Abstract
Sphingomonas sp. strain RB2256 is representative of the ultramicrobacteria that proliferate in oligotrophic marine waters. While this class of bacteria is well adapted for growth with low concentrations of nutrients, their ability to respond to complete nutrient deprivation has not previously been investigated. In this study, we examined two-dimensional protein profiles for logarithmic and stationary-phase cells and found that protein spot intensity was regulated by up to 70-fold. A total of 72 and 177 spots showed increased or decreased intensity, respectively, by at least twofold during starvation. The large number of protein spots (1,500) relative to the small genome size (ca. 1.5 Mb) indicates that gene expression may involve co- and posttranslational modifications of proteins. Rates of protein and RNA synthesis were examined throughout the growth phase and up to 7 days of starvation and revealed that synthesis was highly regulated. Rates of protein synthesis and cellular protein content were compared to ribosome content, demonstrating that ribosome synthesis was not directly linked to protein synthesis and that the function of ribosomes may not be limited to translation. By comparing the genetic capacity and physiological responses to starvation of RB2256 to those of the copiotrophic marine bacterium Vibrio angustum S14 (J. Ostling, L. Holmquist, and S. Kjelleberg, J. Bacteriol. 178:4901–4908, 1996), the characteristics of a distinct starvation response were defined for Sphingomonas strain RB2256. The capacity of this ultramicrobacterium to respond to starvation is discussed in terms of the ecological relevance of complete nutrient deprivation in an oligotrophic marine environment. These studies provide the first evidence that marine oligotrophic ultramicrobacteria may be expected to include a starvation response and the capacity for a high degree of gene regulation.
Most natural aquatic and terrestrial environments are nutrient limited and, as a result, a major portion of the biosphere exists as oligotrophic (nutrient-depleted) habitats (22, 32). Of all the environments on Earth, however, the ocean has the highest cellular production rate (36). The number of prokaryotes in the ocean is estimated to be 1.2 × 1029 (36) and is mainly due to the growth of oligotrophic, extremely small bacteria (i.e., ultramicrobacteria), a unique class of marine bacteria that proliferate by growing slowly, even at nanomolar concentrations of growth substrates (reviewed in reference 32).
Although ultramicrobacteria represent the ocean's main bacterial component in terms of both activity and biomass (32) and although they also predominate in soil ecosystems (19), their physiology has remained largely uncharacterized (19). The lack of knowledge largely relates to the difficulty in isolating them from the environment. In 1990, an important breakthrough was made with the isolation of the ultramicrobacterium Sphingomonas sp. strain RB2256 (herein referred to as RB2256) from Resurrection Bay, Alaska (6, 29). At the time of isolation, it was a numerically dominant bacterium. Morphologically (29) and phylogenetically (M. Vancanneyt, F. Schut, C. Snauwaert, J. Goris, J. Swings, and J. C. Gottschal, submitted for publication) related bacteria have also been isolated as dominant species from the North Sea, and two years after initial isolation, RB2256 was again isolated as one of the dominant species in Resurrection Bay (M. Vancanneyt et al., submitted). Recently, a phylogenetically related strain was also isolated from waters near Japan (M. Eguchi et al., unpublished data).
Characteristics that define RB2256 as a typical or model oligotrophic ultramicrobacterium (15, 18, 32, 33) include a constant ultramicrosize (<0.1 μm3), irrespective of whether it is growing or starved; a mechanism for avoiding predation (ultramicro size), a relatively slow maximum specific growth rate (<0.2 h−1); the ability to utilize low concentrations of nutrients; high-affinity, broad-specificity uptake systems; and the ability to simultaneously take up mixed substrates (8, 10, 29–31). Based on the Michaelis-Menten constant for substrate transport (Kt) and the available concentrations of mixed amino acids in the ocean, RB2256 is predicted to have an in situ doubling time of 12 h to 3 days (6, 29). As the average doubling time for microorganisms in oligotrophic waters is estimated to be 5 to 15 days (14), RB2256 is likely to be a significant contributor to biomass turnover in oligotrophic ocean waters.
A fundamental characteristic that distinguishes RB2256 from typical copiotrophic bacteria is the capacity for copiotrophic bacteria to grow rapidly in the presence of high concentrations of nutrients and to undergo reductive cell division to form resting-stage cells when exposed to nutrient deprivation (22, 34). This may be referred to as a feast-and-famine response. As RB2256 is well suited for growth in low concentrations of nutrients, it may be expected that it would rarely encounter nutrient levels that would cause starvation, and as a result it may not have the genetic potential to respond to complete nutrient deprivation in the same way as copiotrophic bacteria.
The potential for RB2256 to starve may, however, be considered in terms of the total bacterial carbon content versus the amount of carbon available for microbial assimilation. Bacterial growth in the subarctic Pacific Ocean is limited by carbon (20), and approximately 1 to 10% of dissolved organic carbon (DOC) is available for microbial assimilation (22). This mainly consists of dissolved free amino acids and carbohydrates. The greatest proportion of bioavailable carbon in these oligotrophic waters consists of dissolved free amino acids at a concentration of 3 to 191 nM (9, 23) or 0.1 to 8.8 μg of C liter−1 (based on 110 as the average molecular weight for an amino acid and a ratio of 25/60 for conversion to carbon). Assuming an average bacterial cell carbon content of 5.6 × 10−13 g of C μm−3 (4) and an average cell volume of RB2256 of 0.05 μm3 (10), the carbon content is 28 fg of C cell−1. At the upper limit of bioavailable amino acids (8.8 μg of C liter−1), the cell yield is 3 × 105 cells ml−1. As RB2256 was isolated from a 106-fold dilution series of seawater from Resurrection Bay in which the standing bacterial population was approximately 2 × 105 cells ml−1 (32), this would indicate that the cells were likely to be starving at the time of sampling. There are possible limitations associated with this analysis, including uncertainties regarding measurements of bioavailable DOC and diurnal and seasonal variations (23); however, as an approximation, it indicates the potential for bacteria in oligotrophic marine environments to be exposed to starvation.
In order to assess the capacity of RB2256 to respond to starvation, we compared gene expression in logarithmic and starved cells using a two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) analysis of cellular proteins. The generation of high-resolution protein profiles enabled a quantitative assessment of differential gene expression in RB2256 and a comparison of gene expression to that of Vibrio angustum S14 (26), a typical marine copiotrophic bacterium with a well-characterized response to starvation (34). In addition to patterns of whole-cell gene expression, we examined changes in total protein and RNA synthesis that occurred as the cells made the transition from logarithmic growth to starvation. By monitoring the response throughout the growth phase and up to 7 days of starvation, we have gained significant insight into how the cell prepares for and copes with starvation. Our findings demonstrate that RB2256 exhibits a defined response to starvation with a number of characteristics that differ from copiotrophic bacteria. These results expand our view of the physiology of oligotrophic bacteria, and we discuss the potential advantages this may have for bacterial survival under oligotrophic conditions.
MATERIALS AND METHODS
Organism, growth conditions, and media.
Sphingomonas sp. strain RB2256 is a member of the α-proteobacteria. It has recently been taxonomically and phylogenetically characterized, and the species name Sphingomonas alaskensis sp. nov. has been proposed (M. Vancanneyt et al., submitted). RB2256 was maintained in an artificial seawater medium (10) supplemented with 3 mM d-glucose (ASWG medium). Batch cultures were grown at 30°C with orbital shaking at 150 rpm, as described previously (11). Cell viability was monitored by the drop plate method (13) with VNSS solid medium (25).
Determination of glucose concentration in growth medium.
RB2256 cultures were grown in ASWG medium, and samples were filtered through sterile 0.22-μm-pore-size Millex-GV (Millipore) membranes. Filtrates were analyzed with a glucose oxidase-peroxidase kit according to manufacturer's protocols (Sigma Chemical Co., St. Louis, Mo.).
Determination of rates of protein and RNA synthesis during growth and starvation.
The rates of protein and RNA synthesis were determined as the rate of radioactive methionine-cysteine and uridine incorporation into tricholoroaceteic acid (TCA)-insoluble material, respectively (13). Cultures were monitored by measuring the optical density at 433 nm (OD433) (11). Cultures were sampled during logarithmic growth, at the onset of starvation, and 1, 2, 4, and 7 days thereafter and exposed to a final concentration of 0.06 μM [35S]methionine-cysteine (specific activity, 1,175 Ci mmol−1; ICN Pharmaceuticals) or 0.38 μM [3H]uridine (specific activity, 26 Ci mmol−1; Amersham Life Science) at 30°C for up to 60 min. Initial rates were calculated from the linear portion of graphs of incorporation versus time. To determine synthesis rates, 50-μl duplicate samples were removed and added to 800 μl of ice-cold 10% TCA and maintained on ice until all samples were collected. Samples used for determining rates of protein synthesis were heated at 90°C for 15 min, kept on ice for 1 h, and collected by filtration with 0.2-μm-pore-size filters (Millipore). Samples used for the determination of rates of RNA synthesis were filtered without prior heat treatment. Filters for both protein and RNA determinations were washed three times with ice-cold 5% TCA and transferred into scintillation vials, and 10 ml of scintillation cocktail (Wallac Hi-Safe 3) was added. Filters were mixed by vortexing for 40 s prior to measuring radioactivity (Packard Scintillation Counter Series 2000). Rates of protein and RNA synthesis were calculated from the linear slopes of radionuclide incorporation graphs (25). The rate (in disintegrations per minute per cell) was calculated from the slope (disintegrations per minute), the viable cell count (the number of cells per milliliter), and sample volume (in milliliters). The rate was converted to the number of picomoles per minute per cell by incorporating the specific activity (curies per millimole) for each radionuclide and conversion factor (1 Ci = 2.2 × 1012 dpm) and converting to the number of picomoles per minute per milligram of protein by using the measured protein content per cell (see below). Relative rates were calculated as the proportion of the maximum rate during logarithmic growth and are represented as percentages or decimal values.
Determination of cellular protein content.
Cultures were sampled (15-ml samples) during logarithmic growth, at the onset of starvation, and 1, 2, 4, and 7 days thereafter and collected by centrifugation at 2,570 × g at 25°C for 20 min (Hettich Universal 16R). The cell pellet was resuspended in 15 ml of 0.1% sodium dodecyl sulfate by brief vortex mixing, followed by dilution with an equal volume of sterile Milli-Q water. Protein concentrations were determined with a bicinchoninic acid assay kit in accordance with the manufacturer's protocols (Sigma Chemical Co.), with standard curves constructed with bovine serum albumin.
Determination of ribosome content.
Data for ribosomes was determined previously (11).
Separation and analysis of 2D-PAGE of proteins.
Proteins from cells in mid-logarithmic growth (OD433 = 0.17; 1.5 × 108 cells ml−1) or following 24 h of starvation were radioactively labelled with [35S]methionine-cysteine (ICN Pharmaceuticals). Pulse-labelling, sample preparation, and electrophoresis were performed as described previously (12). Images of 2D-PAGE gels were obtained with CS phosphor screens on a Bio-Rad GS525 Molecular Imager. Quantitative analysis of 2D-PAGE gels was performed using Melanie II software (Bio-Rad) with three images generated from two separate experiments for each growth condition. Logarithmic-phase cells were labelled for 60 min (12). Starved cells were labelled for various lengths of time to determine the period of exposure that provided an equivalent level of incorporation per cell. By comparing disintegrations per minute from whole-cell counts with liquid scintillation and total disintegrations per minute from 2D-PAGE gels, equivalent levels of incorporation were found after 12-h labelling of cells starved for 24 h. Each spot was allocated a unique identification number. After initial spot detection, landmarks were manually appointed on each image, and all data were matched automatically with Melanie II software. Each spot that was identified by the spot-finding program was manually examined to confirm that it was a real spot. Anomalous spots, including regions of streaking or smearing and regions at the extremities of the gel, were excluded. Each gel was compared to every other gel to identify spots they had in common. Every match between spots from any two gels was designated a group. For each growth condition, groups were visually examined to ensure that the analogous spot was detected in all three gels. Anomalous spots were excluded or manually adjusted. Groups between different growth conditions were statistically analyzed (Student t test, 95% confidence interval) to determine which groups had intensity differences of at least twofold. Spots which appeared in the three gels from one growth condition (e.g., logarithmic growth) but did not appear in any of the three gels from another growth condition (e.g., starvation) were designated unique to the particular growth condition (e.g., logarithmic growth).
RESULTS
In order to examine the physiological changes that take place in stationary phase, we determined rates of RNA and protein synthesis, protein content per cell, and the glucose concentration of the growth medium throughout the growth phase and starvation and generated two-dimensional protein profiles of logarithmic-phase cells and cells starved for 24 h.
Onset of starvation and glucose utilization.
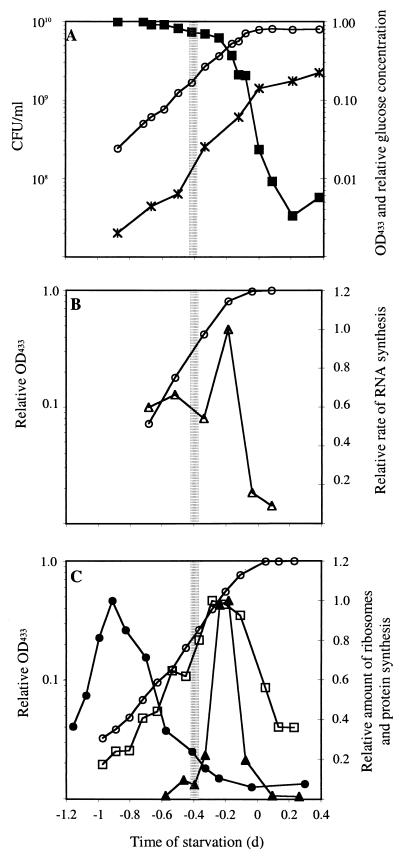
During growth at 30°C in ASWG medium, maximum growth yields corresponded to an OD433 of 0.8 (Fig. 1A). Once this optical density was reached, it did not increase further. In contrast, viable cell numbers increased from 1.4 × 109 CFU ml−1 when OD433 first reached 0.8 to 2.2 × 109 CFU ml−1 over the next 9 h. In association with cell growth, the glucose concentration decreased during log phase to a relative level of 0.023 when the cells reached an OD433 of 0.8. Within the next 5 h, the glucose level continued to fall to a relative level at least as low as 0.003 (the detection limit of the glucose assay).
FIG. 1.
Growth phase-dependent, macromolecular synthesis in RB2256. Cells were grown in ASWG medium. (A) Cell growth is measured as OD433 (○), as the number of CFU per milliliter (∗), and as the concentration of glucose in the medium (■). Curves for optical density and glucose concentration are the averages of two experiments with a maximum standard deviation of less than 20%, and results of a typical experiment are shown for the number of CFU per milliliter. (B) The optical density of cultures (○) and the relative rate of RNA synthesis (▵) are shown. The results of a typical experiment are shown. (C) The optical density of cultures (○), the relative number of ribosomes per cell (●), protein content per cell (□), and rates of protein synthesis (▴) are shown. The vertical hatched bar highlights the logarithmic-phase time point used for 2D-PAGE pulse-labelling experiments (see Fig. 3). Curves for optical density and protein content per cell are results of a typical experiment, rates of protein synthesis are the average of two experiments with a standard deviation of less than 27%, and ribosome numbers are from Fegatella et al. (11). All experiments were performed a minimum of three times.
To determine which point of the growth curve was most appropriately assigned as the onset of starvation, it was necessary to consider the potential growth yield associated with the available carbon. Three millimolar glucose is equivalent to 500 mg of glucose per liter. We have previously shown that 8 mg of DOC per liter yields 2.3 × 107 CFU ml−1 (10). Therefore, at a relative glucose concentration of 0.023 (11.5 mg of glucose per liter or 4.6 mg of DOC per liter), the maximum growth yield would be approximately 1.5 × 107 CFU ml−1. As the number of cells at this stage of growth had already reached 1.4 × 109 CFU ml−1, the concentration of glucose was insufficient to allow cell division, and cells were essentially starved. The onset of starvation for RB2256 was therefore most conveniently defined by the time point at which OD433 first reached maximum levels. The increase in viable count (approximately double), but the absence of increasing optical density in the following 9 h, was likely to reflect final rounds of cell division where dividing cells separated to form single cells. This was consistent with RB2256 cells not undergoing reductive cell division or changing cell volume during starvation (10, 28).
RNA synthesis during growth.
Peak rates of RNA synthesis occurred during late logarithmic phase (Fig. 1B). Prior to the observed increase in the rate of RNA synthesis, rates of RNA synthesis were relatively constant (54 to 66% of maximal rates), and following the peak, synthesis rates decreased rapidly to a relative level of around 16% at the onset of starvation. This indicated that RNA synthesis rates remained at relatively high levels during the most active stages of growth and that synthesis was quickly down-regulated as the cells approached starvation. The highest rates of RNA synthesis were 1.0 × 10−8 pmol of uridine min−1 cell−1.
Protein synthesis during growth.
The rate at which proteins were synthesized increased rapidly during logarithmic phase, peaked around late logarithmic phase, and decreased rapidly to reach a minimal basal level as the cells approached the onset of starvation (Fig. 1C). The rate of protein synthesis increased from a relative level of approximately 2% and decreased from peak levels to 2% at the onset of starvation, demonstrating that protein synthesis was regulated (gene expression and protein turnover) by up to 50-fold throughout the growth phase. Highest rates of protein synthesis were 5.2 × 10−8 pmol of methionine min−1 cell−1 and 284 pmol of methionine min−1 mg of protein−1 (Table 1). The profile describing the protein content per cell had a peak that coincided with the peak in the rate of protein synthesis. However, the profile of the rate of protein synthesis was broad, with relative levels around 1 day prior to starvation of 17% and 56% at the onset of starvation (Fig. 1C). The maximum concentration of protein corresponded to 186 fg cell−1 (Table 1).
TABLE 1.
Protein content and rate of protein synthesis in Sphingomonas sp. RB2256 during logarithmic growth and starvation
| Starvation (days) | Relative rate of synthesis (% of maximum rate) | Rate of protein synthesis (pmol of methionine min−1 cell−1) | Cellular protein content (fg cell−1) | Rate of protein synthesis (pmol of methionine min−1 mg of protein−1) |
|---|---|---|---|---|
| −0.57 | 1.6 | 8.3 × 10−10 | 84 | 10 |
| −0.46 | 9.5 | 4.9 × 10−9 | 116 | 42 |
| −0.39 | 7.1 | 3.6 × 10−9 | 141 | 26 |
| −0.33 | 21.8 | 1.1 × 10−8 | 167 | 68 |
| −0.24 | 97.7 | 5.0 × 10−8 | 186 | 271 |
| −0.18 | 100 | 5.2 × 10−8 | 181 | 284 |
| −0.07 | 19.4 | 1.0 × 10−8 | 164 | 61 |
| 0.09 | 1.6 | 8.1 × 10−10 | 87 | 9 |
| 0.27 | 1.2 | 6.0 × 10−10 | 66 | 9 |
| 0.42 | 0.6 | 2.8 × 10−10 | 60 | 5 |
| 1 | 0.9 | 4.5 × 10−10 | 37 | 12 |
| 4 | 0.3 | 1.3 × 10−10 | 34 | 4 |
| 7 | 0.3 | 1.7 × 10−10 | 34 | 5 |
By using the rates of protein synthesis to calculate the amount of protein synthesized in a fixed time period, the predicted cellular protein content was compared to the experimentally determined protein content. As an example, for the time period between 0.39 and 0.33 day prior to starvation, it was calculated that between 12.6 and 38.9 fg of protein was synthesized. This was derived from the data in Table 1 using the rates of protein synthesis for the two time points (see Materials and Methods). Based on the protein content per cell at 0.39 day prior to starvation, this would lead to a protein content per cell of between 154 and 180 fg cell−1 at 0.33 day prior to starvation. This predicted value compared favorably with the experimentally determined value of 167 fg cell−1.
Ribosome content and protein and RNA synthesis.
The number of ribosomes per cell during logarithmic growth (11) is shown in comparison to rates of protein synthesis and protein content (Fig. 1C). The peak in ribosome content occurred much earlier in the growth phase than the peaks for the protein synthesis and protein content per cell. At the point in the growth phase when the maximum number of ribosomes were present (2,000 per cell [11]), the total protein content was 45 fg cell−1 (data not shown). Assuming that the ribosomes in RB2256 have a mass equivalent to that of Escherichia coli (857,000 [21]), the protein content of the ribosomes at this point was 2.9 fg cell−1 or approximately 6% of total cell protein. This compares to 9% for E. coli at a specific growth rate of 0.6 h−1 (5). By applying this same calculation to time points from the onset to 7 days of starvation (Fig. 2), ribosomal protein represented less than 0.9% of total protein.
FIG. 2.
Macromolecular synthesis in RB2256 during starvation. Conditions for the experiments are as described in the legend to Fig. 1. Cell growth was measured by determining OD433 (○), and protein content per cell (□), relative number of ribosomes per cell (●), relative rate of RNA synthesis (▵), and rates of protein synthesis (▴) are shown.
The maximal rate of RNA synthesis occurred at a later stage in the growth phase than the peak in ribosome synthesis. As the rRNA content was relatively low at this stage of growth, the high level of RNA synthesis may indicate either an elevated synthesis of tRNA or mRNA or a high rate of rRNA turnover.
Macromolecular synthesis during starvation.
During stationary phase, up to 7 days of starvation, rates of RNA (0.3 to 2%) and protein (0.3 to 0.9%) synthesis reached low levels that remained constant throughout the starvation period (Fig. 2). The protein and ribosome contents in the cell also remained constant throughout starvation; however, the relative levels were approximately 25 and 10%, respectively. The protein content has also been found to remain high (50.4 mg of protein liter−1) for glucose-limited chemostat cells that have been starved for up to 29 days (28). These data show that low basal levels of macromolecular synthesis occurred during starvation and that preformed ribosomes and whole cell protein were retained for at least 7 days.
Protein profiles.
High-resolution 2D-PAGE profiles were generated for cells in mid-logarithmic growth (1.5 × 108 CFU ml−1) and after 24 h of starvation. The time point for logarithmic growth corresponded to 0.4 day before the onset of starvation and prior to the major increases in rates of RNA and protein synthesis (Fig. 1). This time was chosen as it was expected to reveal greater differences in expression between logarithmic growth and starvation than if a later time point was chosen for logarithmic growth. These profiles also serve as references maps for future work to examine the potential spectrum of changes in gene expression that may occur throughout the growth phase in association with the cellular levels of ribosomes and rates of protein synthesis.
Radioactive images of equivalent intensity were obtained with logarithmic-phase cells (generation time, ∼3.5 h) that were pulse-labelled for 1 h and 24-h-starved cells that were labelled for 12 h (Fig. 3). Up to 1,500 spots were resolved for logarithmic cells, and only marginally lower numbers (5%) of spots were detected for starved cells. Protein spots from both growth conditions were distributed evenly throughout the resolved pI range from 4 to 7 and a range in molecular mass from 10 to 100 kDa. The similarity in number and distribution of spots between the two growth phase conditions indicated that although rates of protein synthesis were significantly lower during starvation (Fig. 1C and 2) and (as a result) pulse-labelling times were longer, a broad range of proteins continued to be synthesized.
FIG. 3.
Two-dimensional gels of proteins from RB2256. (A and C) Logarithmic-phase (1.5 × 108 cells/ml) cells were labelled with [35S]methionine-cysteine for 60 min, and (B and D) cells starved for 24 h were labelled for 12 h. The molecular mass (in kilodaltons) of broad-range sodium dodecyl sulfate-PAGE standards (Bio-Rad) are indicated on the left of panel A. The molecular mass and pI values of specific spots were assigned with Melanie II software (Bio-Rad) and are shown in Table 3. Spots highlighted with circles and the letter L have ≥10-fold-higher intensities than the gels from logarithmic growth, and those with squares and the letter S have ≥10-fold-higher intensities than gels from starvation cultures. Spots identified with triangles and the letter U are present only in gels from logarithmic growth; some of the more intense spots are highlighted in the figure. Panels C and D are expanded views of sections of panels A and B, respectively, and serve to highlight the differences observable between protein spots from logarithmic-phase and starved cells.
A total of 1,086 spots were matched and analyzed for quantitative and qualitative differences between logarithmic-phase and starved cells. The methods used (12) combined radioactive imaging with phosphor screens, which provided the distinction between spot intensities throughout a 103-fold linear range, with a computer software package that enabled sensitive spot recognition and allowed large numbers of spots to be simultaneously compared. As a result, spots that may otherwise have been unrecognized were detected and quantitated. This was particularly important for determining which spots were unique, in comparison to those spots which were present in both growth conditions but at very different levels.
While the overall pattern was generally similar (i.e., the number and position of spots) between gels from each growth condition (Fig. 3A and B), differences in spot intensity were clearly identifiable. One of these regions is highlighted in Fig. 3C and D. A summary of overall quantitative differences is presented in Table 2. The intensity of 72 spots was increased at least twofold during starvation. Of these, the intensity of 14 spots was increased more than 10-fold. The greatest level of increase was 64-fold (Fig. 3B and D). Relative to the number of spots that have increased intensity during starvation, about 2 1/2 times as many protein spots have decreased intensity during starvation. Most of these were moderately decreased (>2-fold), although seven were decreased greater than 10-fold, and one was decreased 71-fold (Fig. 3A and C). While the numbers of spots between gels from logarithmic growth and starvation were similar, it is noteworthy that 80 protein spots were completely absent from gels of starved cells (i.e., unique to logarithmic growth).
TABLE 2.
Compilation of spot intensities from logarithmic and stationary phase two-dimensional protein gelsa
| Criterion | Intensity increased during starvation | Intensity decreased during starvation |
|---|---|---|
| 2–4 fold | 40 | 68 |
| 5–9 fold | 18 | 22 |
| 10–19 fold | 10 | 6 |
| 20–59 fold | 3 | 0 |
| >60 fold | 1 | 1 |
| Unique | 0 | 80 |
| Total | 72 | 177 |
See Fig. 3.
Specific protein spots are identified in Fig. 3 with intensity that is increased by 10-fold or higher during starvation or logarithmic phase. In addition, a number of spots unique to logarithmic growth have also been identified. A summary describing the characteristics of these specific spots is presented in Table 3. Eight of the 14 spots that have at least 10-fold-higher intensity during starvation were part of protein clusters (i.e., spots S1 to S5 and S6 to S8) (Fig. 3D). The spots in the cluster containing spots S1 to S5 included three rows of spots with up to six or seven closely spaced spots per row. In addition, the cluster was clearly present during logarithmic growth. Most of the spots in the two lower rows have spot intensities at least seven times higher during starvation. The cluster that includes spots S6 to S8 had somewhat different characteristics and appeared to be part of a single row of four spots which were more widely spaced than those in the S1-to-S5 cluster, and these four spots were almost absent during logarithmic growth. This cluster contained the spot (S8) with the highest differential intensity during starvation.
TABLE 3.
Summary of protein characteristics for spots from two-dimensional protein gelsa that have spot intensities varying by ≥10-fold or more between logarithmic and stationary-phase cells
| Criterion | Identificationb | Molecular mass (kDa) | pI | Fold increase or decrease |
|---|---|---|---|---|
| Intensity increased during starvation | S1 | 71 | 5.01 | 11 |
| S2 | 74 | 4.93 | 13 | |
| S3 | 72 | 4.96 | 11 | |
| S4 | 69 | 4.93 | 10 | |
| S5 | 69 | 4.97 | 11 | |
| S6 | 55 | 5.32 | 25 | |
| S7 | 55 | 5.39 | 37 | |
| S8 | 55 | 5.46 | 64 | |
| S9 | 36 | 5.12 | 12 | |
| S10 | 33 | 5.38 | 11 | |
| S11 | 30 | 4.96 | 13 | |
| S12 | 28 | 5.10 | 21 | |
| S13 | 27 | 5.02 | 10 | |
| S14 | 26 | 4.76 | 12 | |
| Intensity decreased during starvation | L1 | 63 | 6.03 | 16 |
| L2 | 52 | 5.71 | 10 | |
| L3 | 33 | 5.91 | 13 | |
| L4 | 32 | 6.15 | 16 | |
| L5 | 30 | 5.95 | 10 | |
| L6 | 26 | 5.08 | 71 | |
| L7 | 23 | 4.52 | 20 | |
| U1 | 52 | 5.89 | ∞ | |
| U2 | 52 | 5.52 | ∞ | |
| U3 | 48 | 5.00 | ∞ | |
| U4 | 40 | 5.37 | ∞ | |
| U5 | 36 | 4.52 | ∞ | |
| U6 | 26 | 4.96 | ∞ |
See Fig. 3.
S, starvation; L, logarithmic phase; U, unique to logarithmic growth.
The genome size of RB2256 is on the order of 1.5 Mb (29). The average sizes of the genes in E. coli (3) and Helicobacter pylori (35) are 1,082 and 1,049 bp, respectively. Assuming a similar relationship for other gram-negative bacteria (1 gene = 1 kb), the genome of RB2256 may encode approximately 1,500 genes. As it is unlikely that RB2256 simultaneously expressed every gene from its genome, it is equally unlikely that each of the 1,500 spots detected on the protein gels represented a unique protein. Due to the similarity of molecular mass and pI between spots from the same cluster (Table 3; Fig. 3), it is possible that the clusters represented posttranslationally or cotranslationally modified versions of the same protein and/or isozymes. Similar features have previously been associated with the glycosylation and phosphorylation of proteins (16). Furthermore, a high level of posttranslational and cotranslational modifications have been detected in the proteomes of E. coli (24) and Mycoplasma (I. Humphrey-Smith, personal communication). It is possible that in addition to regulated synthesis of protein production, modification of preexisting proteins plays an important role in growth-phase-specific physiology of RB2256.
DISCUSSION
A comparison of gene expression and protein synthesis during growth and starvation in RB2256 and V. angustum S14.
Differences in spot intensity of 60- to 70-fold are present between matching spots from protein gels for cells from logarithmic growth and starvation (Table 3), and 80 spots are present only in gels from logarithmic-phase cells (Table 2). These observed changes in spot intensity are indicative of growth phase-specific alterations in gene expression and provide strong support for the presence of a genetically programmed starvation response in RB2256.
We compared protein gels from RB2256 (Fig. 3) with comparable gels (1,700 spots) from V. angustum S14 (26) and attempted to rationalize the types of observed changes with the physiology and genetic characteristics of each bacterium. The total number of spots with higher intensity during starvation and logarithmic growth is 72 and 177, respectively, for RB2256 (Table 2), which compares with 157 and 144, respectively, for V. angustum S14 (26). The most noticeable feature of this comparison is the relatively low number of protein spots that have higher intensity during starvation in RB2256. A contributing factor to this difference may relate to cell division. Unlike copiotrophic microorganisms such as V. angustum S14, RB2256 does not undergo reductive cell division during starvation. As a result, genes involved in cell division are likely to be repressed in RB2256 but would need to be expressed in V. angustum S14 during stationary phase to enable reductive cell division to take place. Consistent with this, a large proportion of the spots in RB2256 that are differentially expressed during stationary phase are spots that are unique to logarithmic phase (Table 2) and are thus only required when the cell is growing.
We have previously shown that, unlike what is observed in V. angustum S14 and many other nondifferentiating heterotrophic bacteria, starvation in RB2256 does not lead to enhanced resistance to hydrogen peroxide or heat stress (10) and that cells starved for 24 h respond to the addition of excess glucose by immediately achieving maximum growth rates (11). These results indicate that genes often associated with stress resistance during starvation may not be induced during starvation in RB2256. In contrast, genes necessary for rapid recovery from starvation may be induced during starvation. It will clearly be valuable to characterize the spots on the protein gels that have been highly regulated during starvation, as this will determine the genetic basis and inferred physiological significance of the starvation response in RB2256. This will be particularly valuable for testing hypotheses regarding the types of catabolic systems and regulatory mechanisms that may be expected to be present in typical oligotrophic bacteria (27).
During starvation, the regulation of protein and RNA synthesis in RB2256 differs markedly from that in V. angustum S14. In RB2256, at the onset of starvation the rate of protein synthesis is 9 pmol of methionine min−1 mg of protein−1, decreasing to a minimum level of 4 pmol min−1 mg of protein−1 throughout 7 days of starvation (Table 1). In V. angustum S14, the rate of synthesis is 4.3 pmol of leucine min−1 mg of protein−1 immediately prior to starvation; however, the rate dramatically decreases to 0.1 and 0.009 pmol of leucine min−1 mg of protein−1 at 1 and 6 days of starvation, respectively (13). These data equate to a 2-fold decrease in specific rates of synthesis in RB2256 compared with a 470-fold decrease in V. angustum S14. A similar trend is observed for rates of RNA synthesis, whereby rates remain largely unchanged throughout starvation in RB2256 (Fig. 2) and abruptly decrease during starvation in V. angustum S14 (13). These responses to starvation further highlight the different physiology of these two classes of bacteria.
Protein synthesis and ribosomes.
We have previously shown that the ribosome content in RB2256 is highly regulated throughout the growth phase, and yet as cells can achieve maximum growth rates with a relative level of 10%, the cells appear to contain a cellular excess of ribosomes (11). To further examine the role of ribosomes in the cell, in this study we compared rates of protein synthesis and cellular protein content to the ribosome content (Fig. 1C). These data showed that the number of ribosomes per cell reached a maximum and then fell to basal levels approximately a half day before the rates of protein synthesis and cellular protein content reached maximum levels. In effect, the highest rate of protein translation occurs when there is a relative level of 10 to 15% of ribosomes (200 to 300 ribosomes cell−1). This indicates that ribosome synthesis is largely uncoupled from protein synthesis and that ribosomes may perform alternative functions during mid-logarithmic growth when the ribosome content greatly exceeds protein synthesis requirements. The ribosomes may function as protein reserves or be bound to and affect the stability of mRNA. These possibilities, however, need to be experimentally investigated.
During stationary phase, rates of protein synthesis do not exceed a relative level of 1%; however, the relative number of ribosomes per cell remains around 10% (11). This indicates that in starvation the number of ribosomes is also in large excess relative to protein synthesis requirements. However, unlike conditions of mid-logarithmic growth, where cells are dividing at maximum specific growth rates, an excess of ribosomes may provide an advantage for starved cells when they encounter a new source of carbon. This is supported by the ability of RB2256 cells that have been starved for 7 days in complex medium to resume growth immediately following the addition of excess glucose (11).
Monitoring starvation and ecological relevance.
In RB2256, maximum rates of RNA synthesis and protein synthesis coincide with the maximum rate of change in glucose concentration of the growth medium (Fig. 1). The rapid decrease in carbon flux during late logarithmic phase may provide a signal to the cell that starvation is imminent. In this way, much of this activity may be aimed at preparing the cell for complete nutrient deprivation.
Cells starved for 7 days in a glucose-minimal medium respond with submaximal rates of growth when subjected to glucose excess, whereas they respond with maximal rates when subjected to carbon in complex medium (11). This outgrowth response of cells in minimal medium is indicative of a repression mechanism, whereby rates of growth are repressed until the biosynthetic capacity of the cell is able to meet the demand necessary for achieving maximum rates of growth. Our present results indicate that the repression mechanism does not involve a typical stringent response (7, 17, 34), as the relaxation phase that normally accompanies a stringent response is not evident in the protein synthesis profile (Fig. 1C). This interpretation will be clarified by evaluating levels of ppGpp(p) throughout the growth phase.
The oligotrophic marine environment contains microzones that may create sustained gradients of nutrients utilizable by marine bacteria, produced by the lysis of or excretion from phytoplankton, the release of intracellular components during predation, or the presence of organic detritus. As a result of bacterial movement relative to the microzones caused by motility and chemotaxis (2) or hydrodynamic forces, bacteria may encounter sharp gradients of nutrients (1). As a result, they may experience periods of nutrient depletion and rapid changes in nutrient concentration that are sufficient to induce a starvation response and rapid increases in nutrients as they encounter relatively enriched microzones. Therefore, a marine bacterium such as RB2256 with an effective means of utilizing oligotrophic levels of nutrients is likely to have an enhanced ability to compete in the environment if it is able to mount an appropriate starvation response and rapid outgrowth response.
Marked changes in gene expression, as observed in protein gels from logarithmic phase and periods of cell starvation, and a high level of regulation of protein and RNA synthesis and ribosome content occur in RB2256. It remains to be determined what contributions are afforded by well-known gene regulatory pathways and alternative mechanisms of controlling gene expression, such as mRNA stability, protein turnover, and co- and posttranslational modification of proteins. However, the implications of these findings strongly argue that in RB2256 and perhaps other oligotrophic ultramicrobacteria, maintaining the normal physiological state of the cell involves a high level of gene regulation. It is not unreasonable to find that a bacterium such as RB2256 that is capable of becoming numerically dominant would have evolved a genotype with sufficient regulatory mechanisms to ensure optimized physiological activity in its native environment.
ACKNOWLEDGMENTS
We acknowledge constructive and critical discussions with M. Eguchi and S. Kjelleberg and thank M. Ostrowski, T. Kolesnikow and S. Rice for critical review of the manuscript. We warmly acknowledge the reviewers for their particularly constructive and detailed critiques during the review process.
This work was supported by the Australian Research Council.
REFERENCES
- 1.Azam F, Ammerman J W. Cycling of organic matter by bacterioplankton in pelagic marine ecosystems: microenvironmental considerations. In: Fasham M J R, editor. Flows of energy and materials in marine ecosystems: theory and practice. New York, N.Y: Plenum Publishing; 1984. pp. 345–360. [Google Scholar]
- 2.Blackburn N, Fenchel T, Mitchell J. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science. 1998;282:2254–2256. doi: 10.1126/science.282.5397.2254. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Bratbak G. Bacterial biovolume and biomass estimations. Appl Environ Microbiol. 1985;49:1488–1493. doi: 10.1128/aem.49.6.1488-1493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 6.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1456–1496. [Google Scholar]
- 8.Cavicchioli R, Fegatella F, Ostrowski M, Eguchi M, Gottschal J C. Sphingomonads from marine environments. J Ind Microbiol Biotechnol. 1999;23:268–272. doi: 10.1038/sj.jim.2900732. [DOI] [PubMed] [Google Scholar]
- 9.Eguchi M, Ishida Y. Oligotrophic properties of heterotrophic bacteria and in situ heterotrophic activity in pelagic seawaters. FEMS Microbiol Ecol. 1990;73:23–30. [Google Scholar]
- 10.Eguchi M, Nishikawa T, MacDonald K, Cavicchioli R, Gottschal J C, Kjelleberg S. Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1996;62:1287–1294. doi: 10.1128/aem.62.4.1287-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fegatella F, Ostrowski M, Cavicchioli R. An assessment of protein profiles from the marine oligotrophic ultramicrobacterium, Sphingomonas sp. strain RB2256. Electrophoresis. 1999;20:2094–2098. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2094::AID-ELPS2094>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Flardh K, Cohen P S, Kjelleberg S. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1992;174:6780–6788. doi: 10.1128/jb.174.21.6780-6788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrman J A, Sleeter T D, Carlson C A, Proctor L M. Dominance of bacterial biomass in the Sargasso Sea and its ecological implications. Mar Ecol Prog Ser. 1989;57:207–217. [Google Scholar]
- 15.Gonzales J M, Sherr E B, Sherr B F. Differential feeding by marine flagellates on growing versus starving, and on motile versus nonmotile, bacterial prey. Mar Ecol Prog Ser. 1993;102:257–267. [Google Scholar]
- 16.Gooley A A, Packer N H. The importance of protein co- and post-translational modifications in proteome projects. In: Wilkins M R, Williams K L, Appel R D, Hochstrasser D F, editors. Proteome research: new frontiers in functional genomics. Berlin, Germany: Springer-Verlag; 1997. pp. 65–89. [Google Scholar]
- 17.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1497–1512. [Google Scholar]
- 18.Hirsch P, Bernhard M, Cohen S S, Ensign J C, Jannasch H W, Koch A L, Marshall K C, Poindexter J S, Rittenberg S C, Smith D C, Veldkamp H. Life under conditions of low nutrient concentrations. In: Shilo M, editor. Strategies of microbial life in extreme environments. Weinheim, Germany: Verlag Chemie; 1979. pp. 357–372. [Google Scholar]
- 19.Janssen P H, Schuhmann A, Mörschel E, Rainey F A. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol. 1997;63:1382–1388. doi: 10.1128/aem.63.4.1382-1388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchman D L. Limitation of bacterial growth by dissolved organic matter in the subarctic Pacific. Mar Ecol Prog Ser. 1990;62:47–54. [Google Scholar]
- 21.Lewin B. Genes IV. New York, N.Y: Oxford University Press; 1990. [Google Scholar]
- 22.Morita R Y. Bacteria in oligotrophic environments, starvation-survival lifestyle. New York, N.Y: Chapman & Hall; 1997. [Google Scholar]
- 23.Munster U. Concentration and fluxes of organic carbon substrates in the aquatic environment. Antonie Leeuwenhoek. 1993;63:243–274. doi: 10.1007/BF00871222. [DOI] [PubMed] [Google Scholar]
- 24.Nouwens A S, Hopwood F G, Traini M, Williams K L, Walsh B J. Proteome approach to the identification of cellular Escherichia coli proteins. In: Charlebois R L, editor. Organization of the prokaryotic genome. Washington, D.C.: American Society for Microbiology; 1999. pp. 331–349. [Google Scholar]
- 25.Nystrom T, Marden P, Kjelleberg S. Relative changes in incorporation rates of leucine and methionine during starvation survival of two bacteria isolated from marine waters. FEMS Microbiol Ecol. 1986;38:285–292. [Google Scholar]
- 26.Ostling J, Holmquist L, Kjelleberg S. Global analysis of the carbon starvation response of a marine Vibrio disrupted in genes homologous to relA and spoT. J Bacteriol. 1996;178:4901–4908. doi: 10.1128/jb.178.16.4901-4908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poindexter J S. Oligotrophy. Fast and famine existence. Adv Microb Ecol. 1981;5:63–89. [Google Scholar]
- 28.Schut F. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1994. [Google Scholar]
- 29.Schut F, de Vries E J, Gottschal J C, Robertson B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schut F, Gottschal J C, Prins R A. Isolation and characterization of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol Rev. 1997;20:363–369. [Google Scholar]
- 31.Schut F, Jansen M, Pedro Gomes T M, Gottschal J C, Harder W, Prins R A. Substrate uptake and utilization by a marine ultramicrobacterium. Microbiology. 1995;141:351–361. doi: 10.1099/13500872-141-2-351. [DOI] [PubMed] [Google Scholar]
- 32.Schut F, Prins R A, Gottschal J C. Oligotrophy and pelagic marine bacteria: facts and fiction. Aquat Microb Ecol. 1997;12:177–202. [Google Scholar]
- 33.Simek K, Chrzanowski H. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl Environ Microbiol. 1992;58:3715–3720. doi: 10.1128/aem.58.11.3715-3720.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasan S, Kjelleberg S. Cycles of famine and feast: the starvation and outgrowth strategies of a marine Vibrio. J Biosci. 1998;23:501–511. [Google Scholar]
- 35.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L X, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L, Lee N, Adams M D, Hickey E K, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 36.Whitman W B, Coleman D C, Weibe W J. Prokaryotes—the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]