Abstract
A novel method, which involves a nested PCR in a single closed tube, was developed for the sensitive detection of Erwinia amylovora in plant material. The external and internal primer pairs used had different annealing temperatures and directed the amplification of a specific DNA fragment from plasmid pEA29. The procedure involved two consecutive PCRs, the first of which was performed at a higher annealing temperature that allowed amplification only by the external primer pair. Using pure cultures of E. amylovora, the sensitivity of the nested PCR in one tube was similar to that of a standard nested PCR in two tubes. The specificity and sensitivity were greater than those of standard PCR procedures that used a single primer pair. The presence of inhibitors in plant material, very common in E. amylovora hosts, is overcome with this system in combination with a simple DNA extraction protocol because it eliminates many of the inhibitory compounds. In addition, it needs a very small sample volume (1 μl of DNA extracted). With 83 samples of naturally infected material, this method achieved better results than any other PCR technique: standard PCR detected 55% of positive samples, two-tube nested PCR detected 71% of positive samples, and nested PCR in a single closed tube detected 78% of positive samples. When analyzing asymptomatic plant material, the number of positive samples detected by the developed nested PCR was also the highest, compared with the PCR protocols indicated previously (17, 20, and 25% of 251 samples analyzed, respectively). This method is proposed for the detection of endophytic and epiphytic populations of E. amylovora in epidemiological studies and for routine use in quarantine surveys, due to its high sensitivity, specificity, speed, and simplicity.
Erwinia amylovora, the causal agent of fire blight, is one of the most destructive plant-pathogenic bacteria, affecting different rosaceous species of economical importance (pear, apple, loquat, and several ornamental species). This pathogen moves from one geographical area to another in very diverse and effective ways (3, 7, 34, 40, 41, 44) and in the last 20 years has undergone a rapid spread to many countries around the world (35, 42; T. van der Zwet, Abstr. 8th Int. Workshop Fire Blight, p. 30, 1998). E. amylovora can survive as an endophyte and an epiphyte (5, 8, 17), and its systemic distribution in plants has been demonstrated (28, 36). This has prompted in the last years an increasing interest for reliable and sensitive methods to analyze potentially infected but symptomless plant material, because the inadvertent introduction of infected plants to pathogen-free areas could result in the unstoppable spread of E. amylovora (10). This in fact might have been the reason for some of the outbreaks in certain Mediterranean countries, which for many years have imported host plants from North European countries where the disease is present.
The already available methods (2, 6, 9, 11, 13, 14, 15, 18, 21, 23, 26, 27, 31, 35) allow reliable detection of the pathogen with a relatively good level of sensitivity in plant material with symptoms, but all have some drawbacks. Isolation takes several days and needs confirmation of the identity of the pathogen by other techniques (6, 15, 21). Serological techniques are not sensitive enough, except the enrichment–enzyme-linked immunosorbent assay (ELISA) method (9), although it requires 3 days to complete and the sensitivity could be affected by other bacteria present in the sample. PCR inhibitors, which are very common in fire blight hosts, present a serious drawback for conventional PCR techniques (13, 23, 27, 32). Furthermore, the actual population of epiphytic and endophytic E. amylovora in symptomless plant material could be well below the detection levels of these techniques. The implementation of methodologies that overcome the above problems is therefore necessary. In countries affected by fire blight, such methods could help to improve the knowledge of the pathogen life cycle under their specific ecological conditions. Additionally, the availability of simple and sensitive protocols to analyze imported material and to perform quarantine surveys is crucial in those countries that are still free of the disease.
The rapidity and sensitivity of detection of this pathogen are desirable characteristics that have been met by the use of a nested-PCR procedure (27). However, the introduction of a second amplification step, and the concomitant manipulation of the previously amplified material, could lead to a significant increase of false positives due to cross-contamination, making this approach too risky for routine analysis. A realistic alternative to avoid the manipulation of the PCR tubes between the first and second round of amplification is the nested PCR in one tube (25, 27, 29, 30).
In this study, we describe the development of a nested PCR in a single closed tube which gives sensitivity levels equal to or higher than those of previous detection methods and saves both time and reagents. This method greatly reduces the cross-contamination risks and, due to the low volume of sample used, is unaffected by the presence of PCR inhibitors. The application of this method to several host plants (apple, loquat, pear, quince, Cotoneaster spp., Crataegus spp., and Pyracantha spp.) and different plant material (flowers, buds, shoots, stems, fruits, and leaves) produced satisfactory results in all cases. Combined with an efficient DNA extraction protocol previously developed in our laboratory (22), this procedure could be used as a rapid and sensitive technique for the routine detection of E. amylovora in plant material.
MATERIALS AND METHODS
Bacterial strains and sensitivity studies.
The E. amylovora strains employed in this study and their origins are listed in Table 1. The specificity tests were carried out with 71 E. amylovora strains, 24 strains from other plant pathogenic species (one Agrobacterium tumefaciens strain, one Agrobacterium vitis strain, one Brenneria nigrifluens strain, one Brenneria rubrifaciens strain, one Brenneria quercina strain, six Pectobacterium carotovorum subsp. carotovorum strains, one Pseudomonas corrugata strain, eight Pseudomonas syringae strains, one Pseudomonas savastanoi pv. savastanoi strain, one Ralstonia solanacearum strain, one Xylophilus ampelinus strain, and one Xanthomonas vesicatoria strain) and 16 strains of saprophytic bacteria isolated from fire blight hosts (5 identified as Pantoea agglomerans strains and 11 identified as Pseudomonas fluorescens strains). All the strains were grown on King's medium B (18) at 25°C for 48 h, and a suspension of each culture (ca. 108 CFU/ml) was prepared for the PCRs in sterile ultrapure water.
TABLE 1.
Summary of specificity assays with several E. amylovora strains using the nested procedure in one closed tube
| E. amylovora straina | Host | Origin | Size of band obtained by PCR amplificationb
|
||
|---|---|---|---|---|---|
| 447 bp | Intermediate | 391 bp | |||
| CFBP 179 | Pyrus communis | United States | + | ||
| CFBP 1430 | Pyrus communis | France | + | ||
| CFBP 2314 | Malus sylvestris | France | + | ||
| CFBP 2582 | Pyrus communis | Sweden | + | ||
| GCCM 841 | Pyrus communis | Cyprus | + | ||
| GCCM 909 | Pyrus communis | Greece | + | ||
| GCCM 1634 | Sorbus sp. | Czechoslovakia | + | ||
| IVIA 1867 | Pyrus communis | Spain | + | ||
| IVIA 1897 | Malus sylvestris | Spain | + | ||
| IVIA 1898 | Sorbus aucuparia | Spain | + | ||
| IVIA 1909 | Pyrus communis | Spain | + | ||
| IVIA 1924 | Pyracantha sp. | Spain | + | ||
| IVIA 1952 | Pyracantha sp. | Spain | + | ||
| IVIA 1961 | Pyrus pyrifolia | Spain | + | ||
| IVIA 1966-1 | Pyracantha sp. | Spain | + | ||
| IVIA 1966-2 | Pyracantha sp. | Spain | + | ||
| IVIA 1966-3 | Pyracantha sp. | Spain | + | ||
| IVIA 1967 | Pyracantha sp. | Spain | + | ||
| NCPPB 311 | Pyrus communis | Canada | + | ||
| NCPPB 595 | Pyrus communis | United Kingdom | + | ||
| NCPPB 683 | Pyrus communis | United Kingdom | + | ||
| NCPPB 1734 | Pyrus communis | Egypt | + | ||
| NCPPB 1819 | Crataegus sp. | United States | + | ||
| NCPPB 1859 | Rubus idaeus | United States | + | ||
| NCPPB 2080 | Pyrus communis | New Zealand | + | ||
| NCPPB 2291 | Rubus idaeus | United States | + | ||
| NCPPB 2292 | Rubus idaeus | United States | + | ||
| NCPPB 2293 | Rubus idaeus | United States | + | ||
| NCPPB 2791 | Pyrus communis | United States | + | ||
| NCPPB 2950 | Rubus sp. | United States | + | ||
| NCPPB 3159 | Malus sylvestris | The Netherlands | + | ||
| NCPPB 3548 | Eriobotrya japonica | Turkey | + | ||
| PMV 6089 | Crataegus sp. | France | + | ||
| PMV 1887 | Cotoneaster lacteus | France | + | ||
| SL2156 | Cotoneaster sp. | Ireland | + | ||
| SL2159 | Sorbus sp. | Ireland | + | ||
| UPN 500 | Crataegus sp. | Spain | + | ||
| UPN 501 | Pyrus communis | Spain | + | ||
| UPN 502 | Pyrus communis | Spain | + | ||
| UPN 503 | Malus sylvestris | Spain | + | ||
| UPN 504 | Pyrus communis | Spain | + | ||
| UPN 505 | Pyrus communis | Spain | + | ||
| UPN 506 | Malus sylvestris | Spain | + | ||
| UPN 507 | Malus sylvestris | Spain | + | ||
| UPN 508 | Pyrus communis | Spain | + | ||
| UPN 509 | Malus sylvestris | Spain | + | ||
| UPN 510 | Pyrus communis | Spain | + | ||
| UPN 511 | Malus sylvestris | Spain | + | ||
| UPN 512 | Malus sylvestris | Spain | + | ||
| UPN 513 | Pyrus communis | Spain | + | ||
| UPN 514 | Pyrus communis | Spain | + | ||
| UPN 515 | Pyrus communis | Spain | + | ||
| UPN 516 | Pyrus communis | Spain | + | ||
| UPN 517 | Pyrus communis | Spain | + | ||
| UPN 518 | Malus sylvestris | Spain | + | ||
| UPN 519 | Malus sylvestris | Spain | + | ||
| UPN 520 | Malus sylvestris | Spain | + | ||
| UPN 521 | Malus sylvestris | Spain | + | ||
| UPN 522 | Malus sylvestris | Spain | + | ||
| UPN 523 | Malus sylvestris | Spain | + | ||
| UPN 524 | Malus sylvestris | Spain | + | ||
| UPN 525 | Pyracantha sp. | Spain | + | ||
| UPN 526 | Pyrus communis | Spain | + | ||
| UPN 527 | Malus sylvestris | Spain | + | ||
| UPN 528 | Malus sylvestris | Spain | + | ||
| UPN 529 | Pyracantha sp. | Spain | + | ||
| UPN 530 | Pyracantha sp. | Spain | + | ||
| UPN 531 | Pyracantha sp. | Spain | + | ||
| UPN 532 | Pyracantha sp. | Spain | + | ||
| UPN 533 | Pyracantha sp. | Spain | + | ||
| UPN 534 | Pyracantha sp. | Spain | + | ||
E. amylovora strains classified in the following collections: CFBP, Collection Française de Bactéries Phytopathogènes, Institut National de la Recherche Agronomique (INRA), Angers, France; GCCM, Greek Coordinated Collections of Microorganisms, Benaki Phytopathological Institute, Athens, Greece; IVIA, Collection of Plant Pathogenic Bacteria, Instituto Valenciano de Investigaciones Agrarias, Moncada, Spain; NCPPB, National Collection of Plant Pathogenic Bacteria, Central Science Laboratory, York, United Kingdom; PMV, Pathologie Moléculaire et Végétale, Institut National Agronomique-INRA, Paris, France; SL, State Laboratory, Bacterial Collection from Republic of Ireland, Abbotstown, Dublin, Ireland; UPN, Collection of Plant Pathogenic Bacteria, Universidad Pública de Navarra, Pamplona, Spain.
+, presence of band.
Serial dilutions ranging from 7 × 107 to 7 CFU/ml were made from a concentrated suspension of E. amylovora strain PMV 6089 (mutant of strain CFBP 1430), and 5 μl from each was used to compare the sensitivity of the different PCRs (Table 2). Similar sensitivity assays were performed with bacterial suspensions added to pear, apple, and Pyracantha extracts obtained from comminuted shoots of greenhouse-grown plants in the buffer described by Gorris et al. (9) (phosphate-buffered saline [pH 7.2] with 2% polyvinylpyrrolidone 10, 1% mannitol, 10 mM ascorbic acid, and 10 mM reduced glutathione). The bacterial suspensions were mixed with the plant extracts to give a final concentration ranging from 5 × 105 to 5 CFU/ml. Bacterial counts were in all cases confirmed by plating 50 μl from each dilution in triplicate on King's medium B. With these samples, a simple DNA extraction protocol was used (22). Briefly, 1 ml of sample was centrifuged at 10,000 × g for 10 min. The pellet was resuspended in 500 μl of extraction buffer (200 mM Tris-HCl [pH 7.5], 250 mM NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate, 2% polyvinylpyrrolidone), vortexed, and left for 1 h at room temperature with continuous shaking. After centrifugation, 450 μl of the supernatant was taken, mixed gently with 450 μl of isopropanol, and left for 1 h at room temperature. The mixture was centrifuged, the supernatant was discarded, and the dried pellet was resuspended in 200 μl of sterile water. Five microliters of DNA extract was used for standard PCRs (2, 11, 23, 27) and for the first round of the two-tube nested-PCR assay (27), while 1 μl was used for the nested PCR in a single closed tube. All the analyses were performed twice.
TABLE 2.
Sensitivities of several sets of primers designed for the detection of E. amylovora by various PCR proceduresa
| Sampleb | Lowest positive dilution (CFU/ml) detected by PCR procedure
|
||||||
|---|---|---|---|---|---|---|---|
| Bereswill et al. (2)c | McManus and Jones (27)c | Maes et al. (23)c | Guilford et al. (11)c | PEANT1-PEANT2d (this work) | S. nestede (27)c | N1Tf (this work) | |
| PMV 6089 + water | 7 × 102 | 7 × 101 | 7 × 102 | 7 × 103 | 7 × 101 | 7 × 10−1 | 7 × 10−1 |
| PMV 6089 + pear | 5 × 102 | 5 × 103 | 5 × 103 | 5 × 104 | 5 × 102 | 5 | 5 |
| PMV 6089 + apple | 5 × 103 | 5 × 102 | 5 × 102 | 5 × 104 | 5 × 102 | 5 | 5 |
| PMV 6089 + Pyracantha | 5 × 103 | 5 × 102 | 5 × 103 | 5 × 104 | 5 × 102 | 5 | 5 |
Samples of plant extracts were analyzed following DNA extraction.
Serial dilutions of E. amylovora strain PMV 6089 in water and in different plant extracts.
Reference of the primers employed on each analysis.
PEANT1-PEANT2, internal primers designed for the nested-PCR method in one tube.
S. nested, standard nested PCR (two tubes).
N1T, nested PCR in a single closed tube.
Naturally infected samples.
From naturally infected plants, we selected 83 samples that included material from parts of the plants without symptoms as well as from organs showing fire blight symptoms (Table 3). Also, 251 samples from symptomless pear, apple, loquat, quince, Pyracantha sp., Cotoneaster sp., and Crataegus sp. plants were obtained from different plots close to others with infected plants where an outbreak was detected, to monitor for potential dissemination of the pathogen (Table 4). The samples, consisting of flowers, buds, leaves, stems, and/or fruits, were prepared according to the European Plant Protection Organization (EPPO) methodology (6) for plants with symptoms using the buffer described previously (9). For symptomless plants, the EPPO method for plants with symptoms was also followed. In all samples, isolation was performed according to the standard procedure (6) on King's medium B (18) and on selective CCT medium (15). Enrichment-ELISA-double-antibody sandwich indirect (DASI) using specific monoclonal antibodies was assayed (9) after an enrichment step in King's medium B and in CCT medium (15, 18), and the DNA was extracted as described above. Greenhouse plant material previously determined to be free of the bacterium was interspersed among the samples to monitor potential cross-contamination during sample preparation. Additionally, up to five negative controls were also placed among the sample tubes during PCR analysis. All the PCR analyses, including the DNA extraction from each sample, were repeated at least twice.
TABLE 3.
Detection of E. amylovora in naturally infected plant material by various PCR procedures
| Referencea (host) | No. of samples | No. of positive samples detected with primer set
|
||||
|---|---|---|---|---|---|---|
| Bereswill et al. (2)b | McManus and Jones (27)b | Maes et al. (23)b | S. nestedc (27)b | N1Td (this work) | ||
| 1899-f (Crataegus azarolus) | 1 | 0 | 0 | 0 | 0 | 1 |
| 1913-b (Crataegus monogyna) | 2 | 0 | 0 | 0 | 0 | 1 |
| 1899-g (Crataegus sp.) | 2 | 1 | 2 | 2 | 2 | 2 |
| 1913-c (Cotoneaster dammeri) | 2 | 0 | 1 | 1 | 1 | 2 |
| 1899-e (Cydonia sp.) | 6 | 0 | 0 | 0 | 0 | 2 |
| 1899-d (Eryobotria japonica) | 1 | 0 | 0 | 0 | 1 | 1 |
| 1899-h (Malus domestica) | 4 | 1 | 1 | 1 | 1 | 1 |
| 1961-b (Malus domestica) | 3 | 3 | 3 | 3 | 3 | 3 |
| 1913-a (Pyracantha sp.) | 2 | 0 | 1 | 0 | 0 | 1 |
| 1952-b (Pyracantha sp.) | 15 | 9 | 9 | 8 | 13 | 13 |
| 1961-c (Pyracantha sp.) | 5 | 4 | 3 | 3 | 5 | 5 |
| 1966-b (Pyracantha sp.) | 7 | 6 | 6 | 6 | 6 | 6 |
| 2112-b (Pyracantha sp.) | 5 | 4 | 3 | 4 | 4 | 4 |
| 1892-b (Pyrus communis) | 11 | 6 | 9 | 6 | 10 | 10 |
| 1961-d (Pyrus communis) | 5 | 4 | 3 | 4 | 4 | 4 |
| 1961-e (Pyrus communis) | 5 | 4 | 2 | 4 | 4 | 4 |
| 1961-f (Pyrus communis) | 1 | 0 | 0 | 0 | 1 | 1 |
| 1961-g (Pyrus communis) | 5 | 1 | 2 | 2 | 3 | 3 |
| 1961-h (Pyrus pyrifolia) | 1 | 1 | 1 | 1 | 1 | 1 |
| Total no. of samples | 83 | 44 | 46 | 45 | 59 | 65 |
Samples with different reference numbers (not including suffix letter) have different origins.
Reference of the primers employed on each analysis.
S. nested, standard nested PCR (two tubes).
N1T, nested PCR in a single closed tube.
TABLE 4.
Detection of E. amylovora in symptomless plant material by various PCR procedures
| Referencea (host) | No. of samples | No. of positive samples detected with each set
|
||||
|---|---|---|---|---|---|---|
| Bereswill et al. (2)b | McManus and Jones (27)b | Maes et al. (23)b | S. nestedc (27)b | N1Td (this work) | ||
| 1898-a (Cotoneaster sp.) | 1 | 1 | 1 | 1 | 1 | 1 |
| 1910-a (Crataegus sp.) | 6 | 0 | 1 | 0 | 1 | 4 |
| 1899-c (Cydonia sp.) | 2 | 0 | 0 | 1 | 0 | 1 |
| 1899-b (Eriobotrya japonica) | 1 | 0 | 0 | 0 | 1 | 1 |
| 1898-b (Malus sp.) | 2 | 1 | 1 | 1 | 1 | 1 |
| 1961-a (Malus sp.) | 8 | 7 | 7 | 7 | 7 | 7 |
| 1898-c (Pyracantha sp.) | 1 | 1 | 1 | 1 | 1 | 1 |
| 1952-a (Pyracantha sp.) | 4 | 3 | 3 | 1 | 4 | 4 |
| 1966-a (Pyracantha sp.) | 5 | 5 | 5 | 2 | 5 | 5 |
| 1967 (Pyracantha sp.) | 2 | 1 | 2 | 2 | 2 | 2 |
| 2112-a (Pyracantha sp.) | 3 | 3 | 3 | 3 | 3 | 3 |
| 2161 (Pyracantha sp.) | 30 | 1 | 1 | 0 | 1 | 1 |
| 1895 (Pyrus communis) | 14 | 0 | 1 | 1 | 2 | 3 |
| 1899-a (Pyrus communis) | 1 | 0 | 1 | 0 | 0 | 1 |
| 1886 (Pyrus sp.) | 4 | 4 | 4 | 4 | 4 | 4 |
| 1892-a (Pyrus sp.) | 1 | 0 | 0 | 0 | 1 | 1 |
| 1910-b (Pyrus sp.) | 9 | 0 | 1 | 0 | 3 | 7 |
| 2083 (Pyrus sp.) | 40 | 0 | 1 | 1 | 1 | 1 |
| 2116 (Pyrus sp.) | 18 | 0 | 8 | 10 | 12 | 13 |
| 2181 (Pyrus sp.) | 99 | 0 | 1 | 1 | 1 | 1 |
| Total no. of samples | 251 | 27 | 42 | 36 | 51 | 62 |
Samples with different reference numbers (not including suffix letter) have different origins.
Reference of the primers employed on each analysis.
S. nested, standard nested PCR (two tubes).
N1T, nested PCR in a single closed tube.
PCR design and comparison of amplifications.
We have designed the nested PCR in a single closed tube considering primers previously described because they have shown a good sensitivity and specificity in this study and in our previous work. The criteria we used for selecting the external and internal primer pairs were (i) the external primer pair should amplify a fragment large enough to permit the design of an appropriate internal couple, (ii) annealing temperatures of the primer pairs should allow for the separation of both PCRs only by this parameter, and (iii) high sensitivity of the primers, to increase as much as possible the detection threshold of the nested PCR in one tube. The standard PCRs were performed as described by Bereswill et al. (2), using primers A and B; by McManus and Jones (27), using primers AJ75-AJ76; by Maes et al. (23), using primers EAF-EAR; and by Guilford et al. (11), using primers EA71-EA72. After some sensitivity assays, we choose as external primers those designed by McManus and Jones (27), which were used at an annealing temperature of 72°C. We then designed as internal pair the primers PEANT1 (5′-TATCCCTAAAAACCTCAGTGC-3′) and PEANT2 (5′-GCAACCTTGTGCCCTTTA-3′), which lie within 844 bases of the fragment from the 29-kb plasmid pEA amplified by the external pair (27). Since PEANT1 and PEANT2 produced amplification products at 56°C but not at 72°C, it was thus possible to separate the activity of the internal and the external primer pairs by modifying the annealing temperature. PCRs were performed in a final volume of 50 μl with the following reagents: 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 3% (vol/vol) formamide, a 200 μM concentration of each deoxynucleoside triphosphate, 0.03 pmol each of external primers AJ75 and AJ76 (27), 10 pmol each of internal primers PEANT1 and PEANT2, and 3 U of Taq polymerase (Gibco BRL). The reaction conditions were a denaturation step of 94°C for 4 min followed by 25 cycles of 94°C for 30 s and 72°C for 1 min. This first round of PCR was followed in the same thermocycler by a second denaturation step of 94°C for 4 min and 40 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s. The PCR products were visualized after electrophoresis on 1.5% agarose gels.
Restriction fragment length polymorphisms and sequencing.
The restriction pattern of the amplification products obtained from the bacterial suspensions was examined with DraI and SmaI (Amersham/Pharmacia Biotech) to confirm their identity. The fragments amplified from strains CFBP 1430 and PVM 6089 with the primers designed by Bereswill et al. (2) were excised from the gel, purified using the Concert Nucleic Acid purification kit (Gibco BRL), and sequenced with the same primers. The resulting sequences were then compared to the corresponding sequence obtained from strain CA11 by McManus and Jones (27) (GenBank accession no. U19245) using the program CLUSTAL W, version 1.5 (39).
RESULTS
Sensitivity and specificity tests.
We compared the sensitivity of the nested PCR in a single closed-tube assay we developed with that of a two-tube nested PCR and other PCR procedures that used a single primer pair. The results of sensitivity assays performed with pure E. amylovora cultures are shown in Table 2. The sensitivity of the nested PCR in a single closed tube and the two-tube nested assays was 7 × 10−1 CFU/ml, while in the best case it was possible to detect only 7 × 10 cfu/ml with the standard PCR procedures. When the assays were carried out with plant extracts spiked with bacteria, the sensitivity levels of the nested procedures were slightly reduced, to 5 CFU/ml, although they were still 100 to 1,000 times more sensitive than the standard one-round PCR assays. For the sensitivity assays with plant material, we tested the effects of different amounts of sample volume, looking for a balance between minimum inhibitory effects of the extract on the PCR and the maximum sensitivity (data not shown). For the standard PCRs, the volume was 5 μl, and for the two-tube nested PCR it was 5 μl in the first round and 2 μl in the second. For the nested PCR in a single closed tube, only 1 μl of sample was necessary to obtain the strongest band signals.
The specificity of the procedure developed in this work was tested using pure cultures of 40 strains from 14 species of phytopathogenic and saprophytic bacteria. No unspecific banding was observed with any of the bacteria analyzed (data not shown), while all of the 71 E. amylovora strains examined produced a single amplification band (Table 1). With only three of these strains the amplified fragment was 447 bp long, as predicted from the sequence obtained from strain CA11 (P. McManus and A. Jones, accession no. U19245) from which the primers were designed, while 63 strains produced 391-bp fragments and bands of intermediate size were amplified from 5 strains (Table 1; Fig. 1). Nonetheless, digestion of the amplicons with DraI or SmaI in all cases produced two fragments whose sizes were as predicted or slightly smaller, supporting the identity of the amplified fragment. To investigate the reasons for the discrepancies in the size of the amplicons, we obtained the nucleotide sequence of the fragments amplified from strains CFBP 1430 and PMV 6089 (391 bp). Both sequences were identical and showed a deletion of 56 nucleotides with respect to the sequence from strain CA11, comprising seven 8-bp tandem repeats (GAATTACA) (Fig. 2).
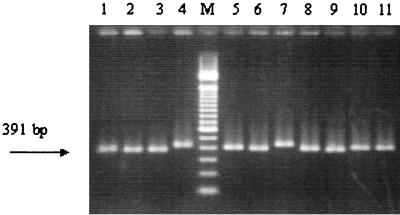
FIG. 1.
Diversity of fragments obtained after amplification following the nested-PCR method in a single closed tube. The sizes vary, including the expected 447 bp (lanes 4 and 7), 391 bp (lanes 1, 2, 3, and 9), and intermediate values (lanes 5, 6, 8, 10, and 11). Numbered lanes contain samples from the NCPPB collection (strain number and country of origin are given in parentheses): lane 1, 2292 (United States); lane 2, 2293 (United States); lane 3, 2950 (United States); lane 4, 311 (Canada); lane 5, 683 (United Kingdom); lane 6, 1734 (Egypt); lane 7, 1819 (United States); lane 8, 2080 (New Zealand); lane 9, 2791 (United States); lane 10, 3159 (The Netherlands); lane 11, 3548 (Turkey); lane M, marker (100-bp DNA ladder; Gibco BRL).
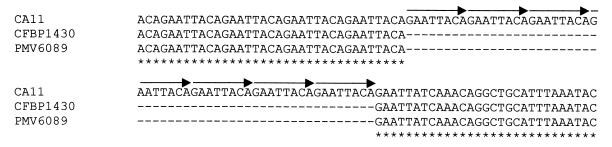
FIG. 2.
Location and extent of the deletion in the amplified fragments. The fragments amplified by the one-round PCR using primers designed by Bereswell et al. (2) from strains CFBP 1430 and PMV 6089 that gave a band of 391 bp by the nested PCR in a single closed tube were sequenced and compared to the corresponding sequence of strain CA11 (447 bp) using the program CLUSTAL W. For simplicity, only the sequence surrounding the 56-bp deletion present in strains CFBP 1430 and PMV 6089 is shown, since the rest was identical for the three strains. The 8-bp repeats are indicated by arrows.
Detection in naturally infected plant material.
To test its suitability for routine analyses, the nested PCR in a single closed tube was compared to standard one-round and two-tube nested procedures (2, 23, 27) using naturally infected plant material. The primers proposed by Guilford et al. (11) were not included in this comparison due to their low sensitivity in bacterial cultures and spiked plant material (Table 2).
We first tested our method with material from plants naturally infected with E. amylovora. All of the PCR procedures tested detected E. amylovora in samples both with and without symptoms, although the nested procedure in a single closed tube allowed the detection of the pathogen in the largest number of samples: 65 positives out of 83 samples versus 46 positives in the best case with standard one-round PCRs and 59 for the nested PCR in two tubes (Table 3). A further advantage of the nested PCR in a single closed tube is its greater specificity and thus a higher reliability for diagnosis, since no spurious bands were observed in any of the samples analyzed in this work. In contrast, using the standard PCR procedures we commonly observed the appearance of several unspecific amplification bands that hampered the interpretation of the results, as shown in Fig. 3. The presence of the pathogen in the positive samples was confirmed by its isolation in culture medium and/or by enrichment-ELISA. The controls employed to monitor the reliability of the sample preparation and the PCRs were all negative.
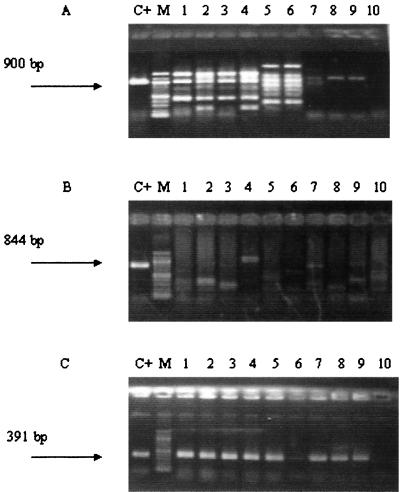
FIG. 3.
Specificity of the nested PCR in a single closed tube compared to that of other PCR methods. Samples were taken from naturally infected plants and analyzed by one-round PCR using the primers described by Bereswill et al. (2) (A), the primers described by McManus and Jones (27) (B), the nested PCR developed in this work (C). Note that the first two pairs of primers produce unspecific amplifications. Samples: 1, Pyrus communis 1892-b.1; 2, Pyracantha sp. 1952-b.5; 3, Pyracantha sp. 1952-b.9; 4, Pyracantha sp. 1952-b.11; 5, Pyrus communis 1961-d.1; 6, Pyrus communis 1961-e.2; 7, Pyrus communis 1961-e.3; 8, Pyrus communis 1961-f; 9, Pyrus communis 1961-g.3; 10, Malus domestica 1899-h. All the samples were positive except number 10. Sample number 6 gave a faint band. C+, positive control; M, marker (100 bp; New England Biolabs). The negative control is not shown in this figure.
Secondly, we tested the validity of our method for routine detection by assaying 251 samples from symptomless plants from areas where fire blight outbreaks were reported. The nested PCR in a single closed tube allowed the detection of the pathogen in 62 samples, the standard one-round PCRs allowed detection in 42 samples, and the two-tube nested PCR allowed detection in 51 samples (Table 4). The samples that were negative for E. amylovora with the single closed-tube method were also negative by the other PCR analysis. Three months after the analysis, six of the plants in which the pathogen was detected by the nested PCR in a single closed tube, but not by the other PCR procedures, developed typical fire blight symptoms and the pathogen could be recovered from affected tissues. Fifty-three out of 62 of the positive samples obtained by the proposed method could be confirmed by other methods like isolation, enrichment-ELISA, and other PCR systems (Table 5), so only nine samples remained with no confirmation.
TABLE 5.
Confirmation by other techniques of some samples positive by nested PCR in one single closed tube
| Species | Reference | Result byd:
|
||
|---|---|---|---|---|
| Other PCRa | E-ELISAb | Isolationc | ||
| Cotoneaster sp. | 1898-2 | + | − | + |
| Crataegus sp. | 1910-11 | + | − | − |
| Cydonia sp. | 1899-5 | − | − | + |
| Eriobotrya japonica | 1899-20 | + | − | − |
| Malus domestica | 1898-11 | + | − | − |
| 1961-7 | + | − | − | |
| 1961-26 | + | + | − | |
| 1961-27 | + | − | − | |
| 1961-29 | + | − | − | |
| 1961-30 | + | − | + | |
| 1961-31 | + | + | + | |
| 1961-32 | + | + | + | |
| Pyracantha sp. | 1898-7 | + | − | − |
| 1952-6 | + | − | − | |
| 1952-12 | + | − | − | |
| 1952-16 | + | − | + | |
| 1952-19 | + | + | + | |
| 1966-1 | + | + | − | |
| 1966-5 | + | − | − | |
| 1966-7 | + | − | − | |
| 1966-8 | + | − | − | |
| 1966-11 | + | + | + | |
| 1967-1 | + | − | − | |
| 1967-2 | + | − | − | |
| 2112-1 | + | + | + | |
| 2112-4 | + | + | + | |
| 2112-10 | + | + | + | |
| 2161-26 | + | − | − | |
| Pyrus sp. | 1886-2 | + | + | + |
| 1886-3 | + | + | − | |
| 1886-4 | + | + | − | |
| 1886-5 | + | + | − | |
| 1892-9 | + | − | − | |
| 1895-3 | + | − | − | |
| 1895-10 | + | − | − | |
| 1899-1 | + | − | − | |
| 1910-3 | + | − | − | |
| 1910-6 | + | − | − | |
| 1910-11 | + | − | − | |
| 2083-39 | + | − | − | |
| 2116-2 | + | − | − | |
| 2116-3 | + | − | − | |
| 2116-5 | + | − | − | |
| 2116-6 | + | − | − | |
| 2116-7 | + | − | − | |
| 2116-8 | + | − | − | |
| 2116-9 | + | − | − | |
| 2116-11 | + | − | − | |
| 2116-13 | + | − | − | |
| 2116-15 | + | + | − | |
| 2116-16 | + | + | − | |
| 2116-18 | + | − | − | |
| 2116-19 | + | − | − | |
DISCUSSION
The importance of controlling the spread of the fire blight is well known in the United States, the European Community, and other countries (17, 42; van der Zwet, Abstr. 8th Int. Workshop Fire Blight). Recent outbreaks in several countries (10, 17; P. Battilani, L. Mazzoli, and U. Mazzuchi, Abstr. 8th Int. Workshop Fire Blight, p. 17, 1998) show how difficult the control of this disease is and how fast it spreads, even when different measures of control are taken (4, 12, 16, 20, 24, 38, 43; Battilani et al., Abstr. 8th Int. Workshop Fire Blight). In addition, no symptoms are observed in winter in deciduous species, and the surveys, made mainly by visual detection of typical lesions, are useless. Apparent healthy plants can carry latent infections (5, 8, 23, 43), and from these E. amylovora could be distributed from nurseries to other parts of the country or other countries, where it will only take favorable conditions for symptoms to develop. As pointed out by other authors, in spite of being a very useful and sensitive technique, PCR is still seriously limited due to inhibition by different compounds (13, 23, 27, 32). In fact, our experience in diagnosing fire blight has shown the importance of this problem, sometimes detecting fewer positive samples by the standard PCR technique than by plating or enrichment-ELISA (data not shown). The nested PCR in a single closed tube developed in this work solves the main drawbacks of this technique, since we overcome the problem of false-negative results by reducing the volume of sample used, thus avoiding plant inhibitors, and by minimizing sample manipulations, which drastically reduces the possibility of cross-contamination. The comparison of the two nested systems with symptomless samples shows the inhibitory effect of the plant material on the PCR. The slightly larger amount of sample volume employed in the two-tube nested procedure (5 μl instead of 1 μl) seems enough to affect the first round of PCR, and thus, the whole nested reaction. The results obtained in the sensitivity assays are concordant with what we expected from the nested technology (27), the two nested systems being 100 to 1,000 times more sensitive than the standard PCR systems. The highest sensitivity of the nested PCR in a singleclosed tube, compared to the other PCR systems, was observed with asymptomatic material, with 62 positive samples versus 51 (two-tube nested PCR) and 42 (standard PCRs) (Table 4). In this assay, some of the samples that were positive by the nested PCR in a single closed tube were confirmed by other techniques, such as isolation, enrichment-ELISA, or other types of PCR (Table 5). Furthermore, 3 months later typical fire blight symptoms appeared in some of the analyzed plants. The bacterium could then be isolated, corroborating the presence of latent infections of E. amylovora, as demonstrated by other authors (5, 8, 23, 43). The use of this highly sensitive method allows a more rapid detection of the pathogen in asymptomatic plants, since it overcomes the need to wait for the results of time-consuming techniques and for the appearance of typical symptoms. Moreover, we do not know much about the survival of the bacterium or after applying different control treatments (4, 16, 17, 24, 43; Battilani et al., Abstr. 8th Int. Workshop Fire Blight). Thus, this system could be useful for monitoring the effectiveness of some of these methods.
The amplification of plasmid sequences for detection of a given pathogen could produce misleading results if (i) plasmidless cells remain virulent or (ii) the plasmid is transferred to other bacterial species (1). Nevertheless, virulent E. amylovora strains without the plasmid have not been found in nature (1, 31), and the transfer of the plasmid to other species or to other genera has not been reported. On the other hand, the advantages of using primers designed to amplify pEA29 sequences are a higher sensitivity and specificity.
The size of the amplified bands was variable in samples from plant material as well as from the E. amylovora collection strains analyzed. This can be explained by the variability in the number of 8-bp repeats in the amplified sequence, which has already been described (33). This was confirmed by the comparison of the sequence from strain CA11 with those of strains CFBP 1430 and PMV 6089, which were used as positive controls. Previous works (1, 19, 27, 33) have reported the amplification of different sized fragments, rather than the 900 bp reported by Bereswill et al. (2), from several strains from the United States and New Zealand (33) and from Europe (19). Nevertheless, these small variations in the size of the amplicons do not compromise the validity of the one-tube nested system for detection. The use of two consecutive and specific amplification reactions greatly reduces the possibility of obtaining false positives, while making it possible to further confirm the identity of the amplified fragment by restriction analysis.
The combination of the nested PCR in a single closed tube with a simple and effective DNA extraction protocol that involves little handling and does not employ toxic compounds such as phenol or chloroform (22) has led to very high levels of sensitivity. The large number of species of naturally infected plant material tested and their different origins show that the method developed here can be of universal use for fire blight detection and epidemiological applications. The probability of contamination by amplicons under the system presented here is as low as that with standard PCRs, although the sensitivity is at least as good as that of the two-tube nested PCR, thus allowing the implementation of the one-tube nested approach for routine detection. As far as we know, this is the first development of such a methodology for the detection of a bacterial plant pathogen, and the features it presents could be applied to other plant-pathogenic bacteria.
ACKNOWLEDGMENTS
We thank J. Laurent, J. P. Paulin, D. Stead, N. Alivizatos, D. Hayes, D. Berra, M. Borruel, and J. Murillo for kindly providing some of the E. amylovora strains employed and J. Cubero and B. Lastra for critical reading of the manuscript. We give special thanks to J. Murillo for extensive revision of our English and useful comments.
We are grateful to the Subdirección General de Sanidad Vegetal, MAPA, Madrid, Spain, CICYT project AGF98 0402CO302, and SMT project 4-CT98 2252 for funding.
REFERENCES
- 1.Bereswill S, Bugert P, Bruchmuller I, Geider K. Identification of the fire blight pathogen, Erwinia amylovora, by PCR assays with chromosomal DNA. Appl Environ Microbiol. 1995;61:2636–2642. doi: 10.1128/aem.61.7.2636-2642.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bereswill S, Pahl A, Bellemann P, Berger F, Zeller W, Geider K. Sensitive and species-specific detection of Erwinia amylovora by PCR analysis. Appl Environ Microbiol. 1992;58:3522–3526. doi: 10.1128/aem.58.11.3522-3526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark G G, Hickey K D, Travis J W. Fireblight management: evaluation of the role of aphids in transmission of bacteria and development of a computerized management system for growers. Penn Fruit News. 1991;71:43–44. [Google Scholar]
- 4.Covey R P, Fischer W R. Timely cutting of fire blight infections reduces losses. Acta Hortic. 1990;273:351–353. [Google Scholar]
- 5.Crepel C, Geenen J, Maes M, Bonn W G. The latent survival of Erwinia amylovora in hibernating shoots. Acta Hortic. 1996;411:21–25. [Google Scholar]
- 6.European Plant Protection Organization. Quarantine procedure no. 40. Erwinia amylovora. Sampling and test methods. Bull EPPO. 1992;22:225–231. [Google Scholar]
- 7.Ficke W, Ehrig F, Nachtigall M, Naumann K, Richter K, Schaefer H J, Zielke R. Possibilities for detecting the causal agent of fireblight (Erwinia amylovora (Burrill) Winslow et al.) in the air space of orchards. Zentbl Mikrobiol. 1990;145:121–133. [Google Scholar]
- 8.Ge Q, van der Zwet T, Bonn W G. Persistence and recovery of endophytic Erwinia amylovora in apparently healthy apple tissues. Acta Hortic. 1996;411:29–33. [Google Scholar]
- 9.Gorris M T, Cambra M, Llop P, López M M, Lecomte P, Chartier R, Paulin J P, Bonn W G. A sensitive and specific detection of Erwinia amylovora based on the ELISA-DASI enrichment method with monoclonal antibodies. Acta Hortic. 1996;411:41–46. [Google Scholar]
- 10.Griffo R, Tartaglia A, Capriolo F, Adamo F. Fire blight in Campania. Inform Agrar. 1998;54:73–75. [Google Scholar]
- 11.Guilford P J, Taylor R K, Clark R G, Hale C N, Forster R L S, Bonn W G. PCR-based techniques for the detection of Erwinia amylovora. Acta Hortic. 1996;411:53–56. [Google Scholar]
- 12.Hasler T, Vogelsanger J, Schoch B, Bonn W G. Disinfection of fire blight contaminated tools. Acta Hortic. 1996;411:369–371. [Google Scholar]
- 13.Henson J M, French R. The polymerase chain reaction and plant disease diagnosis. Annu Rev Phytopathol. 1993;31:81–109. doi: 10.1146/annurev.py.31.090193.000501. [DOI] [PubMed] [Google Scholar]
- 14.Hutschemakers J, Verhoyen M, Bazin H. Production of rat monoclonal antibodies specific to Erwinia amylovora. Acta Hortic. 1987;217:71–76. [Google Scholar]
- 15.Ishimaru C, Klos E J. New medium for detecting Erwinia amylovora and its use in epidemiological studies. Phytopathology. 1984;74:1342–1345. [Google Scholar]
- 16.Keck M, Chartier R, Zislavsky W, Lecomte P, Paulin J P. Heat treatment of plant propagation material for the control of fire blight. Plant Pathol. 1995;44:124–129. [Google Scholar]
- 17.Keck M, Reich H, Chartier R, Paulin J P, Bonn W G. First record of fire blight (Erwinia amylovora) in Austria. Preliminary experiments on the survival on fruit boxes. Acta Hortic. 1996;411:9–11. [Google Scholar]
- 18.King E O, Ward M, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 19.Lecomte P, Manceau C, Paulin J P, Keck M. Identification by PCR analysis on plasmid pEA29 of isolates of Erwinia amylovora responsible for an outbreak in Central Europe. Eur J Plant Pathol. 1997;103:91–98. [Google Scholar]
- 20.Lecomte P, Paulin J P. Chemical control of bacterial blight: balance sheet and prospects after ten years of experimentation in France. Fruit Belge. 1992;60:204–216. [Google Scholar]
- 21.Lelliot R A, Stead D E. Methods for the diagnosis of bacterial diseases of plants. Methods Plant Pathol. 1987;2:1–216. [Google Scholar]
- 22.Llop P, Caruso P, Cubero J, Morente C, López M M. A simple extraction procedure for efficient routine detection of pathogenic bacteria in plant material by polymerase chain reaction. J Microbiol Methods. 1999;37:23–31. doi: 10.1016/s0167-7012(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 23.Maes M, Garbeva P, Crepel C. Identification and sensitive endophytic detection of the fire blight pathogen Erwinia amylovora with 23S ribosomal DNA sequences and the polymerase chain reaction. Plant Pathol. 1996;45:1139–1149. [Google Scholar]
- 24.Manceau C, Gardan L, Paulin J P. Use of antibiotics to control fire blight in France: environmental hazards and established legislation. Acta Hortic. 1987;217:195–202. [Google Scholar]
- 25.Mathis A, Weber R, Kuster H, Speich R. Reliable one-tube nested PCR for detection and SSCP-typing of Pneumocystis carinii. J Eukaryot Microbiol. 1996;43:7S. doi: 10.1111/j.1550-7408.1996.tb04948.x. [DOI] [PubMed] [Google Scholar]
- 26.McLaughlin R J, Chen T A, Wells J M. Monoclonal antibodies against Erwinia amylovora: characterization and evaluation of a mixture for detection by enzyme-linked immunosorbent assay. Phytopathology. 1989;79:610–613. [Google Scholar]
- 27.McManus P S, Jones A L. Detection of Erwinia amylovora by nested PCR and PCR-dot-blot and reverse blot hybridizations. Phytopathology. 1995;85:618–623. [Google Scholar]
- 28.Momol M, Norelli J, Piccioni D, Momol E A, Gustafson H L, Cummins J N, Aldwinckle H S. Internal movement of Erwinia amylovora through symptomless apple scion tissues into the rootstock. Plant Dis. 1998;82:646–650. doi: 10.1094/PDIS.1998.82.6.646. [DOI] [PubMed] [Google Scholar]
- 29.Niepold F, Schober B. Application of the one-tube PCR technique in combination with a fast DNA extraction procedure for detecting Phytophtora infestans in infected potato tubers. Microb Res. 1997;152:345–351. [Google Scholar]
- 30.Orou A, Fechner B, Menzel H J, Utermann G. Automatic separation of two PCRs in one tube by annealing temperature. Trends Genet. 1995;11:127–128. doi: 10.1016/s0168-9525(00)89021-6. [DOI] [PubMed] [Google Scholar]
- 31.Paulin, J. P. In J. Vanneste (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora, part II. The pathogen, in press.
- 32.Pulawska J, Maes M, Deckers T, Sobiczewski P. Proceedings of the 49th International Symposium on Crop Protection. Vol. 62. 1997. The influence of pesticide contamination on detection of epiphytic Erwinia amylovora using PCR; pp. 959–962. , part IV. [Google Scholar]
- 33.Schnabel E L, Jones A L. Instability of a pEA29 marker in Erwinia amylovora previously used for strain classification. Plant Dis. 1998;82:1334–1336. doi: 10.1094/PDIS.1998.82.12.1334. [DOI] [PubMed] [Google Scholar]
- 34.Seidel M, Steffen E, Seidel D, Walter A. Survival of Erwinia amylovora (Burrill) Winslow et al. on bird feet. Arch Phytopathol Plant Protection. 1994;29:25–27. [Google Scholar]
- 35.Sobiczewski P, Deckers T, Puławska J. Fire blight (Erwinia amylovora): some aspects of epidemiology and control. Skierniewice, Poland: Research Institute of Pomology and Floriculture; 1997. [Google Scholar]
- 36.Steiner P W, Suleman P. The systemic nature of fire blight: implications for control. Penn Fruit News. 1993;73:16–22. [Google Scholar]
- 37.Thomson S V, Gouk S C, Popay A J. Proceedings of the Forty-Fifth New Zealand Plant Protection Conference. 1992. The effect of rain on the development of Erwinia amylovora and E. herbicola populations on apple flowers; pp. 301–303. Wellington, New Zealand. [Google Scholar]
- 38.Thomson S V, Bonn W G. Solarization of pear and apple trees to eradicate bacteria in fire blight cankers. Acta Hortic. 1996;411:337–339. [Google Scholar]
- 39.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Wael L, de Greef M, van Laere O. The honeybee as a possible vector of Erwinia amylovora (Burr.) Winslow et al. Acta Hortic. 1990;273:107–113. [Google Scholar]
- 41.van der Zwet T. The various means of dissemination of the fire blight bacterium Erwinia amylovora. Bull EPPO. 1994;24:209–214. [Google Scholar]
- 42.van der Zwet T. Present worldwide distribution of fire blight. Acta Hortic. 1996;411:7–8. [Google Scholar]
- 43.van der Zwet T, Beer S V. A practical guide to integrated disease management. U.S. Department of Agriculture, Wash: U.S. Department of Agriculture Agriculture Information Bulletin 631; 1995. Fire blight: its nature, prevention and control; pp. 1–91. ington, D.C. [Google Scholar]
- 44.van der Zwet T, Bell R L. Survival of Erwinia amylovora on apple and pear pollen. Acta Hortic. 1993;338:111. [Google Scholar]