Abstract
In vitro enzyme-based ATP regeneration systems are important for improving yields of ATP-dependent enzymatic reactions for preparative organic synthesis and biocatalysis. Several enzymatic ATP regeneration systems have been described but have some disadvantages. We report here on the use of polyphosphate:AMP phosphotransferase (PPT) from Acinetobacter johnsonii strain 210A in an ATP regeneration system based on the use of polyphosphate (polyP) and AMP as substrates. We have examined the substrate specificity of PPT and demonstrated ATP regeneration from AMP and polyP using firefly luciferase and hexokinase as model ATP-requiring enzymes. PPT catalyzes the reaction polyPn + AMP → ADP + polyPn−1. The ADP can be converted to ATP by adenylate kinase (AdK). Substrate specificity with nucleoside and 2′-deoxynucleoside monophosphates was examined using partially purified PPT by measuring the formation of nucleoside diphosphates with high-pressure liquid chromatography. AMP and 2′-dAMP were efficiently phosphorylated to ADP and 2′-dADP, respectively. GMP, UMP, CMP, and IMP were not converted to the corresponding diphosphates at significant rates. Sufficient AdK and PPT activity in A. johnsonii 210A cell extract allowed demonstration of polyP-dependent ATP regeneration using a firefly luciferase-based ATP assay. Bioluminescence from the luciferase reaction, which normally decays very rapidly, was sustained in the presence of A. johnsonii 210A cell extract, MgCl2, polyPn=35, and AMP. Similar reaction mixtures containing strain 210A cell extract or partially purified PPT, polyP, AMP, glucose, and hexokinase formed glucose 6-phosphate. The results indicate that PPT from A. johnsonii is specific for AMP and 2′-dAMP and catalyzes a key reaction in the cell-free regeneration of ATP from AMP and polyP. The PPT/AdK system provides an alternative to existing enzymatic ATP regeneration systems in which phosphoenolpyruvate and acetylphosphate serve as phosphoryl donors and has the advantage that AMP and polyP are stabile, inexpensive substrates.
Enzyme-catalyzed phosphoryl transfer reactions (i.e., those which form or cleave P—O bonds) represent a viable alternative to multistep chemical phosphorylations in preparative organic synthesis (7, 10, 20). Many useful phosphorylating enzymes require nucleoside triphosphates as cofactors. ATP is the most important biological phosphate donor and is a required cofactor for numerous enzymatic reactions in both anabolic and catabolic metabolism, specifically in the formation of P—O bonds. Cofactor provision for ATP-dependent enzymes can be accomplished by their direct addition in stoichiometric amounts or by the inclusion of a cofactor regeneration system. Direct cofactor addition is not only costly but can unfavorably alter reaction equilibrium, lead to an accumulation of inhibitory cofactor by-products, and complicate recovery of end products (20). For preparative-scale organic synthesis, these problems have been overcome by the development of enzymatic systems for ATP regeneration.
Whitesides et al. and others have described several enzymatic systems for the regeneration of ATP from ADP (4). The two most common systems are based on acetyl phosphate/acetate kinase (AcP/AcK) and phosphoenolpyruvate/pyruvate kinase (PEP/PK). Both systems involve the transfer of a phosphoryl group from a high-energy donor to ADP but have characteristics (discussed later) that favor particular applications. Both AcK and PK have broad substrate specificities, allowing the regeneration of other nucleoside or 2′-deoxynucleoside triphosphates (i.e., GTP, UTP, CTP, and 2′-dATP) (20). Disadvantages of both systems relate to the stability, expense, or requisite chemical syntheses (for large-scale application) of the high-energy phosphoryl donors and product inhibition, in the case of PK, by pyruvate. Despite minor drawbacks, these enzymatic ATP regeneration systems have facilitated the study of numerous kinase and synthetase reactions and led to improved chemoenzymatic synthesis of valuable phosphorylated compounds (5, 6, 9, 11, 12, 19, 21).
ATP regeneration from ADP using polyphosphate/polyphosphate kinase (polyP/PPK) has also been demonstrated (13) and was recently applied in a practical synthesis of the oligosaccharide, N-acetyllactosamine (14). The latter work showed PPK from Escherichia coli to phosphorylate not only ADP but also other nucleoside diphosphates to their corresponding triphosphates using polyP as a phosphoryl donor. Although PPK is not yet commercially available, the low cost of production and high stability of polyP make it an attractive alternative to the phosphoryl donors in the systems mentioned above (3).
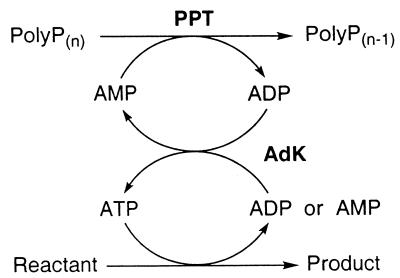
While each system described above recycles ATP from ADP, many ATP-dependent enzymes generate AMP rather than ADP and some produce adenine. The regeneration of ATP from AMP can be achieved by including adenylate kinase (AdK), while ATP regeneration from adenosine requires inclusion of both adenosine kinase and AdK in the above regenration systems (7, 11). Thus, efficient systems for the enzymatic regeneration of ATP from AMP could benefit these scenarios. The focus of the present work was to examine the potential of polyphosphate:AMP phosphotransferase (PPT) from Acinetobacter johnsonii strain 210A (1) for in vitro ATP regeneration from polyP and AMP (16). PPT catalyzes the reaction polyPn + AMP → ADP + polyPn − 1, and the ADP can be converted to ATP by AdK (or PPK) (Fig. 1). The substrate specificity of PPT with other nucleoside and 2′-deoxynucleoside monophosphates was determined with partially purified preparations and the ability of PPT/AdK to regenerate ATP from AMP using polyP as a phosphoryl donor was demonstrated with luciferase and hexokinase as model ATP-dependent enzymes.
FIG. 1.
General scheme showing enzymatic ATP regeneration from AMP and polyP by the PPT/AdK system. Note that AMP released by certain ATP-dependent enzymes requires the activity of both PPT and AdK to regenerate ATP.
MATERIALS AND METHODS
Strain cultivation and fermentation conditions.
A. johnsonii strain 210A was grown in the basic medium with trace metals (BM) described by van Groenestijn (17) with 5.67 g of sodium acetate trihydrate per liter as a carbon source. Sufficient biomass for initial studies was obtained by culturing strain 210A in batch mode in a 30-liter fermentor vessel containing 25 liters of BM with butyrate (2.29 g of butyric acid per liter [adjusted to pH 7.0]) and inoculated (3% vol/vol) with an acetate-grown preculture. The culture was grown at 25°C with agitation at 230 rpm, 15 liters of air/min (pH 7.0), and a maintained pO2 of >80%. After 22 h of growth, the culture was cooled (10°C), stirred at 120 rpm with 10 liters of air/min, and harvested by centrifugation to give a 120-g wet-cell weight. In order to identify carbon sources which lead to both high cell yields and PPT activity, strain 210A was grown in 200-ml batch cultures (30°C, 200 rpm) on BM containing 20 mM acetate, crotonate, or dl-lactate. Cells were also grown in complex Luria-Bertani (LB) medium alone and LB medium supplemented with 40 mM dl-lactate or 0.8 g of potassium phosphate per liter. Large-scale cultivation of strain 210A was in a 1-m3 fermentor containing 700 liters of BM with 75 mM dl-lactate (pH 7.2) and inoculation with a 24-liter preculture grown for 22 h. The culture was grown at 25°C with agitation at 100 to 150 rpm for 15 h (A546 = 11.2), cooled to 18°C, and harvested using a Sharples continuous centrifuge. The final cell yield was approximately 11.5 g (wet weight) per liter. Culture purity was assessed by plating to rich (LB) medium and by microscopic examination at each stage of fermentation. Cells were frozen at −80°C until use.
Preparation of cell extracts and a PPT active fraction.
Strain 210A cells were suspended 1:1 (wt/vol) in breakage buffer (50 mM Tris-HCl, 4 mM EDTA, pH 6.8) containing 0.5 mM Perfabloc (AEBSF; Boehringer Mannheim) and DNase I (Fluka Chemie AG) at 50 μg/ml. Cells were disrupted by passage of the suspension twice through a chilled French pressure cell at 20,000 lb/in2. The cell homogenate was centrifuged at 18,000 × g for 1 h to remove cell debris, and the resulting crude extract was either frozen at −70°C or immediately ultracentrifuged at 150,000 × g (4°C, 1.5 h). The soluble fraction, designated the high-speed supernatant (HSS), was further fractionated. Ammonium sulfate (AS) precipitation was done by addition of a saturated solution and/or finely ground solid crystalline material to the HSS with stirring on a 4°C ice bath. AS cuts were made at 30, 40, 50, 60, and 75% saturation (percentages assume additive volume), and precipitates were resuspended in 0.5 volume of 0.05 M Tris-HCl (pH 7.5). These solutions were used directly in the experiments described here or concentrated and desalted using a stirred ultrafiltration cell equipped with a YM30 regenerated-cellulose membrane (Amicon bioseparations; Millipore Corp., Bedford, Mass.).
Assays.
PPT was measured spectrophotometrically essentially as described by Bonting et al. (method A) (1), except that the assay buffer was 50 mM Tris-HCl (pH 7.6). The 1.0-ml assay mixtures contained 8 mM MgCl2, 5 mM glucose, 0.4 mM NADP+, 2 U of yeast hexokinase (HK), 1 U of glucose 6-phosphate (G6P) dehydrogenase, 1 U of AdK, 0.2 mg of polyPn = 35, and 1 mM AMP (added to initiate the reaction). The reduction of NADP+ by G6P dehydrogenase was monitored at 340 nm (Jasco V-550) at room temperature (22 ± 2°C). As noted previously, the initial rate of NADP+ reduction in this coupled-enzyme assay is nonlinear and usually required at least 5 min to reach linear rates corresponding to ADP formation (16). Since the assay required 0.5 to 1.0 mg of crude cell extract protein for measurable activity, more-sensitive luciferase-based assays of PPT activity were also used to monitor enzyme activity in a microtiter plate (MTP) format.
PPT activity was measured by two methods with firefly luciferase-based ATP Bioluminescence Assays (CLSII and HSII Kits; Boehringer Mannheim) following enzymatic conversion of ADP to ATP. In the first method (assay 1), 1.0-ml reaction mixtures contained, in 50 mM Tris-HCl (pH 7.6), 3 mM MgCl2, 0.2 mg of polyPn = 35, cell extract, and 2 mM AMP (added to initiate the reaction). The mixtures were incubated at room temperature, and at predetermined time intervals (10 to 30 min), subsamples (typically 50 μl) were added to 5 ml of breakage buffer and boiled for 5 min. These solutions were treated with 2 mM PEP and 1 U of PK per ml for 30 min at 35°C. The concentration of ATP, which is directly proportional to the amount of ADP formed by PPT, was measured in MTP wells containing 50 μl each of assay sample (or ATP standard) and CLSII luciferase reagent using a MicroLumat LB-96P Luminometer (EG&G Berthold).
The second method (assay 2) was developed to minimize sample manipulation steps, increase sensitivity, and provide a rapid in vitro assay of ATP regeneration activity in real time. This method takes advantage of the natural decay kinetics of bioluminescence in the HSII ATP Assay Kit (1 and 10 μM ATP standards show >97% and >93% relative light unit [RLU] decay by 30 min). Complete reaction mixtures (0.1 ml) were set up on ice and contained MgCl2, polyPn = 35, and AMP in Tris-HCl (pH 7.6) at the concentrations listed for assay 1 and AdK at 1 U/ml. Activity was measured immediately in MTP wells as described above, except that the HSII ATP Assay Kit luciferase reagent was used. Assay constituents were systematically eliminated or serially diluted to experimentally determine required components.
To determine whether AdK or PPK is responsible for converting ADP to ATP in strain 210A, 0.1-ml reaction mixtures were set up with ADP as the substrate using strain 210A crude cell extract, the HSS, and the AS 50% cut and containing the concentrations of polyPn = 35, MgCl2, and Tris-HCl listed for assay 1. Reactions were set up with and without polyP to assess the contributions of AdK and PPK (polyP dependent) to the formation of ATP, which was measured over a 3-h period using the HSII ATP Assay Kit luciferase reagent.
Protein concentrations were estimated by the method of Bradford (2), using protein dye reagent from Bio-Rad, and are relative to the standard, bovine serum albumin.
HK coupled to the PPT/AdK system for ATP regeneration.
HK reaction mixtures with crude cell extract contained the following components (where indicated; see Table 3) in a final volume of 1.0 ml: 2 U of yeast HK, 50 mM glucose, 3 mM MgCl2, 5 mM AMP or ATP, 0.5 mg of polyPn = 35, 1 U of AdK, and 84 mU of PPT in 50 mM Tris-HCl (pH 7.5). Rates of G6P formation were measured in 1.0-ml reaction mixtures containing 5 U of HK, 6 mM MgCl2, 100 mM glucose, 1 U of AdK, 1 mg of polyPn = 35, 5 mM AMP, and different concentrations of PPT from a 40 to 60% AS cut (concentrated by ultrafiltration; 13.6 mg of protein/ml) in 50 mM Tris-HCl (pH 7.5). In control reaction mixtures the AMP, polyP, and PPT were omitted and 5 mM ATP was provided with and without AdK. Reaction mixtures were incubated at 30°C with horizontal shaking (175 rpm). At predetermined intervals, aliquots of the reaction mixtures were boiled for 5 min to inactivate enzymes and centrifuged for 10 min (14,000 rpm, Microfuge) and the supernatants were retained for G6P analysis. G6P concentrations were measured by monitoring the G6P dehydrogenase-coupled reduction endpoint of NADP+ at 340 nm with a UV/Vis spectrophotometer. The 1.0-ml assay mixtures contained 6 mM MgCl2, 40 mM NADP+, and 0.5 U of G6P dehydrogenase in Tris-HCl (pH 7.6).
TABLE 3.
HK-catalyzed formation of G6P with and without ATP regeneration by PPT/AdK from strain 210A cell extract
| Reaction componentsa | G6P concn (mM)b |
|---|---|
| ATP (control) | 5.8 |
| CE, AdK, polyP, AMP | 5.0 |
| CE, polyP, AMP | 4.5 |
| CE, AdK, polyP, ATP | 11.7 |
| CE, AdK, AMP | 0.5 |
In addition to HK, Mg+2, and glucose. CE, cell extract.
G6P was measured as described in Materials and Methods after incubation of HK reaction mixtures for 18 h at 30°C with shaking (150 rpm).
Substrate specificity.
The ability of PPT to phosphorylate AMP and several related nucleoside phosphates was examined in reaction mixtures (1.0 ml) containing in Tris-HCl (pH 7.6), 3 mM MgCl2, 0.4 mg of polyPn = 35, 0.5 mM nucleoside monophosphate (AMP, 2′-dAMP, GMP, CMP, UMP, and IMP), and 0.23 mg of protein from a 40 to 50% AS cut with PPT activity. Reactions were also conducted with 1.0 mM AMP and 2′-dAMP. At predetermined intervals, 0.1-ml subsamples were boiled for 5 min, diluted 1:10 in the high-pressure liquid chromatography (HPLC) mobile phase, and filtered (0.45-μm pore size). Sample analysis was performed on a Gynkotek HPLC system (M480G pump, Gina 50T autosampler, and UVD340S photodiode array detector; Gynkotek GmbH) equipped with a Nucleosil 100-5 C18 column (250 by 4.6 mm [inside diameter]; Macherey-Nagel). An isocratic mobile phase of 0.08 M sodium dihydrogen phosphate in methanol (77:23, vol/vol) with 5 mM tetrabutylammonium hydrogen sulfate (pH 6.0) operated at a flow rate of 0.7 ml/min, and compounds were detected at 260 nm. Under these conditions, the retention times of the nucleoside mono-, di-, or triphosphates were as follows: AMP, ADP, and ATP, 5.5, 7.2, and 10.4 min; 2′-dAMP, 2′-dADP, and 2′-dATP, 6.5, 8.9, and 12.3 min; GMP and GDP, 4.4 and 5.6 min; CMP and CDP, 4.2 and 5.3 min; IMP and IDP, 4.4 and 5.5 min; UMP and UDP, 4.4 and 5.7 min. Concentrations of analytes were determined from standard curves prepared using authentic compounds.
RESULTS
Expression of PPT.
To determine suitable conditions for large-scale cultivation of strain 210A and expression of PPT, the specific activity in crude cell extracts was measured using the spectrophotometric assay after growth on different substrates (200-ml scale). The specific activity of PPT in cells cultivated on butyrate was 68 nmol/min/mg of protein, which agreed well with the activity reported previously for butyrate-grown cells of strain 210A (1). Almost identical PPT activities were observed in cells grown on crotonate, acetate, dl-lactate, and LB medium (Table 1). The activity of PPT was stable, as no difference was observed in LB medium-grown cultures assayed at 19 and 31 h. The determination of conditions allowing both high cell yield and PPT activity facilitated studies demonstrating the utility of PPT as a functional biocatalyst for ATP-recycling systems and for direct phosphorylation reactions.
TABLE 1.
Expression of polyP:AMP phosphotransferase in crude cell extracts prepared from A. johnsonii sp. 210A grown on different media
| Growth condition | Cultivation time (h) | Growth rate (h−1) | Sp act (nmol/min/mg)a |
|---|---|---|---|
| Minimal mediab | |||
| Butyrate | 23 | 0.20 | 68 |
| Crotonate | 23 | 0.17 | 70 |
| Acetate | 18 | 0.30 | 64 |
| dl-Lactate | 18 | 0.33 | 70 |
| Complex media | |||
| LB | 16.5 | NDd | 74 |
| LB + KH2PO4 at 0.8 g/liter | 16.5 | ND | 73 |
| LB + dl-lactatec | 19 | 0.40 | 63 |
| LB + dl-lactatec | 31 | 0.40 | 61 |
| LB + KH2PO4 + dl-lactatec | 25 | ND | 66 |
Reported are averages of duplicate determinations using the spectrophotometric assay. Standard deviations were less than 10%.
Defined media (see Materials and Methods) containing 20 mM carbon source in 200-ml volumes.
Media contained 40 mM dl-lactate.
ND, not determined.
Regeneration of ATP by PPT/AdK.
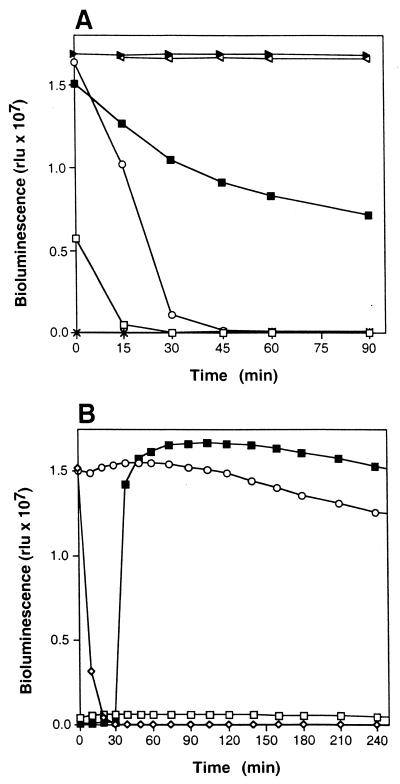
ATP regeneration was initially demonstrated using strain 210A cell extract and a commercially available firefly luciferase-based ATP assay (HSII Kit; Boehringer Mannheim). Figure 2A shows that bioluminescence, which normally decays very rapidly with ATP standards (>90% RLU decay by 30 min), could be sustained in the presence of cell extract, MgCl2, AdK, polyP, and AMP (complete reaction mixture components). Reaction mixtures in which AdK was omitted also showed high levels of sustained bioluminescence, indicating that cell extracts were not limited for AdK activity. Reduced levels of bioluminescence were observed when AMP was omitted, and complete reaction mixtures lacking polyP showed no bioluminescence (Fig. 2A). In the latter case, high levels of sustained luciferase activity were observed immediately following polyP addition at 30 min (Fig. 2B) and demonstrate the polyP-dependent nature of the PPT-based ATP regeneration system. Luciferase activity in complete reaction mixtures was sustained for >24 h (data not shown).
FIG. 2.
Regeneration of ATP from polyP and AMP as demonstrated by sustained bioluminescence from firefly luciferase (assay 2). (A) Effect of assay constituents on ATP regeneration activity. Complete reaction mixtures contained the following, except where otherwise indicated: cell extract protein at 0.5 mg/ml, 3 mM MgCl2, AdK at 1 U/ml, polyP at 0.2 g/liter, and 2 mM AMP. Symbols: □, 1.0 μM ATP; ○, 10 μM ATP; ▸, complete reaction mixture; ■, complete reaction mixture without AMP; ◃, complete reaction mixture without AdK; ×, complete reaction mixture without polyP. (B) Demonstration of the polyP requirement for ATP regeneration (assay constituents as defined for panel A). Symbols: ◊, 10 μM ATP; ○, complete reaction mixture; □, complete reaction mixture without polyP; ■, complete reaction mixture without polyP but with polyP at 0.2 g/liter added at 30 min.
AS precipitation.
AS precipitation was an effective method for the concentration and recovery of PPT activity from the 150,000 × g HSS. Recovery of PPT activity and protein in a typical AS precipitation series is shown in Table 2. Based on activity, PPT in the HSS was recovered in the 50% AS cut with a purification factor of 2 to 5. The most reproducible precipitation of PPT was achieved when a saturated AS solution was used to make cuts of up to 60%. When the procedure was scaled to volumes greater than 100 ml, PPT activity was distributed in approximately equal amounts in both the 40 to 50% and 50 to 60% AS cuts (data not shown).
TABLE 2.
Protein and polyP:AMP phosphotransferase activity distribution following AS precipitation of strain 210A HSS
| AS (% saturation) | Protein concn (mg/ml) | Relative sp act (RLU/mg of protein)a | Relative activity recovery (%) |
|---|---|---|---|
| 0b | 8.6 | 1,620 | 100 |
| 30 | 2.1 | 37 | 2.2 |
| 40 | 6.5 | 146 | 9.0 |
| 50 | 3.5 | 3,124 | 193 |
| 60 | 2.6 | 139 | 8.5 |
| 70 | 1.3 | 2 | 0.1 |
PPT activity was measured using assay 2 (see Materials and Methods). The relative specific activity reported represents 10−2 times the actual RLU magnitude measured at 30 min.
100,000 × g HSS.
Involvement of AdK or PPK in ATP formation.
To distinguish between AdK- and PPK-catalyzed formation of ATP, reactions were set up containing various strain 210A preparations of cell extract (crude, HSS, 50% AS cut) and ADP, with and without polyPn = 35. Initial rates of luciferase activity, measured as bioluminescence within 10 min, indicated only slightly higher levels of ATP formed in the presence of polyP (results not shown). However, by 105 min, the magnitude of bioluminescence (RLU) and the decay rate of luciferase activity were identical for reaction mixtures with and without polyP, suggesting that the formation of ATP from ADP was polyP independent and catalyzed by AdK.
Substrate specificity.
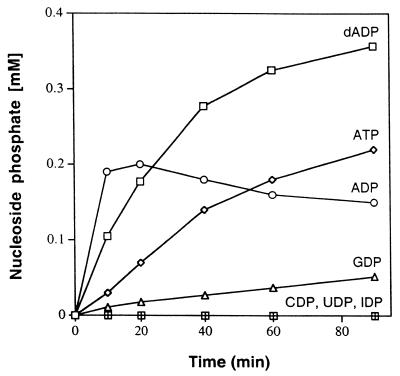
The potential of PPT to phosphorylate other nucleoside monophosphate substrates was examined using the 40 to 50% AS cut and measuring the formation of nucleoside di- and triphosphates by ion pair reverse-phase C18 HPLC with UV detection at 260 nm. Reaction mixtures containing PPT activity from the 40 to 50% AS cut (0.23 mg of protein/ml) efficiently converted 0.5 mM AMP to ADP (and ATP by AdK activity present in the preparation) but failed to phosphorylate CMP, UMP, or IMP (Fig. 3). Under identical conditions, GMP and 2′-dAMP were converted to GDP and 2′-dADP, respectively. The rate of GMP phosphorylation was 4% of that measured for AMP. The specific activity of PPT in these reaction mixtures (based on AMP depletion) was 76 nmol/min/mg.
FIG. 3.
Substrate specificity of PPT. Reaction mixtures contained different nucleoside monophosphates (AMP, 2′-dAMP, GMP, CMP, UMP, and IMP) and were incubated with the 50% AS cut (0.23 mg of protein per ml; see Materials and Methods). The formation of the corresponding nucleoside di- and triphosphates was monitored by HPLC.
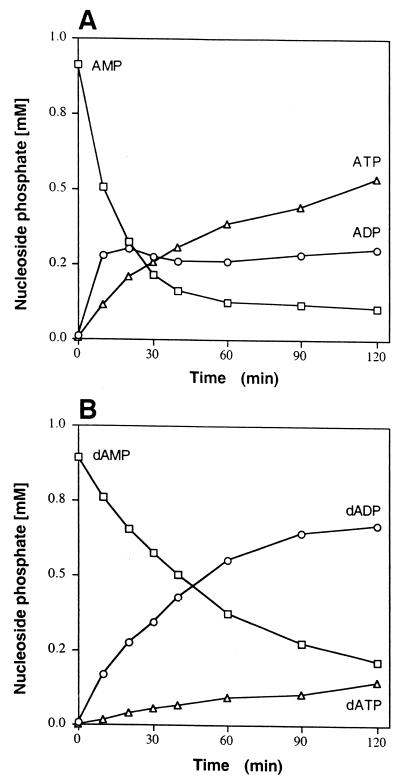
The kinetics of AMP and 2′-dAMP phosphorylation were examined in reaction mixtures containing a 40 to 50% AS cut and a 1.0 mM nucleotide substrate. AMP was rapidly phosphorylated to ADP, which underwent subsequent phosphorylation to ATP by the action of the AdK in the same preparation (Fig. 4A). ATP levels continued to rise (0.5 mM at 120 min) until AMP was depleted and equilibrium between ADP and ATP was established (16). Under the same conditions, 2′-dAMP was phosphorylated to 2′-dADP at 46% of the initial rate measured for AMP phosphorylation (Fig. 4B). Unlike ADP, however, 2′-dADP was not rapidly phosphorylated to dATP, suggesting that 2′-dADP is a relatively poor substrate for AdK (Fig. 4B). This appears to contradict a previous report of 2′-dADP phosphorylation by AdK (or PK) (12).
FIG. 4.
Course of AMP (A) and 2′-dAMP (B) conversion to the corresponding nucleoside diphosphates (○) and triphosphates (▵) by the PPT/AdK system. Reaction mixtures were as described in the legend to Fig. 3 but contained 1.0 mM substrate and 0.46 mg of protein of a 50% AS cut per ml.
HK reactions with ATP formed via PPT/AdK.
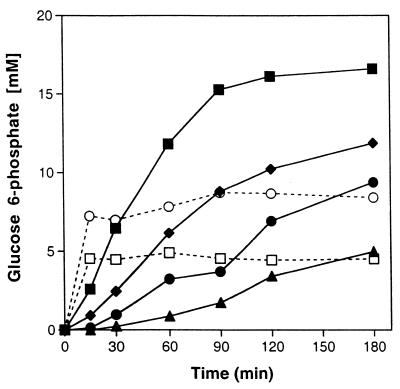
The applicability of the PPT/AdK system for biocatalysis was shown using strain 210A cell extract with HK as a model ATP-requiring enzyme. G6P was measured in reaction mixtures containing ATP or cell extract, polyP, and AMP. Results demonstrated G6P formation by HK with ATP provided from polyP and AMP by PPT/AdK (Table 3, reaction 2). Identical reaction mixtures lacking added AdK indicated that sufficient AdK activity was present in the cell extract to allow ATP recycling. Elevated G6P could be achieved by adding AdK to facilitate regeneration of added ATP (Table 3, reaction 4). Generation of ATP from AMP was shown to be polyP dependent, since only very low concentrations of G6P were detected when polyP was omitted (Table 3, reaction 5). HK was also used to examine ATP regeneration from polyP and 5 mM AMP with different concentrations of PPT protein activity precipitating between 40 and 60% AS saturation. In reaction mixtures containing 1.36 mg of protein per ml, the formation of over 16 mM G6P represented a threefold product excess relative to the added AMP concentration (Fig. 5). Plotting of the initial rates of G6P formation versus protein concentration results in a linear relationship (data not shown) and a calculated product formation rate of 164 nmol of G6P/min/mg of protein. While G6P formation was more rapid in the control reaction mixtures provided with 5 mM ATP, product formation was stoichiometric in both the absence and the presence of AdK, where final measured G6P concentrations were about 5 and 8 mM, respectively (Fig. 5).
FIG. 5.
HK-catalyzed formation of G6P in reaction mixtures with ATP supplied from 5 mM AMP and polyPn = 35 using AdK with different amounts of PPT protein. PPT concentrations, represented as total protein from a 40 to 60% AS cut, are 1.36 (■), 0.68 (⧫), 0.34 (●), and 0.136 (▴) mg/ml. Control reaction mixtures contained 5 mM ATP (dashed lines) with (○) and without (□) AdK.
DISCUSSION
The results of the present study provide information on the expression and substrate specificity of PPT and its potential application for the regeneration of ATP from AMP and polyP. Work conducted with crude extracts and partially purified preparations demonstrated the ability of PPT, when coupled with AdK, to regenerate ATP and sustain activities of the ATP-requiring enzymes luciferase and HK. The main advantage of using PPT with a kinase such as AdK (or PPK) is the ability to use AMP as the initial acceptor and polyP as the phosphoryl donor. Both substrates for the system are stable, commercially available, and at least 10-fold less expensive than corresponding substrates of systems based on PEP/PK or AcP/AcK. The reaction catalyzed by PPT is unidirectional and has an apparent half-saturation (Km) value of 0.26 mM for AMP (1), which is within the range of Km values for ADP measured with PK and AcK (see below). The Km of 0.8 μM for polyP suggests that the phosphoryl donor is tightly bound to PPT and efficiently used until it is completely degraded (1).
The PEP/PK system is considered the most efficient enzymatic ATP regeneration system when a strong, stable phosphorylating agent is required. The stability of PEP (half-life, 1,000 h at 25°C, pH 7) and its strength as a phosphoryl donor make it suitable for slow and thermodynamically unfavorable phosphorylation reactions (4). PK has a half-saturation constant (Km) for ADP of 0.1 mM, which is smaller than that of AcK (Km [MgADP] = 0.4 mM) and allows ATP regeneration at low concentrations of ADP. The AcP/AcK system is widely used for large-scale ATP regeneration, since AcP can be prepared relatively easily (20). Although AcP is a weaker phosphoryl donor than PEP, the major drawback is its relative instability in solution (half-life, 21 h at 25°C, pH 7) (4). Thus, an ATP regeneration system based on PPT and adenylate (or polyP) kinase is attractive since it uses polyP and AMP as the initial, stable, low-cost substrates.
Physiological-expression studies using cell extracts prepared after batch growth of strain 210A on different carbon sources showed that the specific activity of PPT was in the range of 60 to 70 nmol/min/mg. These activity levels are consistent with those reported previously for butyrate-grown batch cultures (1) and slightly higher than the activities reported for strain 210A under continuous-culture conditions (15). The results indicating that PPT activity in strain 210A was unchanged when it was batch cultured on carbon sources supporting different specific growth rates are in contrast to the finding that PPT activity is inversely related to growth rate under continuous-culture conditions (15). The finding that dl-lactate allowed a high growth rate and expression of PPT led to its selection as the carbon source for large-scale cultivation of strain 210A. The use of dl-lactate also eliminated the odor emission associated with butyrate. On the basis of the conditions examined, the results suggest that PPT activity in strain 210A was not inducible to levels higher than those previously measured and that it was independent of growth rate in batch culture.
In vitro ATP regeneration from AMP and polyP was initially demonstrated using strain 210A cell extracts and a firefly luciferase ATP assay. Results showed that bioluminescence from the luciferase reaction, which normally decays within 30 min for ATP standards (Fig. 2A), could be sustained for more than 24 h in the presence of strain 210A cell extract, MgCl2, polyP, and AMP. Sufficient AdK activity was present in the cell extract to convert ADP to ATP. Since firefly luciferase releases AMP from ATP, this assay was able to demonstrate the application of the PPT/AdK system for the regeneration of ATP exclusively through AMP.
Recent studies using acetate-limited chemostat cultures of A. johnsonii 210A suggested the involvement of PPK in polyP synthesis (18). Therefore, the possibility was considered that PPK functioning in the reverse direction could be responsible for the conversion of ADP to ATP in our studies. Assays to delineate the involvement of AdK or PPK in the formation of ATP indicated that ATP levels produced in the presence of polyP were only slightly higher than those produced without polyP, a result that is likely due to the PPT recycling the AMP released by luciferase. The finding that ATP formation from ADP was not polyP dependent indicated that AdK catalyzed the conversion of ADP to ATP in strain 210A cell-free preparations (Fig. 1). Unamended polyP levels in these preparations did not support PPT activity with AMP as the phosphoryl acceptor.
AS precipitation provided a scalable step for enrichment of PPT from the 150,000 × g HSS and can be followed by other chromatographic methods. PPT activity from Corynebacterium xerosis was similarly recovered between 45 and 75% AS saturation in a partial purification described previously (8). Streptomycin sulfate has been used to precipitate PPT from crude cell extracts with high efficiency (1); however, the procedure is dependent on the polyP concentration of the cell extract and is not easily scaled up. The protein precipitating between 40 and 60% AS saturation reproducibly provided an active PPT fraction suitable for substrate specificity and ATP regeneration studies.
Analysis of PPT substrate specificity showed that AMP and 2′-dAMP were the only nucleoside monophosphates accepted for efficient phosphorylation. GMP was also phosphorylated but at much slower rates (Fig. 3). Relative to that of AMP, the rates of PPT-catalyzed phosphorylation of GMP and 2′-dAMP were 4 and 46%, respectively. Our results obtained with ribonucleoside monophosphates are consistent with those reported for a 60-fold-purified preparation of PPT from C. xerosis which also failed to phosphorylate GMP, UMP, CMP, and IMP (8). The relatively restricted substrate spectrum for PPT contrasts with that of PPK from E. coli, which accepts ADP, GDP, CDP, and UDP as substrates with kination (phosphorylation) efficiencies ranging from 31 to 52% (14). The substrate specificity assay results suggest that use of PPT will be restricted to regeneration of ATP and not other nucleoside phosphates. However, the finding that PPT phosphorylates 2′-dAMP to 2′-dADP suggests potential utility in the enzymatic preparation of 2′-dADP or 2′-dATP (12).
HK catalyzed the formation of G6P and was used to demonstrate the applicability of the PPT/AdK system for ATP regeneration from polyP and AMP (Table 3 and Fig. 5). HK converts ATP to ADP, which can be recycled by AdK to ATP and AMP. The continuously formed AMP can be converted to ADP by PPT, preventing buildup of AMP and completing the cofactor-recycling process. HK has been used as a model ATP-requiring biocatalyst to demonstrate the utility of other ATP regeneration systems (13), some of which have enabled studies of its versatility (9). Reactions conducted with partially purified preparations of PPT (40 to 60% AS cut) formed G6P at a rate of 164 nmol/min/mg (Fig. 5), which appears to be a productivity similar to that of the polyP/PPK system for ATP regeneration from ADP (13). The utility of the PPT-based ATP regeneration system is apparent from the higher maximum G6P yields compared to ATP-containing control reaction mixtures (Fig. 5). Since the PPT preparation contained ammonium ions, which can sequester magnesium ions from solution (7), partial inhibition of HK, adenylate kinase, or PPT itself cannot be ruled out.
In conclusion, PPT appears to be a promising biocatalyst, when coupled with AdK activity, for the regeneration of ATP from AMP and polyP. The restricted substrate specificity of PPT indicates that its use for cofactor recycling, when coupled with AdK, may be limited to ATP (regeneration). The identification of 2′-dAMP as a substrate suggests the potential utility of PPT as a biocatalyst for direct phosphorylation reactions. The use of PPT/AdK to supply energy for luciferase and HK activities demonstrated the ability to recycle AMP through ADP to ATP. For both activities, ATP regeneration was strictly dependent on polyP as the phosphoryl donor. The present work provides a basis for the development of PPT as a functional biocatalyst for direct phosphorylation reactions and use in ATP regeneration to enhance the productivity of biotechnologically relevant ATP-requiring enzymes. Purification of the protein and cloning of the corresponding gene are the necessary steps for complete characterization of this interesting enzyme.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Swiss National Science Foundation (Swiss Priority Program in Biotechnology, project 5002-46093).
We thank Andreas Schmid and Hans-Juergen Feiten for assistance with cultivation and harvesting of strain 210A on a 700-liter scale; Roland Wohlgemuth (Fluka Chemie, AG) for providing several nucleotides; A. Schmid, R. Wohlgemuth, and Hans-Peter Kohler for helpful discussions; and Paolo Landini for critical reading of the manuscript.
REFERENCES
- 1.Bonting C F C, Kortstee G J J, Zehnder A J B. Properties of polyphosphate:AMP phosphotransferase of Acinetobacter strain 210A. J Bacteriol. 1991;173:6484–6488. doi: 10.1128/jb.173.20.6484-6488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;161:280–290. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 3.Butler L. A suggested approach to ATP regeneration for enzyme technology applications. Biotechnol Bioeng. 1977;19:591–593. doi: 10.1002/bit.260190415. [DOI] [PubMed] [Google Scholar]
- 4.Crans D C, Kazlauskas R J, Hirschbein B L, Wong C-H, Abril O, Whitesides G M. Enzymatic regeneration of adenosine 5′-triphosphate: acetyl phosphate, phosphoenolpyruvate, methoxycarbonyl phosphate, dihydroxyacetone phosphate, 5-phospho-α-D-ribosyl pyrophosphate, uridine-5′-diphosphoglucose. Methods Enzymol. 1987;136:263–280. doi: 10.1016/s0076-6879(87)36027-6. [DOI] [PubMed] [Google Scholar]
- 5.Crans D C, Whitesides G M. Glycerol kinase: synthesis of dihydroxyacetone phosphate, sn-glycerol-3-phosphate, and chiral analogues. J Am Chem Soc. 1985;107:7019–7027. [Google Scholar]
- 6.Crans D C, Whitesides G W. Glycerol kinase: substrate specificity. J Am Chem Soc. 1985;107:7008–7018. [Google Scholar]
- 7.Davies H G, Green R H, Kelly D R, Roberts S M. Biotransformations in preparative organic chemistry—the use of isolated enzymes and whole cell systems in synthesis. San Diego, Calif: Academic Press; 1989. pp. 82–86. [Google Scholar]
- 8.Dirheimer G, Ebel J-P. Caracterisation d'une polyphosphate-AMP-phosphotransferase dans Corynebacterium xerosis. C R Acad Sci Paris. 1965;260:3787–3790. [PubMed] [Google Scholar]
- 9.Drueckhammer D G, Wong C-H. Chemoenzymatic synthesis of fluoro sugar phosphates and analogues. J Org Chem. 1985;50:5912–5913. [Google Scholar]
- 10.Faber K. Biotransformations in organic chemistry. New York, N.Y: Springer-Verlag; 1992. pp. 100–106. [Google Scholar]
- 11.Gross A, Abril O, Lewis J M, Geresh S, Whitesides G M. Practical synthesis of 5-phospho-d-ribosyl-α-1-pyrophosphate (PRPP): enzymatic routes from ribose 5-phosphate or ribose. J Am Chem Soc. 1983;105:7428–7435. [Google Scholar]
- 12.Ladner W E, Whitesides G M. Enzymatic synthesis of deoxyATP using DNA as starting material. J Org Chem. 1985;50:1076–1079. [Google Scholar]
- 13.Murata K, Uchida T, Kato J, Chibata I. Polyphosphate kinase: distribution, some properties and its application as an ATP regeneration system. Agric Biol Chem. 1988;52:1471–1477. [Google Scholar]
- 14.Noguchi T, Shiba T. Use of Escherichia coli polyphosphate kinase for oligosaccharide synthesis. Biosci Biotechnol Biochem. 1998;62:1594–1596. doi: 10.1271/bbb.62.1594. [DOI] [PubMed] [Google Scholar]
- 15.van Groenestijn J W, Bentvelsen M M A, Deinema M H, Zehnder A J B. Polyphosphate-degrading enzymes in Acinetobacter spp. and activated sludge. Appl Environ Microbiol. 1989;55:219–223. doi: 10.1128/aem.55.1.219-223.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Groenestijn J W, Deinema M H, Zehnder A J B. ATP production from polyphosphate in Acinetobacter strain 210A. Arch Microbiol. 1987;148:14–19. [Google Scholar]
- 17.van Groenestijn J W, Zuidema M, van de Worp J J M, Deinema M H, Zehnder A J B. Influence of environmental parameters on polyphosphate accumulation in Acinetobacter sp. Antonie van Leeuwenhoek. 1989;55:67–82. doi: 10.1007/BF02309620. [DOI] [PubMed] [Google Scholar]
- 18.van Niel E W J, de Best J H, Kets E P W, Bonting C F C, Kortstee G J J. Polyphosphate formation by Acinetobacter johnsonii 210A: effect of cellular energy status and phosphate-specific transport system. Appl Microbiol Biotechnol. 1999;51:639–646. [Google Scholar]
- 19.Walt D R, Findeis M A, Rios-Mercadillo V M, Auge J, Whitesides G M. An efficient chemical and enzymatic synthesis of nicotinamide adenine dinucleotide (NAD+) J Am Chem Soc. 1984;106:234–239. [Google Scholar]
- 20.Whitesides G M. Formation and cleavage of P—O bonds. In: Drauz K, Waldman H, editors. Enzyme catalysis in organic synthesis, a comprehensive handbook. II. Weinheim, Germany: VCH Publishers; 1995. pp. 505–527. [Google Scholar]
- 21.Wong C-H, Whitesides G M. Synthesis of sugars by aldolase-catalyzed condensation reactions. J Org Chem. 1983;48:3199–3205. [Google Scholar]