Abstract
Extrachromosomal circular DNA (eccDNA) of chromosomal origin is found in many eukaryotic species and cell types, including cancer, where eccDNAs with oncogenes drive tumorigenesis. Most studies of eccDNA employ short-read sequencing for their identification. However, short-read sequencing cannot resolve the complexity of genomic repeats, which can lead to missing eccDNA products. Long-read sequencing technologies provide an alternative to constructing complete eccDNA maps. We present a software suite, Construction-based Rolling-circle-amplification for eccDNA Sequence Identification and Location (CReSIL), to identify and characterize eccDNA from long-read sequences. CReSIL’s performance in identifying eccDNA, with a minimum F1 score of 0.98, is superior to the other bioinformatic tools based on simulated data. CReSIL provides many useful features for genomic annotation, which can be used to infer eccDNA function and Circos visualization for eccDNA architecture investigation. We demonstrated CReSIL’s capability in several long-read sequencing datasets, including datasets enriched for eccDNA and whole genome datasets from cells containing large eccDNA products. In conclusion, the CReSIL suite software is a versatile tool for investigating complex and simple eccDNA in eukaryotic cells.
Keywords: CRESIL, eccDNA, long-read sequence, bioinformatic tool
Introduction
Extrachromosomal circular DNA (eccDNA) of chromosomal origin is a common byproduct of mutation found across the eukaryotic kingdoms from plants to fungi and animals [1–3]. EccDNA can range in size from a hundred base pairs to several megabases and thereby often capture genes or parts of genes [4–7]. A lack of centromeres on eccDNA means that these elements segregate unfaithfully and tend to accumulate in subpopulations of cells that can thrive on a high copy number when genes included in eccDNA are expressed, providing a selective advantage [6]. This is particularly evident in the unicellular yeast and cancer cells, where eccDNA can greatly affect the cell phenotype and disease progression [2, 8–10]. Similarly, yeast cells grown under nutrient-limiting conditions can obtain a selective advantage when transporter genes are trapped and accumulate in clonal sub-populations [5, 11]. An example of this is when yeast cells grow with limited glucose, and clones of yeast cells with eccDNA harboring circularized glucose transporter genes [HXT6/7circle] accumulate in the population [5]. Many tumor cells accumulate oncogenes on circular DNA [6, 10, 12], suggesting circular DNA plays a role in tumorigenesis. These structures are often complex and comprise several fragments, such as enhancers and several oncogenes, indicating that eccDNA can evolve to become more transcriptionally efficient [13, 14]. Besides, small eccDNAs (200–3000 nt) can express small non-coding RNA, which act as transcriptional regulators without a canonical promoter [9]. These recent advances in our understanding of eccDNA are mainly obtained through short-read sequencing characterization with tools such as AmpliconArchitect [15], Circle-Map [16], ECCsplorer [17], or eccDNA_finder [18].
However, the identification of eccDNA from short reads is limited by the capacity to identify repetitive regions of genomes such as centromeres and complex eccDNA composed of several fragments. This problem can be solved with long-read sequencing technologies such as Pacific Bioscience Technology (PacBio) and Oxford Nanopore Technology (ONT) because the long reads span longer stretches, thereby covering elements in repeats and multiple fragments in complex eccDNA. Identification is further aided by amplifying the eccDNA with rolling circle amplification (RCA) [19–21]. This simple, yet powerful approach allows for reads that have often traveled around the circle several times, generating concatemeric tandem copies (CTC). Still, bioinformatic tools to identify eccDNA from RCA long-read sequences are scarce, and the available tools have never been compared. CIDER-Seq2 is designed to characterize virus genomes [22, 23], while ecc_finder is made for both short and long-read sequences [18]. This tool was extensively tested in short-read sequences; however, the performance evaluation in long-read datasets are limited. Wang et al. presented the eccDNA_RCA_Nanopore software [24], which only focuses on sequences that contain CTC and ignores the remaining 52% of the sequenced reads, which could be important products derived from incomplete RCA of large-size eccDNA or DNA breakages during the experimental procedures. We have recently presented two tools for mapping eccDNA: NanoCircle and CReSIL [25]. NanoCircle identifies complex and simple circles from sequence reads that span the junction site in the circular DNA but often fails to assemble complex circles correctly. CReSIL is designed to construct eccDNA de novo; however, advancing computational workflow and additional features that aid deep investigations of the identified eccDNAs has been needed to uncover the complete architecture of eccDNA sequences, reflecting their biological function.
Here, we present the bioinformatics software suite, Construction-based Rolling-circle-amplification for eccDNA Sequence Identification and Location (CReSIL), to accurately construct and represent eccDNA from long-read sequences. We benchmarked it to other known tools for identifying eccDNA from long-read sequences and found that it is superior to all other available tools. CReSIL provides useful features to investigate genomic annotations, sequence variations of eccDNA, and visualization of eccDNA architecture. We demonstrated the capability of CReSIL to identify eccDNA molecules from many human and mouse samples enriched for eccDNA and showed how CReSIL could be used to identify eccDNA from whole genome long-read sequencing (WGLS) datasets.
Material and methods
The methodological details are described in the Supplementary Materials and Methods.
Results
EccDNA enrichment workflow and CReSIL computational workflow for eccDNA identification
We designed a computational workflow to identify eccDNA from long-read sequence samples enriched for eccDNA (Figure 1.0). Enrichment was obtained by removing the chromosomal DNA and mitochondrial DNA from eccDNA. In this procedure, the mitochondrial DNA was linearized using either rare cutting restriction enzymes DNA or CRISPR-Cas9 [26] specifically designed to target mtDNA; after which the linear chromosomal and mitochondrial DNA was digested using Exonuclease V. We next amplified the eccDNA molecules using RCA, which allows for DNA sequences that spanned the eccDNAs more than once. The RCA approach produced hyperbranched DNA products that required de-branching with the T7 endonuclease before sequencing library preparation, obtaining long reads and minimizing pore clogging during DNA sequencing that would otherwise result in ghost sequences (i.e. improperly generated sequences that are unmappable or have similarities to the known sequence but in the opposite orientation). ONT sequencing was next performed to generate long-read sequences used as the input for the CReSIL 2.0 bioinformatic analysis.
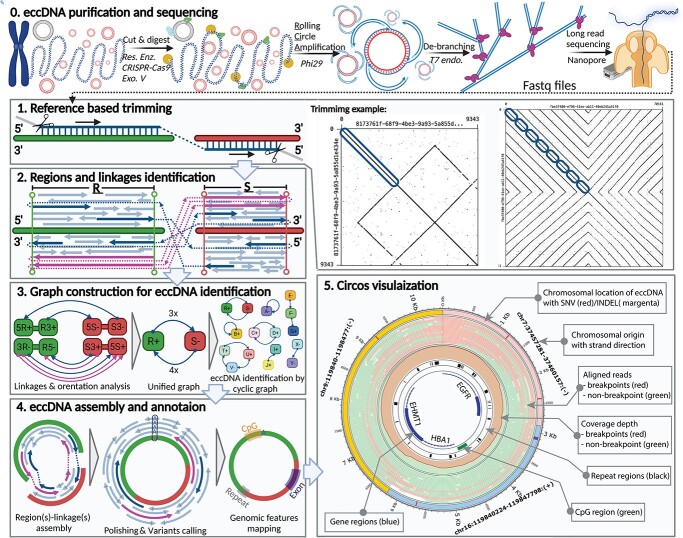
Figure 1.
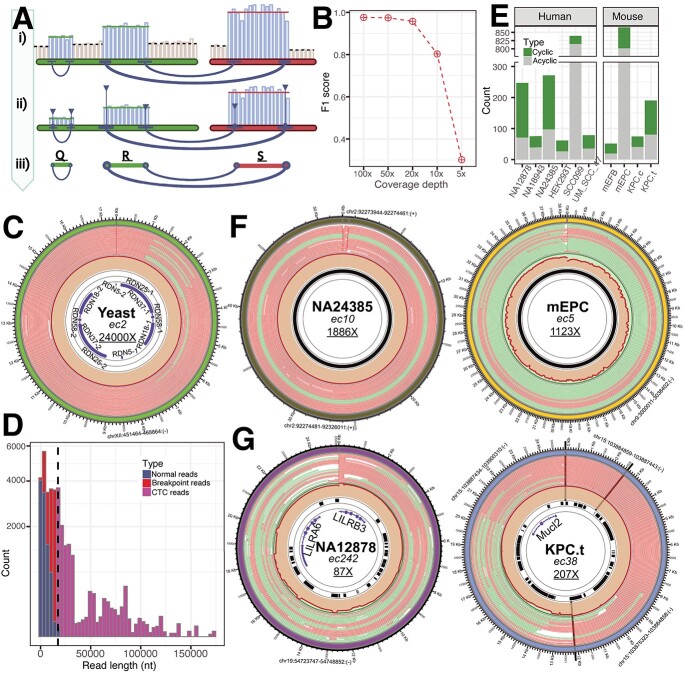
EccDNA identification by long-read sequencing. Step 0. The experimental workflow begins with purified genomic DNA; chromosomal DNA (blue), mitochondrial DNA (magenta), eccDNA (red), restriction enzyme (green), CRISPR-Cas9 (cyan), and exonuclease V (yellow). Step 1. A read (left panel) can be aligned on regions of 2 chromosomes (green and red) of the reference genome (blue) with breakpoint reads (dashed line) linking the aligned chromosomal regions and trimmed unmapped portions (gray). Self-read dot plots (right panels) showing read regions that align to the reference genome (blue ovals); reads without CTCs (left dot plot) and with CTCs (right dot plot). Step 2. Merged regions R and S with the reference sequences (green and red bars) and the breakpoint event (dashed lines) that links the two regions; arrows for non-breakpoint reads (light blue), breakpoint reads (blue), and reads with CTCs (magenta) point in the direction of the aligned orientation on the plus strand of the reference sequence (arrows above the bars) or the minus strand (arrows below the bars). Step 3. The information of the read alignments was converted to directed graphs. Step 4. The reads were assembled using our developed regions/linkages algorithm; the assembled sequences were polished, variants were identified, and eccDNA was annotated with genomic features such as exon, repeat, and CpG. Step 5. Circos visualization to present the eccDNA architecture.
Even though the de-branching was performed, we observed ghost sequences in the long-read data as examples of dot plots illustrated in Figure 1.1. Therefore, Step 1 of CReSIL was the preprocessing of the raw reads by reference-based trimming, which is essential in avoiding false positives from ghost reads. First, the reads were aligned on the reference genome. Next, high confidence mapped region(s) of the reads were obtained, and the rest were trimmed out. For example, the read without CTC (left dot plot of Figure 1.1) has a sequence length of 9343 nucleotides (nt); we found a 3514 nt region at the beginning of the read that was mapped to the reference genome. CReSIL trimmed the rest of the read, which was assumed to be a ghost sequence of self-inverted repeats followed by an unmapped sequence. An example of a CTC read (right dot plot of Figure 1.1) was a sequence of 70 161 nt. We found that the first half of this read contained eight consecutive sequence copies that could be mapped on the same region of the reference genome. The second half of the read was a sequence of self-inverted repeats, which was assumed to be a ghost sequence that CReSIL discarded. The reference alignment results of the individual reads were used for the next step.
In Step 2 (Figure 1.2), chromosomally aligned locations of the trimmed reads were aggregated to identify merged regions representative of the chromosomal origins of eccDNA. On the individual merge regions, we typically observed three types of aligned reads: (1) reads aligned only to one region (normal reads), (2) reads aligned on multiple regions without CTC (breakpoint reads without CTC), and (3) reads aligned on multiple regions with CTC (CTC reads). CReSIL recorded the aligned reads of the identified individual merged region, the linkages of the identified merged region(s), the orientation of the reads (indicated by arrows direction), and the aligned strand orientation of the chromosomal regions. The last two types contain breakpoint(s), which is critical to identify linkages that connect region(s) together.
In Step 3 (Figure 1.3), CReSIL formulated the recorded regions and linkages information into graph representations. First, CReSIL constructed directed graphs with the information of regions, terminals, and strands; therefore, an individual region contained four nodes and multiple edges derived from linkages (Figure 1.3. left panel). After the linkages and orientation analysis based on read alignment orientation and strand, the graphs were unified (see Supplementary Figure S1 for methodological details), resulting in one node representing one region and the weight of the edges representing the number of linkages (Figure 1.3. middle panel). From this point, CReSIL identified high-confidence eccDNA from cyclic graphs and low-confidence eccDNA (possible translocation) from acyclic graphs.
In Step 4 (Figure 1.4), CReSIL performed region(s) and linkage(s) assembly based on the graphs (Figure 1.4. left panel). All the reads belonging to an individual graph were assembled and polished to generate a consensus sequence. During polishing, CReSIL also identified mutations present in the eccDNA (Figure 1.4. middle panel). Moreover, CReSIL annotated some selected genomic features on the identified eccDNA (e.g. exon, intron, repeat, CpG).
In Step 5 (Figure 1.5), CReSIL generated an input file of selected eccDNA for visualization to illustrate eccDNA architecture using the Circos software [27]. Each plot contains information about the chromosomal origin of eccDNA, any single nucleotide variation (SNV) or an insertion–deletion mutation (INDEL), a read alignment, coverage of breakpoint reads, non-breakpoint reads, and selected genomic features.
CReSIL outperformed in eccDNA detection compared with other tools
To evaluate the performance of CReSIL in comparison with other tools such as eccDNA_RCA_nanopore [24], ecc_finder [18], or the long-read de novo assembler Flye [28], we generated a synthetic eccDNA dataset of 1300 true positives, which mimic long reads derived from eccDNA purification and RCA amplification (Figure 1.0), and 1300 true negatives by randomly selecting chromosomal regions of the human genome (hg19). We simulated eccDNA following the size and region distribution previously reported by Henriksen et al. [25] (Figure 2A). The size distribution of the simulated eccDNA true positive and true negative datasets was similar (Figure 2A), with a minimum size of approximately 500 nt, a maximum size of approximately 34 000 nt, and a median size of approximately 6200 nt. The simulated sets contained both simple and complex (>1 region) eccDNAs with a similar distribution of chromosomal-containing regions (Figure 2B) and varying depth coverage from 3× to 100×. We used PBSIM2 [29] software to simulate long reads based on the simulated eccDNA sets varying depth coverage from 3× to 100×. Considering the true positive dataset, we found that in the lower sequencing depth, the number of simulated eccDNA that contained CTC reads was reduced (Figure 2C) due to the lower probability of generating CTC reads by PBSIM2 mimicked the incomplete circular amplification and/or DNA breakage events.
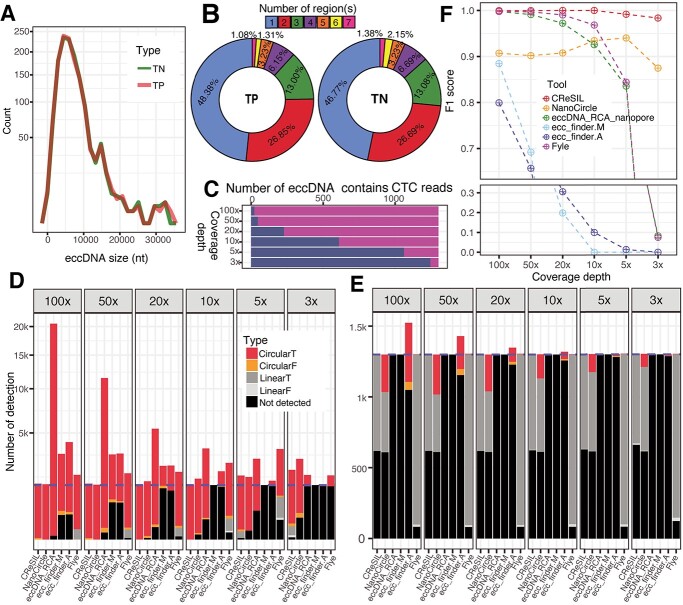
Figure 2.
Evaluation of eccDNA detection performance of CReSIL and its comparison with the other tools. A) Frequency polygon plot showing the size distribution of simulated eccDNA of true positive (TP, orange line) and true negative (TN, green line) datasets. B) Donut plots showing the percent distribution of the number of chromosomal regions of the individual simulated eccDNA datasets. C) Bar plots showing the number of simulated eccDNA that contains CTC reads (magenta) and non-CTC reads (dark blue) across different sequencing depths. D) Stacked bar plots showing the number of eccDNA detected for the true positive dataset by different tools. The eccDNA detection results are classified into five categories; CircularT (red) = circular sequences with 95% reciprocal overlaps and 90% identity with the simulated eccDNA sequences, CircularF (orange) = circular sequences without the criteria, LinearT (dark gray) = linear sequences with the criteria, LinearF (light gray) = linear sequences without the criteria, and not detected (black) = the tool cannot detect; the number of true positive eccDNA (blue dashed line). E) Stacked bar plots presenting the number of eccDNA detected for the true negative dataset. See the color code for panel D, the number of true negative eccDNA (blue dashed line). F) Point and line plots showing the performance (F1 score) of eccDNA detection of the individual tools across different sequencing depths.
The identification of eccDNA results based on synthetic datasets derived from CReSIL and the other tools were separately summarized for true positive (Figure 2D) and true negative (Figure 2E) datasets. CReSIL and NanoCircle correctly detected almost every true positive dataset except low coverage of 5× and 3× (Figure 2D). At 100×, eccDNA_RCA_nanopore detected a large amount of eccDNAs (approximately 20 000 eccDNAs), which is due to high redundancy derived from individual CTC reads. Because eccDNA_RCA_nanopore is designed to detect eccDNA derived from CTC reads only, it missed the detection of eccDNA whose simulated reads contained no CTC reads. We also compared our software to ecc_finder in two modes, assembly (ecc_finder.A), and mapping (ecc_finder.M). The eccDNA detection results from the two modes revealed a high fraction of true positives that were not detected at high depth coverage of 100x and 50x, and missed detection of almost every eccDNA at a low sequencing depth of 10×, 5×, and 3×. The de novo assembly tool, Flye, performed well in detecting eccDNA at 100× (missed detection of 48 eccDNAs), yet the performance of Flye dropped when the sequencing depth was reduced. Considering the true negative datasets (Figure 2E), almost every tool controlled the false positive rate very well, except ecc_finder assembly mode (ecc_finder.A), which produced a high number of false positives at 100×, which was reduced as the sequencing depth decreased. NanoCircle has a similar number of false positives across all sequencing depths, which are derived from complex circles only. We next calculated the F1 score to compare the eccDNA detection performance across the tools at the various coverage depths (Figure 2F, the precision and recall of each tool and scenario are provided in Supplementary Table S4.). All tools performed very well at 100×, but F1 scores dramatically dropped when sequencing depth was reduced, except CReSIL, which maintained an F1 score of 0.98 at 3×. This demonstrates that CReSIL outperforms other available tools in eccDNA detection from long reads derived from eccDNA enrichment and RCA amplification.
Identification and characterization of eccDNA from eccDNA enrichment samples using CReSIL
To demonstrate the capabilities of CReSIL on real data, we next performed eccDNA enrichment and long-read sequencing following the experimental workflow shown in Figure 1.0 to generate high-quality datasets of three human multiple myeloma cancer cell lines (APR1, EJM, and JJN3), two mouse tissues (pancreas and cortex), and three mouse cells (MLOY4 osteocyte-like cells, 5TMG1 multiple myeloma cells, and E0771 breast cancer cells). In addition, we included in the analysis published eccDNA enrichment long-read sequencing data of a human sperm dataset from Henriksen et al. [25] and the mouse embryonic stem cell mESC dataset from Wang et al. [24].
The datasets ranged from 0.25 to 4.16 million reads after trimming (Figure 3A) and CResIL detected a variety of reads containing CTC with debranching, ranging from 9.4 to 53.7%, and only 1.8% CTC reads in samples that had not been debranched before sequencing. This indicated the importance of T7 endonuclease treatment before sequencing. After trimming, CReSIL kept a high fraction of reads for the following steps, averaging 76% for the CTC reads and 98% for the normal reads (Figure 3B).
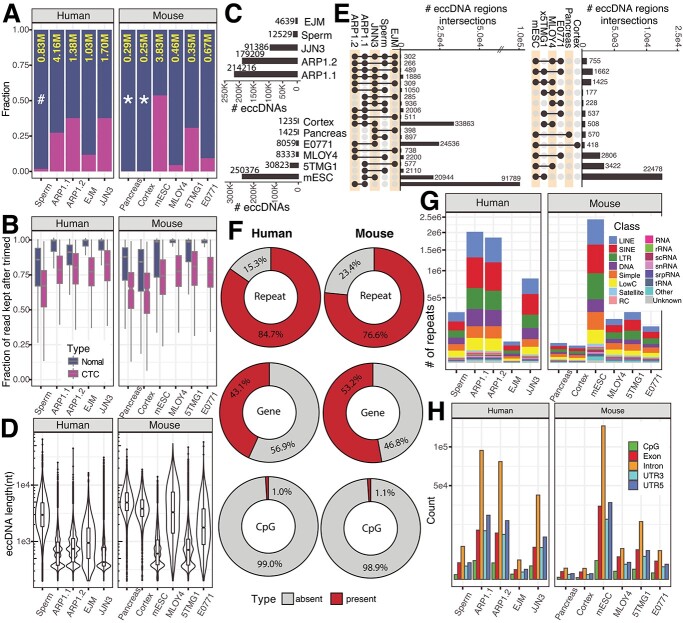
Figure 3.
Identification of eccDNA from long-read sequencing of eccDNA enrichment samples derived from human and mouse cells. A) Stacked bar plot showing the fraction of non-CTC reads (dark blue) and CTCs (magenta) across five human cell samples and six mouse cell samples; the total number of high-quality reads after the CReSIL trimming step (yellow), in a million read units; * = datasets prepared by primer-free-based RCA; the rest were prepared by primer-based RCA; # = sample prepared for sequencing without debranching. B) Box-whiskers plots showing the fraction of reads used for eccDNA identification that was kept after trimming of normal (dark blue) and CTC (magenta) reads. C) Bar plots summarizing the number of eccDNA in human (top panel) and mouse (bottom panel) datasets D) Violin boxplots showing the distributions of the length of identified eccDNA. E) Upset plots showing overlaps (right panels), only overlapped numbers over 100 are shown. F) Donut charts showing the percentages of eccDNA harbored repeats, genes, and CpG islands. G) Stacked bar plots showing the frequency of different classes of repeats harbored in identified eccDNA. H) Bar plots showing the frequency of CpG, exon, intron, 3’UTR, and 5’UTR harbored in identified eccDNA.
In human datasets (Figure 3C top panel), CReSIL identified a high number of eccDNAs in cancer cell lines, over 200 000 eccDNAs for both replicates of APR1, and over 90 000 eccDNAs for JJN3 cells. CReSIL also identified over 12 000 eccDNAs for sperm and over 4600 eccDNAs for EJM cells. In mouse datasets (Figure 3D bottom panel), CReSIL identified over 250 000 eccDNAs from mESC, over 30 000 for 5TMG1, over 8000 eccDNAs for MLOY4 and E0771 cells, and less than 1500 eccDNAs for the two tissues. Overall, for the human and mouse datasets, most of the identified eccDNAs were formed from a single region in the genome (simple eccDNA), and ~1.5% were complex eccDNAs made of several fragments of chromosomal DNA (see example Figure 1 step 5 and Supplementary Table S5). The length size distribution of the identified eccDNA for each dataset is shown in Figure 3D. The smallest size of eccDNA is 200 nt, which is the selected minimum region length cut-off for graph construction. We recorded the biggest eccDNA size to approximately 65 000 nt in sperm for the human dataset and approximately 58 000 nt in E0771 for the mouse dataset. Next, we checked the overlaps among the identified eccDNA for each organism as shown in the upset plots (Figure 3E). We observed a relatively low number of overlaps comparing the number of identified eccDNAs. This is consistent with the random biogenesis of eccDNA proposed by Møller et al. [30, 31] and Wang et al. [24]. However, we found over 28 000 specifically recurrent eccDNAs between replicates of the ARP1 cell line (~15%), indicating a cell-specific population of eccDNA that possibly benefits the ARP1 cell line (Figure 3E left panel).
Characterizing genes and genomic content harbored by eccDNA is essential for understanding the function of eccDNAs in tumor cells [13, 32] or other cells where eccDNA can provide a selective advantage [5, 11]. CReSIL also provides a function for genomic annotations of individual eccDNAs. We annotated three types of genetic elements, repeats, genes, and CpG islands, on the identified eccDNA. CReSIL mapped the repeats on a high percentage of identified eccDNA of in the human datasets (84.7%) and in the mouse datasets (76.6%) (Figure 3E), which is similar to previous reports using short-read sequences to identify eccDNA [30, 33]. Moreover, we found that 43.1% of the identified eccDNAs in the human dataset and 53.2% in the mouse dataset harbored genes or parts of genes (Figure 3E middle panel). We observed a very small amount, less than 1.5% of the identified eccDNA, harbored CpG islands (Figure 3E bottom panel). Repetitive sequences were commonly found in eccDNA reported in many studies [30, 33, 34] and could promote circles forming through microhomology [34–36]. The 16 different repeat classes were distributed across individual datasets (Figure 3F). Long interspersed nuclear elements, short interspersed nuclear elements, and long terminal repeats were the most frequently found repeat classes identified in the eccDNAs. Of the repeat types, the introns were most frequently found in identified eccDNA harboring genes or part of genes. We also observed a high frequency of exons in the identified eccDNAs (Figure 3G).
Because CReSIL identifies eccDNA by aggregating all reads in a sample and calculates the coverage depth of individually identified eccDNA, we can use this information to rank the relative abundance of eccDNA molecules qualitatively. Based on the hexbin plots (Figure 4A), we did not observe a correlation between the size of the identified eccDNA and their coverage depth for both the human and mouse datasets. Interestingly, we found that the most frequent coverage depth of the identified eccDNA is approximately 10x for both datasets. Next, we used CReSIL to generate consensus sequences of identified eccDNA and called genetic variations for individually identified eccDNA. At the variant quality score cut-off of 20, we found 35.6% of the identified eccDNA for the human dataset and 32.0% for the mouse dataset have variant(s) (Figure 4B). Single nucleotides variation events gave the highest fraction of the variants, except for mouse pancreas and cortex eccDNA, in which deletion events gave the highest fraction of the variants (Figure 4C). Interestingly, we observed a low fraction of insertion events across all datasets.
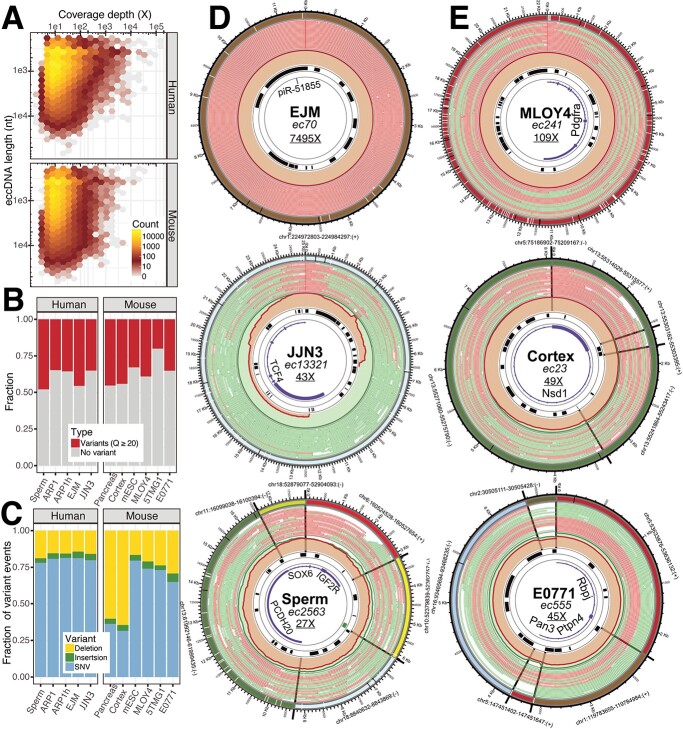
Figure 4.
Identification of eccDNA from long-read sequencing of eccDNA enrichment samples derived from human and mouse cells. A) Hexagonal bin plots showing no correlation between the length of the identified eccDNA with their coverage depth. B) Bar plots showing the fraction of the identified eccDNA containing variants (SNVs and/or INDELs) based on a quality cut-off of 20. C) Bar plots showing the distribution of deletions, insertions, and SNVs of the identified eccDNA. D) Circos plots visualizing the architecture of three selected human eccDNA datasets. E) Circos plots visualizing the architecture of three selected mouse eccDNA datasets (see Figure 1.5 for lane annotation; data set information (bold), eccDNA name (italics), coverage depth (underline).
CReSIL also provides functions to make Circos plots visualize the architecture of identified eccDNA. We arbitrarily selected six eccDNAs from the human (Figure 4D) and mouse datasets (Figure 4E). Of the human eccDNAs, identified in the EJM dataset, ec70 has a size of approximately 11 500 nt and has an extremely high coverage depth of almost 7500×, indicating a very high abundance of this eccDNA molecule. Ec70 harbored only a short non-coding RNA, piR-51,855. Most of the reads contributing to the identification of ec70 were CTC reads. In contrast, identified in the JJN3 dataset, ec13321 has a size of approximately 25 000 nt, which is based on non-CTC reads with the strong support of breakpoint reads. Ec13321 had 43× coverage depth and harbored five exons of the transcription factor 4 gene. Lastly, identified from the sperm dataset, ec2563 is a complex eccDNA that comprises six regions of different chromosomes with a total length of approximately 18 000 nt and harbors a CpG island and the parts of three genes, insulin growth factor 2 receptor, sex-determining region Y–box transcription factor 6, and protocadherin 20, which came from different chromosomal origins. Of the mouse eccDNAs, ec241 was identified in the MLOY4 dataset and is approximately 22 000 nt with a high coverage depth of 109×. Interestingly, ec241 contains five exons of the platelet-derived growth factor receptor alpha gene, a well-known marker for mesenchymal lineage cells [37], reflecting the origin of the osteocyte-like MLOY4 cells. Identified in the cortex dataset, ec23 is a complex eccDNA with a length of approximately 8000 nt and four regions from chromosome 13. Interestingly, based on the chromosomal coordinates, ec23 began at chr13:55303162 and ended at chr13:55315577 (12 416 nt). Ec23 showed that four regions within the chromosomal coordinates were combined to generate an eccDNA with a size of 8068 nt, which is a substantial deletion (a total of 4348 nt). Ec23 had 49× coverage depth and harbored 1 exon of the nuclear receptor binding SET domain protein 1 gene, which is known to be associated with Sotos syndrome [38] (caused by mutations in this gene). Finally, identified in the E0771 dataset, ec555 is a complex eccDNA of five regions from four different chromosomes and has a size of approximately 6600 nt, with 45× coverage depth. Ec555 harbored the parts of three genes, recombination signal binding protein for immunoglobulin kappa J region, protein tyrosine phosphatase non-receptor type 4, and poly(A) specific ribonuclease subunit. The reads used to construct the molecule derived from CTC and normal reads with a similar fraction of half.
Identification of eccDNA from long-read whole genome sequencing datasets by CReSIL
Turner et al. reported that approximately 50% of the focal copy number amplification in tumors derives from eccDNA molecules [6]. The structure can be constructed by their sequence architecture and the AmpliconArchitect algorithm and, in some cases, validated by long-read sequences, as reported by Deshpande et al. [15]. Therefore, we extended CReSIL’s capability to identify eccDNA molecules from WGLS by implementing an additional workflow (Figure 5A) to identify focal regions and their linkages and then fed it to the graph construction algorithm of CReSIL. Beginning with the read alignment result, CReSIL calculated the coverage depth of small windows of 10 nt across the reference chromosomes to ensure that small-sized eccDNAs were captured. The average coverage depth of individual chromosomes was recorded to estimate the backgrounds, which was used to subtract the coverage depth of all windows with respect to their chromosomal location. In parallel, CReSIL identified the breakpoint positions from the read alignment result. After the subtraction, the focal amplified regions were uncovered by aggregation of the windows with depth higher than the background. The focal amplified regions were refined by the breakpoint locations. The refined regions and the linkages information were converted in the graph construction (Figure 1.3) of the CReSIL workflow. Then, the eccDNAs were identified through the CReSIL workflow.
Figure 5.
Identification of eccDNA from WGLS of yeast human and mouse cells. A) Scheme showing CReSIL extension workflow to identify eccDNA from WGLS dataset; the sequencing depth on a small window of focal amplified regions (blue bars) was higher than the background (gray), and the identified focal regions (horizontal lines) are shown with the breakpoint reads generating linkages between the regions (dark blue curved lines); also shown are the number of breakpoints reads (thicker lines = more reads) and the number breakpoints reads at that location (higher vertical lines = more reads). B) Dot-line plot of F1 scores showing the performance of CReSIL in identifying eccDNA from WGLS synthetic data by mixing 20× of human genome reads with different coverage of simulated eccDNA reads of the true positive and true negative set. C) Circos plot showing the identified yeast eccDNA of the known circular rDNA. D) Histogram showing the distribution of normal reads (dark blue), breakpoint reads (red), and CTC reads (magenta) with the size of identified eccDNA of rDNA (vertical dashed line). E) Bar plots showing the number of identified eccDNA from the WGLS datasets. Green represents circular, and gray represents non-circular. F) Circos plots showing examples of identified eccDNA with high coverage depth on centromere regions of human (left) and mouse (right) datasets. All are satellite repeats. G) Circos plots showing examples of identified eccDNA containing gene(s) of human (left) and mouse (right) datasets (see Figure 1.5 for lane annotation; dataset information (bold), the eccDNA name (italic), and coverage depth (underline).
We assessed the capability of CReSIL to identify eccDNA from WGLS using this workflow. We used PBSIM2 to randomly generate long-read datasets of the human genome at 20× coverage depth and mixed them with the simulated eccDNA true positive and true negative datasets with varying coverage depths. CReSIL exhibited a very good performance, with an F1 score > 0.98 (Figure 5B) when the coverage depth of simulated eccDNA was 100×, 50×, and 20×. The F1 score dropped approximately to 0.8 at 10×, and the performance was lower at 5× (score of ~0.3).
We also tested CReSIL’s workflow on the generated WGLS (native DNA) of Saccharomyces cerevisiae dataset of 4.3 Gb, corresponding to 356× genome coverage depth. CReSIL identified only 2 eccDNAs with a very high coverage depth. Interestingly, one of them is the well-known circular ribosomal DNA (rDNA) molecule in yeast. CReSIL identified an eccDNA with a size of approximately 17 000 nt, covering the whole RDN gene clusters of RDN37, RDN25, RDN18, RDN58, and RDN5 (two clusters based on the reference genome), with an extremely high coverage depth of 24 000× (Figure 5C). This result agrees with the recent studies reporting accumulation (> 99% in aged cells) of circular rDNA in yeast based detected using the CIRCLE-Seq method [5, 39]. We explored the reads of the eccDNA and found that approximately 9600 reads were normal reads, approximately 6100 reads were normal breakpoint reads, and approximately 10 000 were breakpoint reads with CTC reads. We found CTC at various lengths (Figure 5D), indicating the size variation of circular rDNA. Interestingly, we found the longest CTC read, whose length was over 150 000 nt, covered 13 rounds of RDN gene clusters.
Furthermore, we applied CReSIL on publicly available WGLS datasets of human and mouse cells to screen for eccDNA. CReSIL identified much smaller amounts of cyclic eccDNAs (less than 200 in the individual dataset) from WGLS (Figure 5E) than from the eccDNA enrichment approach. The high fraction of acyclic regions was possibly derived from other structural variants. Most of the identified eccDNAs that had high coverage depth mostly originated from satellite repeat locations, such as the centromere region, and from variations of the centromere telomere [25] (Figure 5F). We found an eccDNA with a size of approximately 52 000 nt of alpha-like repeats (ALR)/alpha repeats in the class of satellite repeats identified from the human B-Lymphocyte cell NA24385 dataset located on the centromere region of human chromosome 2. Another example found in the mouse dataset of mouse embryonic placenta, ec5, whose size is approximately 38 000 nt (Figure 5F), contained GSAT_MM repeat in the class of satellite repeats located on the centromere region of mouse chromosome 9. We also found that the identified eccDNA harbored whole gene(s) (Figure 5G). LILRA6, encoding leukocyte immunoglobulin-like receptor subfamily A member 6, and LILRB3, encoding leukocyte immunoglobulin-like receptor subfamily B member 3 were harbored in ec242, with a length of approximately 25 000 nt and coverage depth of 87×, identified in B-Lymphocyte cell NA12878 (Figure 5G). In another example in the mouse dataset of pancreatic cancer cell lines under treatment, we found the whole gene of Mucl2, encoding mucin-like protein 2, harbored in ec38, with a length of approximately 24 500 nt and 207× coverage depth. These examples showed the importance of eccDNA in focal amplification regions that possibly serve a niche of cells.
Discussion
We presented a bioinformatic software suite package, CReSIL, for accurately identifying eccDNA molecules from long-read sequencing data derived from either eccDNA enrichment or WGLS. We incorporated useful features to assess the function of eccDNA molecules and visualization for the deep investigation of eccDNA architecture. Compared with other bioinformatic tools for identifying eccDNA, CReSIL maintains the highest precision and a performance with the lowest F1 score of 0.98 at coverages as low as 3×, which is better than the other tested tools (NanoCircle, eccDNA_RCA_nanopore, ecc_finder, and Flye). NanoCircle has good precision and good control of false positives for simple eccDNA. However, NanoCircle has difficulty detecting false positives derived from complex eccDNAs. EccDNA_RCA_nanopore was designed to identify eccDNA at the read level and strictly based on CTC reads; therefore, the tool missed the detection of eccDNAs that are only represented by non-CTC reads in the sequencing library. Based on the real-world samples (Figure 3A), the fraction of CTC reads across samples was highly diverse, depending on sample preparation. Therefore, eccDNA_RCA_nanopore missed detecting several eccDNA that had not completed circular amplification by RCA in one amplicon or DNA breakage events. Because eccDNA_RCA_nanopore provides the result of individual reads, the user needs to write a custom script to reduce the redundancy of the result. Flye in metagenomic mode, which was used in the development of CReSIL software [25], performed very well when assembling eccDNAs at high sequencing depths (F1 score > 0.93 for 10×–100×. Yet, the F1 score dropped to 0.73 at 5x and 3x [Figure 2F]). This is caused by Flye’s design, which is made to assemble genomes. Therefore, small-sized eccDNAs with low-sequencing depth are difficult to capture.
Besides high accuracy, CReSIL provides many useful features to study eccDNA. CReSIL generates consensus sequences and variants of individual eccDNAs that can be used to infer potential functional effects of eccDNA when variations on the chromosomes are generated. Genes or gene fragments harbored in eccDNAs can be expressed and alter protein levels, resulting in growth advantages for the cells carrying them [13, 32]. Moreover, eccDNA is also thought to be involved in stable chromosomal gene duplication events when eccDNAs reintegrate into the genome [5, 6]. Furthermore, repeats such as alpha-satellite harbored in eccDNA could contribute to centromere variation [25]. Thus, identifying the genetic elements of eccDNA is important to understanding the biological impact of eccDNA. To aid the user in investigating eccDNA architecture, CReSIL provides a function to generate the config. file, populate the chromosomal origin information and genomic annotations and plot it in Circos for visualization.
Detection of eccDNA from WGLS is challenging [40]; however, it is possible by leveraging the higher copy number of eccDNA over the genomic background (Figure 5). Using CReSIL, we found that WGLS data can be used as the basis for identification of eccDNA when the circular DNA is amplified over the general background level of genomic DNA. This is especially relevant in tumor data where oncogenes are often amplified on large eccDNAs [6, 13, 32]. This opens the possibility that WGLS of tumors can be used to identify eccDNA with effects on tumorigenesis. Thus, biobank WGLS from tumors can become an important resource for identifying such eccDNAs.
CReSIL has many advantages over the other tools, including high accuracy of eccDNA identification, complete construction and assembly of eccDNA molecules, and features to identify genetic variation and gene annotation with eccDNA architecture visualization. CReSIL relies on the reference genome read alignment result, enabling the construction of linkages among regions. Therefore, CReSIL cannot capture the eccDNA that originated outside the reference genome sequences. This is the main limitation of CReSIL. In the future, unmapped reads, especially unmapped CTC reads, can be an additional step to identify eccDNA outside the reference sequences. We used 200 nt region cutoff for graph construction. Therefore, eccDNAs with less than 200 nt cannot be detected. Nevertheless, this limitation can be overcome by lowering the parameter of aligned region length for the graph construction algorithm to identify smaller eccDNAs. However, the quality of alignments of the short region needs to be assessed to ensure the reliability of the results.
In summary, the CReSIL suite software solves major problems in the identification and analysis of eccDNA. It provides a tool for accurate and sensitive identification of eccDNA from long-read sequences, thereby allowing identification of eccDNA from repeat regions of the human genomes and mapping complex eccDNAs made of several fragments from long reads. CReSIL provides a versatile tool to study the immense diversity and impact of eccDNAs created from any part of a genome from existing WGS data and data enriched for eccDNA.
Key Points
CReSIL provides a robust and comprehensive bioinformatic workflow to identify eccDNA form long-read sequences derived from standard eccDNA enrichment, Circle-Seq, with useful features and visualization for deeper investigation of eccDNA molecules.
Comparative analysis of CReSIL and other bioinformatics tools for eccDNA identification from long-read sequences was performed, showing that CReSIL performs best.
Extension of CReSIL enables direct identification of eccDNA from whole genome long-read sequencing.
Supplementary Material
Acknowledgments
We thank Thidathip Wonsurawat for the technical discussion and evaluation on DNA debranching. We thank Taylor Wadley for technical assistance on yeast sequencing. We thank Henriette Pilegaard for mice housing, ethical permits and administration.
Author Biographies
Visanu Wanchai is a postdoctoral researcher at the Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, USA 72211. Research interest includes developing and applying bioinformatics tools and databases for biological sequence analysis.
Piroon Jenjaroenpun is a lecturer at the Division of Bioinformatics and Data Management for Research, Department of Research and Development, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand, 10700. Research interest includes developing and applying bioinformatics tools for biological sequence analysis.
Thongpan Leangapichart has a PhD in Microbiology. Research interest includes pathogen and eccDNA identification.
Gerard Arrey is a PhD student. Research interest includes identifying and characterizing eccDNA in mouse and yeast models.
Charles M Burnham is a PhD student. Research interest includes the identification of eccDNA in breast cancer.
Maria C Tümmler is a master’s student. Research interests include the development of experimental methods to identify eccDNA
Jesus Delgado-Calle is a professor in the Department of Physiology and Cell Biology, College of Medicine, University of Arkansas for Medical Sciences, USA 72211. Research interest includes bone and cancer biology.
Birgitte Regenberg is a professor in the Department of Biology, University of Copenhagen, Denmark, 2100. Her research interest focuses on genetic mechanisms for cell–cell variation in eukaryotic cells and their impact on diseases and chromosome evolution, with a particular interest in eccDNA.
Intawat Nookaew is a professor in the Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, USA 72211. Research interest focuses on the development, application and translation of bioinformatics and systems biology.
Contributor Information
Visanu Wanchai, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas, United States of America.
Piroon Jenjaroenpun, Division of Bioinformatics and Data Management for Research, Research Group and Research Network Division, Research Department, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand.
Thongpan Leangapichart, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas, United States of America.
Gerard Arrey, Ecology and Evolution, Department of Biology, University of Copenhagen, Copenhagen, Denmark.
Charles M Burnham, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas, United States of America.
Maria C Tümmler, Ecology and Evolution, Department of Biology, University of Copenhagen, Copenhagen, Denmark.
Jesus Delgado-Calle, Department of Physiology and Cell Biology, College of Medicine, Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences, Little Rock, Arkansas, United States of America.
Birgitte Regenberg, Ecology and Evolution, Department of Biology, University of Copenhagen, Copenhagen, Denmark.
Intawat Nookaew, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas, United States of America.
Software availability
CReSIL 2.0 is freely available at https://github.com/visanuwan/cresil under an MIT license.
Author contribution
I.N. designed and conceived the project. V.W. and P.J. developed and implemented the CReSIL software. V.W., PJ, and I.N. performed computational analysis. T.L. and C.M.B performed eccDNA enrichment and sequencing of cell line samples under the supervision of I.N. Cell culture experiments were performed in J.D.C. laboratory. G.A. and M.C.T. performed eccDNA enrichment and sequencing of mouse tissues under the supervision of B.R. I.N. wrote the manuscript, and all authors edited the manuscript. All authors have read and approved the final version.
Funding
National Institute of General Medical Sciences of the National Institutes of Health (award P20GM125503) support to I.N. National Institute of General Medical Sciences of the National Institutes of Health (T32GM106999) support to C.M.B. National Cancer Institute of the National Institutes of Health (R37 CA251763) support to J.D.C. Novo Nordisk Foundation (NNF18OC0053139 and NNF21OC0072023) to B.R. and G.A and the VILLUM Foundation (00023247) to G.A. and B.R., European Union’s Horizon 2020 research and innovation action under the FET-Open Programme (899417—CIRCULAR VISION) to B.R., and Innovation Fund Denmark under the Grand Solutions programme (8088-00049B CARE DNA) to B.R. G.A. also received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no 801199.
Competing interests
None of the authors have any competing interests.
Data availability
All data generated in this study will be available in the Sequence Read Archives database under bioproject number PRJNA806866.
The simulated data will be available at the CReSIL project site. The sample data to test CReSIL is available at https://uams.app.box.com/folder/158081132175?s=exs4yue7v8fe3h5414hnh6j6mruarz71
References
- 1. Paulsen T, Kumar P, Koseoglu MM, et al. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet 2018;34:270–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zuo S, Yi Y, Wang C, et al. Extrachromosomal circular DNA (eccDNA): from chaos to function. Front Cell Dev Biol 2022;9:792555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peng H, Mirouze M, Bucher E. Extrachromosomal circular DNA: a neglected nucleic acid molecule in plants. Curr Opin Plant Biol 2022;69:102263. [DOI] [PubMed] [Google Scholar]
- 4. Kanda T, Otter M, Wahl GM. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci 2001;114:49–58. [DOI] [PubMed] [Google Scholar]
- 5. Prada-Luengo I, Moller HD, Henriksen RA, et al. Replicative aging is associated with loss of genetic heterogeneity from extrachromosomal circular DNA in Saccharomyces cerevisiae. Nucleic Acids Res 2020;48:7883–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turner KM, Deshpande V, Beyter D, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 2017;543:122–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yi E, Gujar AD, Guthrie M, et al. Live-cell imaging shows uneven segregation of extrachromosomal DNA elements and transcriptionally active extrachromosomal DNA hubs in cancer. Cancer Discov 2022;12:468–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li R, Wang Y, Li J, et al. Extrachromosomal circular DNA (eccDNA): an emerging star in cancer. Biomark Res 2022;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paulsen T, Shibata Y, Kumar P, et al. Small extrachromosomal circular DNAs, microDNA, produce short regulatory RNAs that suppress gene expression independent of canonical promoters. Nucleic Acids Res 2019;47:4586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noer JB, Horsdal OK, Xiang X, et al. Extrachromosomal circular DNA in cancer: history, current knowledge, and methods. Trends Genet 2022;38:766–81. [DOI] [PubMed] [Google Scholar]
- 11. Gresham D, Usaite R, Germann SM, et al. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A 2010;107:18551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. deCarvalho AC, Kim H, Poisson LM, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 2018;50:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morton AR, Dogan-Artun N, Faber ZJ, et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell 2019;179:1330–1341.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helmsauer K, Valieva ME, Ali S, et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat Commun 2020;11:5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deshpande V, Luebeck J, Nguyen ND, et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun 2019;10:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prada-Luengo I, Krogh A, Maretty L, et al. Sensitive detection of circular DNAs at single-nucleotide resolution using guided realignment of partially aligned reads. BMC Bioinformatics 2019;20:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mann L, Seibt KM, Weber B, et al. ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data. BMC Bioinformatics 2022;23:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang P, Peng H, Llauro C, et al. ecc_finder: a robust and accurate tool for detecting extrachromosomal circular DNA from sequencing data. Front Plant Sci 2021;12:743742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dean FB, Hosono S, Fang L, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A 2002;99:5261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Esteban JA, Salas M, Blanco L. Fidelity of phi 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem 1993;268:2719–26. [PubMed] [Google Scholar]
- 21. de Paz AM, Cybulski TR, Marblestone AH, et al. High-resolution mapping of DNA polymerase fidelity using nucleotide imbalances and next-generation sequencing. Nucleic Acids Res 2018;46:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mehta D, Hirsch-Hoffmann M, Were M, et al. A new full-length circular DNA sequencing method for viral-sized genomes reveals that RNAi transgenic plants provoke a shift in geminivirus populations in the field. Nucleic Acids Res 2019;47:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mehta D, Cornet L, Hirsch-Hoffmann M, et al. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq. Nat Protoc 2020;15:1673–89. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Wang M, Djekidel MN, et al. eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature 2021;599:308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henriksen RA, Jenjaroenpun P, Sjostrom IB, et al. Circular DNA in the human germline and its association with recombination. Mol Cell 2021;82:209–217.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng W, Arrey G, Zole E, et al. Targeted removal of mitochondrial DNA from mouse and human extrachromosomal circular DNA with CRISPR-Cas9. Comput Struct Biotechnol J 2022;20:3059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009;19:1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kolmogorov M, Yuan J, Lin Y, et al. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 2019;37:540–6. [DOI] [PubMed] [Google Scholar]
- 29. Ono Y, Asai K, Hamada M. PBSIM2: a simulator for long-read sequencers with a novel generative model of quality scores. Bioinformatics 2021;37:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moller HD, Mohiyuddin M, Prada-Luengo I, et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun 2018;9:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moller HD, Ramos-Madrigal J, Prada-Luengo I, et al. Near-random distribution of chromosome-derived circular DNA in the condensed genome of pigeons and the larger, more repeat-rich human genome. Genome Biol Evol 2020;12:3762–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koche RP, Rodriguez-Fos E, Helmsauer K, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat Genet 2020;52:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dillon LW, Kumar P, Shibata Y, et al. Production of extrachromosomal MicroDNAs is linked to mismatch repair pathways and transcriptional activity. Cell Rep 2015;11:1749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shibata Y, Kumar P, Layer R, et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 2012;336:82–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones RS, Potter SS. L1 sequences in HeLa extrachromosomal circular DNA: evidence for circularization by homologous recombination. Proc Natl Acad Sci U S A 1985;82:1989–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Misra R, Matera AG, Schmid CW, et al. Recombination mediates production of an extrachromosomal circular DNA containing a transposon-like human element, THE-1. Nucleic Acids Res 1989;17:8327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kfoury Y, Scadden DT. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell 2015;16:239–53. [DOI] [PubMed] [Google Scholar]
- 38. Oishi S, Zalucki O, Vega MS, et al. Investigating cortical features of Sotos syndrome using mice heterozygous for Nsd1. Genes Brain Behav 2020;19:e12637. [DOI] [PubMed] [Google Scholar]
- 39. Moller HD, Parsons L, Jorgensen TS, et al. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci U S A 2015;112:E3114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mouakkad-Montoya L, Murata MM, Sulovari A, et al. Quantitative assessment reveals the dominance of duplicated sequences in germline-derived extrachromosomal circular DNA. Proc Natl Acad Sci U S A 2021;118:e2102842118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study will be available in the Sequence Read Archives database under bioproject number PRJNA806866.
The simulated data will be available at the CReSIL project site. The sample data to test CReSIL is available at https://uams.app.box.com/folder/158081132175?s=exs4yue7v8fe3h5414hnh6j6mruarz71