Abstract
Increasing rates of serious multi-drug resistant (MDR) Pseudomonas aeruginosa infections have been reported globally, including in Saudi Arabia. This retrospective study investigates the epidemiological, microbiological, and clinical characteristics of multi-resistant P. aeruginosa (n3579 clinical isolates) in King Fahad Medical City, Riyadh, Saudi Arabia (2019–2021). Information on antimicrobial susceptibility and medical history was collected from the hospital database. P. aeruginosa infections occurred in 55.6% of males and 44.4% of females, and P. aeruginosa was more prevalent in children than in adults. Our analysis showed that P. aeruginosa had the highest sensitivity to amikacin (92.6%) and greatest resistance to aztreonam (29.8%), imipenem (29.5%), ceftazidime (26.1%), meropenem (25.6%), and cefepime (24.3%). MDR and extensively drug resistant (XDR) strains were more prevalent in male than female patients. Female patients showed higher rates of infection with pan-drug resistant (PDR) strains. Respiratory samples contained the majority of resistant isolates. Septic shock and liver disease were strongly correlated with mortality in the ICU patient group after analysing the relative risk associated with mortality. Our study emphasises the threat of multi-resistant P. aeruginosa in Saudi Arabia (and potentially the Middle East) and highlights important sources and contexts of infection that inhibit its effective control and clinical management.
Keywords: Pseudomonas aeruginosa, respiratory, intensive care unit, nosocomial infection, multi-drug resistant, pan-drug resistant, COVID-19
1. Introduction
Pseudomonas aeruginosa is an important Gram-negative opportunistic pathogen found in many environmental settings and can be isolated from different living sources, such as plants, animals, and humans. This organism can persist in both community and hospital settings under conditions of limited nutrition [1]. Community-acquired infections (CAI) are less prevalent than nosocomial infections [2]. The prevalence rate of P. aeruginosa isolates in patients with community-acquired pneumonia (CAP) is 3.8% in Europe, 4.3% in North America, 5.2% in Asia, 4.9% in South America, and 5.5% in Africa [3]. P. aeruginosa is implicated in various nosocomial infections, including ventilator-associated pneumonia (VAP), bloodstream infections (BSI), urinary tract infections (UTI), and wound infections. It is the fourth most frequently isolated nosocomial pathogen and the second leading contributor to pneumonia, with mortality rates of 27–48% in critically ill patients [4]. Therefore, it is considered as one of the most life-threating bacteria [5].
The emergence of antibiotic resistance in P. aeruginosa has become a global public health concern because of the lack of therapeutic alternative treatments available [6], and its ability to metabolize various antibiotics, which promotes infectivity potential [7]. P. aeruginosa displays intrinsic and acquired resistance to many antimicrobial agents [8]. According to the European Centre for Disease Prevention and Control’s annual report, 18.7% of all P. aeruginosa isolates were carbapenem resistant and 13.4% were resistant to three or more antimicrobial classes [9]. P. aeruginosa can be classified into phenotypes based on drug resistance patterns. Multi-drug resistant (MDR) strains are resistant to three or more anti-pseudomonal drugs, including aminoglycosides, carbapenems, cephalosporins, fluoroquinolones, and penicillin/-lactamase. Extensively drug-resistant (XDR) strains are susceptible to only one or two anti-pseudomonal drugs. Pan-drug resistant (PDR) strains are resistant to all anti-pseudomonal drugs [10].
Infections caused by MDR P. aeruginosa strains are associated with high morbidity and mortality rates, as well as increased durations of hospital stays and overall costs of treatment [11,12]. Several factors are associated with MDR P. aeruginosa infection, including previous antimicrobial use, prolonged antibiotic treatment, history of chronic obstructive pulmonary disease (COPD), long hospital stay or admission to the intensive care unit (ICU), history of P. aeruginosa infection, use of invasive indwelling devices (e.g., tracheostomy tubes or urethral catheters), and previous surgery [13].
A recent increase in the prevalence of MDR and XDR P. aeruginosa strains in hospitalised patients has been reported across the globe [6]. In Saudi Arabia, MDR P. aeruginosa was most commonly isolated from patients in the ICU [14]. Each year, 13–19% of hospital-acquired infections (HAIs) in the United States are caused by MDR P. aeruginosa [15]. MDR and XDR isolates are highly prevalent in Europe, particularly in Greece [16]. In a multinational prospective cohort study of 236 ICUs from 77 hospitals in 10 Middle Eastern countries, age, length of stay, central-line days, use of mechanical ventilators, central-line blood stream infection, ventilator associated pneumonia, female gender, and hospitalization were found to be associated with mortality among ICU-admitted patients [17].
The aim of this retrospective study was to examine the epidemiological, microbiological, and clinical characteristics of MDR P. aeruginosa isolated at the King Fahad Medical City (KFMC) to inform the development of updated clinical treatment strategies in Saudi Arabia.
2. Methods
2.1. Study Design
This retrospective study considered data from January 2019 to December 2021 collected at KFMC, Riyadh, Saudi Arabia. A total of 3579 P. aeruginosa isolates from various clinical samples were analysed. The clinical history of 255 adult ICU patients was also included in the study.
2.2. Data Collection
P. aeruginosa isolates were collected from a variety of sources, including blood (central and peripheral lines), the respiratory system (sputum, endotracheal, BAL, nasopharyngeal, throat swab, ear, and eye), urine (mid-stream urine and in-and-out catheters), miscellaneous sources (abscess, wound, tissue, and body fluid), and cerebrospinal fluid (CSF). The inclusion criteria were the same as in the previously published study by Hafiz et al. (2022) [18]. Any bacteria other than P. aeruginosa were excluded from this study. ICU patient data (collected from the KFMC database) were included based on their clinical histories as follows: (1) clinical symptoms, such as fever or gastrointestinal tract (GIT) symptoms; (2) wound or urinary tract infections; (3) respiratory diseases, such as COPD, acute respiratory distress syndrome (ARDS), and pulmonary oedema; (4) pneumonia, including ventilator-associated pneumonia (VAP), CAP, and hospital-acquired pneumonia (HAP); (5) mechanical ventilation used at some point; (6) chronic diseases, such as renal disease, heart disease, liver disease, brain disorders, malignancy, diabetes mellitus (DM), asthma, or hypertension (HTN); (6) septicaemia; (7) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection; (8) clinical outcomes for the patient, and additional notes, if present.
2.3. P. aeruginosa Identification and Antimicrobial Susceptibility Testing
For complete identification and sensitivity testing, isolates were confirmed to be Pseudomonas spp. using a BD Phoenix automated microbiological system (Becton Dickinson Diagnostic Systems, Sparks, MD, USA). Only data of patients with definitively identified P. aeruginosa isolates were included in this study. The following antibiotics were tested for antimicrobial sensitivity testing (AST): ceftazidime, cefepime, ciprofloxacin, piperacillin-tazobactam, gentamicin, amikacin, imipenem, meropenem, colistin, levofloxacin, and aztreonam. The E test was used to confirm the presence of resistant isolates. The results were interpreted and reported in accordance with the CLSI-M100, 32nd edition, and classified as susceptible (S), intermediate (I), or resistant (R). According to the International Consensus, P. aeruginosa isolates were classified based on their antibiotic resistance [19].
2.4. Statistical Analysis
All data were analysed using SPSS software (IBM SPSS, Armonk, NY, USA). Graphs were generated using GraphPad Prism software (v9.4.1; GraphPad Software, San Diego CA, USA). We compared clinical characteristics among adult ICU-admitted patients over one year using a univariate chi-squared test, with significance set at p < 0.05. Similarly, a univariate analysis was performed to compare the outcomes and risk factors associated with mortality among ICU patients. Relative risk (RR) was calculated to determine how the risk of death increased among patients with respiratory infections, and the results were reported as RR and 95% confidence intervals. Antimicrobial susceptibility test results were presented as percentages.
2.5. Ethical Consideration
The study protocol was approved by the Ethical Research Committee of King Fahad Medical City. Consent was acquired from the KFMC in accordance with the ICH GCP guidelines (IRB log number: 21-063E).
3. Results
3.1. Demographic Characteristics of Patients with Multi-Resistant P. aeruginosa
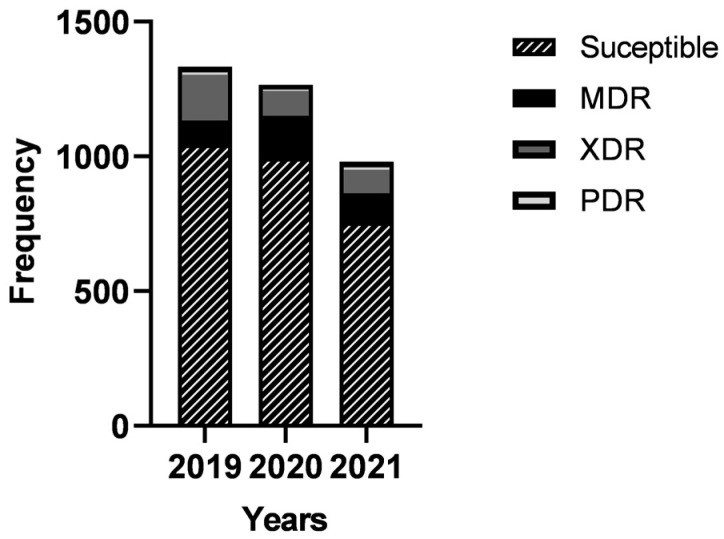
Out of the 3579 P. aeruginosa isolates, 422 (11.8%) were XDR, 348 (9.7%) were MDR, and 11 (0.3%) were PDR. MDR isolates increased after 2019, whereas the percentages of XDR and PDR isolates decreased (Figure 1).
Figure 1.
Distribution of multi-resistant P. aeruginosa isolates for the period 2019–2021.
Most MDR and XDR P. aeruginosa isolates (61.8 and 61.1%, respectively) were isolated from male patients, while female patients were infected with more PDR isolates (Table 1). The majority of the MDR isolates (38.8%, 26/67) among paediatrics occurred in infants < 1 year of age, whereas the highest number of XDR isolates (36.8%, 25/68) occurred in paediatric children. PDR isolates were only found in infants, with 100% (2/2) of the paediatric population. Among adult patients, those aged 65–84 years had the highest incidence of MDR (39.5%, 111/281) and XDR isolates (34.5%, 89/258). Additionally, 44.4% (4/11) of PDR isolates occurred in adults aged 45–64 years, 33.3% (3/11) in adults aged 65–84 years, and only two isolates in adults aged 19–44 years. No PDR isolates were observed in adults aged 85 years and above.
Table 1.
Distribution of multi-resistant P. aeruginosa isolates according to gender, age, and sample source and site.
| Characteristic | Total n (%) (Total = 3579) |
P. aeruginosa Isolates | p-Value | |||
|---|---|---|---|---|---|---|
|
P. aeruginosa n (%) (Total = 2798) |
MDR n (%) (Total = 348) |
XDR n (%) (Total = 422) |
PDR n (%) (Total = 11) |
|||
| Gender | ||||||
| Male | 1990 (55.6) | 1513 (54.1) | 215 (61.8) | 258 (61.1) | 4 (36.4) | 0.002 ** |
| Female | 1589 (44.4) | 1285 (45.9) | 133 (38.2) | 164 (38.2) | 7 (63.6) | |
| Age | ||||||
| Paediatric infant (<1 year) | 236 (26.2) | 185 (24.2) | 26 (38.8) | 23 (33.8) | 2 (100) | 0.001 ** |
| Paediatric child (1–10 years) | 466 (51.7) | 417 (54.6) | 24 (35.8) | 25 (36.8) | 0 (0) | |
| Paediatric adolescent (11–18 years) | 199 (22.1) | 162 (21.2) | 17 (25.4) | 20 (29.4) | 0 (0) | |
| Adult (19–44 years) | 658 (24.6) | 491 (24.1) | 59 (21) | 83 (32.2) | 2 (22.2) | 0.022 * |
| Adult (45–64 years) | 889 (33.2) | 695 (34.2) | 83 (29.5) | 80 (31) | 4 (44.4) | |
| Adult (65–84 years) | 968 (36.1) | 728 (35.8) | 111 (39.5) | 89 (34.5) | 3 (33.3) | |
| Adult (≥85 years) | 163 (6.1) | 120 (5.9) | 28 (10) | 6 (2.3) | 0 (0) | |
| Source | ||||||
| Miscellaneous | 927 (25.9) | 738 (26.4) | 79 (22.7) | 108 (25.6) | 2 (18.2) | <0.001 ** |
| Blood | 284 (7.9) | 232 (8.3) | 24 (6.9) | 25 (5.9) | 3 (27.3) | |
| Respiratory | 1633 (45.6) | 1194 (42.7) | 205 (58.9) | 228 (54) | 6 (54.5) | |
| Urine | 719 (20.1) | 622 (22.2) | 39 (11.2) | 58 (13.7) | 0 (0) | |
| CSF | 16 (0.4) | 12 (0.4) | 1 (0.3) | 3 (0.7) | 0 (0) | |
* Statistically significant; the number of * represents the degree of significance. CSF, cerebrospinal fluid; MDR, multi-drug resistant; XDR, extensively drug-resistant; PDR, pan-drug resistant.
The most common sources of MDR and XDR isolates were from respiratory samples (58.9 and 54%, respectively), followed by miscellaneous sources (Table 1). In contrast, a relatively small number of isolates were obtained from urine, blood, and CSF samples. Regarding PDR P. aeruginosa, 54.4% (6/11) were isolated from respiratory sources, 27.3% (3/11) from blood, and 18.2% (2/11) from miscellaneous sources. No PDR isolates were obtained from urine and CSF samples.
3.2. Antimicrobial Susceptibility of Multi-Resistant P. aeruginosa Isolates
Different isolates showed varying rates of resistance to the respective antibiotics (Table 2). Among MDR and XDR P. aeruginosa isolates, the highest rates of resistance were observed against cephalosporins (cefepime and ceftazidime), carbapenems (imipenem and meropenem), and aztreonam. On the other hand, these isolates showed lower rates of resistance to amikacin and colistin. XDR P. aeruginosa isolates were highly resistant to ciprofloxacin (90.8%) and levofloxacin (91.2%). PDR P. aeruginosa isolates showed 100% resistance to all antibiotics tested.
Table 2.
Comparison of antimicrobial susceptibility among multi-resistant P. aeruginosa isolates.
| Antibiotics | Total n (%) (Total = 3579) |
P. aeruginosa Isolates | p-Value | |||
|---|---|---|---|---|---|---|
|
P. aeruginosa n (%) (Total = 2798) |
MDR n (%) (Total = 348) |
XDR n (%) (Total = 422) |
PDR n (%) (Total = 11) |
|||
| Ceftazidime | ||||||
| Sensitive | 2529 (70.7) | 2498 (89.3) | 25 (7.2) | 6 (1.4) | 0 (0) | <0.001 ** |
| Intermediate | 117 (3.3) | 78 (2.8) | 31 (8.9) | 8 (1.9) | 0 (0) | |
| Resistant | 933 (26.1) | 222 (7.9) | 292 (83.9) | 408 (96.7) | 11 (100) | |
| Cefepime | ||||||
| Sensitive | 2617 (73.1) | 2567 (91.7) | 42 (12.1) | 8 (1.9) | 0 (0) | <0.001 ** |
| Intermediate | 92 (2.6) | 33 (1.2) | 39 (11.2) | 20 (4.7) | 0 (0) | |
| Resistant | 870 (24.3) | 198 (7.1) | 267 (76.7) | 394 (93.4) | 11 (100) | |
| Ciprofloxacin | ||||||
| Sensitive | 2783 (77.8) | 2554 (91.3) | 218 (62.6) | 11 (2.6) | 0 (0) | <0.001 ** |
| Intermediate | 159 (4.4) | 95 (3.4) | 36 (10.3) | 28 (6.6) | 0 (0) | |
| Resistant | 637 (17.8) | 149 (5.3) | 94 (27) | 383 (90.8) | 11 (100) | |
| Piperacillin-Tazobactam | ||||||
| Sensitive | 2573 (71.9) | 2516 (89.9) | 44 (12.6) | 13 (3.1) | 0 (0) | <0.001 ** |
| Intermediate | 394 (11) | 178 (6.4) | 114 (32.8) | 102 (24.2) | 0 (0) | |
| Resistant | 612 (17.1) | 104 (3.7) | 190 (54.6) | 307 (72.7) | 11 (100) | |
| Gentamicin | ||||||
| Sensitive | 3085 (86.2) | 2709 (96.8) | 274 (78.7) | 102 (24.2) | 0 (0) | <0.001 ** |
| Intermediate | 77 (2.2) | 38 (1.4) | 12 (3.4) | 27 (6.4) | 0 (0) | |
| Resistant | 417 (11.7) | 51 (1.8) | 62 (17.8) | 293 (69.4) | 11 (100) | |
| Amikacin | ||||||
| Sensitive | 3313 (92.6) | 2776 (99.2) | 317 (91.1) | 220 (52.1) | 0 (0) | <0.001 ** |
| Intermediate | 66 (1.8) | 7 (0.3) | 9 (2.6) | 50 (11.8) | 0 (0) | |
| Resistant | 200 (5.6) | 15 (0.5) | 22 (6.3) | 152 (36) | 11 (100) | |
| Imipenem | ||||||
| Sensitive | 2365 (66.1) | 2270 (81.1) | 85 (24.4) | 10 (2.4) | 0 (0) | <0.001 ** |
| Intermediate | 158 (4.4) | 139 (5) | 16 (4.6) | 3 (0.7) | 0 (0) | |
| Resistant | 1056 (29.5) | 389 (13.9) | 247 (71) | 409 (96.9) | 11 (100) | |
| Meropenem | ||||||
| Sensitive | 2555 (71.4) | 2446 (87.4) | 99 (28.4) | 10 (2.4) | 0 (0) | <0.001 ** |
| Intermediate | 108 (3) | 88 (3.1) | 15 (4.3) | 5 (1.2) | 0 (0) | |
| Resistant | 916 (25.6) | 264 (9.4) | 234 (67.2) | 407 (96.4) | 11 (100) | |
| Colistin | ||||||
| Sensitive | 3234 (90.4) | 2546 (91) | 308 (88.5) | 380 (90) | 0 (0) | <0.001 ** |
| Intermediate | 305 (8.5) | 241 (8.6) | 33 (9.5) | 31 (7.3) | 0 (0) | |
| Resistant | 40 (1.1) | 11 (0.4) | 7 (2) | 11 (2.6) | 11 (100) | |
| Levofloxacin | ||||||
| Sensitive | 2587 (72.3) | 2412 (86.2) | 155 (44.5) | 20 (4.7) | 0 (0) | <0.001 ** |
| Intermediate | 261 (7.3) | 179 (6.2) | 65 (18.7) | 17 (4) | 0 (0) | |
| Resistant | 729 (20.4) | 207 (6.4) | 126 (36.2) | 385 (91.2) | 11 (100) | |
| Aztreonam | ||||||
| Sensitive | 2124 (59.3) | 2043 (73) | 54 (15.5) | 27 (6.4) | 0 (0) | <0.001 ** |
| Intermediate | 381 (10.6) | 306 (10.9) | 40 (11.5) | 35 (8.3) | 0 (0) | |
| Resistant | 1065 (29.8) | 442 (15.8) | 253 (72.7) | 359 (85.1) | 11 (100) | |
* Statistically significant; the number of * represents the degree of significance.
3.3. Clinical Characteristics of ICU Patients Infected with Multi-Resistant P. aeruginosa
During the study period, 255 multi-resistant P. aeruginosa isolates were identified from adult patients admitted to the ICU (Table 3). The percentage of ICU patients infected with multi-resistant P. aeruginosa was higher for males (61.6%) than that for females (38.4%). The majority of the infected ICU patients (44.3%) were aged 65–84 years. The highest number of multi-resistant P. aeruginosa isolates were from respiratory sources (69.8%). Several clinical variables differed between years and were significantly higher in 2019 than in 2020 or 2021, including the incidences of cancer (p = 0.006) and liver disease (p = 0.001). Conversely, the incidences of pneumonia (p < 0.001), mechanical ventilation (p < 0.001), and COVID-19 (p < 0.001) were significantly higher in 2021 than in 2019 or 2020. Regarding clinical outcomes, mortality rates among infected adults in ICU were high overall (55.7%, 142/255), though mortality rates were highest in 2019. A relatively small number (31/255) of patients were transferred to other hospitals and 32.2% (82/255) were treated.
Table 3.
Characteristics of ICU patients with multi-resistant P. aeruginosa infection for the period 2019–2021.
| Characteristic | Total n (%) (Total = 255) |
Years | p-Value | ||
|---|---|---|---|---|---|
| 2019 n (%) (Total = 114) |
2020 n (%) (Total = 78) |
2021 n (%) (Total = 63) |
|||
| Gender | |||||
| Male | 157 (61.6) | 66 (57.9) | 52 (66.7) | 39 (61.9) | 0.470 |
| Female | 98 (38.4) | 48 (42.1) | 26 (33.3) | 24 (38.1) | |
| Age | |||||
| Adult (19–44 years) | 54 (21.2) | 33 (28.9) | 11 (14.1) | 10 (15.9) | 0.256 |
| Adult (45–64 years) | 72 (28.2) | 29 (25.4) | 24 (30.8) | 19 (30.2) | |
| Adult (65–84 years) | 113 (44.3) | 46 (40.4) | 38 (48.7) | 29 (46) | |
| Adult (⇒85 years) | 16 (6.3) | 6 (5.3) | 5 (6.4) | 5 (7.9) | |
| Source of specimen | |||||
| Miscellaneous | 31 (12.2) | 12 (10.5) | 14 (17.9) | 5 (7.9) | 0.003 ** |
| Blood | 26 (10.2) | 12 (10.5) | 3 (3.8) | 11 (17.5) | |
| Respiratory | 178 (69.8) | 75 (65.8) | 56 (71.8) | 47 (74.6) | |
| Urine | 20 (7.8) | 15 (13.2) | 5 (6.4) | 0 (0) | |
| Clinical presentation/infection | |||||
| Fever | 34 (13.4) | 18 (15.8) | 12 (15.4) | 4 (6.3) | 0.182 |
| Sepsis | 25 (9.8) | 9 (7.9) | 7 (9) | 9 (14.3) | 0.375 |
| Septic shock | 40 (15.7) | 22 (19.3) | 5 (6.4) | 13 (20.6) | 0.025 * |
| Respiratory | 195 (76.5) | 102 (89.5) | 63 (80.8) | 30 (47.6) | <0.001 ** |
| GIT | 8 (3.1) | 5 (4.4) | 2 (2.6) | 1 (1.6) | 0.558 |
| UTI | 32 (12.5) | 16 (14) | 12 (15.4) | 4 (6.3) | 0.222 |
| WI | 46 (18) | 24 (21.1) | 12 (15.4) | 10 (15.9) | 0.529 |
| Pneumonia | 117 (45.9) | 27 (23.7) | 38 (48.7) | 52 (82.5) | <0.001 ** |
| Underlying disease | |||||
| Kidney disease | 90 (35.3) | 42 (36.8) | 28 (35.9) | 20 (31.7) | 0.787 |
| Heart disease | 101 (39.6) | 44 (38.6) | 35 (44.9) | 22 (34.9) | 0.465 |
| Liver disease | 32 (12.5) | 26 (22.8) | 4 (5.1) | 2 (3.2) | <0.001 ** |
| Brain disorder | 68 (26.7) | 34 (29.8) | 24 (30.8) | 10 (15.9) | 0.082 |
| Cancer | 33 (12.9) | 23 (20.2) | 4 (5.1) | 6 (9.5) | 0.006 ** |
| DM | 136 (53.3) | 56 (49.1) | 44 (56.4) | 36 (57.1) | 0.478 |
| Asthma | 8 (3.1) | 4 (3.5) | 2 (2.6) | 2 (3.2) | 0.934 |
| HTN | 152 (59.6) | 55 (48.2) | 54 (69.2) | 43 (68.3) | 0.004 ** |
| Risk factors | |||||
| Mechanical ventilation | 93 (36.5) | 23 (20.2) | 24 (30.8) | 46 (73) | <0.001 ** |
| COVID-19 | 37 (14.5) | 0 (0) | 14 (17.9) | 23 (36.5) | <0.001 ** |
| Outcome | |||||
| Died | 142 (55.7) | 71 (62.3) | 35 (44.9) | 36 (57.1) | 0.002 ** |
| Treated | 82 (32.2) | 35 (30.7) | 24 (30.8) | 23 (36.5) | |
| Transferred | 31 (12.2) | 8 (7) | 19 (24.4) | 4 (6.3) | |
* Statistically significant; the number of * represents the degree of significance. GIT, gastrointestinal tract; UTI, urinary tract infection; WI, wound infection; DM, diabetes mellitus; HTN, hypertension; COVID-19, coronavirus disease 2019.
3.4. Clinical Outcomes and Factors Associated with Mortality Rates of ICU Patients Infected with Multi-Resistant P. aeruginosa
Mortality rates were higher among males than females (61.1% and 38.9%, respectively; Table 4). Mortality rates were highest in patients aged 65–84 years (38.9%), followed by those aged 45–64 years (25.7%). The occurrence of septic shock (p = 0.001), kidney disease (p = 0.038), liver disease (p = 0.006), cancer (p = 0.035), and mechanical ventilation (p = 0.016) was significantly higher in dead patients than that in live patients, while fever (p = 0.028) and brain disorders (p = 0.002) were significantly higher in live patients. A considerable proportion of ICU patients had a respiratory infection (51.1%). Septic shock (RR = 2.458, CI 95% = 1.080–5.593, p = 0.032) and liver disease (RR = 3.107, CI 95% = 1.053–9.162, p = 0.039) were the factors most significantly associated with mortality (Table 5).
Table 4.
Comparison of demographic and clinical characteristics among dead and alive ICU patients infected with multi-resistant P. aeruginosa.
| Characteristic | Outcome | p-Value | |
|---|---|---|---|
| Alive n (%) (Total = 113) |
Dead n (%) (Total = 142) |
||
| Year | |||
| 2019 | 43 (38.1) | 71 (50) | 0.056 |
| 2020 | 43 (38.1) | 35 (24.6) | |
| 2021 | 27 (23.9) | 36 (25.4) | |
| Gender | |||
| Male | 69 (61.1) | 88 (62) | 0.882 |
| Female | 44 (38.9) | 54 (38) | |
| Age | |||
| Adult (19–44 years) | 32 (28.3) | 22 (15.5) | 0.075 |
| Adult (45–64 years) | 29 (25.7) | 43 (30.3) | |
| Adult (65–84 years) | 44 (38.9) | 69 (48.6) | |
| Adult (≥85 years) | 8 (7.1) | 8 (5.6) | |
| Respiratory culture | |||
| Positive | 87 (48.9) | 91 (51.1) | 0.026 * |
| Clinical presentation/infection | |||
| Fever | 21 (18.6) | 13 (9.2) | 0.028 * |
| Sepsis | 9 (8) | 16 (11.3) | 0.378 |
| Septic shock | 8 (7.1) | 32 (22.5) | 0.001 ** |
| Respiratory | 80 (70.8) | 115 (81) | 0.057 |
| GIT | 5 (4.4) | 3 (2.1) | 0.293 |
| UTI | 15 (13.3) | 17 (12) | 0.755 |
| WI | 23 (20.4) | 23 (16.2) | 0.391 |
| Pneumonia | 56 (49.6) | 61 (43) | 0.293 |
| Underlying disease | |||
| Kidney | 32 (28.3) | 58 (40.8) | 0.038 * |
| Heart | 46 (40.7) | 55 (38.7) | 0.749 |
| Liver | 7 (6.2) | 25 (17.6) | 0.006 ** |
| Brain disorder | 41 (36.3) | 27 (19) | 0.002 ** |
| Cancer | 9 (8) | 24 (16.9) | 0.035 * |
| DM | 53 (46.9) | 83 (58.5) | 0.066 |
| Asthma | 6 (5.3) | 2 (1.4) | 0.076 |
| HTN | 70 (61.9) | 82 (57.7) | 0.497 |
| Risk factors | |||
| Mechanical ventilation | 23 (28.3) | 61 (43) | 0.016 * |
| COVID-19 | 15 (3.3) | 22 (15.5) | 0.617 |
Univariate analysis (Fisher’s exact test, p < 0.05). * Statistically significant; the number of * represents the degree of significance. GIT, gastrointestinal tract; UTI, urinary tract infection; WI, wound infection; DM, diabetes mellitus; HTN, hypertension; COVID-19, corona virus disease 2019.
Table 5.
Relative risk associated with mortality among adult ICU patients infected with multi-resistant P. aeruginosa isolates.
| Variable | RR | CI 95% | p-Value |
|---|---|---|---|
| Septic shock | 2.458 | 1.080–5.593 | 0.032 * |
| Kidney disease | 1.287 | 0.851–1.946 | 0.232 |
| Liver disease | 3.107 | 1.053–9.162 | 0.039 * |
| Cancer | 1.673 | 0.738–3.789 | 0.217 |
| Mechanical ventilation | 1.219 | 0.828–1.796 | 0.314 |
* Statistically significant; CI, confidence interval; RR, relative risk. RR < 1 indicates no association with mortality, RR >1 indicates a strong association with mortality.
4. Discussion
P. aeruginosa is commonly associated with HAIs. The emergence and spread of MDR/XDR P. aeruginosa strains are a global public health concern. The prevalence of MDR P. aeruginosa has increased globally, comprising 30% of reported P. aeruginosa infections in some countries [20]. In Saudi Arabia, P. aeruginosa is an increasingly frequent nosocomial pathogen, accounting for 11% of all hospital-acquired infections [21]. In this study, we analysed the epidemiological, microbiological, and clinical characteristics of P. aeruginosa isolates. Respiratory samples were the most common source of MDR (58.9%) and XDR (54%) isolates, consistent with previous observations [14,21,22].
XDR P. aeruginosa was the most prevalent, followed by MDR and PDR strains. The overall prevalence rate of MDR isolates during the three years was 9.7%, which was higher than the previously reported rate of [23]. This increase coincides with the COVID-19 pandemic (2020–2021), which is consistent with the trends in the prevalence of other MDR bacteria [24,25,26]. Tiri et al., showed that the incidence of carbapenem-resistant Enterobacterales colonisation increased from 6.7% in 2019 to 50% in 2020 [24]. Similarly, a study from Jizan, Saudi Arabia, reported a significant increase in the prevalence of carbapenem-resistant enterobacterales (22.4%) during the pandemic period compared to the pre-pandemic period (5.4%) [25]. Additionally, the latest national report from the Centers for Disease Control and Prevention (CDC) reported that, from 2019 to 2020, there was a 35% increase in hospital-onset MDR P. aeruginosa infections [26]. This increased incidence could be attributed to the increased selection of resistant strains, owing to the high consumption of antibiotics to treat secondary bacterial infections associated with SARS-CoV-2 infection.
We found that the highest number of P. aeruginosa isolates, including MDR and XDR isolates, was collected from patients aged between 65 and 84 years, consistent with the findings of previous studies [6,22]. Older adults are generally at a higher risk of developing nosocomial infections, especially MDR infections, owing to their diminished immune response and increased prevalence of comorbidities, such as cardiovascular disease and diabetes [27]. Additionally, we found that MDR and XDR P. aeruginosa isolates were more predominant in males (61%) than females (38.2%). This is consistent with the findings of both national and international studies, showing that the prevalence and drug resistance of Gram-negative pathogens, including P. aeruginosa, are higher in males than females [14,22,28,29]. Males are generally more susceptible to bacterial infections due to physiological factors related to sex chromosomes and hormones. The variation between males and females can also be linked to behavioural differences, such as adherence to treatment or smoking rates [28]. Interestingly, we found that PDR isolates were more prevalent in females (63.6%) than males (36.4%), though the causative factors are unclear.
Globally, the overuse of antibiotics has changed the antimicrobial resistance profile of P. aeruginosa, resulting in an increase in the prevalence of carbapenemase- or β-lactamase-producing strains and the development of resistance to colistin. We assessed the resistance profiles of P. aeruginosa isolates against commonly used antibiotics and observed high resistance rates against monobactams (aztreonam [29.8%]), carbapenems (imipenem [29.5%] and meropenem [25.6%]), and cephalosporins (cefepime [24.3%] and ceftazidime [26.1%]). Overall, XDR P. aeruginosa isolates showed higher resistance rates to carbapenems and cephalosporins than the MDR isolates. We found that amikacin and colistin were the most effective antibiotics, with high susceptibility rates of 92.6% and 90.4%, respectively, which is consistent with the findings of previous studies [30,31,32,33,34].
In Saudi Arabia, the overall level of cephalosporin resistance in P. aeruginosa is low compared with resistance rates in neighbouring countries (96% and 86% in Qatar and Bahrain, respectively) [23]. A recent nationally-representative survey of P. aeruginosa clinical isolates collected in March 2018 and April 2019 reported resistance rates of 14.17% and 8.53% to ceftazidime and cefepime, respectively, with the highest resistance levels observed in Riyadh (19.14% and 10.49%, respectively) [35]. In our study, the resistance rates against cephalosporin were higher than those previously reported [23,35]. We found that the overall resistance rates among all P. aeruginosa isolates were 26.1% and 24.4% against ceftazidime and cefepime, respectively. For XDR and MDR isolates, the resistance rates against ceftazidime were 96% and 83%, respectively, whereas the resistance rates against cefepime were 93% and 76%, respectively. This increased resistance rate may be associated with selective pressure exerted by the overuse of broad-spectrum antibiotics, including cephalosporins, to treat or prevent secondary infections in COVID-19 patients [36].
Our study reported high resistance rates against carbapenems, including imipenem and meropenem, of up to 96% among MDR and XDR isolates. This result is of particular concern as carbapenems are the “last resort” drugs to treat severe nosocomial infections caused by MDR Gram-negative bacteria, including P. aeruginosa [37]. In Saudi Arabia, the prevalence of infections caused by carbapenemase-producing organisms, mainly Enterobacterales, Acinetobacter spp., and Pseudomonas spp., has increased in recent years [38,39]. Most of these infections occur in hospitalised patients and are associated with high carbapenem resistance rates. It has been shown that P. aeruginosa isolates from UTI samples have a concerning resistance rate of 50% against meropenem and 52.4% against imipenem [23,39]
The emergence and spread of carbapenem resistance has led to the reuse of colistin (a last-resort drug) to treat carbapenem-resistant Gram-negative bacteria, including P. aeruginosa [6,40]. We observed a comparatively lower resistance rate against colistin (1.1%). Colistin remains highly effective in the Middle East and North Africa Region, reaching 100% in most countries where some MDR P. aeruginosa isolates remain susceptible to colistin [30,41]. The prevalence of colistin resistance has recently increased in Saudi Arabia, reaching 8% and 30% [14,42]. Although low, the level of colistin resistance reported in this study should serve as a warning against the mismanagement of this drug in clinical practice. Therefore, continued surveillance of drug resistance is essential to control the spread of colistin-resistant pathogens.
In this study, 255 multi-resistant P. aeruginosa isolates were obtained from adult ICU patients, mainly from respiratory sources (69.8%). Most ICU patients (44.3%) were aged 65-84 years. ICU patients are at high risk of HAIs [43]. In the European Union countries, P. aeruginosa is the most common culprit of ICU-acquired pneumonia, UTIs, and bloodstream infections [44]. A comprehensive surveillance study performed in 14 countries of the Middle East demonstrated that, after Escherichia coli (7%) and Klebsiella pneumoniae (8%), P. aeruginosa (5%) was the most common pathogen responsible for peripheral venous catheters-related bloodstream infections (PVCR-BSIs) in ICU patients [45]. In Saudi Arabia, the prevalence of MDR P. aeruginosa in ICU patients has remained high over the past few decades (30.6% in 2004–2009 and 35.7% in 2016) [30]. Moreover, a prospective cohort study found that P. aeruginosa was responsible for 21.7% of VAP among adult patients in ICU [46]. Several studies reported a high prevalence of antimicrobial-resistant bacterial infections among COVID-19 patients admitted to ICU [24,47]. A recent study from Makkah, Saudi Arabia, showed that a large percentage of bacteria (57.1–76.2%) that infected ICU COVID-19 patients were resistant to multiple antibiotics, including Ampicillin, Ciprofloxacin, Levofloxacin, Imipenem, and Moxifloxacin [48]. We reported an increase in the prevalence of MDR P. aeruginosa among ICU COVID-19 patients from 17.9% in 2020 to 36.5% in 2021. Infections with MDR P. aeruginosa are often associated with severe adverse clinical outcomes, including an increase in the duration of hospital stay, as well as mortality and morbidity rates [6,49]. In our study, several co-morbidities, including UTI, pneumonia, and septic shock, were identified in ICU patients infected with MDR P. aeruginosa, with the most common comorbidities being respiratory tract infections (76%) and pneumonia (45%). Our analysis also showed that mechanical ventilation was strongly associated with MDR P. aeruginosa infections in ICU patients during 2020/2021. Mechanical ventilators are widely used in ICUs and are a well-known source of nosocomial infections, mainly VAP [50]. Balkhy et al., showed that, in an adult ICU in Riyadh, VAP-associated P. aeruginosa had 13–31% resistance rates to carbapenems, third-generation cephalosporins, fluoroquinolones, aminoglycosides, and antipseudomonal penicillins [51]. Mechanical ventilators are also essential for the respiratory support of critically ill patients with COVID-19, which may explain the increased rates of pneumonia cases in our study during 2020/2021.
The high mortality rates of ICU patients infected with MDR P. aeruginosa are a major concern. Mortality rates associated with MDR P aeruginosa can be up to four times higher than those associated with non-MDR P aeruginosa [52]. Our results demonstrated an overall high mortality rate among ICU patients during the three years (55.7%, 142/255). We observed a decline in the mortality rate from 62.3% in 2019 to 44.9% and 57.1% in 2020 and 2021, respectively. Overall, mortality rates in our study were significantly higher than those reported in USA and Europe (35.7%) [20]. As expected, mortality rates in ICU patients aged 65–84 years old were the highest (38.9%). Furthermore, the mortality rate was significantly higher in patients with septic shock, kidney disease, liver disease, cancer, and those on mechanical ventilation. An alarmingly high percentage of ICU patients had a respiratory infection (51.1%). With further analysis of the relative risk associated with mortality in the ICU patient group, septic shock and liver disease were significantly associated with mortality. The liver plays an essential role in clearing pathogenic bacteria, and there is a high association between liver dysfunction and sepsis. Pre-existing liver dysfunction is a risk factor for developing sepsis and septic shock and is strongly associated with mortality in patients with sepsis. Additionally, sepsis induces liver dysfunction, which is considered a late manifestation of sepsis-induced multiple organ dysfunction syndrome [53]. Our result suggests that patients with pre-existing liver disease and sepsis are at particularly high risk of death due to infection with MDR P. aeruginosa.
Our study design has several strengths. The sampling effort comprised a range of diverse sample sources from various sites that enabled an analysis of the type of infection associated with different sources. Additionally, the large sample size of ICU patients allowed us to make confident conclusions. However, the study’s main limitation is that it only considered data from a single tertiary institution. A large-scale multi-centre surveillance system in Riyadh and other cities in Saudi Arabia is needed to fully elucidate the threat of P. aeruginosa, among other drug-resistant pathogens.
5. Conclusions
Although P. aeruginosa can still be treated with anti-pseudomonal drugs, trends in its resistance profile require a large-scale monitoring effort in Saudi Arabia. Our study and the data from recent studies reported an increase in antimicrobial resistance during the COVID-19 pandemic. The observed increase in resistance could be attributed to the extensive and inappropriate use of antibiotics as preventive and therapeutic management of COVID-19, which can increase the selective pressure for antimicrobial resistance [47]. We did not address in our study the antibiotic consumption during the pandemic, nevertheless, we believe that the local pattern of antibiotic prescription during the pandemic is similar to the global pattern, which showed an increase in antibiotic use during the pandemic [54,55]. Our study provides well-supported evidence for MDR P. aeruginosa being a high-risk pathogen in Saudi Arabia, especially against older adults and ICU patients.
Acknowledgments
The authors would like to express their thanks to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project no. (IFKSURG-2-490) for funding this project. Furthermore, we express our gratitude to the Microbiology Department at King Fahad Medical City in Riyadh for supporting data collection, as well as to Abeer Al Mazyed, who assisted with data collection. Additionally, the authors thank Prince Naif Health Research Center, Investigator Support Unit for the language editing service provided.
Author Contributions
T.A.H. theorised and designed the study, reviewed the literature, acquired the data, wrote the original draft, and critically revised the manuscript. E.A.B.E. contributed to the conceptualisation and design of the study, interpreted the results, wrote the manuscript, performed critical revisions, and edited the final version of the manuscript. A.S.A. managed the data collection and analysis. Z.S.A. interpreted the results and edited the final version of the manuscript. S.R.A. contributed to the discussion. M.A.M. critically commented on and edited the final version of the manuscript. F.A. and N.A.A. contributed to the study design, managed the data collection, and interpreted the results. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of King Fahad Medical City, Saudi Arabia (IRB log number: 21-063E).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data available in the KFMC Institute data system and could be available for public upon special request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was funded by the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project no. (IFKSURG-2-490).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Arber W. Horizontal gene transfer among bacteria and its role in biological evolution. Life. 2014;4:217–224. doi: 10.3390/life4020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sando E., Suzuki M., Ishida M., Yaegashi M., Aoshima M., Ariyoshi K., Morimoto K. Definitive and indeterminate Pseudomonas aeruginosa infection in adults with community-acquired pneumonia: A prospective observational study. Ann. Am. Thorac. Soc. 2021;18:1475–1481. doi: 10.1513/AnnalsATS.201906-459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Restrepo M.I., Babu B.L., Reyes L.F., Chalmers J.D., Soni N.J., Sibila O., Faverio P., Cilloniz C., Rodriguez-Cintron W., Aliberti S. Burden and risk factors for Pseudomonas aeruginosa community-acquired pneumonia: A multinational point prevalence study of hospitalised patients. Eur. Respir.J. 2018;52:1701190. doi: 10.1183/13993003.01190-2017. [DOI] [PubMed] [Google Scholar]
- 4.Afshari A., Pagani L., Harbarth S. Year in review 2011: Critical care–infection. Crit. Care. 2012;16:242. doi: 10.1186/cc11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moradali M.F., Ghods S., Rehm B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horcajada J.P., Montero M., Oliver A., Sorlí L., Luque S., Gómez-Zorrilla S., Benito N., Grau S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019;32:e00031-19. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sousa T., Hébraud M., Dapkevicius M.L., Maltez L., Pereira J.E., Capita R., Alonso-Calleja C., Igrejas G., Poeta P. Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021;22:12892. doi: 10.3390/ijms222312892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang W., Yoon S.S. Virulence characteristics and an action mode of antibiotic resistance in multidrug-resistant Pseudomonas aeruginosa. Sci. Rep. 2019;9:487. doi: 10.1038/s41598-018-37422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Centre for Disease Prevention and Control . Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2021. ECDC; Stockholm, Sweden: 2022. [Google Scholar]
- 10.Gill J.S., Arora S., Khanna S.P., Kumar K.H. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level intensive care unit. J. Glob. Infect. Dis. 2016;8:155. doi: 10.4103/0974-777X.192962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales E., Cots F., Sala M., Comas M., Belvis F., Riu M., Salvadó M., Grau S., Horcajada J.P., Montero M.M., et al. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health Serv. Res. 2012;12:122. doi: 10.1186/1472-6963-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaye K.S., Pogue J.M. Infections caused by resistant gram-negative bacteria: Epidemiology and management. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2015;35:949–962. doi: 10.1002/phar.1636. [DOI] [PubMed] [Google Scholar]
- 13.Alhussain F.A., Yenugadhati N., Al Eidan F.A., Al Johani S., Badri M. Risk factors, antimicrobial susceptibility pattern and patient outcomes of Pseudomonas aeruginosa infection: A matched case-control study. J. Infect. Public Health. 2021;14:152–157. doi: 10.1016/j.jiph.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Ibrahim M.E. High antimicrobial resistant rates among gram-negative pathogens in intensive care units: A retrospective study at a tertiary care hospital in Southwest Saudi Arabia. Saudi Med.J. 2018;39:1035. doi: 10.15537/smj.2018.10.22944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raman G., Avendano E.E., Chan J., Merchant S., Puzniak L. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control. 2018;7:1–14. doi: 10.1186/s13756-018-0370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez A., Gato E., Pérez-Llarena J., Fernández-Cuenca F., Gude M.J., Oviaño M., Pachón M.E., Garnacho J., González V., Pascual Á., et al. High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob.Chemother. 2019;74:1244–1252. doi: 10.1093/jac/dkz030. [DOI] [PubMed] [Google Scholar]
- 17.Rosenthal V.D., Jin Z., Memish Z.A., Daboor M.A., Ruzzieh M.A.A., Hussien N.H., Guclu E., Olmez-Gazioglu E., Ogutlu A., Agha H.M., et al. Risk factors for mortality in ICU patients in 10 middle eastern countries: The role of healthcare-associated infections. J. Crit. Care. 2022;72:154149. doi: 10.1016/j.jcrc.2022.154149. [DOI] [PubMed] [Google Scholar]
- 18.Hafiz T.A., Aldawood E., Albloshi A., Alghamdi S.S., Mubaraki M.A., Alyami A.S., Aldriwesh M.G. Stenotrophomonas maltophilia Epidemiology, Resistance Characteristics, and Clinical Outcomes: Understanding of the Recent Three Years’ Trends. Microorganisms. 2022;10:2506. doi: 10.3390/microorganisms10122506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 20.Micek S.T., Wunderink R.G., Kollef M.H., Chen C., Rello J., Chastre J., Antonelli M., Welte T., Clair B., Ostermann H., et al. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: Impact of multidrug resistance. Crit. Care. 2015;19:219. doi: 10.1186/s13054-015-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yezli S., Shibl A.M., Livermore D.M., Memish Z.A. Prevalence and antimicrobial resistance among Gram-negative pathogens in Saudi Arabia. J. Chemother. 2014;26:257–272. doi: 10.1179/1973947814Y.0000000185. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed O.B. Incidence and antibiotic susceptibility pattern of pseudomonas aeruginosa isolated from inpatients in two Tertiary Hospitals. Clin. Microbiol. Open Access. 2016;5:2. [Google Scholar]
- 23.Al-Orphaly M., Hadi H.A., Eltayeb F.K., Al-Hail H., Samuel B.G., Sultan A.A., Skariah S. Epidemiology of Multidrug-Resistant Pseudomonas aeruginosa in the Middle East and North Africa Region. Msphere. 2021;6:e00202-21. doi: 10.1128/mSphere.00202-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiri B., Sensi E., Marsiliani V., Cantarini M., Priante G., Vernelli C., Martella L.A., Costantini M., Mariottini A., Andreani P., et al. Antimicrobial Stewardship Program, COVID-19, and Infection Control: Spread of Carbapenem-Resistant Klebsiella Pneumoniae Colonization in ICU COVID-19 Patients. What Did Not Work? J. Clin. Med. 2020;9:2744. doi: 10.3390/jcm9092744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.AlDiba M., Daghriri A.M.H., Jamali E.M.E., Alzahrani A., Alsharif A.B., Almudeer H.G., Khobrani H., Hakami N.T.M., Ghannam W.M., Alasy H.M. Prevalence of Antimicrobial Resistance of Common Bacterial Isolates before and during COVID-19 Pandemic in Armed Forces Hospital Jazan, Saudi Arabia. Eur. J. Med. Health Sci. 2021;3:31–38. doi: 10.24018/ejmed.2021.3.5.1047. [DOI] [Google Scholar]
- 26.Centers for Disease; National Center for Emerging and Zoonotic Infectious Diseases; Division of Healthcare Quality Promotion, editor. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. The Centers for Disease Control and Prevention (CDC); Hyattsville, MD, USA: 2022. [DOI] [Google Scholar]
- 27.Augustine S., Bonomo R.A. Taking stock of infections and antibiotic resistance in the elderly and long-term care facilities: A survey of existing and upcoming challenges. Eur. J. Microbiol. Immunol. 2011;1:190–197. doi: 10.1556/EuJMI.1.2011.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dias S.P., Brouwer M.C., Beek D.V.D. Sex and Gender Differences in Bacterial Infections. Infect. Immun. 2022;90:e00283-22. doi: 10.1128/iai.00283-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGregor J.C., Elman M.R., Bearden D.T., Smith D.H. Sex- and age-specific trends in antibiotic resistance patterns of Escherichia coli urinary isolates from outpatients. BMC Fam. Pract. 2013;14:25. doi: 10.1186/1471-2296-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azim N.S.A., Al-Harbi M.A., Al-Zaban M.I., Nofal M.Y., Somily A.M. Prevalence and Antibiotic Susceptibility among Gram Negative Bacteria Isolated from Intensive Care Units at a Tertiary Care Hospital in Riyadh, Saudi Arabia. J. Pure Appl. Microbiol. 2019;13:201–208. doi: 10.22207/JPAM.13.1.21. [DOI] [Google Scholar]
- 31.Galani I., Papoutsaki V., Karantani I., Karaiskos I., Galani L., Adamou P., Deliolanis I., Kodonaki A., Papadogeorgaki E., Markopoulou M., et al. In vitro activity of ceftolozane/tazobactam alone and in combination with amikacin against MDR/XDR Pseudomonas aeruginosa isolates from Greece. J. Antimicrob. Chemother. 2020;75:2164–2172. doi: 10.1093/jac/dkaa160. [DOI] [PubMed] [Google Scholar]
- 32.del Barrio-Tofiño E., Zamorano L., Cortes-Lara S., López-Causapé C., Sánchez-Diener I., Cabot G., Bou G., Martínez-Martínez L., Oliver A., GEMARA-SEIMC/REIPI Pseudomonas Study Group Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J. Antimicrob. Chemother. 2019;74:1825–1835. doi: 10.1093/jac/dkz147. [DOI] [PubMed] [Google Scholar]
- 33.Said K.B., Al-Jarbou A.N., Alrouji M. Surveillance of antimicrobial resistance among clinical isolates recovered from a tertiary care hospital in Al Qassim, Saudi Arabia. Int. J. Health Sci. 2014;8:3–12. doi: 10.12816/0006066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alnasser AH A., Al-Tawfiq J.A., Ahmed HA A., Alqithami SM H., Alhaddad ZM A., Rabiah AS M., Albrahim M.A.A., Al Kalif M.S.H., Barry M., Temsah M.-H., et al. Public knowledge, attitude and practice towards antibiotics use and antimicrobial resistance in Saudi Arabia: A web-based cross-sectional survey. J. Public Health Res. 2021;10:2276. doi: 10.4081/jphr.2021.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doumith M., Alhassinah S., Alswaji A., Alzayer M., Alrashidi E., Okdah L., Aljohani S., Balkhy H.H., Alghoribi M.F., NGHA AMR Surveillance Group Genomic Characterization of Carbapenem-Non-susceptible Pseudomonas aeruginosa Clinical Isolates From Saudi Arabia Revealed a Global Dissemination of GES-5-Producing ST235 and VIM-2-Producing ST233 Sub-Lineages. Front. Microbiol. 2022;12:4155. doi: 10.3389/fmicb.2021.765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beović B., Doušak M., Ferreira-Coimbra J., Nadrah K., Rubulotta F., Belliato M., Berger-Estilita J., Ayoade F., Rello J., Erdem H. Antibiotic use in patients with COVID-19: A ‘snapshot’ Infectious Diseases International Research Initiative (ID-IRI) survey. J. Antimicrob. Chemother. 2020;75:3386–3390. doi: 10.1093/jac/dkaa326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Codjoe F.S., Donkor E.S. Carbapenem Resistance: A Review. Med.Sci. 2018;6:1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moghnieh R.A., Kanafani Z.A., Tabaja H.Z., Sharara S.L., Awad L.S., Kanj S.S. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. Lancet Infect. Dis. 2018;18:e379–e394. doi: 10.1016/S1473-3099(18)30414-6. [DOI] [PubMed] [Google Scholar]
- 39.Alotaibi F. Carbapenem-Resistant Enterobacteriaceae: An update narrative review from Saudi Arabia. J. Infect. Public Health. 2019;12:465–471. doi: 10.1016/j.jiph.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 40.Falagas M.E., Kasiakou S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005;40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 41.Ramadan R.A., Gebriel M.G., Kadry H.M., Mosallem A. Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: Characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infect. Drug Resist. 2018;11:1261–1269. doi: 10.2147/IDR.S170233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saeed N.K., Kambal A.M., El-Khizzi N.A. Antimicrobial-resistant bacteria in a general intensive care unit in Saudi Arabia. Saudi Med. J. 2010;31:1341. [PubMed] [Google Scholar]
- 43.Blot S., Ruppé E., Harbarth S., Asehnoune K., Poulakou G., Luyt C.-E., Rello J., Klompas M., Depuydt P., Eckmann C., et al. Healthcare-associated infections in adult intensive care unit patients: Changes in epidemiology, diagnosis, prevention and contributions of new technologies. Intensive Crit. Care Nurs. 2022;70:103227. doi: 10.1016/j.iccn.2022.103227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.European Centre for Disease Prevention and Control . ECDC. Annual Epidemiological Report for 2017. ECDC; Stockholm, Sweden: 2019. Healthcare-Associated Infections Acquired in Intensive Care Units. [Google Scholar]
- 45.Rosenthal V.D., Belkebir S., Zand F., Afeef M., Tanzi V.L., Al-Abdely H.M., El-Kholy A., AlKhawaja S.A.A., Demiroz A.P., Sayed A.F., et al. Six-year multicenter study on short-term peripheral venous catheters-related bloodstream infection rates in 246 intensive units of 83 hospitals in 52 cities of 14 countries of Middle East: Bahrain, Egypt, Iran, Jordan, Kingdom of Saudi Arabia, Kuwait, Lebanon, Morocco, Pakistan, Palestine, Sudan, Tunisia, Turkey, and United Arab Emirates-International Nosocomial Infection Control Consortium (INICC) findings. J. Infect. Public Health. 2020;13:1134–1141. doi: 10.1016/j.jiph.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 46.El-Saed A., Balkhy H.H., Al-Dorzi H.M., Khan R., Rishu A.H., Arabi Y. Acinetobacter is the most common pathogen associated with late-onset and recurrent ventilator-associated pneumonia in an adult intensive care unit in Saudi Arabia. Int. J. Infect. Dis. 2013;17:e696–e701. doi: 10.1016/j.ijid.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Langford B.J., So M., Simeonova M., Leung V., Lo J., Kan T., Raybardhan S., E Sapin M., Mponponsuo K., Farrell A., et al. Antimicrobial resistance in patients with COVID-19: A systematic review and meta-analysis. Lancet Microbe. 2023;4:e179–e191. doi: 10.1016/S2666-5247(22)00355-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabrah A., Bahwerth F., Alghamdi S., Alkhotani A., Alahmadi A., Alhuzali M., Aljerary I., Alsulami A. Antibiotics Usage and Resistance among Patients with Severe Acute Respiratory Syndrome Coronavirus 2 in the Intensive Care Unit in Makkah, Saudi Arabia. Vaccines. 2022;10:2148. doi: 10.3390/vaccines10122148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maraolo A.E., Cascella M., Corcione S., Cuomo A., Nappa S., Borgia G., De Rosa F.G., Gentile I. Management of multidrug-resistant Pseudomonas aeruginosa in the intensive care unit: State of the art. Expert Rev. Anti Infect. Ther. 2017;15:861–871. doi: 10.1080/14787210.2017.1367666. [DOI] [PubMed] [Google Scholar]
- 50.Blot S., Koulenti D., Dimopoulos G., Martin C., Komnos A., Krueger W.A., Spina G., Armaganidis A., Rello J. Prevalence, Risk Factors, and Mortality for Ventilator-Associated Pneumonia in Middle-Aged, Old, and Very Old Critically Ill Patients*. Crit. Care Med. 2014;42:601–609. doi: 10.1097/01.ccm.0000435665.07446.50. [DOI] [PubMed] [Google Scholar]
- 51.Balkhy H.H., El-Saed A., Maghraby R., Al-Dorzi H.M., Khan R., Rishu A.H., Arabi Y.M. Drug-resistant ventilator associated pneumonia in a tertiary care hospital in Saudi Arabia. Ann. Thorac. Med. 2014;9:104–111. doi: 10.4103/1817-1737.128858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aloush V., Navon-Venezia S., Seigman-Igra Y., Cabili S., Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob. Agents Chemother. 2006;50:43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan J., Li S., Li S. The role of the liver in sepsis. Int. Rev. Immunol. 2014;33:498–510. doi: 10.3109/08830185.2014.889129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langford B.J., So M., Raybardhan S., Leung V., Soucy J.-P.R., Westwood D., Daneman N., MacFadden D.R. Antibiotic prescribing in patients with COVID-19: Rapid review and meta-analysis. Clin. Microbiol. Infect. 2021;27:520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nandi A., Pecetta S., Bloom D.E. Global antibiotic use during the COVID-19 pandemic: Analysis of pharmaceutical sales data from 71 countries, 2020–2022. Eclinicalmedicine. 2023;57:101848. doi: 10.1016/j.eclinm.2023.101848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available in the KFMC Institute data system and could be available for public upon special request.