Abstract
Bluetongue virus (BTV) is an arbovirus that is transmitted between domestic and wild ruminants by Culicoides spp. Its worldwide distribution depends on competent vectors and suitable environmental ecosystems that are becoming affected by climate change. Therefore, we evaluated whether climate change would influence the potential distribution and ecological niche of BTV and Culicoides insignis in Peru. Here, we analyzed BTV (n = 145) and C. insignis (n = 22) occurrence records under two shared socioeconomic pathway scenarios (SSP126 and SSP585) with five primary general circulation models (GCMs) using the kuenm R package v.1.1.9. Then, we obtained binary presence–absence maps and represented the risk of transmission of BTV and niche overlapping. The niche model approach showed that north and east Peru presented suitability in the current climate scenario and they would have a decreased risk of BTV, whilst its vector would be stable and expand with high agreement for the five GCMs. In addition, its niche overlap showed that the two niches almost overlap at present and would completely overlap with one another in future climate scenarios. These findings might be used to determine the areas of highest priority for entomological and virological investigations and surveillance in order to control and prevent bluetongue infections in Peru.
Keywords: Peru, bluetongue virus, Culicoides insignis, ecological niche, climate change
1. Introduction
Bluetongue (BT) is a vector-borne disease caused by bluetongue virus (BTV) and is transmitted by Culicoides spp. (Diptera; Ceratopogonidae) to ruminants [1]. BTV belongs to the Orbivirus genus within the Reoviridae family and is a non-enveloped virus with 10 segments of double-stranded RNA (dsRNA) [1]. Despite there being twenty-seven identified serotypes, eight additional potential serotypes have been suggested [1,2,3]. BTV can affect several domestic (cattle, sheep and goats) [4,5] and wild ruminants (bighorn sheep, deer and pronghorn antelope), as well as some camelids (alpaca) [6,7]. In general, BT severe illness is mainly restricted to certain breeds of sheep and white-tailed deer; so, they can manifest many of the signs including fever, nasal discharge, hyperemia and edema of the lips, ears, face and submaxillary region, erosions of the oral mucosa, reduced milk yield and abortions [1,8,9]. Cattle play an important role in BTV epidemiology because of prolonged and persistent viremia from 7 to 63 days without clinical signs; so, they are considered to be reservoirs [5]. In addition, cattle enhance the spread of BTV as Culicoides spp. preferentially feed on them [9,10].
BTV is listed in the notifiable terrestrial and aquatic animal diseases of the World Organization for Animal Health (WOAH founded OIE) [11]. It has a negative economic impact on countries and also on the health of threatened wild populations [12]. BTV affects the national and international trade of animals; for instance, loss costs amounted to EUR 40 million during a 2007 outbreak in Germany with fatality rates up to 13.1% for cattle and 41.5% for sheep [13,14]. Regarding South America, BTV has been reported in Ecuador, Argentina, Colombia, French Guiana, Brazil and Peru [15]. In Brazil, BTV outbreaks have been reported since 2001 in sheep and goats that showed clinical signs, where some of them died [16]; furthermore, BTV caused significant mortality (18.4%) in 2014 [17,18]. In addition, dwarf brocket deer (Mazama nana) were affected by hemorrhagic disease during a BTV outbreak in 2015–2016 [19]. Conversely, Peru has not reported any clinical evidence of BT, although infection and seropositive animals have been documented in tropical regions [20,21].
The worldwide distribution of BTV depends on competent Culicoides vectors and suitable environmental ecosystems [22]. Culicoides sonorensis (With and Jones 1957) is the main vector of BTV in North America [23], while Culicoides insignis (Lutz 1913) is the main form in the southeastern United States of America, South America (SA) and the Caribbean [23,24]; Culicoides pusillus (Lutz 1913) could also be involved with BTV in SA [25,26]. Biting midges are blood-sucking insects that are often associated with farm environments close to muddy areas or along the margins of vegetated ponds among other areas [23,26,27]. Regarding C. insignis, it is significantly more abundant in livestock farms compared with both poultry and sylvatic habitats [28]. In addition, temperatures between 20 and 25 °C are associated with an abundance that refers to spring and summer seasons, and variables such as precipitation, wind speed and rainfall affected its highest abundances [29]. These variables, as well as agricultural and animal husbandry patterns, have been related to the presentation of BT infection [22,23].
Understanding the factors that are related to the geographic distribution of diseases is possible with ecological niche modeling (ENM) and species distribution modeling (SDM) [30,31,32]. Basically, SDM is the process of estimating the actual or potential distribution area, or set of suitable habitats for a species, based on its observed presences and (sometimes) absences [33]. In addition, ENM can estimate the Grinnellian fundamental niche that represents the environmental conditions needed for long-term population persistence [30,33]. However, certain assumptions should be considered, such as biotic interactions that could be neglected which denote “the Eltonian noise hypothesis” under the biotic, abiotic and movement (BAM) framework [33,34]. In addition, forecasting distributions and distributional changes in diverse spatial scales is possible with ENM approaches, but such forecasts should come from analyses of extensive data [30].
ENM and SDM have already been used to predict the potential distribution of BTV in the current [31,35] and future climate scenarios [31]. SDMs have also been generated for some Culicoides spp., such as Culicoides imicola (Kieffer 1913), Culicoides varipennis (Wirth and Williams 1957), C. sonorensis, C. insignis and others [31,36,37,38], highlighting abiotic (precipitation, temperature and seasonality) and biotic factors (sheep distribution or livestock density) as being the most important predictors for BTV and Culicoides spp. [31,35,36,37]. Therefore, it would be possible to determine the areas of the highest priority for the control and prevention of BTV in Peru using these tools, hence reducing personnel and equipment costs. These analysis approaches provide an opportunity to identify the factors that affect the spatial and temporal distribution of the vectors and hosts of BTV, thus promoting knowledge of the epidemiology and dynamics of virus transmission to domestic and wild ruminants. For this reason, our main aim was to evaluate whether climate change would influence the potential distribution and ecological niche of BTV and C. insignis in Peru.
2. Materials and Methods
2.1. Occurrence Records
BTV occurrence records (n = 513 individual animals) were obtained from ruminants’ serum (cattle, goats and sheep) via field sampling using competitive ELISA for antibody detection from 24 departments of Peru via Servicio Nacional de Sanidad Agraria (SENASA) from June 2017 to October 2019, and the virology laboratory at Universidad Nacional Mayor de San Marcos (UNMSM) was also used for occurrences (n = 52) from July 2021 to July 2022. Culicoides insignis collections were performed in different farms of Peru using mini-CDC light traps from July 2021 to July 2022, with 15 occurrences registered. We also acquired one C. insignis record from the scientific literature [39] and 13 from speciesLink (https://specieslink.net/, accessed on 16 September 2022). The geographic coordinates of each location were obtained using a GPS unit; however, for obtaining records from the database, we assigned them based on the consultation of online gazetteer data (www.gpsvisualizer.com (accessed on 16 September 2022)).
2.2. Occurrence Records Cleaning
BTV occurrence records were filtered to eliminate duplicates or georeferencing errors, and we also reduced the data to avoid problems with spatial autocorrelation; so, we rarefied based on 20 km buffer that was proposed by Samy and Peterson [31] using the spThin R package [40]. After these data cleaning steps, we had 145 and 13 occurrence records from 2017 to 2019 and 2021 to 2022 data, respectively (Table S1). Then, we randomly split the 145 records into two sets for model calibration (80%) and evaluation (20%). The 13 occurrences represented BTV independent data for model validation (Table S1). Similarly, C. insignis occurrence records (n = 29) were filtered and rarefied based on a 2 km distance that represented the average flight distance for Culicoides [41]. At the end, we had data for C. insignis from 22 occurrences (Table S1).
2.3. Calibration Area
The accessible area (M) was delimited based on our occurrence data. Therefore, we created the virus’ calibration area taking the elevation at which seropositive animals for BTV were found; hence, we used a DEM raster from Worldclim at a 30 arc-second and maximum elevation of 2888 m above sea level. For C. insignis, however, we relied on a map of the terrestrial ecoregions of the world [42]; therefore, each occurrence record was overlaid on the ecoregion map to shape its “M” area.
2.4. Environmental Data
Bioclimatic variables were gathered from CHELSEA V.2.1 [43] (https://chelsa-climate.org/ (accessed on 16 September 2022)) in the form of 19 bioclimatic data layers at a 30 arc-second (~1 km) of spatial resolution. Both current (1981–2010) and future climate scenarios (2071–2100) were downloaded under two shared socioeconomic pathway scenarios (SSP126 and SSP585), to represent an optimistic scenario that assumed climate protection measures and the dramatic scenario of the range of pathways with fossil-fueled development, respectively [44]. In addition, five primary general circulation models (GCMs) were used in our prediction model (GFDL-ESM4, IPSL-CM6A-LR, MPI-ESM1-2-HR, MRI-ESM2-0 and UKESM1-0-LL), as recommended by the literature [43].
We used four and five sets of bioclimatic variables for BTV and C. insignis, respectively (Tables S2 and S3). Following these criteria, we obtained (a) Set 1: 19 variables from CHELSA v.2.1; (b) Set 2: jackknife processes in Maxent to select variables that contributed most to models (>80%); (c) Set 3: pairwise Pearson’s correlation coefficients (r), thereby eliminating one variable per pair with the value of |r| being less than 0.80; (d) Set 4: variables with VIFs (variance inflation factors) less than 10 [45] and (e) Set 5 (only for C. insignis): variables used for the construction of ENMs of other species of medical importance such as sandflies and culicids [46].
2.5. Ecological Niche Modeling and Model Transfers
The maximum entropy (Maxent v.3.4.4) algorithm was used to model an ecological niche using the kuenm R package v.1.1.9 [40]. We combined four and five sets of environmental variables for BTV and C. insignis, respectively, 17 values of regularization multiplier (0.1–1 with intervals of 0.1; 2–6 with intervals of 1, 8 and 10) and 7 possible combinations of 3 feature classes (linear = l, quadratic = q and product = p) [40]. The candidate model performance and best model selection were based on the significance of the partial receiver operating characteristic (pROC), with 500 iterations and 50% of the data for bootstrapping, prediction ability (omission rate, E = 5%), model fit and complexity AICc (Akaike’s information criteria), delta AICCc and weight AICCc [40].
The final model creation and its evaluation for each species was obtained using 10 replicates via bootstrap with raw outputs; these were transferred from the accessible area “M” to the projection area “G”—Peru. Then, the type of model output for BTV and C. insignis (free extrapolation, extrapolation and clamping, and no extrapolation) was selected based on the two important biological variables (Bio 1: mean annual air temperature and Bio 12: annual precipitation amount); so, they were plotted in a two-dimensional space, along with the occurrences, to determine their position in the environmental space (Figures S3 and S4) [47]. After that, the median of the GCMs (GFDL-ESM4, IPSL-CM6A-LR MPI-ESM1-2-HR, MRI-ESM2-0 and UKESM1-0-LL) was used to reclassify the maps into binaries for generating a consensus of each future climate scenario (SSP126 and SSP585). It was based on a threshold that omitted all regions with habitat suitability below the suitability value, using the 5 % omission error rate threshold (based on the lowest training presence threshold approach), assuming that this value refers to the percentage of data that may include errors misrepresenting the species’ environment [48].
Furthermore, the identification of changes in suitable areas and suitability in projections was calculated and represented by agreement maps of changes (stable, gain and loss); therefore, we compared five GCMs against the current projection and quantified the agreement of the gain and loss of suitable areas, as well as the stability of suitable and un-suitable conditions using the function “kuenm_projchanges” of the kuenm R package v.1.1.9 [40]. Next, the degree of extrapolation (i.e., prediction areas where environmental conditions differ from those represented within the training data [49]) was computed via a multivariate environmental similarity surface (MESS) using the kuenm R package v.1.1.9 [40]. We binarized with 10% of threshold using the average map of each SSP-GCM replicate. These were used to cut their respective maps of BTV or C. insignis in current and future climate scenarios (SSP126 and SSP585). In addition, the final model for BTV in the projection area was evaluated after being created; for this step, independent data from samples of 2021–2022 were used.
2.6. Ecological Niche Overlap
The environmental spaces were visualized using the software Niche Analyst (NicheA) version 3.0 [50], available at http://nichea.sourceforge.net/ (accessed on 20 October 2022). A three-dimensional ellipsoid of environmental distribution was created for BTV and C. insignis in a virtual space of bio-climatic conditions, represented by three principal components (PCAs). The PCAs of the 19 bioclimatic variables from CHELSA V.2.1 were represented by PC1, PC2 and PC3, and we quantified similarity between the niches in terms of overlap using NicheA, whilst also estimating the Jaccard index for each one [51].
2.7. Risk Map Design
We used binary maps of the raw outputs of the best final models cut with MESS and then we classified the data into three categories: (1) low risk, which refers to areas not suitable for either BTV or C. insignis; (2) moderate risk, which means suitable areas only for BTV or C. insignis; and (3) high risk, which refers to suitable areas for both. In addition, the percent of the number of grids was calculated for each category and represented in a pie chart.
3. Results
3.1. Model Calibration Results
We obtained 476 and 595 candidate models for BTV and C. insignis in the calibration area. Then, three and six models were found as the final model candidates for BTV and C. insignis, respectively. Afterward, we selected the best model for each one based on the lowest ∆AICc. Therefore, the models that worked better with the variables in Set 4 were selected for both species (Table 1). The final model validation for BTV in the projection area of the current climate scenario showed a pROC equal to 0 and a mean AUC ratio of 1.32.
Table 1.
Description of metrics of the selected models for each species.
| Species | Occurrence Records | Model Settings | Set of Variables | p-ValuepROC | E% | ΔAICc | Parameters |
|---|---|---|---|---|---|---|---|
| Bluetongue virus | 145 | RM = 0.8; FC = lp | Set 4: Bio 2, Bio 3, Bio 4, Bio 8, Bio 15, Bio 18 and Bio 19 | 0 | 0.17 | 0 | 14 |
| Culicoides insignis | 22 | RM = 0.8; FC = q | Set 4: Bio 2, Bio 3, Bio 4, Bio 8, Bio 13, Bio 15, Bio 18 and Bio 19 | 0 | 0.00 | 0 | 5 |
RM: regularization multiplier; FC: feature classes (linear = l, quadratic = q, product = p); pROC: partial receiver operating characteristic; E%: omission rate at 5%; ΔAICc: delta Akaike information criterion corrected for small sample sizes. Bio 2: mean diurnal air temperature range; Bio 3: isothermality; Bio 4: temperature seasonality; Bio 8: mean daily mean air temperatures of the wettest quarter; Bio 13: precipitation amount of the wettest month; Bio 15: precipitation seasonality; Bio 18: mean monthly precipitation amount of the warmest quarter; Bio 19: mean monthly precipitation amount of the coldest quarter.
3.2. Potential Suitable Areas for Bluetongue Virus and C. insignis under Current and Two Future Climate Scenarios (SSP126 and SSP585)
The current distribution area of BTV in Peru is mostly concentrated in the east and north of the country. The BTV location mainly covers eleven of twenty-four political divisions and the ecoregions that presented suitability for the occurrences were Ucayali moist forests, Eastern Cordillera Real montane forests, Southwest Amazon moist forests, Marañón dry forests, Peruvian Yungas, Sechura Desert, Napo moist forests, Central Andean wet puna and Tumbes–Piura dry forests. In addition, the suitable area in Peru accounts for 77.42% in the current climate scenario (Figure 1A), and it would be decreased to 31.42% (Figure 1B) or 12.57% (Figure 1C) depending on whether the future climate scenarios were SSP126 or SSP585 by 2071–2100, respectively.
Figure 1.
Suitable and unsuitable areas for the potential distribution of bluetongue virus in Peru under (A) the current climate scenario, and (B) the climate change in 2071–2100 in the future climate scenarios of SSP126 and (C) SSP585. Occurrence records (purple points).
Regarding C. insignis, its current distribution mostly covers eastern Peru and eight of the twenty-four political divisions. In the case of the ecoregions, suitability is possibly found in Ucayali moist forests, Eastern Cordillera Real montane forests, Southwest Amazon moist forests, Peruvian Yungas and Marañón dry forests (Figure 2A). Conversely to BTV, the suitable area for C. insignis in the current climate scenario would be increased from 66.33% to 87.02% in the SSP126 future climate scenario, or to 75.80% in the SSP585 by 2071–2100.
Figure 2.
Suitable and unsuitable areas for the potential distribution of Culicoides insignis in Peru under (A) the current climate scenario, and (B) the climate change in 2071–2100 in the future climate scenarios of SSP126 and (C) SSP585. Occurrence records (red points).
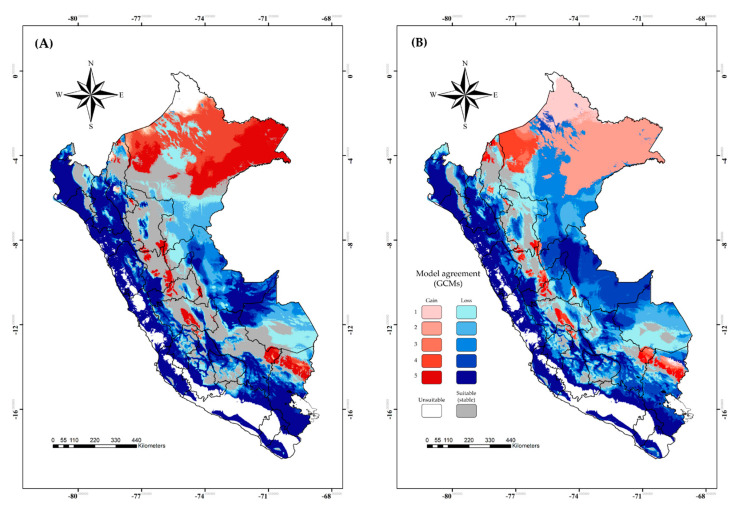
Additionally, future model transfers showed more stable suitable areas for C. insignis (i.e., suitable in current and future climate scenarios) than BTV across the Peruvian east (Figure 3 and Figure 4). The areas of range reduction (loss) were higher in BTV and mainly on the coast and the eastern zone (Figure 3A,B), whereas there was no evidence of a reduction in SSP126 for C. insignis (Figure 4A), but SSP585 showed few reductions in the center of Peru (Figure 4B). A range expansion (gain) was evidenced for C. insignis with high agreement for the five GCMs; however, BTV distribution showed that the gain of suitable areas was concentrated in the northern parts of Peru (Figure 3A,B). In general, we noted greater agreement among the five GCMs in terms of losses for BTV and gains for C. insignis in both SSP126 and SSP585 scenarios.
Figure 3.
Comparison of binary models considering agreement among five GCMs for the potential geographic distribution of bluetongue virus based on the current and future climate in two distinct scenarios: (A) SSP126 and (B) SSP585. A blue scale means an agreement level of five GCMs which are no longer suitable, while a red scale denotes them being newly suitable. Gray color means suitable in the current climate scenario and for all GCMs, while white denotes being unsuitable in the current climate scenario and for all GCMs.
Figure 4.
Comparison of binary models considering agreement between five GCMs for the potential geographic distribution of Culicoides insignis based on the current and future climate in two distinct scenarios: (A) SSP126 and (B) SSP585. A blue scale means an agreement level of five GCMs which are no longer suitable, while a red scale denotes them being newly suitable. Gray color means suitable in the current climate scenario and for all GCMs, while white denotes unsuitable in the current climate scenario and for all GCMs.
3.3. Uncertainty in Current and Future Climate Scenarios
MESS analysis detected environmental areas that were similar and different between model training and model projection data. For BTV, different environmental areas were shown in the middle that represented the Andean highlands of Peru (Figure 1A); however, future climate scenarios (SSP126 and SSP585) showed areas of no extrapolation in the coast and some parts of the eastern area (Figure 1B,C). Regarding C. insignis, MESS results exhibited high agreement in non-extrapolative areas among future scenarios, with most of these areas concentrated on Peru’s coast. In addition, sources of variability results indicated no variation coming from GCMs, SSPs or replicates (Figures S1 and S2), with the only sources of variation documented for C. insignis coming from replicates (Figure S2C).
3.4. Risk Map in Current and Future Climate Scenarios
The BTV risk map in Peru showed that high risk areas are mainly in the east of the country (current climate scenario) and represent the highest coverage of Peruvian territory (Figure 5A). However, the high risk would decrease gradually from current to SSP126 and SPP585 future climate scenarios, except for the northeastern corner of Peru (Figure 5B,C), and it would increase the moderate risk by 2071–2100 in both scenarios. All of the scenarios would show low-risk areas that are lower than the others, as represented in the top of each map as a pie chart (Figure 5).
Figure 5.
Risk map of bluetongue virus transmission by Culicoides insignis in Peru based on current (A) and future climate in two distinct scenarios (B) SSP126 and (C) SSP585. Colors represent different risk levels.
3.5. Ecological Niche Overlap
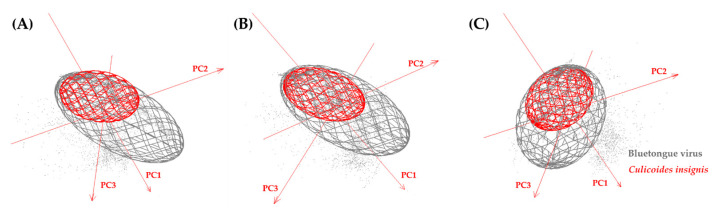
Minimum-volume ellipsoid (MVE) niche estimations showed that the two niches almost overlap in the current climate scenario (Figure 6A) and would completely overlap with one another in future climate scenarios (Figure 6B,C). The MVE reconstructions demonstrate that the niche would undergo an important shift in the SSP585 scenery. Additionally, the degree of overlap using the Jaccard similarity index was generally low. Values were 0.29 and 0.21 in the scenarios SSP585 and SSP126, respectively, and 0.12 in the current climate scenario. Regarding niche amplitudes, BTV had a higher niche compared to C. insignis in all of the scenarios (Figure 6).
Figure 6.
Minimum-volume ellipsoid (MVE) niche estimations for bluetongue virus and Culicoides insignis in three-dimensional environmental space in Peru based on the current (A) and future climate in two distinct scenarios: (B) SSP126 and (C) SSP585. Gray points represent environmental background conditions.
4. Discussion
The species distribution modeling for BTV and C. insignis in the current climate scenario allowed potential areas that mainly cover the east of the country to be identified. Peru is located in the Neotropic and its eastern region is part of the Amazon basin, with the main biome being tropical and subtropical moist broadleaf forest [42], where BTV infection is considered to be endemic because of the presence of its Culicoides vector [1]. This area has a temperature that promotes faster rates of virogenesis with more rapid BTV replication [52,53] and it would also decrease the intervals between feedings, thereby increasing the chances for potential transmission. In addition, egg and larval development of Culicoides spp will be faster and the higher temperatures may shorten the lifespan of the female [23,54,55]. The BTV potential current distribution was also described by Samy and Peterson [31] but the focus was worldwide. Their results were different from our results as they showed that BTV would be present along the Andes region of Peru (center), possibly because they did not use any occurrences from Peru with most of them being from Europe and the United States. Regarding C. insignis, its potential current distribution was evaluated in previous studies that were modeled for northwestern Argentina [56] and Florida [36]. Therefore, this study represents the first potential distribution of C. insignis in the northwest of South America.
Furthermore, the suitable areas for BTV and C. insignis were mainly represented by the ecoregions of Ucayali moist forests and Southwest Amazon moist forests. These ecoregions are part of the rainforest of Peru where the altitude is above 500 masl and it has a tropical climate, with high temperatures and rainfall throughout the year [57]. The average annual rainfall is between 1000 and 3000 mm, with temperatures ranging from 22 °C in the eastern Andes to 31 °C in the Amazon [58]. The Peruvian Amazon is home to numerous Culicoides species, which prefer moist regions, grass and decomposing material such as the remains of banana trees [59]. This part of the planet showed 64% more deforestation in 2020 than in 2019 as registered by Brazilian satellites [60]. This increases the likelihood of environmental degradation, heightens population health risks and causes ecosystem loss on a local, regional and international level. Therefore, these suitable areas should be monitored by SENASA to establish passive surveillance programs for BTV and its vectors.
On the one hand, the suitable area for C. insignis would increase in both future climate scenarios with high agreement among the five models. The Culicoides’ response to climatic variables is different between midge species and midge species groups [61,62]. In addition, the emergence of BTV in Europe has been particularly attributed to climate change because of the range of expansion of C. imicola [23,63,64], and these findings could also be possible due the more intense sampling and the use of more effective traps because of the BTV outbreaks. However, there are no studies of C. insignis adaptation to changing climate, but there is evidence of records located outside of this historical range [36]. In our results, C. insignis would invade the central Andes, where it could find naive populations and cause a massive outbreak and the subsequent death of cattle, similar to that seen for Culicoides obsoletus (Meigen 1918) in the western Palearctic region in 2006–2008 [65]. In addition, Peru is one of the 20 nations that are most susceptible to climate change and its consequences can already be seen in many places [66]. For example, temperature records from 1981 to 2010 showed a warming trend of more than 0.2 °C per decade in the central Andes [67]. Furthermore, C. insignis is a versatile midge; hence, it may utilize more diverse larval development sites including mangrove swamps, sugarcane fields and a variety of streams, springs, ponds and ditches of both natural and human origin [23].
On the other hand, our results showed that a BTV-suitable area would decrease in both future climate scenarios and the MESS analysis cut the coastal zone of Peru despite the presence of occurrence records in the current climate scenario. The coastal zone is represented by the Sechura desert, Peruvian coastal desert and the Atacama desert, which are nearly devoid of rain and below 1000 masl, with a coastal topography that is low and flat [68]. These areas are environmentally different from those where BTV positive cases are frequently observed in Peru [20,21,69]. In addition, the occurrence records were obtained via serological analysis that does not have 100% sensitivity and specificity [70]. Additionally, the occurrence records from the coastal zone could come from migrations as a result of the movement of animals in the country.
The current risk map in Peru showed high overlap between BTV and C. insignis areas. These areas represent endemic spaces of the country for BTV where its presence has been documented at the serological and molecular level [20,21,69]. The high risk would decrease from current to future climate scenarios, except for the northeastern corner of Peru where it would be possible because a new competent vector could emerge similar to the case of C. obsoletus in the northern region of Europe [71]. The moderate risk would remain latent in the future climate conditions and it would cover a larger part of the country. Surveillance programs into C. insignis and the host have to be performed because BTV is a double-stranded RNA virus with 10 segments, so reassortment plays an important role in the overall genetic diversification of BTV [72]. In addition, previous studies have documented that reassortment in some Culicoides was reportedly higher (42% in C. sonorensis) than in vertebrate hosts (5% in sheep) [73,74,75]. Furthermore, we consider the importance of adding a density layer of cattle, goat and sheep to improve the prediction of BTV and C. insignis distribution, as this species is the most abundant and frequently associated with livestock facilities [10]. In addition, it is important to establish more exhaustive sampling and use more effective capture traps for an adequate estimate of the biodiversity of Culicoides associated with livestock.
In the environmental space, BTV and vector populations appear to share similar ecological niches, but BTV has a higher niche compared to C. insignis in all of the scenarios. This might be due to the potential presence of another competent vector. Currently, C. insignis is probably the only competent vector of BTV in South America [76], and it has been recorded predominantly in the eastern part of Peru [39,77]. Although C. pusillus has also been recorded in the country [77], its role as a competent vector of BTV remains unknown [22,26]. In addition, BTV transmission is not only restricted to being vector-borne, as it can also be transmitted by direct contact with the atypical BTV serotypes [78,79]. Moreover, BTV is usually transmitted for more than one species in one continent such as C. imicola and C. obsoletus in Europe [41,80], Culicoides brevitarsis (Kieffer 1917), Culicoides fulvus (Sen and Das Gupta 1959), Culicoides wadai (Kitaoka 1980), C. obsoletus, C. imicola and Culicoides pulicaris (Linnaeus 1758) in Asia [41,80] C. sonorensis, Culicoides stellifer (Coquillett 1901) and C. insignis in North America [23,41,76,81]. For these reasons, medical veterinary entomology has to be developed in Peru and should incorporate the ecological dimension into management and arbovirus/vector control strategies to determine the mechanisms involved in BTV transmission.
5. Conclusions
We presented ecological niche models for BTV and its main vector C. insignis in Peru. This modeling strategy enabled us to evaluate the effect of climate change in BTV and C. insignis distribution for the period 2071–2100, that would remain with a moderate risk (green) in much of Peru. These findings might be used to determine the areas of the highest priority for entomological and virological investigations and surveillance in order to control and prevent bluetongue infections in Peru.
Acknowledgments
The authors sincerely thank “Servicio Nacional de Sanidad Agraria del Perú—SENASA” for their significant contributions to the sample provision.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/v15040892/s1: Figure S1: Model variability for bluetongue virus: median of variance coming from GCMs (A), SSPs (B) and replicates (C) in future climate scenarios; Figure S2: Model variability for Culicoides insignis: median of variance coming from GCMs (A), SSPs (B) and replicates (C) in future climate scenarios; Figure S3: Scatterplot of bluetongue virus in an environmental space. Gray points are the projection area (Peru) while purple circles are used for occurrences in the ENM model; Figure S4: Scatterplot of Culicoides insignis in an environmental space. Gray points are the projection area (Peru) while red circles are used for occurrences in the ENM model; Table S1: Occurrence records of bluetongue virus and Culicoides insignis; Table S2: Set of bioclimatic variables used for construction of the ENM for bluetongue virus; Table S3: Set of bioclimatic variables used for construction of the ENM for Culicoides insignis.
Author Contributions
Conceptualization, D.A.N.M., H.R.G. and M.A.-S.; methodology, D.A.N.M., H.R.H., R.V.B. and M.A.-S.; software, M.A.-S. and D.A.N.M.; validation, D.A.N.M. and M.A.-S.; formal analysis, D.A.N.M., M.R., E.V.G. and W.V.A.; investigation, D.A.N.M. and M.A.-S.; resources, D.A.N.M.; data curation, D.A.N.M., H.R.H. and R.V.B.; writing—original draft preparation, D.A.N.M.; writing—review and editing, M.A.-S., W.A.C. and H.R.G.; visualization, D.A.N.M.; supervision, M.A.-S.; project administration, H.R.G.; funding acquisition, H.R.G. and D.A.N.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used for this study are included in the supplementary information files.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Programa Nacional de Investigación Científica y Estudios Avanzados (PROCIENCIA)—CONCYTEC en el marco de la convocatoria de Proyectos de Investigación Básica 2019-01 [Contrato N° 356-2019-FONDECYT]. And, this research was supported by Universidad Nacional Mayor de San Marcos—RR N° 05753-R-21 and project number A21081401.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.MacLachlan N.J., Mayo C.E., Daniels P.W., Savini G., Zientara S., Gibbs E.P.J. Bluetongue. Rev. Sci. Tech. 2015;34:329–340. doi: 10.20506/rst.34.2.2360. [DOI] [PubMed] [Google Scholar]
- 2.Ries C., Sharav T., Tseren-Ochir E.O., Beer M., Hoffmann B. Putative Novel Serotypes ‘33’ and ‘35’ in Clinically Healthy Small Ruminants in Mongolia Expand the Group of Atypical BTV. Viruses. 2021;13:42. doi: 10.3390/v13010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bumbarov V., Golender N., Jenckel M., Wernike K., Beer M., Khinich E., Zalesky O., Erster O. Characterization of Bluetongue Virus Serotype 28. Transbound. Emerg. Dis. 2020;67:171–182. doi: 10.1111/tbed.13338. [DOI] [PubMed] [Google Scholar]
- 4.MacLachlan N.J. The Pathogenesis and Immunology of Bluetongue Virus Infection of Ruminants. Comp. Immunol. Microbiol. Infect. Dis. 1994;17:197–206. doi: 10.1016/0147-9571(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 5.MacLachlan N.J. Bluetongue: Pathogenesis and Duration of Viraemia. Vet. Ital. 2004;40:462–467. [PubMed] [Google Scholar]
- 6.Coetzee P., van Vuuren M., Venter E.H., Stokstad M. A Review of Experimental Infections with Bluetongue Virus in the Mammalian Host. Virus Res. 2014;182:21–34. doi: 10.1016/j.virusres.2013.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera N.A., Varga C., Ruder M.G., Dorak S.J., Roca A.L., Novakofski J.E., Mateus-Pinilla N.E. Bluetongue and Epizootic Hemorrhagic Disease in the United States of America at the Wildlife-Livestock Interface. Pathogens. 2021;10:915. doi: 10.3390/pathogens10080915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacLachlan N.J., Drew C.P., Darpel K.E., Worwa G. The Pathology and Pathogenesis of Bluetongue. J. Comp. Pathol. 2009;141:1–16. doi: 10.1016/j.jcpa.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Saminathan M., Singh K.P., Khorajiya J.H., Dinesh M., Vineetha S., Maity M., Rahman A.F., Misri J., Malik Y.S., Gupta V.K., et al. An Updated Review on Bluetongue Virus: Epidemiology, Pathobiology, and Advances in Diagnosis and Control with Special Reference to India. Vet. Q. 2020;40:258–321. doi: 10.1080/01652176.2020.1831708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayllón T., Nijhof A.M., Weiher W., Bauer B., Allène X., Clausen P.H. Feeding Behaviour of Culicoides spp. (Diptera: Ceratopogonidae) on Cattle and Sheep in Northeast Germany. Parasit. Vectors. 2014;7:34. doi: 10.1186/1756-3305-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen S.S., Bicout D.J., Calistri P., Canali E., Drewe J.A., Garin-Bastuji B., Gonzales Rojas J.L., Gortázar C., Herskin M., Michel V., et al. Assessment of Listing and Categorisation of Animal Diseases within the Framework of the Animal Health Law (Regulation (EU) No 2016/429): Antimicrobial-Resistant Escherichia Coli in Dogs and Cats, Horses, Swine, Poultry, Cattle, Sheep and Goats. EFSA J. 2022;20:7311. doi: 10.2903/j.efsa.2022.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orange J.P., Dinh E.T.N., Goodfriend O., Citino S.B., Wisely S.M., Blackburn J.K. Evidence of Epizootic Hemorrhagic Disease Virus and Bluetongue Virus Exposure in Nonnative Ruminant Species in Northern Florida. J. Zoo. Wildl. Med. 2021;51:745–751. doi: 10.1638/2019-0174. [DOI] [PubMed] [Google Scholar]
- 13.Conraths F.J., Gethmann J.M., Staubach C., Mettenleiter T.C., Beer M., Hoffmann B. Epidemiology of Bluetongue Virus Serotype 8, Germany. Emerg. Infect. Dis. 2009;15:433–443. doi: 10.3201/eid1503.081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gethmann J., Probst C., Conraths F.J. Economic Impact of a Bluetongue Serotype 8 Epidemic in Germany. Front. Vet. Sci. 2020;7:65. doi: 10.3389/fvets.2020.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legisa D., Santos M.J.D. Bluetongue Virus in South America: Current Status Based on Phylogenetic Analysis. J. Gen. Virol. 2021;102:1–10. doi: 10.1099/jgv.0.001561. [DOI] [PubMed] [Google Scholar]
- 16.Clavijo A., Sepulveda L., Riva J., Pessoa-Silva M., Tailor-Ruthes A., Lopez J.W. Isolation of Bluetongue Virus Serotype 12 from an Outbreak of the Disease in South America. Vet. Rec. 2002;151:301–302. doi: 10.1136/vr.151.10.301. [DOI] [PubMed] [Google Scholar]
- 17.Balaro M.F.A., dos Santos Lima M., del Fava C., de Oliveira G.R., Pituco E.M., Brandão F.Z. Outbreak of Bluetongue Virus Serotype 4 in Dairy Sheep in Rio de Janeiro, Brazil. J. Vet. Diagn. Investig. 2014;26:567–570. doi: 10.1177/1040638714538020. [DOI] [PubMed] [Google Scholar]
- 18.Guimarães L.L.B., Rosa J.C.C., Matos A.C.D., Cruz R.A.S., Guedes M.I.M.C., Dorella F.A., Figueiredo H.C.P., Pavarini S.P., Sonne L., Lobato Z.I.P., et al. Identification of Bluetongue Virus Serotypes 1, 4, and 17 Co-Infections in Sheep Flocks during Outbreaks in Brazil. Res. Vet. Sci. 2017;113:87–93. doi: 10.1016/j.rvsc.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Baldini M.H.M., Rosa J.C.C., Matos A.C.D., Cubas Z.S., Guedes M.I.M.C., de Moraes W., de Oliveira M.J., Felippi D.A., Lobato Z.I.P., de Moraes A.N. Multiple Bluetongue Virus Serotypes Causing Death in Brazilian Dwarf Brocket Deer (Mazama Nana) in Brazil, 2015–2016. Vet. Microbiol. 2018;227:143–147. doi: 10.1016/j.vetmic.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Jurado J., Navarro D., Ramírez M., Santiago M.A., Rivera H. Detection of Antibodies against Bluetongue Virus in Sheep from Two Locations in Junín, Peru. Rev. Inv. Vet. Peru. 2020;31:e17850. doi: 10.15381/rivep.v31i2.17850. [DOI] [Google Scholar]
- 21.Navarro D., Rojas M., Jurado J., Manchego A., Ramírez M., Castillo A., Rivera H. Molecular Detection of Bluetongue Virus in Culicoides insignis and Sheep of Pucallapa, Peru. Rev. Inv. Vet. Peru. 2019;30:465–476. doi: 10.15381/rivep.v30i1.15690. [DOI] [Google Scholar]
- 22.MacLachlan N.J. Bluetongue: History, Global Epidemiology, and Pathogenesis. Prev. Vet. Med. 2011;102:107–111. doi: 10.1016/j.prevetmed.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Purse B.V., Carpenter S., Venter G.J., Bellis G., Mullens B.A. Bionomics of Temperate and Tropical Culicoides Midges: Knowledge Gaps and Consequences for Transmission of Culicoides-Borne Viruses. Annu. Rev. Entomol. 2015;60:373–392. doi: 10.1146/annurev-ento-010814-020614. [DOI] [PubMed] [Google Scholar]
- 24.Vigil S.L., Ruder M.G., Shaw D., Wlodkowski J., Garrett K., Walter M., Corn J.L. Apparent Range Expansion of Culicoides (Hoffmania) insignis (Diptera: Ceratopogonidae) in the Southeastern United States. J. Med. Entomol. 2018;55:1043–1046. doi: 10.1093/jme/tjy036. [DOI] [PubMed] [Google Scholar]
- 25.Mo C.L., Thompson L.H., Homan E.J., Oviedo M.T., Greiner E.C., González J., Sáenz M.R. Bluetongue Virus Isolations from Vectors and Ruminants in Central America and the Caribbean. Interamerican Bluetongue Team. Am. J. Vet. Res. 1994;55:211–215. [PubMed] [Google Scholar]
- 26.Ronderos M.M., Greco N.M., Spinelli G.R. Diversity of Biting Midges of the Genus Culicoides Latreille (Diptera: Ceratopogonidae) in the Area of the Yacyretá Dam Lake between Argentina and Paraguay. Mem. Inst. Oswaldo Cruz. 2003;98:19–24. doi: 10.1590/S0074-02762003000100003. [DOI] [PubMed] [Google Scholar]
- 27.Erram D., Blosser E.M., Burkett-Cadena N. Habitat Associations of Culicoides Species (Diptera: Ceratopogonidae) Abundant on a Commercial Cervid Farm in Florida, USA. Parasit. Vectors. 2019;12:367. doi: 10.1186/s13071-019-3626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González M.A., Bravo-Barriga D., Rodríguez-Sosa M.A., Rueda J., Frontera E., Alarcón-Elbal P.M. Species Diversity, Habitat Distribution, and Blood Meal Analysis of Haematophagous Dipterans Collected by CDC-UV Light Traps in the Dominican Republic. Pathogens. 2022;11:714. doi: 10.3390/pathogens11070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrasco D., Felippe-Bauer M.L., Dumont L.F., D’incao F. Abundance of Culicoides (Diptera, Ceratopogonidae) Species in Salt Marshes of the Patos Lagoon Estuary, Rio Grande Do Sul, Brazil: Influence of Climatic Variables. Panam. J. Aquat. Sci. 2014;9:8–20. [Google Scholar]
- 30.Escobar L.E. Ecological Niche Modeling: An Introduction for Veterinarians and Epidemiologists. Front. Vet. Sci. 2020;7:519059. doi: 10.3389/fvets.2020.519059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samy A.M., Peterson A.T. Climate Change Influences on the Global Potential Distribution of Bluetongue Virus. PLoS ONE. 2016;11:e0150489. doi: 10.1371/journal.pone.0150489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escobar L.E., Peterson A.T. Spatial Epidemiology of Bat-Borne Rabies in Colombia. Rev. Panam. Salud Publica. 2013;34:135–136. [PubMed] [Google Scholar]
- 33.Soberón J., Nakamura M. Niches and Distributional Areas: Concepts, Methods, and Assumptions. Proc. Natl. Acad. Sci. USA. 2009;106:19644–19650. doi: 10.1073/pnas.0901637106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson E.E., Escobar L.E., Zambrana-Torrelio C. An Ecological Framework for Modeling the Geography of Disease Transmission. Trends Ecol. Evol. 2019;34:655–668. doi: 10.1016/j.tree.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J., Gao X., Liu B., Chen H., Xiao J., Wang H. Epidemiology and Spatial Distribution of Bluetongue Virus in Xinjiang, China. PeerJ. 2019;7:e6514. doi: 10.7717/peerj.6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloyer K.E., Burkett-Cadena N.D., Yang A., Corn J.L., Vigil S.L., McGregor B.L., Wisely S.M., Blackburn J.K. Ecological Niche Modeling the Potential Geographic Distribution of Four Culicoides Species of Veterinary Significance in Florida, USA. PLoS ONE. 2019;14:e0206648. doi: 10.1371/journal.pone.0206648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thameur B.H., Soufiène S., Ammar H.H., Hammami S. Spatial Distribution and Habitat Selection of Culicoides imicola: The Potential Vector of Bluetongue Virus in Tunisia. Onderstepoort J. Vet. Res. 2021;88:1861. doi: 10.4102/ojvr.v88i1.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guichard S., Guis H.L.N., Tran A., Garros C., Balenghien T., Kriticos D.J. Worldwide Niche and Future Potential Distribution of Culicoides imicola, a Major Vector of Bluetongue and African Horse Sickness Viruses. PLoS ONE. 2014;9:e0119323. doi: 10.1371/journal.pone.0112491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navarro D., Rivera H., Caceres A., Rondón J. Morphological Identification of Culicoides spp. Described as Orbivirus Transmitters Trapped in Sheep Farms in Pucallpa, Peru. Rev. Inv. Vet. Peru. 2018;29:302–309. doi: 10.15381/rivep.v29i1.14203. [DOI] [Google Scholar]
- 40.Cobos M.E., Peterson A.T., Barve N., Osorio-Olvera L. Kuenm: An R Package for Detailed Development of Ecological Niche Models Using Maxent. PeerJ. 2019;7:e6281. doi: 10.7717/peerj.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellor P.S. The replication of bluetongue virus in Culicoides vectors. Curr. Top. Microbiol. Inmmunol. 1990;162:143–161. doi: 10.1007/978-3-642-75247-6_6. [DOI] [PubMed] [Google Scholar]
- 42.Olson D.M., Dinerstein E., Wikramanayake E.D., Burgess N.D., Powell G.V.N., Underwood E.C., D’Amico J.A., Itoua I., Strand H.E., Morrison J.C., et al. Terrestrial ecoregions of the world: A new map of life on Earth. Bioscience. 2001;51:933–938. doi: 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2. [DOI] [Google Scholar]
- 43.Karger D.N., Conrad O., Böhner J., Kawohl T., Kreft H., Soria-Auza R.W., Zimmermann N.E., Linder H.P., Kessler M. Climatologies at High Resolution for the Earth’s Land Surface Areas. Sci. Data. 2017;4:170122. doi: 10.1038/sdata.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riahi K., van Vuuren D.P., Kriegler E., Edmonds J., O’Neill B.C., Fujimori S., Bauer N., Calvin K., Dellink R., Fricko O., et al. The Shared Socioeconomic Pathways and Their Energy, Land Use, and Greenhouse Gas Emissions Implications: An Overview. Glob. Environ. Chang. 2017;42:153–168. doi: 10.1016/j.gloenvcha.2016.05.009. [DOI] [Google Scholar]
- 45.Estrada-Peña A., Estrada-Sánchez A., Estrada-Sánchez D., de la Fuente J. Assessing the Effects of Variables and Background Selection on the Capture of the Tick Climate Niche. Int. J. Health Geogr. 2013;12:43. doi: 10.1186/1476-072X-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moo-Llanes D., Ibarra-Cerdeña C.N., Rebollar-Téllez E.A., Ibáñez-Bernal S., González C., Ramsey J.M. Current and Future Niche of North and Central American Sand Flies (Diptera: Psychodidae) in Climate Change Scenarios. PLoS Negl. Trop. Dis. 2013;7:e2421. doi: 10.1371/journal.pntd.0002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owens H.L., Campbell L.P., Dornak L.L., Saupe E.E., Barve N., Soberón J., Ingenloff K., Lira-Noriega A., Hensz C.M., Myers C.E., et al. Constraints on Interpretation of Ecological Niche Models by Limited Environmental Ranges on Calibration Areas. Ecol. Modell. 2013;263:10–18. doi: 10.1016/j.ecolmodel.2013.04.011. [DOI] [Google Scholar]
- 48.Peterson A.T., Campbell L.P., Moo-Llanes D.A., Travi B., González C., Ferro M.C., Ferreira G.E.M., Brandão-Filho S.P., Cupolillo E., Ramsey J., et al. Influences of Climate Change on the Potential Distribution of Lutzomyia longipalpis Sensu Lato (Psychodidae: Phlebotominae) Int. J. Parasitol. 2017;47:667–674. doi: 10.1016/j.ijpara.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Elith J., Kearney M., Phillips S. The Art of Modelling Range-Shifting Species. Methods Ecol. Evol. 2010;1:330–342. doi: 10.1111/j.2041-210X.2010.00036.x. [DOI] [Google Scholar]
- 50.Qiao H., Peterson A.T., Campbell L.P., Soberón J., Ji L., Escobar L.E. NicheA: Creating Virtual Species and Ecological Niches in Multivariate Environmental Scenarios. Ecography. 2016;39:805–813. doi: 10.1111/ecog.01961. [DOI] [Google Scholar]
- 51.Mammola S. Assessing Similarity of N-Dimensional Hypervolumes: Which Metric to Use? J. Biogeogr. 2019;46:2012–2023. doi: 10.1111/jbi.13618. [DOI] [Google Scholar]
- 52.Kopanke J., Lee J., Stenglein M., Carpenter M., Cohnstaedt L.W., Wilson W.C., Mayo C. Exposure of Culicoides sonorensis to Enzootic Strains of Bluetongue Virus Demonstrates Temperature-and Virus-Specific Effects on Virogenesis. Viruses. 2021;13:1016. doi: 10.3390/v13061016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullens B.A., Gerry A.C., Lysyk T.J., Schmidtmann E.T. Environmental Effects on Vector Competence and Virogenesis of Bluetongue Virus in Culicoides: Interpreting Laboratory Data in a Field Context. Vet. Ital. 2004;40:160–166. [PubMed] [Google Scholar]
- 54.Mayo C., McDermott E., Kopanke J., Stenglein M., Lee J., Mathiason C., Carpenter M., Reed K., Perkins T.A. Ecological Dynamics Impacting Bluetongue Virus Transmission in North America. Front. Vet. Sci. 2020;7:186. doi: 10.3389/fvets.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veronesi E., Venter G.J., Labuschagne K., Mellor P.S., Carpenter S. Life-History Parameters of Culicoides (Avaritia) imicola Kieffer in the Laboratory at Different Rearing Temperatures. Vet. Parasitol. 2009;163:370–373. doi: 10.1016/j.vetpar.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 56.Aybar C.A.V., Juri M.J.D., de Grosso M.S.L., Spinelli G.R. Spatial and Temporal Distribution of Culicoides insignis and Culicoides paraensis in the Subtropical Mountain Forest of Tucumn, Northwestern Argentina. Fla. Entomol. 2011;94:1018–1025. doi: 10.1653/024.094.0440. [DOI] [Google Scholar]
- 57.Brack A., Mendiola C. Ecología del Perú. 2nd ed. Publisher; Bruño, Peru: 2004. pp. 83–254. [Google Scholar]
- 58.Climate Change Knowledge Portal for Develolpment Practitionersand Policy Makers. [(accessed on 2 January 2023)]. Available online: https://climateknowledgeportal.worldbank.org/country/peru/climate-data-historical.
- 59.Mercer D.R., Spinelli G.R., Watts D.M., Tesh R.B. Biting Rates and Developmental Substrates for Biting Midges (Diptera: Ceratopogonidae) in Iquitos, Peru. J. Med. Entomol. 2003;40:807–812. doi: 10.1603/0022-2585-40.6.807. [DOI] [PubMed] [Google Scholar]
- 60.Lowe R., Lee S., Martins Lana R., Torres Codeço C., Castro M.C., Pascual M. Emerging Arboviruses in the Urbanized Amazon Rainforest. BMJ. 2020;371:m4385. doi: 10.1136/bmj.m4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kluiters G., Sugden D., Guis H., McIntyre K.M., Labuschagne K., Vilar M.J., Baylis M. Modelling the Spatial Distribution of Culicoides Biting Midges at the Local Scale. J. Appl. Ecol. 2013;50:232–242. doi: 10.1111/1365-2664.12030. [DOI] [Google Scholar]
- 62.Calvete C., Estrada R., Miranda M.A., Borrás D., Calvo J.H., Lucientes J. Ecological Correlates of Bluetongue Virus in Spain: Predicted Spatial Occurrence and Its Relationship with the Observed Abundance of the Potential Culicoides spp. Vector. Vet. J. 2009;182:235–243. doi: 10.1016/j.tvjl.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Guis H., Caminade C., Calvete C., Morse A.P., Tran A., Baylis M. Modelling the Effects of Past and Future Climate on the Risk of Bluetongue Emergence in Europe. J. R. Soc. Interface. 2012;9:339–350. doi: 10.1098/rsif.2011.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elbers A.R.W., Koenraadt C.J.M., Meiswinkel R. Mosquitoes and Culicoides Biting Midges: Vector Range and the Influence of Climate Change. Rev. Sci. Tech. 2015;34:123–137. doi: 10.20506/rst.34.1.2349. [DOI] [PubMed] [Google Scholar]
- 65.Carpenter S., Groschup M.H., Garros C., Felippe-Bauer M.L., Purse B.V. Culicoides Biting Midges, Arboviruses and Public Health in Europe. Antiviral Res. 2013;100:102–113. doi: 10.1016/j.antiviral.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 66.Altea L. Perceptions of Climate Change and Its Impacts: A Comparison between Farmers and Institutions in the Amazonas Region of Peru. Clim. Dev. 2020;12:134–146. doi: 10.1080/17565529.2019.1605285. [DOI] [Google Scholar]
- 67.Vuille M., Franquist E., Garreaud R., Lavado Casimiro W.S., Cáceres B. Impact of the Global Warming Hiatus on Andean Temperature. J. Geophys. Res. 2015;120:3745–3757. doi: 10.1002/2015JD023126. [DOI] [Google Scholar]
- 68.Philip W., Rundel P.W., Dillon M.O. Ecological Patterns in the Bromeliaceae of the Lomas Formations of Coastal Chile. Plant Syst. Evol. 1998;212:261–278. [Google Scholar]
- 69.Escano J., Navarro D., Jurado J., Ara M., Mantilla J., Ramírez M., Rivera H. Seroprevalencia del Virus de Lengua Azul en Cabras (Capra hircus) de la Región Norte Del Perú. Rev. Inv. Vet. Perú. 2022;33:e24096. doi: 10.15381/rivep.v33i6.24096. [DOI] [Google Scholar]
- 70.Vandenbussche F., Vanbinst T., Verheyden B., van Dessel W., Demeestere L., Houdart P., Bertels G., Praet N., Berkvens D., Mintiens K., et al. Evaluation of Antibody-ELISA and Real-Time RT-PCR for the Diagnosis and Profiling of Bluetongue Virus Serotype 8 during the Epidemic in Belgium in 2006. Vet. Microbiol. 2008;129:15–27. doi: 10.1016/j.vetmic.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 71.Meiswinkel R., van Rijn P., Leijs P., Goffredo M. Potential new Culicoides vector of bluetongue virus in northern Europe. Vet. Rec. 2007;161:564–565. doi: 10.1136/vr.161.16.564. [DOI] [PubMed] [Google Scholar]
- 72.van den Bergh C., Coetzee P., Venter E.H. Reassortment of Bluetongue Virus Vaccine Serotypes in Cattle. J. S. Afr. Vet. Assoc. 2018;89:1649. doi: 10.4102/jsava.v89i0.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaw A.E., Ratinier M., Nunes S.F., Nomikou K., Caporale M., Golder M., Allan K., Hamers C., Hudelet P., Zientara S., et al. Reassortment between Two Serologically Unrelated Bluetongue Virus Strains Is Flexible and Can Involve Any Genome Segment. J. Virol. 2013;87:543–557. doi: 10.1128/JVI.02266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Batten C.A., Maan S., Shaw A.E., Maan N.S., Mertens P.P.C. A European Field Strain of Bluetongue Virus Derived from Two Parental Vaccine Strains by Genome Segment Reassortment. Virus Res. 2008;137:56–63. doi: 10.1016/j.virusres.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 75.Dal Pozzo F., Martinelle L., Thys C., Sarradin P., de Leeuw I., van Campe W., de Clercq K., Thiry E., Saegerman C. Experimental Co-Infections of Calves with Bluetongue Virus Serotypes 1 and 8. Vet. Microbiol. 2013;165:167–172. doi: 10.1016/j.vetmic.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 76.Tanya V.N., Greiner E.C., Gibbs E.P.J. Evaluation of Culicoides insignis (Diptera: Ceratopogonidae) as a Vector of Bluetongue Virus. Vet. Microbiol. 1992;32:1–14. doi: 10.1016/0378-1135(92)90002-B. [DOI] [PubMed] [Google Scholar]
- 77.Felippe-Bauer L.M., Cáceres A., Santos da Silva C., Valderrama-Bazan W., Gonzales-Perez A., Martins Costa J. New Records of Culicoides Latreille (Diptera: Ceratopogonidae) from Peruvian Amazonian Region. Biota. Neotrop. 2008;8:33–38. doi: 10.1590/S1676-06032008000200002. [DOI] [Google Scholar]
- 78.Batten C., Darpel K., Henstock M., Fay P., Veronesi E., Gubbins S., Graves S., Frost L., Oura C. Evidence for Transmission of Bluetongue Virus Serotype 26 through Direct Contact. PLoS ONE. 2014;9:e96049. doi: 10.1371/journal.pone.0096049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van der Sluijs M.T.W., de Smit A.J., Moormann R.J.M. Vector Independent Transmission of the Vector-Borne Bluetongue Virus. Crit. Rev. Microbiol. 2016;42:57–64. doi: 10.3109/1040841X.2013.879850. [DOI] [PubMed] [Google Scholar]
- 80.Sick F., Beer M., Kampen H., Wernike K. Culicoides Biting Midges—Underestimated Vectors for Arboviruses of Public Health and Veterinary Importance. Viruses. 2019;11:376. doi: 10.3390/v11040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGregor B.L., Shults P.T., McDermott E.G. A Review of the Vector Status of North American Culicoides (Diptera: Ceratopogonidae) for Bluetongue Virus, Epizootic Hemorrhagic Disease Virus, and Other Arboviruses of Concern. Curr. Trop. Med. Rep. 2022;9:130–139. doi: 10.1007/s40475-022-00263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used for this study are included in the supplementary information files.