) (lane 2), or EDTA (1 mM) (lane 3); lane 4 shows the control supernatant. An equal volume of each sample was electrophoresed.
TABLE 3.
Liberation of fatty acid-containing material from serovar Typhimurium SL696 by EDTA, lactic acid, and HCl
| Fatty acid | Amt (μg) of fatty acid in 8.6 ml of cell-free supernatant after treatment with:
|
|||
|---|---|---|---|---|
| None (control) | EDTA (1 mM) | Lactic acid (5 mM, pH 3.5) | HCl (pH 3.5) | |
| C12:0a | 1.4 | 2.4 | 2.8 | 2.0 |
| C14:0a | 0.9 | 4.1 | 4.7 | 2.6 |
| C14:0(3-OH)a | 0.8 | 6.1 | 8.6 | 4.6 |
| C16:0 | 3.4 | 6.8 | 10.7 | 7.2 |
| C16:1 | 2.3 | 5.5 | 8.6 | 6.0 |
| C18:1 | 2.5 | 3.1 | 5.5 | 3.1 |
| Total | 11.3 | 28.0 | 40.9 | 25.5 |
The percentages of LPS-specific fatty acids for the four treatment groups in columns 2 through 5 are 27, 45, 39, and 36%, respectively.
DISCUSSION
The results presented here permit the conclusion that lactic acid is a potent OM-disintegrating agent, as evidenced by its ability to cause LPS release and to sensitize bacteria to detergents or lysozyme. Increase of the uptake of the hydrophobic probe NPN further suggests a permeabilizing action for lactic acid. Whereas acidity as adjusted by hydrochloric acid also brought about effects indicative of OM disruption, the direct effect of lactic acid as measured by the NPN uptake method was always stronger than that observed in HCl-treated bacteria at the same pH. Our data are thus in accord with and offer a potential mechanism for earlier findings that organic acids, including lactic acid, cause sublethal injury for gram-negative bacteria, as indicated by their decreased viability on bile salt-containing agar (23, 25, 26).
Disruption of the OM by acids can possibly involve the action of both dissociated and undissociated forms. Our finding that hydrochloric acid causes significant OM damage at pH 4 shows that the disintegration of the LPS layer can be caused by a fully dissociable acid. The additional OM-disintegrating effect demonstrated here for lactic acid is likely due to the action of undissociated lactic acid molecules; at pH 4 ca. 40% and at pH 3.6 ca. 60% of lactic acid are present in the undissociated form. This conclusion is further supported by our finding that the dissociated potassium lactate at neutral conditions had no permeabilizing activity. Although the addition of MgCl2 to the NPN assay system with 5 mM lactic acid challenge resulted in reduced NPN uptake for P. aeruginosa and E. coli especially in the presence of KCN, these effects cannot be regarded as indicative of chelation of cations from the OM, since similar effects were also observed with HCl and E. coli. A more likely mechanism than chelation would be protonation of anionic components such as carboxyl and phosphate groups and the consequent weakening of molecular interactions between OM components. It is plausible that rather than interacting directly with the acid molecules, MgCl2 stabilizes the OM, making it more resistant to acid challenge. Instead, excess Mg2+ with each bacterial species expectedly abolished the NPN uptake induced by EDTA, which is believed to act solely by chelation.
The permeabilizing capacity of lactic acid has a number of important consequences. Above all, lactic acid should be able to potentiate the apparent antimicrobial activity of other components against gram-negative bacteria. In natural situations such as in fermented low-pH products obtained by lactic acid starter culture bacteria, numerous metabolites are present that are too lipophilic or too large to effectively penetrate the intact gram-negative bacterial OM but that could possibly do so in the presence of lactic acid. Beside well-recognized lactic acid bacterial antimicrobial factors such as diacetyl (24), hydrogen peroxide, lactoperoxidase systems, and reuterin (5), a plethora of cryptic antimicrobials acting in synergy with lactic acid could theoretically exist. In fact, culture supernatant of Lactobacillus plantarum was recently shown to contain small-molecular-mass substances acting together with lactic acid against the gram-negative target organism Pantoea agglomerans (19). There are also indications (3, 4) that high concentrations of lactic acid sensitizes gram-negative bacteria to bacteriocins such as nisin. The sublethal injury caused by lactic acid could play a major role in such sensitizing, along with providing an acidic milieu required for the chemical stability of nisin (6).
Despite the neutral pH conditions of the large intestine, which probably are not favorable for the permeabilizing action of lactic acid in general, it could be assumed that probiotic lactic acid bacterial strains might, however, be beneficial in combating gram-negative pathogens in the large intestine. This could happen through local production of relevant concentrations of lactic acid in microenvironments, with inhibition of harmful gram-negative strains by the combined action of lactic acid and bile salts; the latter possess detergent-like action against which many enteric gram-negative pathogens exhibit resistance (18). Another interesting feature is that lactobacilli and lactic acid have been reported to suppress the gastric pathogen Helicobacter pylori (1, 16, 17). H. pylori is naturally adapted to an acid environment (9), and it would be of interest to investigate the permeability properties of the OM of this pathogen in response to challenge with different acids.
A gradual increase in acidity can allow induced tolerance to acid to occur (acid habituation), and this will permit the organisms to survive subsequent exposures which could be lethal to nonhabituated cells (2, 20, 21, 27). The effect of such an adaptive response on the permeability properties of gram-negative bacteria should be examined, especially with enteric pathogens such as E. coli O157:H7.
ACKNOWLEDGMENTS
We thank Päivi Lepistö and Anna-Liisa Ruskeepää for excellent technical assistance.
This work was supported by the Academy of Finland (project 44163) and by the European Commission (project NISINPLUS, FAIR-CT96-1148).
REFERENCES
- 1.Aiba Y, Suzuki N, Kabir A M A, Takagi A, Koga Y. Lactic acid-mediated suppression of Helicobacter pylori by the oral administration of Lactobacillus salivarius as a probiotic in a gnotobiotic murine model. Am J Gastroenterol. 1998;93:2097–2101. doi: 10.1111/j.1572-0241.1998.00600.x. [DOI] [PubMed] [Google Scholar]
- 2.Baik H S, Bearson S, Dunbar S, Foster J W. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology. 1996;142:3195–3200. doi: 10.1099/13500872-142-11-3195. [DOI] [PubMed] [Google Scholar]
- 3.Cutter C N, Siragusa G R. Population reductions of gram-negative pathogens following treatments with nisin and chelators under various conditions. J Food Prot. 1995;58:977–983. doi: 10.4315/0362-028X-58.9.977. [DOI] [PubMed] [Google Scholar]
- 4.Cutter C N, Siragusa G R. Treatments with nisin and chelators to reduce Salmonella and Escherichia coli on beef. J Food Prot. 1995;58:1028–1030. doi: 10.4315/0362-028X-58.9.1028. [DOI] [PubMed] [Google Scholar]
- 5.Daeschel M A, Penner M H. Hydrogen peroxide, lactoperoxidase systems, and reuterin. In: Ray B, Daeschel M, editors. Food preservatives of microbial origin. Boca Raton, Fla: CRC Press; 1992. pp. 155–175. [Google Scholar]
- 6.De Vuyst L, Vandamme E J. Nisin, a lantibiotic produced by Lactococcus lactis subsp. lactis: properties, biosynthesis, fermentation and application. In: De Vuyst L, Vandamme E J, editors. Bacteriocins of lactic acid bacteria. Glasgow, United Kingdom: Blackie A&P; 1994. pp. 151–221. [Google Scholar]
- 7.Dickson J S, Anderson M E. Microbiological decontamination of food animal carcasses by washing and sanitising systems: a review. J Food Prot. 1992;55:133–140. doi: 10.4315/0362-028X-55.2.133. [DOI] [PubMed] [Google Scholar]
- 8.Doores S. Organic acids. In: Davidson P M, Branen A L, editors. Antimicrobials in foods. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 95–136. [Google Scholar]
- 9.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer G G, Dilts B D. Lactic acid inhibition of the growth of spoilage bacteria and cold tolerant pathogens on pork. Int J Food Microbiol. 1995;25:141–151. doi: 10.1016/0168-1605(94)00088-n. [DOI] [PubMed] [Google Scholar]
- 11.Helander I M, Mäkelä P H, Westphal O, Rietschel E T. Lipopolysaccharides. In: Meyers R A, editor. Encyclopedia of molecular biology and molecular medicine. Vol. 3. Weinheim, Germany: VCH; 1996. pp. 462–471. [Google Scholar]
- 12.Helander I M, Alakomi H-L, Latva-Kala K, Koski P. Polyethyleneimine is an effective permeabilizer of gram-negative bacteria. Microbiology. 1997;143:3193–3199. doi: 10.1099/00221287-143-10-3193. [DOI] [PubMed] [Google Scholar]
- 13.Helander I M, von Wright A, Mattila-Sandholm T. Potential of lactic acid bacteria and novel antimicrobials against gram-negative bacteria. Trends Food Sci Technol. 1997;8:146–150. [Google Scholar]
- 14.Helander I M, Alakomi H-L, Latva-Kala K, Mattila-Sandholm T, Pol I, Smid E J, Gorris L G M, von Wright A. Characterization of the action of selected essential oil components on gram-negative bacteria. J Agric Food Chem. 1998;46:3590–3595. [Google Scholar]
- 15.Helander I M, Mattila-Sandholm T. Fluorometric assessment of gram-negative bacterial permeabilization. J Appl Microbiol. 2000;88:213–219. doi: 10.1046/j.1365-2672.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- 16.Kabir A M A, Aiba Y, Takagi A, Kamiya S, Miwa T, Koga Y. Prevention of Helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut. 1997;41:49–55. doi: 10.1136/gut.41.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Midolo P D, Lambert J R, Hull R, Luo F, Grayson M L. In vitro inhibition of Helicobacter pylori NCTC 11637 by organic acids and lactic acid bacteria. J Appl Bacteriol. 1995;79:475–479. doi: 10.1111/j.1365-2672.1995.tb03164.x. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido H. Outer membrane. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 29–47. [Google Scholar]
- 19.Niku-Paavola M-L, Laitila A, Mattila-Sandholm T, Haikara A. New types of antimicrobial compounds produced by Lactobacillus plantarum. J Appl Microbiol. 1999;86:29–35. doi: 10.1046/j.1365-2672.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- 20.Nojoumi S A, Smith D G, Rowbury R J. Tolerance to acid in pH 5.0-grown organisms of potentially pathogenic gram-negative bacteria. Lett Appl Microbiol. 1995;21:359–363. doi: 10.1111/j.1472-765x.1995.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 21.Park Y-K, Bearson B, Bang S H, Bang I S, Foster J W. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol Microbiol. 1996;20:605–611. doi: 10.1046/j.1365-2958.1996.5441070.x. [DOI] [PubMed] [Google Scholar]
- 22.Podolak R K, Zayas J F, Kastner C L, Fung D Y C. Inhibition of Listeria monocytogenes and Escherichia coli O157:H7 on beef by application of organic acids. J Food Prot. 1996;59:370–373. doi: 10.4315/0362-028X-59.4.370. [DOI] [PubMed] [Google Scholar]
- 23.Przybylski K S, Witter L D. Injury and recovery of Escherichia coli after sublethal acidification. Appl Environ Microbiol. 1979;37:261–265. doi: 10.1128/aem.37.2.261-265.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray B. Diacetyl of lactic acid bacteria as a food biopreservative. In: Ray B, Daeschel M, editors. Food preservatives of microbial origin. Boca Raton, Fla: CRC Press; 1992. pp. 137–153. [Google Scholar]
- 25.Ray B, Sandine W E. Acetic, propionic, and lactic acids of starter culture bacteria as biopreservatives. In: Ray B, Daeschel M, editors. Food preservatives of microbial origin. Boca Raton, Fla: CRC Press; 1992. pp. 103–136. [Google Scholar]
- 26.Roth L A, Keenan D. Acid injury in Escherichia coli. Can J Microbiol. 1971;17:1005–1008. doi: 10.1139/m71-160. [DOI] [PubMed] [Google Scholar]
- 27.Ryu J-H, Deng Y, Beuchat L. Behavior of acid-adapted and unadapted Escherichia coli O157:H7 when exposed to reduced pH achieved with various organic acids. J Food Prot. 1999;62:451–455. doi: 10.4315/0362-028x-62.5.451. [DOI] [PubMed] [Google Scholar]
- 28.Salminen S, Deighton M A, Benno Y, Gorbach S L. Lactic acid bacteria in health and disease. In: Salminen S, von Wright A, editors. Lactic acid bacteria: microbiology and functional aspects. New York, N.Y: Marcel Dekker, Inc.; 1998. pp. 211–253. [Google Scholar]
- 29.Smulders F J M, Greer G G. Integrating microbial decontamination with organic acids in HACCP programmes for muscle foods: prospects and controversies. Int J Food Microbiol. 1998;44:149–169. doi: 10.1016/s0168-1605(98)00123-8. [DOI] [PubMed] [Google Scholar]
- 30.Träuble H, Overath P. The structure of Escherichia coli membranes studied by fluorescence measurements of lipid phase transition. Biochim Biophys Acta. 1973;307:491–512. doi: 10.1016/0005-2736(73)90296-4. [DOI] [PubMed] [Google Scholar]
- 31.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Netten P, Huis In't Veld J H J, Mossel D A A. An in-vitro meat model to assess the immediate bactericidal effect of lactic acid decontamination of meat. J Appl Bacteriol. 1994;76:49–54. doi: 10.1111/j.1365-2672.1994.tb04414.x. [DOI] [PubMed] [Google Scholar]
- 33.van Netten P, Mossel D A A, Huis In't Veld J H J. Lactic acid decontamination of fresh pork carcasses: a pilot plant study. Int J Food Microbiol. 1995;21:1–15. doi: 10.1016/0168-1605(94)00039-9. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson R G, Gemski P, Jr, Stocker B A D. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972;70:527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- 35.Zähringer U, Lindner B, Rietschel E T. Molecular structure of lipid A, the endotoxic center of bacterial lipopolysaccharides. Adv Carbohydr Chem Biochem. 1994;50:211–276. [PubMed] [Google Scholar]