Abstract
Nine novel bacterial strains were isolated from the feces of cats and sheep in 2019 and 2020 in Beijing, China. Cells were 1–3 μm long and ≤0.5 μm wide, Gram-stain negative, microaerobic, motile, oxidase positive, and urease negative. Phylogenetic analyses based on 16S rRNA gene sequences indicated that these nine isolates belong to the genus Campylobacter but formed two robust clades that were clearly separate from the currently recognized species and, respectively, isolated from the cat and sheep. Both these strains shared low 16S rRNA gene sequence similarity, dDDH relatedness, and ANI values with their closest species C. upsaliensis CCUG 14913T and C. lanienae NCTC 13004T, and against each other, which are below the cut-off values generally recognized for isolates of the same species. The genomic DNA G + C contents of type strains XJK22-1T and SYS25-1T were 34.99 mol% and 32.43 mol%, respectively. Electron microscopy showed that these cells were spiral shaped, with bipolar single flagella. Based on results from genotypic, phenotypic, phylogenetic, and phylogenomic analyses, these nine strains represent two novel species within the genus Campylobacter, for which the names Campylobacter felis sp. nov. (Type strain XJK22-1T = GDMCC 1.3684T = JCM 35847T) and Campylobacter ovis sp. nov. (Type strain SYS25-1T = GDMCC 1.3685T) are proposed.
Keywords: Campylobacter felis, Campylobacter ovis, novel species, genomic characteristics, phylogenetic analyses
1. Introduction
The genus Campylobacter belongs to the family Campylobacteraceae and the order Campylobacterales, which currently contains 37 and 13 validly described species and subspecies, respectively, and 2 not validly described species (https://lpsn.dsmz.de/genus/campylobacter) (accessed on 15 February 2023). Members of the Campylobacter genus are morphologically diverse, and can be spiral, curved, or rod shaped. These bacteria are nutritionally fastidious and grow under strictly anaerobic or microaerobic conditions. They naturally colonize humans, other mammals, birds, reptiles, shellfish, etc. [1,2,3].
Campylobacter is among the four main causes of gastroenteritis worldwide [4]. Most reported Campylobacter infections are caused by C. jejuni, which is a leading cause of bacterial gastroenteritis in humans worldwide [2] and whose antecedent infection could trigger a Guillain–Barré Syndrome (GBS) outbreak [5], and, to a lesser extent, C. coli, which accounts for 1–25% of all Campylobacter-related diarrheal diseases [2]. However, the other emerging Campylobacter pathogens are gaining increasing recognition as important pathogens in humans and animals [6]. In addition, with the continuous progress of isolation and culture technology, more and more novel Campylobacter species have been identified in recent years [7,8,9,10,11,12,13,14]. Domestic animals, especially pets, have close contact with humans. Additionally, Campylobacter spp. infections in humans caused by pets have been reported several times in recent years [15,16,17,18,19]. A total of 117 of 121 patients with Campylobacter infections reported contact with a dog in the week before symptom onset in 2016–2020 [20].
In this study, taxonomic and genomic characteristics of the novel Campylobacter-like isolates were described, and the phylogenetic relationships between the isolated strains and their closest relatives were also clarified. Based on polyphasic taxonomic analyses, these novel isolates are proposed as two novel Campylobacter species, designated Campylobacter felis sp. nov. (XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1) and Campylobacter ovis sp. nov. (SYS25-1T, SYS28-3, and S13-1).
2. Materials and Methods
2.1. Sampling, Isolation and Culturing
During the investigation of the Campylobacter spp. diversity in both healthy animals and animals with diarrhea, which included animals such as dogs, cats, sheep, and pigs that are in close contact with humans, isolation was carried out using the Campylobacter isolation kit incorporating a membrane filter method (ZC-CAMPY-002, Qingdao Sinova Biotechnology Co., Ltd., Qingdao, China). Briefly, 0.4 mL stool specimen suspension was transferred into a 4 mL enrichment buffer, which was provided in the kit. The principal component of the enrichment buffer was the modified Preston broth, which was described in the manual book. The enriched suspension was incubated at 37 °C for 24 h in a microaerophilic atmosphere consisting of 5% O2, 10% CO2, and 85% N2. About 300 μL cultured enrichment suspension was then spotted on the surface of the filter pasted on the double medium plates, which contained Karmali and Columbia agar, respectively, with 5% defibrinated sheep blood. The medium plates were incubated in a microaerophilic atmosphere at 37 °C for 48 h [21]. The suspected monoclonal colonies were selected and purified and were subjected to preliminary characterization by PCR amplification and sequencing of 16S rRNA gene sequence analysis [22], and subsequently conserved at −80 °C in BHI with 20% (v/v) glycerol for further identification. The exact 16S rRNA gene PCR amplification primer sequences were listed in Table 1.
Table 1.
Primers and probes used in this study.
| Target | Sequence(5′–3′) a | Target Gene/Region | Reference |
|---|---|---|---|
| Campylobacter spp. | 27F: AGAGTTTGATCCTGGCTCAG | 16S rRNA gene | [8] |
| 1492R: CGGTTACCTTGTTACGACTT | |||
| C. felis sp. nov. | F: GCGCCATCTTGGACGAGTAT | Putative hydrolase YxeP gene | This study |
| R: GGGCAGGGCGTCCATATC | |||
| P: FAM-CGCGAAGGAGGACGCAGGGA-BHQ1 | |||
| C. ovis sp. nov. | F: TGAAGCTGGAGAAAGTGGCC | Hypothetical protein gene | This study |
| R: TCCTATTATGGCGCCAGCTG | |||
| P: FAM-CAACCCTAAGTAGCGGAAGCGGTGG-BHQ1 |
a FAM—6-carboxyfluorescein.
2.2. Morphological, Physiological and Biochemical Characteristics
For the study of the morphological and biochemical characteristics, cells were cultivated and harvested in the late-exponential growth phase. Gram staining was conducted using a Gram-staining kit (Baso, Zhuhai, China) [23] and observed under a light microscope. The morphological characteristics of these two putative novel species’ type strains were determined using transmission electron microscopy. Fresh cells were gently suspended in 0.1 M phosphate-buffered saline (PBS) to an OD600 of 1 and collected via gentle centrifugation. The pellet was gently resuspended in a 2% (v/v) glutaraldehyde solution for fixation. Fixation was completed by incubating strains for 1 h on the grid. All samples were stained with 2% (w/v) uranyl acetate for 1 min and examined using a Hitachi H7700 transmission electron microscope (Eclipse Ci-L, NIKON, Tokyo, Japan) at 80 kV.
The catalase activity was evaluated using a 3% (v/v) H2O2 solution for bubble production. The general phenotypic traits of Campylobacter spp., oxidase, catalase, hydrolysis of hippurate, and indoxyl acetate were evaluated using the Campylobacter Biochemical Assay kit (ZC-CAMPY-010, Qingdao Sinova Biotechnology Co., Ltd., Qingdao, China). Further biochemical characteristics were obtained using the identification system of API Campy, strictly following the manufacturers’ instructions (bio-Mérieux, Lyon, France). Biochemical tests were carried out to characterize the physiology and chemotaxonomy of the isolates. C. jejuni ATCC 33560T, C. coli ATCC 33559T, and C. upsaliensis CCUG 14913T were used as controls.
2.3. Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) for 11 antimicrobials (erythromycin, azithromycin, nalidixic acid, ciprofloxacin, gentamicin, streptomycin, chloramphenicol, florfenicol, tetracycline, telithromycin, and clindamycin) at concentrations ranging from 0.02 to 256 μg mL−1 were determined for all isolates using the agar dilution method (ZC-AST-001, Qingdao Sinova Biotechnology Co., Ltd., Qingdao, China) and the gradient strip diffusion method (E-test, bio Mérieux, Nürtingen, Germany) following the manufacturer’s instructions, as previously reported [24,25]. The MIC was read as the lowest concentration without visible growth. Type strain C. jejuni ATCC 33560T was used as a control.
2.4. Species-Specific PCR
To define a diagnostic method for the rapid detection and identification of these two putative new species, specific TaqMan real-time PCR primers targeting different genes were designed. The exact primer and probe sequences are listed in Table 1. The PCR conditions were as follows: initial denaturation at 94 °C for 30 s, followed by 40 cycles of 94 °C for 5 s and 60 °C for 30 s. The positive and negative results refer to the cycle threshold (Ct) value of the real-time PCR that, according to the manual, Ct < 35 with the typical S curve, is determined as positive; Ct > 40 or no typical S curve is determined as negative, and 35 ≤ Ct ≥ 40 should be repeated 3 times and determined as weakly positive after the third repeat. To assess the sensitivity and specificity of this qPCR method, some species of Campylobacter strains which were stored in our laboratory, including C. coli, C. concisus, C. fetus subsp. fetus, C. gracilis, C. helveticus, C. hyointestinalis subsp. hyointestinalis, C. jejuni subsp. doylei, C. jejuni subsp. jejuni, C. lari subsp. lari, C. mucosalis, C. rectus, C. showae, and C. upsaliensis, were used as reference strains. Several kinds of other genus bacteria stored in our laboratory were used as negative controls, such as Arcobacter butzleri, Arcobacter skirrowii, Helicobacter pylori, Escherichia coli, etc.
2.5. Genome Extraction and Sequencing
After culturing, the DNA for genome sequence was extracted using the QIAamp DNA Mini Kit (Qiagen, German) according to the manufacturer’s instructions for sequencing. Then, the NanoDrop 2000 spectrophotometer (Thermo Fisher, Massachusetts, USA) was used to measure the concentration and purity of DNAs. The quality requirements were a concentration ≥ 20 ng/μL and a total amount > 2 μg. The purity requirement was as follows: OD260/OD280 value should be between 1.6 and 1.8. The DNA sequencing was performed by an Illumina PE150 platform (Illumina Inc., San Diego, CA, USA) at the Novogene Corporation (Beijing, China) with a depth of 100× coverage. To sequence the genomes, a 350 bp paired-end library was constructed and then 150 bp reads were generated. FastQC v0.11.8 and fastp v0.23.2 were applied to evaluate and improve the quality of the raw sequence data, respectively. Low-quality reads were removed if the quality scores of ≥3 consecutive bases were ≤Q30. The clean reads were assembled by SOAPdenovo v2.40.
2.6. Genomic Analysis
The genome was predicted and annotated using the Prokka pipeline [26] and tRNA-scan tool [27]. Phage Search Tool (PHAST) web server (http://phaster.ca/) (accessed on 29 January 2023)and phiSpy v4.2.21 [28] were used to search for phage sequences. The antimicrobial resistance genes were predicted using the Comprehensive Antibiotic Resistance Database (CARD) [29] and the ResFinder v4.2.21. The virulence genes of all the genomes were detected on VFanalyzer (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi?func=VFanalyzer) (accessed on 7 September 2022) [30]. The digital DNA-DNA hybridization (dDDH) relatedness was calculated and compared using the Genome-to-Genome Distance Calculator 3.0 (https://ggdc.dsmz.de/) (accessed on 15 February 2023) [31]. The average nucleotide identity (ANI) values were determined by pyani 0.2.10 [32].
2.7. Phylogenetic and Phylogenomic Analysis
To determine the phylogenetic positions of strains, 16S rRNA gene PCR amplification was performed with primers 27F and 1492R, as previously reported. Each almost-complete sequence of the 16S rRNA gene PCR product was purified, sub-cloned into the pMD18-T vector for 30 min at 16 °C, transformed into Escherichia coli DH5α, and the inserted 16S rRNA gene fragment was obtained from a single colony after lysis and sequenced. The newly generated 16S rRNA gene sequences were compared with other Campylobacter species by EzBioCloud’s identification service to locate their taxonomic position [33]. Multiple sequence alignment of the 16S rRNA gene sequences of the type strains in the genus Campylobacter was performed using the MAFFT 7.471 software [34] and phylogenetic analysis using the software package MEGA X [35], by the neighbor-joining (NJ) [36], maximum parsimony (MP) [37] and maximum likelihood (ML) [38] algorithms with a bootstrap analysis of 1000 replicates [39] and strain Arcobacter butzleri ATCC 49616T was used as an outgroup.
The protein sequences of core genes of genomes from the isolates and the other Campylobacter species were extracted using the CD-HIT v4.8.1 [40] based on 40% protein sequence similarity and aligned to reconstruct a phylogenomic tree using FastTree v2.1.11 [41]. The multiple sequence alignment of the core genomes of the genus Campylobacter was also performed using the MAFFT software. Then, the phylogenomic tree was visualized by Dendroscope 3.8.3 [42], and modified with Interactive Tree of Life (https://itol.embl.de/) (accessed on 28 February 2023).
2.8. Accession Numbers
The GenBank/EMBL/DDBJ accession numbers for the nearly full-length 16S rRNA gene and the draft genome sequences of these 9 isolates were submitted to NCBI (https://www.ncbi.nlm.nih.gov/) (accessed on 9 August 2022). In addition to these 9 isolates, the other 10 temporarily undefined genomes in NCBI which belonged to Campylobacter ovis sp. nov. and genomes from the other type strains of Campylobacter genus were downloaded from NCBI. There were no genomes found in NCBI belonging to Campylobacter felis sp. nov. More information about the genomes of novel species used in this study was listed in Table 2.
Table 2.
Background of strains Campylobacter felis sp. nov. and Campylobacter ovis sp. nov.
| Isolate | Species | Isolation Date | Isolation Country: City | Host | Source | 16S rRNA Gene Accesion Number | Accession Number |
|---|---|---|---|---|---|---|---|
| XJK22-1 | C. felis | 2019 | China: BeiJing | cat | feces | OP278862 | JANURX000000000 |
| XJK33-1 | C. felis | 2019 | China: BeiJing | cat | feces | OP278861 | JANURU000000000 |
| XJK49-2 | C. felis | 2019 | China: BeiJing | cat | feces | OP278863 | JANURW000000000 |
| XJK56-3 | C. felis | 2019 | China: BeiJing | cat | feces | OP278858 | JANURS000000000 |
| XJK62-3 | C. felis | 2019 | China: BeiJing | cat | feces | OP278859 | JANURT000000000 |
| XJK7-1 | C. felis | 2019 | China: BeiJing | cat | feces | OP278860 | JANURV000000000 |
| S13-1 | C. ovis | 2020 | China: BeiJing | Sheep | feces | OP278865 | JANURP000000000 |
| SYS25-1 | C. ovis | 2019 | China: BeiJing | Sheep | feces | OP278866 | JANURR000000000 |
| SYS28-3 | C. ovis | 2019 | China: BeiJing | Sheep | feces | OP278867 | JANURQ000000000 |
| RM8835 | C. ovis | 2009 | USA: California | Alpaca | feces | OP821422 | MJLL01000000 |
| RM8965 | C. ovis | 2009 | USA: California | Goat | feces | OP821424 | MJLM01000000 |
| RM8966 | C. ovis | 2009 | USA: California | Goat | feces | OP821425 | MJLN01000000 |
| RM9262 | C. ovis | 2009 | USA: California | Alpaca | feces | OP821428 | MJLQ01000000 |
| S0112 | C. ovis | 2013 | UK: Scotland | Sheep | feces | OP821431 | MJLS01000000 |
| RM8970 | C. ovis | 2009 | USA: California | Goat | feces | OP821426 | MJLO01000000 |
| RM9263 | C. ovis | 2009 | USA: California | Alpaca | feces | OP821429 | MJLR01000000 |
| RM9261 | C. ovis | 2009 | USA: California | Alpaca | feces | OP821427 | MJLP01000000 |
| RM12175 | C. ovis | 2010 | USA: California | Alpaca | feces | OP821430 | CP018793 |
| RM8964 | C. ovis | 2009 | USA: California | Goat | feces | OP821423 | CP018791 |
3. Results and Discussion
3.1. Isolation and Phenotypic Characterization
Six isolates (XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1) and three other isolates (S13-1, SYS25-1T, and SYS28-3) were isolated from fecal samples of asymptomatic carriers in cat and sheep, respectively.
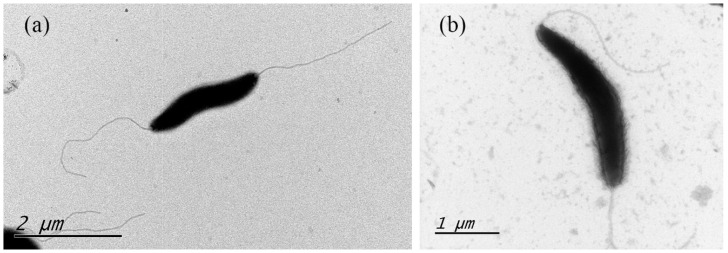
These Gram-negative, microaerobic, motile, spiral-shaped cells with bipolar single flagellum ranged from 1.8 to 2.2 μm for type strain XJK22-1T and 2.1 to 2.5 μm for type strain SYS25-1T (Figure 1). Colonies were circular, 2–3 mm in diameter, smooth, and gray after 2 days of growth on Karmali agar with 5% defibrinated sheep blood. The cells appeared coccoid after 5–6 days of incubation or when exposed to air.
Figure 1.
Transmission electron microscope image of the novel Campylobacter strains from 48 h culture. (a) C. felis strain XJK22-1T. (b) C. ovis strain SYS25-1T.
Like most other Campylobacter species, the isolates were positive for oxidase and negative for urease activities. The catalase was negative for species of type strain XJK22-1T and positive for species of type strain SYS25-1T. While most Campylobacter species are unable to hydrolyze hippurate and able to hydrolyze indoxyl acetate and reduce nitrate, two-thirds (n = 6, XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1) of the new species isolates were able to hydrolyze hippurate and indoxyl acetate positive and could reduce nitrate, and the other one-third (n = 3, S13-1, SYS25-1T, and SYS28-3) could not hydrolyze hippurate and indoxyl acetated, among which only two isolates could reduce nitrate (n = 2, S13-1 and SYS25-1T). There are only five other species of the genus Campylobacter, C. avium, C. curvus, C. jejuni, C. hepaticus, and C. geochelonis that can hydrolyze hippurate [11].
All nine isolates were initially identified as C. upsaliensis and C. lanienae according to the results of the sequencing of the 16S rRNA gene. Thus, it was not unexpected that the results of the standard biochemical of strain XJK22-1T and SYS25-1T showed a strong similarity between the composite phenotypic profile observed from the isolates and the phenotypic profile reported previously for C. upsaliensis CCUG 14913T and C. lanienae NCTC 13004T, respectively (Table 3). Nevertheless, these six strains (XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1) could be unambiguously distinguished from C. upsaliensis by the ability to hydrolyze hippurate, which may challenge the discriminability of C. jejuni to distinguish from other Campylobacter species by hydrolyzing hippurate. Meanwhile, the other three strains (S13-1, SYS25-1T, and SYS28-3) could be distinguished from C. lanienae by the phenotypic characteristic of positive for GGT (gamma-glutamyltransferase) and negative for reduction of TTC (Triphenyltetrazolium chloride). These partial results preliminarily supported the theory that these nine strains were two novel Campylobacter species.
Table 3.
Phenotypic characteristics of Campylobacter felis sp. nov. strains, Campylobacter ovis sp. nov. strains, and the type strains of related species.
| Species | Isolate | Catalase | Oxidase | URE | NIT | EST | HIP | GGT | TTC | PyrA | ArgA | AspA | PAL | H2S |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. felis | XJK22-1 | − | + | − | + | + | + | − | − | − | + | + | + | − |
| C. felis | XJK33-1 | − | + | − | + | + | + | − | − | − | + | + | + | − |
| C. felis | XJK49-2 | − | + | − | + | + | + | − | − | − | + | + | + | − |
| C. felis | XJK56-3 | − | + | − | + | + | + | − | − | − | + | + | + | − |
| C. felis | XJK62-3 | − | + | − | + | + | + | − | − | − | + | + | + | − |
| C. felis | XJK7-1 | − | + | − | + | + | + | − | − | − | + | + | + | − |
| C. ovis | S13-1 | + | + | − | + | − | − | + | − | − | + | − | + | − |
| C. ovis | SYS25-1 | + | + | − | + | − | − | + | − | − | + | − | + | − |
| C. ovis | SYS28-3 | + | + | − | − | − | − | + | − | − | + | − | + | − |
| C. upsaliensis | CCUG 14913 | − | + | − | + | + | − | − | − | − | + | + | + | − |
| C. coli | ATCC 33559 | + | + | − | − | + | − | − | − | − | + | − | + | − |
| C. concisus | ATCC 33237 | − | − | − | − | − | − | − | − | + | − | + | ||
| C. gracilis | ATCC 33236 | − | − | + | − | + | − | − | + | + | − | + | ||
| C. helveticus | CPD4-1 | − | + | − | + | + | + | + | − | − | − | − | − | − |
| C. hyointestinalis | ATCC 35217 | + | + | − | − | − | − | + | − | − | − | − | + | − |
| C. jejuni subsp. doylei | ATCC 49349 | − | − | + | + | + | + | + | − | − | + | − | ||
| C. jejuni subsp. jejuni | ATCC 33560 | + | + | − | − | + | + | − | − | − | − | − | + | − |
| C. lari | ATCC 35221 | + | + | − | + | − | − | + | − | − | + | − | − | − |
| C. rectus | ATCC 33238 | − | − | + | − | − | − | − | + | + | + | − | ||
| C. showae | ATCC 51146 | − | − | + | − | + | − | − | + | + | − | + | ||
| C. jejuni subsp. jejuni | NCTC 11168 | − | + | + | + | − | − | + | − | − | + | − | ||
| C. upsaliensis a | CCUG 14913 | − | + | − | + | + | − | − | − | − | + | + | + | − |
| C. lanienae a | CCUG 44467 | + | + | − | + | +/− | − | − | + | − | +/− | − | + | − |
a These data are referenced from BacDave (https://bacdive.dsmz.de/) (accessed on 15 February 2023).
3.2. Phylogenetic and Phylogenomic Analysis
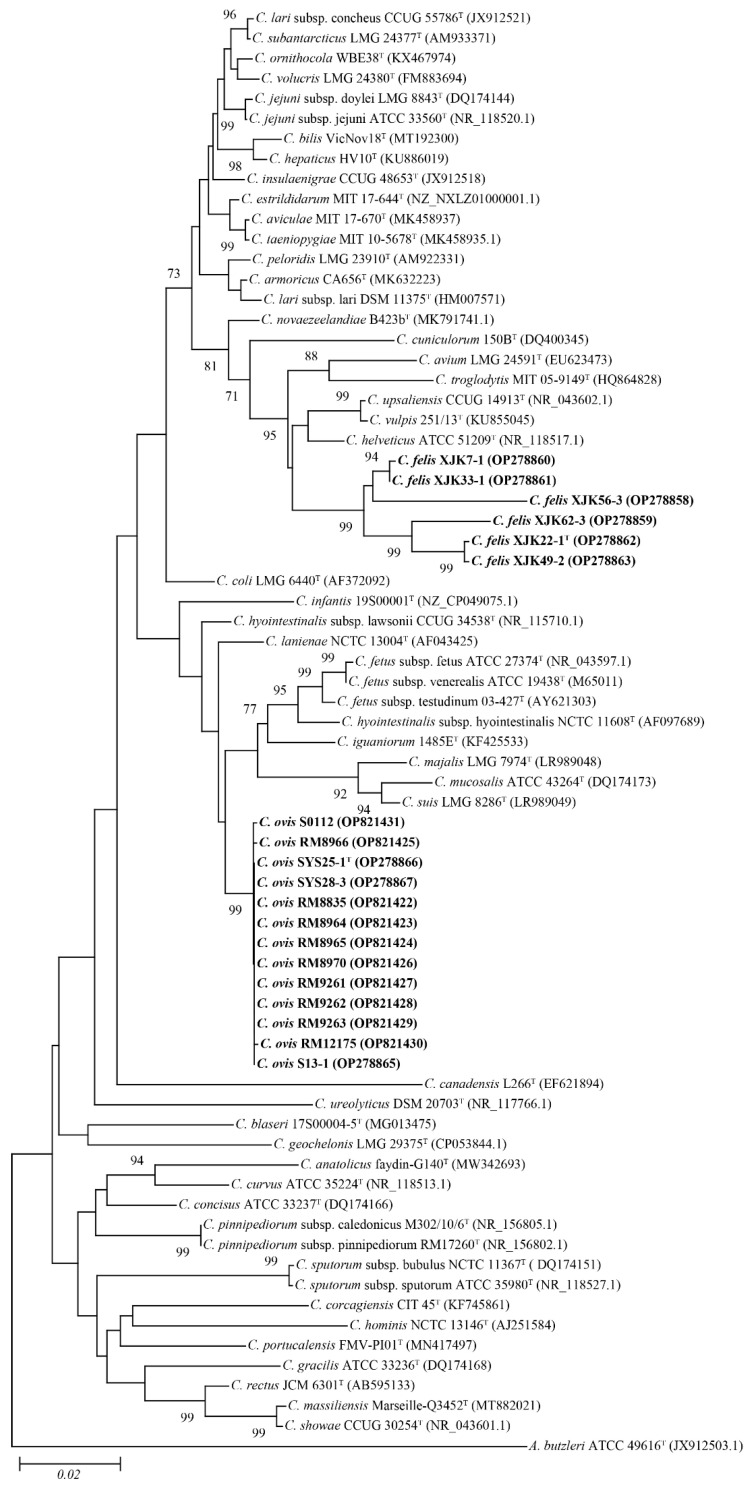
The comparison against the EzTaxon-e database of near full-length 16S rRNA gene sequences (1474–1478 bp) revealed that our 9 isolates and the other 10 strains were most closely related to the representatives of the genus Campylobacter (Domain, Bacteria; Phylum, Pseudomonadota; Class, Epsilonproteobacteria; Order, Campylobacterales; Family, Campylobacteraceae). Strains XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1 were closest to C. upsaliensis CCUG 14913T (96.66% of 16S rRNA gene identity of strain XJK22-1T), whereas strains S13-1, SYS25-1T, SYS28-3, and the other 10 strains were closest to C. lanienae NCTC 13004T (98.47% of 16S rRNA gene identity of strain SYS25-1T). The similarity between strains XJK22-1T and SYS25-1T was 90.70%. These values were lower than 98.70%, which was the generally accepted threshold for species [43], suggesting that these 19 strains should belong to the genus Campylobacter and represent two novel species.
The NJ phylogenetic tree (Figure 2) based on the nearly complete 16S rRNA gene sequences also revealed that these 19 strains belong to the genus Campylobacter and form two independent clusters. Strains, XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1, were grouped into a cluster and closest to C. upsaliensis CCUG 14913T, C. vulpis 251/13T, C. helveticus ATCC 51209T, whereas strains S13-1, SYS25-1T, SYS28-3, and the other 10 strains were grouped into another cluster and closest to C. lanienae NCTC 13004T, C. infantis 19S00001T, C. hyointestinalis subsp. lawsonii CCUG 34538T, which was similar to the topological results obtained from the ML and MP trees (Supplementary data Figures S1 and S2).
Figure 2.
Neighbor-joining phylogenetic tree based on nearly complete 16S rRNA gene showing the relationships between our isolates and the type strains of the genus Campylobacter. Bootstrap values (>70%) based on 1000 replicates are shown at branch nodes, with Arcobacter butzleri ATCC 49616T as an outgroup. Bar—0.02 changes per nucleotide position. Novel strains are highlighted in bold.
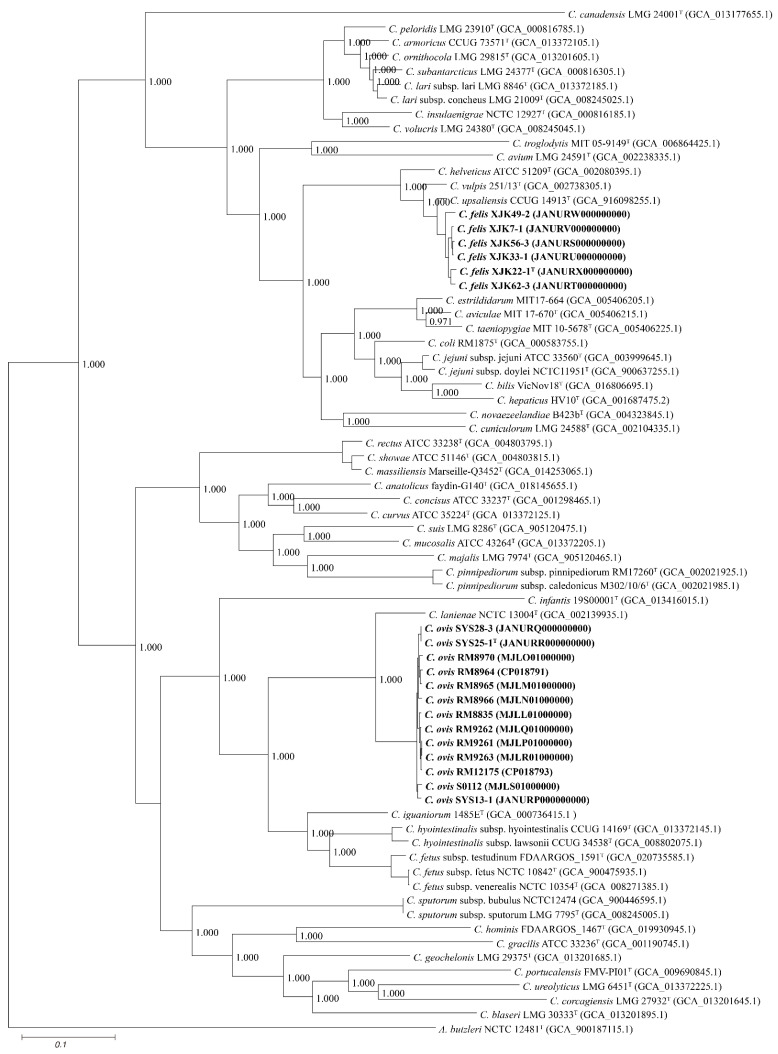
Based on 40% protein identity, orthologous groups of 332 core genes shared by our 9 isolates and all available genomes of the genus Campylobacter were extracted and used to build a phylogenomic tree (Figure 3).
Figure 3.
Neighbor-joining phylogenomic tree based on 332 core genes of the genus Campylobacter. The outgroup is Arcobacter butzleri ATCC 49616T. Novel strains are highlighted in bold.
This robust pangenomic tree revealed that strains XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1 were grouped with C. upsaliensis CCUG 14913T and strains S13-1, SYS25-1T, and SYS28-3 were grouped with C. lanienae NCTC 13004T, a result identical to that of phylogenetic trees based on 16S rRNA gene sequences, further proving that the isolates belong to the genus Campylobacter.
3.3. Genome Characteristics
The draft genome of strain XJK22-1T (1.70 Mb) was predicted to contain 1747 coding genes and carried 2 rRNA genes and 43 tRNA genes, whereas SYS25-1T (1.58 Mb) was predicted to contain 1567 coding genes and carried 1 CRISPR/Cas loci, which were obviously different between two type strains, 1 rRNA gene, and 38 tRNA genes. The genomic DNA G + C content of type strain XJK22-1T was 34.99 mol%, which is slightly higher than the most closely related bacterium, C. upsaliensis CCUG 14913T (34.73%). Meanwhile, strain SYS25-1T was 32.43 mol%, which is lower than the most closely related bacterium, C. lanienae NCTC 13004T (34.60%), and within the range of DNA base compositions previously reported for the members in the genus Campylobacter (29–47 mol% G + C) [44]. This further confirms that these nine strains are two novel Campylobacter species. More genomic characteristics (G + C content, CDS, size, etc.) are listed in Table 4.
Table 4.
Genomes characteristics of strains Campylobacter felis sp. nov. and Campylobacter ovis sp. nov.
| Strain | Contigs | Bases | GC Content | CDS | rRNA | CRISPR | tRNA |
|---|---|---|---|---|---|---|---|
| XJK22-1 | 42 | 1,700,455 | 34.99% | 1747 | 2 | - | 43 |
| XJK33-1 | 53 | 1,666,017 | 35.05% | 1707 | 2 | - | 43 |
| XJK49-2 | 37 | 1,640,989 | 35.10% | 1711 | 4 | - | 44 |
| XJK56-3 | 39 | 1,664,700 | 35.03% | 1693 | 2 | - | 42 |
| XJK62-3 | 42 | 1,750,246 | 34.90% | 1841 | 2 | - | 43 |
| XJK7-1 | 51 | 1,668,760 | 35.04% | 1710 | 3 | - | 44 |
| S13-1 | 51 | 1,442,012 | 32.69% | 1454 | 3 | 1 | 37 |
| SYS25-1 | 10 | 1,580,362 | 32.43% | 1567 | 1 | 1 | 38 |
| SYS28-3 | 17 | 1,590,016 | 32.43% | 1576 | 2 | 1 | 39 |
| RM8835 | 49 | 1,694,176 | 32.21% | 1722 | 2 | 2 | 39 |
| RM8965 | 28 | 1,502,748 | 32.45% | 1499 | 2 | 1 | 38 |
| RM8966 | 62 | 1,608,325 | 32.33% | 1601 | 2 | 3 | 39 |
| RM9262 | 86 | 1,707,487 | 32.11% | 1738 | 2 | 2 | 33 |
| S0112 | 18 | 1,533,040 | 32.28% | 1545 | 2 | 2 | 39 |
| RM8970 | 44 | 1,496,869 | 32.43% | 1493 | 2 | 1 | 32 |
| RM9263 | 59 | 1,632,618 | 32.31% | 1680 | 2 | 2 | 39 |
| RM9261 | 66 | 1,634,929 | 32.31% | 1673 | 2 | 2 | 39 |
| RM12175 | 3 | 1,612,610 | 32.40% | 1645 | 6 | 3 | 40 |
| RM8964 | 2 | 1,754,294 | 32.08% | 1771 | 6 | 1 | 41 |
The dDDH scores within each strain pair were 73.80–94.00% (XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1) and 75.00–99.60% (strains S13-1, SYS25-1T, SYS28-3, and the other 10 strains), which were well above 70%, the threshold for species demarcation. In contrast, the scores of these 19 strains with their closest species were below 70%. Meanwhile, the ANI values within each strain pair were 96.92–99.26% (strains XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1) and 97.09–99.91% (strains S13-1, SYS25-1T, SYS28-3, and the other 10 strains), in contrast to below 95%, the cutoff for species demarcation, between our isolates and all established species of Campylobacter (Table 5 and Table S1). Based on the gold standard for the delineation of bacterial species [45], these results suggested that strains XJK22-1T and SYS25-1T represented two novel species of the genus Campylobacter.
Table 5.
ANI (lower diagonal) and dDDH (upper diagonal) among the novel Campylobacter strains and other Campylobacter species.
| (A) Campylobacter felis sp. nov. Strains with Their Closely Related Campylobacter Species. | |||||||||
| XJK22-1 | XJK33-1 | XJK49-2 | XJK56-3 | XJK62-3 | XJK7-1 | C. helveticus | C. upsaliensis | C. vulpis | |
| XJK22-1 | 80.50% | 73.80% | 79.30% | 82.20% | 80.70% | 31.90% | 57.70% | 39.60% | |
| XJK33-1 | 97.70% | 74.40% | 94.00% | 78.90% | 93.60% | 31.60% | 58.10% | 39.00% | |
| XJK49-2 | 97.01% | 97.10% | 74.60% | 74.30% | 74.40% | 31.30% | 57.30% | 38.90% | |
| XJK56-3 | 97.60% | 99.26% | 96.98% | 78.80% | 91.30% | 31.50% | 58.30% | 38.90% | |
| XJK62-3 | 97.94% | 97.60% | 96.92% | 97.60% | 79.30% | 32.90% | 57.50% | 38.70% | |
| XJK7-1 | 97.78% | 99.18% | 97.01% | 98.98% | 97.62% | 31.70% | 58.10% | 39.00% | |
| C. helveticus | 86.55% | 86.60% | 86.45% | 86.59% | 87.21% | 86.61% | 29.90% | 28.50% | |
| C. upsaliensis | 94.45% | 94.71% | 94.54% | 94.74% | 94.58% | 94.71% | 85.53% | 40.20% | |
| C. vulpis | 89.92% | 89.74% | 89.70% | 89.74% | 89.64% | 89.77% | 84.51% | 89.90% | |
| (B) Campylobacter ovis sp. nov. strains with their closely related Campylobacter species. | ||||||||||||||||
| S13-1 | SYS25-1 | SYS28-3 | RM8835 | RM8965 | RM8966 | RM9262 | S0112 | RM8970 | RM9263 | RM9261 | RM12175 | RM8964 | C. hyointestinalis subsp. lawsonii | C. infantis | C. lanienae | |
| S13-1 | 75.10% | 75.00% | 75.20% | 74.20% | 74.30% | 75.60% | 78.30% | 74.70% | 75.30% | 75.30% | 75.30% | 74.30% | 19.40% | 21.50% | 25.10% | |
| SYS25-1 | 97.14% | 99.60% | 77.30% | 81.50% | 80.70% | 78.10% | 74.80% | 80.60% | 78.10% | 78.10% | 77.40% | 79.60% | 19.50% | 18.60% | 25.00% | |
| SYS28-3 | 97.09% | 99.91% | 77.10% | 81.30% | 80.50% | 78.00% | 74.60% | 80.50% | 78.10% | 78.00% | 77.30% | 79.50% | 19.70% | 19.30% | 25.10% | |
| RM8835 | 97.15% | 97.34% | 97.36% | 76.90% | 76.70% | 88.20% | 75.40% | 77.50% | 86.00% | 86.00% | 84.00% | 75.10% | 19.70% | 20.90% | 25.20% | |
| RM8965 | 96.98% | 97.84% | 97.86% | 97.41% | 80.80% | 77.60% | 73.70% | 83.30% | 77.60% | 77.60% | 77.30% | 83.00% | 19.40% | 21.60% | 25.10% | |
| RM8966 | 96.97% | 97.80% | 97.79% | 97.31% | 97.83% | 77.90% | 74.10% | 80.80% | 77.70% | 77.60% | 77.30% | 81.20% | 19.70% | 19.90% | 25.20% | |
| RM9262 | 97.14% | 97.43% | 97.44% | 98.61% | 97.44% | 97.34% | 75.60% | 78.20% | 93.70% | 93.70% | 92.60% | 75.80% | 20.30% | 22.10% | 25.20% | |
| S0112 | 97.51% | 97.10% | 97.12% | 97.18% | 96.99% | 97.07% | 97.24% | 75.00% | 75.00% | 75.00% | 75.30% | 74.10% | 19.80% | 21.50% | 25.00% | |
| RM8970 | 97.03% | 97.75% | 97.78% | 97.36% | 98.02% | 97.79% | 97.47% | 97.11% | 78.30% | 78.20% | 78.00% | 83.60% | 19.30% | 20.60% | 25.10% | |
| RM9263 | 97.15% | 97.45% | 97.44% | 98.37% | 97.48% | 97.36% | 99.25% | 97.16% | 97.54% | 100.00% | 95.30% | 75.80% | 20.00% | 22.00% | 25.10% | |
| RM9261 | 97.10% | 97.38% | 97.40% | 98.33% | 97.46% | 97.31% | 99.22% | 97.14% | 97.44% | 99.99% | 95.30% | 75.70% | 20.00% | 22.00% | 25.10% | |
| RM12175 | 97.15% | 97.49% | 97.53% | 98.18% | 97.47% | 97.41% | 99.16% | 97.22% | 97.51% | 99.44% | 99.43% | 77.20% | 20.10% | 23.50% | 25.20% | |
| RM8964 | 97.06% | 97.70% | 97.72% | 97.16% | 98.02% | 97.84% | 97.31% | 97.06% | 98.04% | 97.27% | 97.27% | 97.41% | 19.60% | 23.50% | 25.20% | |
| C. hyointestinalis subsp. lawsonii | 72.25% | 72.22% | 72.34% | 72.39% | 72.21% | 72.36% | 72.44% | 72.25% | 72.13% | 72.29% | 72.31% | 72.34% | 72.17% | 21.30% | 22.80% | |
| C. infantis | 68.44% | 68.23% | 68.40% | 68.26% | 68.33% | 68.24% | 68.32% | 68.38% | 68.39% | 68.26% | 68.24% | 68.34% | 68.35% | 68.09% | 20.90% | |
| C. lanienae | 82.30% | 82.36% | 82.43% | 82.44% | 82.43% | 82.34% | 82.32% | 82.27% | 82.51% | 82.32% | 82.32% | 82.34% | 82.44% | 74.01% | 68.58% | |
Note: The sequence used is the same as in phylogenomic analysis.
3.4. Antibiotic Resistance and Pathogenicity
Antibiotic resistance demonstrated that the strains XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1 were resistant to three types of antibiotics, macrolides (erythromycin (MIC, ≥64 μg mL−1) and azithromycin (MIC, ≥64 μg mL−1)), quinolones (nalidix acid (MIC, ≥32 μg mL−1)), aminoglycosides (streptomycin (MIC, ≥64 μg mL−1)), and yet strains S13-1, SYS25-1T, and SYS28-3 were resistant to quinolones (nalidix acid (MIC, ≥16 μg mL−1) and ciprofloxacin (MIC, ≥8 μg mL−1)). Part of our results is consistent with a previous report showing that Campylobacter species are highly resistant to quinolones [25,46].
In genomes of XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1, the aminoglycoside antibiotic resistance gene APH(2”)-If was found in two out of six strains (33.33%, XJK49-2 and XJK62-3). Point mutations in the 23S ribosomal RNA domain, Cjej_23S_ERY (A2075G), which confer resistance to macrolide antibiotics, was identified in all six strains (100.00%), and the prevalence of Cjej_gyrA_FLO (T86I) conferring resistance to fluoroquinolones was found among these six strains (100.00%). Meanwhile, in genomes of S13-1, SYS25-1T, SYS28-3, and the other 10 strains, the prevalence of Cjej_gyrA_FLO (T86I) was also identified in 2 of 13 strains (15.38%, SYS25-1T and SYS28-3), and the tetracycline and fluoroquinolone antibiotic resistance gene adeF was found in 11 of 13 strains (84.62%, S13-1, SYS25-1, SYS28-3, RM8835, RM8965, RM9262, S0112, RM9263, RM9261, RM12175, RM8964). These antibiotic resistance genes were consistent with their resistance phenotype, respectively.
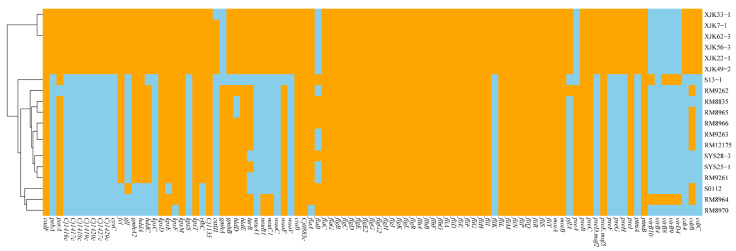
In genomes of these strains, numerous Campylobacter virulence-associated genes were detected, which could encode genes related to adherence, colonization and immune evasion, invasion, motility and export apparatus, secretion system, and toxins. The species of type stains of XJK22-1T and SYS25-1T had significantly different virulence-associated gene profiles. The species of type stain XJK22-1T have more Campylobacter virulence-associated genes than the species of SYS25-1T. The species of XJK22-1T have the complete cytolethal distending toxin, while the species of type strain SYS25-1T only have an incomplete type Ⅳ secretion system (T4SS). The details of the virulence genes were presented in Figure 4.
Figure 4.
Heatmap of the distribution of virulence genes. Orange indicates the presence of the virulence genes, and sky blue indicates the absence of the virulence genes.
3.5. Specific Real-Time PCR
These nine strains were simultaneously tested using Taqman real-time PCR to identify the species of Campylobacter. Amplification of six strains, XJK22-1T, XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1, was positive for Campylobacter felis sp. nov. specific qPCR, whereas the other Campylobacter species and blank control were qPCR negative. Similarly, S13-1, SYS25-1T, and SYS28-3 were positive for Campylobacter ovis sp. nov. specific qPCR, and the other Campylobacter species and blank control were qPCR negative. This suggests that these two specific real-time PCR are robust and can be used to rapidly discriminate between these two novel species and other Campylobacter species strains.
4. Conclusions
A polyphasic approach, including DNA sequencing and analysis (16S rRNA and whole-genome sequencing), electron microscopy, and a wide range of biochemical tests, as suggested by On et al. [47], provided sufficient evidence to distinguish these nine isolates from their closest related type strains and to confirm that they represent two novel species. With XJK22-1T and SYS25-1T as the type strains, we suggest the names Campylobacter felis sp. nov. and Campylobacter ovis sp. nov. for the two novel members of the genus Campylobacter.
4.1. Description of Campylobacter felis sp. nov.
Campylobacter felis (fe’lis. L. gen. n. felis of a Cat)
The cells are Gram negative, mesophilic, motile, and spiral shaped with sizes between 1.8–2.2 μm after 48 h of growth on Karmali or Columbia agar with 5% defibrinated sheep blood in a microaerophilic atmosphere at 37 °C. The colonies are wet, flat, gray, circular, and smooth, with sizes between 1.0 and 1.3 mm after 2 days of growth, but they may vary in size and morphology after a long incubation. No hemolysis on blood agar is observed. Fresh cells are motile and have long bipolar single flagella.
These six strains are negative for catalase, urease, GGT, TTC, and pyroglutamyl-peptidase I, and cannot produce H2S. On the other hand, they are positive for oxidase, reduced nitrate, L-arginine arylamidase, L-aspartic acid arylamidase, and alkaline phosphatase, and all can hydrolyze indoxyl acetate and hippurate. All six strains are sensitive to chloramphenicol, florfenicol, and tetracycline and resistant to erythromycin, azithromycin, nalidixic acid, and streptomycin.
The type strain XJK22-1T (=GDMCC 1.3684T = JCM 35847T), which was isolated from the feces of cats in 2019 and 2020 in Beijing, China, has a DNA G + C content of 34.99 mol%. The other strains XJK33-1, XJK49-2, XJK56-3, XJK62-3, and XJK7-1 are also classified in this species.
4.2. Description of Campylobacter ovis sp. nov.
Campylobacter ovis (o’vis. L. gen. n. ovis of a Sheep)
The cells are Gram negative, mesophilic, motile, and spiral or S-shaped with sizes between 2.1–2.5 μm after 48 h of growth on Karmali or Columbia agar with 5% defibrinated sheep blood in a microaerophilic atmosphere at 37 °C. The colonies are wet, flat, gray, circular, and smooth, with sizes between 1.0 and 1.3 mm after 2 days of growth, but they may vary in size and morphology after a long incubation period. No hemolysis on blood agar is observed. Fresh cells are motile and have long bipolar single flagella.
These three strains are negative for urease, TTC, pyroglutamyl-peptidase I, and L-aspartic acid arylamidase and cannot hydrolyze indoxyl acetate and hippurate or produce H2S. On the other hand, they are positive for catalase, oxidase, GGT, L-arginine arylamidase, and alkaline phosphatase and are variable for the reduction of nitrate. All three strains are resistant to nalidixic acid and sensitive to erythromycin, azithromycin, gentamicin, streptomycin, chloramphenicol, florfenicol, tetracycline, telithromycin, and clindamycin.
The type strain SYS25-1T (=GDMCC 1.3685T), which was isolated from the feces of sheep in 2019 and 2020 in Beijing, has a DNA G + C content of 32.43 mol%. The other strains, S13-1 and SYS28-3, are also classified in this species.
5. Limitations
In this study, we utilized a polyphasic approach to isolate and identify two novel Campylobacter species, which we have named Campylobacter felis sp. nov. and Campylobacter ovis sp. nov. Although their pathogenicity is currently unknown, studies indicate that further investigation into the potential health implications of these novel bacteria is necessary. Such research may aid in the management of diseases that could potentially be caused by these novel Campylobacter species.
Acknowledgments
We thank our colleagues from the Chinese Center for Disease Control and Prevention.
Abbreviations
ANI—average nucleotide identity; dDDH—digital DNA–DNA hybridization; ML—maximum likelihood; MP—maximum parsimony; NJ—neighbor-joining; GBS—Guillain–Barré Syndrome.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11040971/s1. Figure S1: Maximum-likelihood phylogenetic tree based on nearly complete 16S rRNA gene showing the relationships between our isolates and the type strains of the genus Campylobacter. Bootstrap values (>70%) based on 1000 replicates are shown at branch nodes, with Arcobacter butzleri ATCC 49616T as an outgroup. Novel strains are highlighted in bold. Figure S2: Maximum-parsimony phylogenetic tree based on nearly complete 16S rRNA gene showing the relationships between our isolates and the type strains of the genus Campylobacter. Bootstrap values (>70%) based on 1000 replicates are shown at branch nodes, with Arcobacter butzleri ATCC 49616T as an outgroup. Novel strains are highlighted in bold. Table S1: ANI (lower diagonal) and dDDH (upper diagonal) among the novel Campylobacter strains and other Campylobacter species.
Author Contributions
Conceptualization, H.W. and M.Z.; Methodology, X.C. and X.Z.; Software, G.Z.; Validation, Y.L. and Y.G.; Resources, Z.S. and J.Z.; Writing—Original Draft Preparation, H.W.; Writing—Review and Editing, M.Z. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Sponsored by the National Key Research and Development Program of China (2021YFC2301000) and the Sanming Project of Medicine in Shenzhen (SZSM201803081).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hamidian M., Sanaei M., Bolfion M., Dabiri H., Zali M.R., Walther-Rasmussen J. Prevalence of putative virulence markers in Campylobacter jejuni and Campylobacter coli isolated from hospitalized children, raw chicken, and raw beef in Tehran, Iran. Can. J. Microbiol. 2011;57:143–148. doi: 10.1139/W10-089. [DOI] [PubMed] [Google Scholar]
- 2.Man S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011;8:669–685. doi: 10.1038/nrgastro.2011.191. [DOI] [PubMed] [Google Scholar]
- 3.Karmali M.A., Penner J.L., Fleming P.C., Williams A., Hennessy J.N. The serotype and biotype distribution of clinical isolates of Campylobacter jejuni and Campylobacter coli over a three-year period. J. Infect. Dis. 1983;147:243–246. doi: 10.1093/infdis/147.2.243. [DOI] [PubMed] [Google Scholar]
- 4.Costa D., Iraola G. Pathogenomics of Emerging Campylobacter Species. Clin. Microbiol. Rev. 2019;32:e00072-18. doi: 10.1128/CMR.00072-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M., Li Q., He L., Meng F., Gu Y., Zheng M., Gong Y., Wang P., Ruan F., Zhou L., et al. Association study between an outbreak of Guillain-Barre syndrome in Jilin, China, and preceding Campylobacter jejuni infection. Foodborne Pathog. Dis. 2010;7:913–919. doi: 10.1089/fpd.2009.0493. [DOI] [PubMed] [Google Scholar]
- 6.Kaakoush N.O., Castaño-Rodríguez N., Mitchell H.M., Man S.M. Global Epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 2015;28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phung C., Scott P.C., Dekiwadia C., Moore R.J., Van T.T.H. Campylobacter bilis sp. nov., isolated from chickens with spotty liver disease. Int. J. Syst. Evol. Microbiol. 2022;72:005314. doi: 10.1099/ijsem.0.005314. [DOI] [PubMed] [Google Scholar]
- 8.Lynch C., Peeters C., Walsh N., McCarthy C., Coffey A., Lucey B., Vandamme P. Campylobacter majalis sp. nov. and Campylobacter suis sp. nov., novel Campylobacter species isolated from porcine gastrointestinal mucosa. Int. J. Syst. Evol. Microbiol. 2022;72:005510. doi: 10.1099/ijsem.0.005510. [DOI] [PubMed] [Google Scholar]
- 9.Parisi A., Chiara M., Caffara M., Mion D., Miller W.G., Caruso M., Manzari C., Florio D., Capozzi L., D’Erchia A.M., et al. Campylobacter vulpis sp. nov. isolated from wild red foxes. Syst. Appl. Microbiol. 2021;44:126204. doi: 10.1016/j.syapm.2021.126204. [DOI] [PubMed] [Google Scholar]
- 10.Aydin F., Abay S., Kayman T., Karakaya E., Mustak H.K., Mustak I.B., Bilgen N., Goncuoglu M., Duzler A., Guran O., et al. Campylobacter anatolicus sp. nov., a novel member of the genus Campylobacter isolated from feces of Anatolian Ground Squirrel (Spermophilus xanthoprymnus) in Turkey. Syst. Appl. Microbiol. 2021;44:126265. doi: 10.1016/j.syapm.2021.126265. [DOI] [PubMed] [Google Scholar]
- 11.Silva M.F., Pereira G., Carneiro C., Hemphill A., Mateus L., Lopes-da-Costa L., Silva E. Campylobacter portucalensis sp. nov., a new species of Campylobacter isolated from the preputial mucosa of bulls. PLoS ONE. 2020;15:e0227500. doi: 10.1371/journal.pone.0227500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant E., Shen Z., Mannion A., Patterson M., Buczek J., Fox J.G. Campylobacter taeniopygiae sp. nov., Campylobacter aviculae sp. nov., and Campylobacter estrildidarum sp. nov., Novel Species Isolated from Laboratory-Maintained Zebra Finches. Avian Dis. 2020;64:457–466. doi: 10.1637/aviandiseases-D-20-00019. [DOI] [PubMed] [Google Scholar]
- 13.Boukerb A.M., Penny C., Serghine J., Walczak C., Cauchie H.M., Miller W.G., Losch S., Ragimbeau C., Mossong J., Megraud F., et al. Campylobacter armoricus sp. nov., a novel member of the Campylobacter lari group isolated from surface water and stools from humans with enteric infection. Int. J. Syst. Evol. Microbiol. 2019;69:3969–3979. doi: 10.1099/ijsem.0.003836. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert M.J., Zomer A.L., Timmerman A.J., Spaninks M.P., Rubio-Garcia A., Rossen J.W., Duim B., Wagenaar J.A. Campylobacter blaseri sp. nov., isolated from common seals (Phoca vitulina) Int. J. Syst. Evol. Microbiol. 2018;68:1787–1794. doi: 10.1099/ijsem.0.002742. [DOI] [PubMed] [Google Scholar]
- 15.Goyal D., Watkins L.K.F., Montgomery M.P., Jones S.M.B., Caidi H., Friedman C.R. Antimicrobial susceptibility testing and successful treatment of hospitalised patients with extensively drug-resistant Campylobacter jejuni infections linked to a pet store puppy outbreak. J. Glob. Antimicrob. Resist. 2021;26:84–90. doi: 10.1016/j.jgar.2021.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma B., Thille K., Belmar V.M., Thomas R.N., Sharma R.N. Molecular detection and genetic characterization of Arcobacter butzleri isolated from red-footed pet tortoises suspected for Campylobacter spp. from Grenada, West Indies. PLoS ONE. 2020;15:e0230390. doi: 10.1371/journal.pone.0230390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph L.A., Francois Watkins L.K., Chen J., Tagg K.A., Bennett C., Caidi H., Folster J.P., Laughlin M.E., Koski L., Silver R., et al. Comparison of Molecular Subtyping and Antimicrobial Resistance Detection Methods Used in a Large Multistate Outbreak of Extensively Drug-Resistant Campylobacter jejuni Infections Linked to Pet Store Puppies. J. Clin. Microbiol. 2020;58:e00771-20. doi: 10.1128/JCM.00771-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dipineto L., Borrelli L., Pace A., Romano V., D’Orazio S., Varriale L., Russo T.P., Fioretti A. Campylobacter coli infection in pet birds in southern Italy. Acta Vet. Scand. 2017;59:6. doi: 10.1186/s13028-016-0271-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bojanić K., Midwinter A.C., Marshall J.C., Rogers L.E., Biggs P.J., Acke E. Isolation of Campylobacter spp. from Client-Owned Dogs and Cats, and Retail Raw Meat Pet Food in the Manawatu, New Zealand. Zoonoses Public Health. 2017;64:438–449. doi: 10.1111/zph.12323. [DOI] [PubMed] [Google Scholar]
- 20.Francois Watkins L.K., Laughlin M.E., Joseph L.A., Chen J.C., Nichols M., Basler C., Breazu R., Bennett C., Koski L., Montgomery M.P., et al. Ongoing Outbreak of Extensively Drug-Resistant Campylobacter jejuni Infections Associated with US Pet Store Puppies, 2016-2020. JAMA Netw. Open. 2021;4:e2125203. doi: 10.1001/jamanetworkopen.2021.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Zhang S., He M., Zhang Y., Fu Y., Liang H., Jing H., Li Y., Ma H., Zhang M. Prevalence and Molecular Characterization of Campylobacter spp. Isolated from Patients with Diarrhea in Shunyi, Beijing. Front. Microbiol. 2018;9:52. doi: 10.3389/fmicb.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank J.A., Reich C.I., Sharma S., Weisbaum J.S., Wilson B.A., Olsen G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Austrian R. The Gram stain and the etiology of lobar pneumonia, an historical note. Bacteriol. Rev. 1960;24:261–265. doi: 10.1128/br.24.3.261-265.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou G., Liang H., Gu Y., Ju C., He L., Guo P., Shao Z., Zhang J., Zhang M. Comparative genomics of Helicobacter pullorum from different countries. Gut. Pathog. 2020;12:56. doi: 10.1186/s13099-020-00394-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Gu Y., Lv J., Liang H., Zhang J., Zhang S., He M., Wang Y., Ma H., French N., et al. Laboratory Study on the Gastroenteritis Outbreak Caused by a Multidrug-Resistant Campylobacter coli in China. Foodborne Pathog. Dis. 2020;17:187–193. doi: 10.1089/fpd.2019.2681. [DOI] [PubMed] [Google Scholar]
- 26.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 27.Lowe T.M., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akhter S., Aziz R.K., Edwards R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012;40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.V., Cheng A.A., Liu S., et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B., Zheng D., Jin Q., Chen L., Yang J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier-Kolthoff J.P., Carbasse J.S., Peinado-Olarte R.L., Göker M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022;50:D801–D807. doi: 10.1093/nar/gkab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pritchard L., Glover R.H., Humphris S., Elphinstone J.G., Toth I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Analytical. Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 33.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 37.Fitch W.M. Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology. Syst. Zool. 1971;20:406–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- 38.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 40.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price M.N., Dehal P.S., Arkin A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huson D.H., Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 43.Rossi-Tamisier M., Benamar S., Raoult D., Fournier P.E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Pt 6Int. J. Syst. Evol. Microbiol. 2015;65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
- 44.Debruyne L., Broman T., Bergstrom S., Olsen B., On S.L.W., Vandamme P. Campylobacter subantarcticus sp. nov., isolated from birds in the sub-Antarctic region. Pt 4Int. J. Syst. Evol. Microbiol. 2010;60:815–819. doi: 10.1099/ijs.0.011056-0. [DOI] [PubMed] [Google Scholar]
- 45.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M., Gu Y., He L., Ran L., Xia S., Han X., Li H., Zhou H., Cui Z., Zhang J. Molecular typing and antimicrobial susceptibility profiles of Campylobacter jejuni isolates from north China. Pt 10J. Med. Microbiol. 2010;59:1171–1177. doi: 10.1099/jmm.0.022418-0. [DOI] [PubMed] [Google Scholar]
- 47.On S.L.W., Miller W.G., Houf K., Fox J.G., Vandamme P. Minimal standards for describing new species belonging to the families Campylobacteraceae and Helicobacteraceae: Campylobacter, Arcobacter, Helicobacter and Wolinella spp. Int. J. Syst. Evol. Microbiol. 2017;67:5296–5311. doi: 10.1099/ijsem.0.002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.