Abstract
Putative Pseudomonas spp. isolated predominantly from raw and processed milk were characterized by automated ribotyping and by biochemical reactions. Isolates were biochemically profiled using the Biolog system and API 20 NE and by determining the production of proteases, lipases, and lecithinases for each isolate. Isolates grouped into five coherent clusters, predominated by the species P. putida (cluster A), P. fluorescens (cluster B), P. fragi (as identified by Biolog) or P. fluorescens (as identified by API 20 NE) (cluster C), P. fragi (as identified by Biolog) or P. putida (as identified by API 20 NE) (cluster D), and P. fluorescens (cluster E). Isolates within each cluster also displayed similar enzyme activities. Isolates in clusters A, C, and D were generally negative for all three enzyme activities; isolates in cluster B were predominantly positive for all three enzyme activities; and isolates in cluster E were negative for lecithinase but predominantly positive for protease and lipase activities. Thus, only isolates from clusters B and E produced enzyme activities associated with dairy product flavor defects. Thirty-eight ribogroups were differentiated among the 70 isolates. Ribotyping was highly discriminatory for dairy Pseudomonas isolates, with a Simpson's index of discrimination of 0.955. Isolates of the same ribotype were never classified into different clusters, and ribotypes within a given cluster generally showed similar ribotype patterns; thus, specific ribotype fragments may be useful markers for tracking the sources of pseudomonads in dairy production systems. Our results suggest that ribogroups are generally homogeneous with respect to nomenspecies and biovars, confirming the identification potential of ribotyping for Pseudomonas spp.
Phenotypic microbiological techniques have proven useful for quantifying and describing bacteria causing fluid dairy product spoilage; however, precise location of the sources of these spoilage organisms in the processing environment or on the farm requires reliable, differential strain identification strategies. Currently available phenotypic speciation strategies for the most common dairy product spoilers, i.e., Pseudomonas spp. (26) and Bacillus spp., frequently yield inconclusive results. Further, the simple identification of the same genus and species by standard methods in both environmental samples and in the finished product does not unequivocally establish a causal relationship. As the ability to sensitively discriminate among strains within a given species is essential for tracking specific microbial contamination sources, development of analytical methods that allow the characterization and precise identification of microorganisms is necessary to improve product quality and safety assurance programs.
Pseudomonas spp. are important bacterial contributors to spoilage of conventionally pasteurized fluid milk products (30). These psychrotolerant organisms contribute to milk spoilage in two different ways. First, they produce the majority of lipolytic and proteolytic enzymes secreted into raw milk during preprocessing storage. Many of these enzymes can survive pasteurization (72°C for 15 s) and even ultra-high-temperature treatments (138°C for 2 s or 149°C for 10 s) and can thus reduce the sensory quality and shelf life of processed fluid milk products (20, 30). Second, postpasteurization contamination contributes most of the microorganisms, primarily Pseudomonas spp., that cause spoilage of conventionally pasteurized milk during refrigerated storage (7, 13, 24, 29). As most fluid milk products manufactured in the United States are not currently aseptically packaged, the possibility exists for the entry of contaminating microbes into the milk at various points after the heating unit. Therefore, determination of specific contamination sources in individual dairy plants is necessary to reduce or eliminate postprocessing contamination (28).
Although most Pseudomonas spp. are not considered to be human pathogens, several species of this group are associated with human and animal infections (9). Pseudomonas cepacia (recently renamed Burkholderia cepacia) has been isolated from infected human tissues, but members of this species are so difficult to identify by phenotypic means that isolates frequently have been incorrectly assigned to other genera (25). P. aeruginosa has been recognized as an infectious agent transmitted by food and water (23). This organism is an opportunistic pathogen affecting primarily immunocompromised people and those suffering from cystic fibrosis. For this reason, current legislation in several countries demands that bottled water products test free of P. aeruginosa (23). The lack of robust identification tools for these organisms can lead to the misidentification of nonpathogenic Pseudomonas spp. as pathogenic species, potentially forcing costly and unnecessary food product recalls (23). As P. aeruginosa has been isolated from milk (35), and as the dairy industry is likely to face increased domestic and international demand for products free of bacterial contaminants (10), development of reliable tools to identify and track spoilage strains and pathogens will help the industry meet future product quality and safety challenges.
Various phenotypic and molecular methods have been developed and used for subtyping bacterial isolates. Phenotypic subtyping methods include biochemical characterization (biotyping), bacteriocin typing, bacterial fatty acids profiling, and multilocus enzyme electrophoresis. Molecular subtyping methods include pulsed-field gel electrophoresis (PFGE), PCR-based typing methods, DNA sequence-based typing, and ribotyping. Ribotyping, which is broadly applicable for typing bacterial species, is based on restriction digestion of bacterial chromosomal DNA, followed by Southern hybridization with a ribosomal operon probe (6, 17). Ribotyping can be performed in a classical multistep manual Southern blot format, but an automated ribotyping system is also commercially available (5). To date, however, many applications of molecular subtyping, including ribotyping, have focused on differentiation of medically important bacterial species. These typing methods also have tremendous potential for other applications, including those of the food and dairy industry. Development of correlations between genetic types and spoilage potentials of dairy and food microflora will allow application of molecular subtyping methods for rapid tracking of dairy spoilage organisms to source.
The goal of this project was to establish a taxonomic, molecular, and phenotypic framework for species and strain identification of dairy pseudomonads to ultimately facilitate tracking spoilage organism sources in dairy and food production systems.
MATERIALS AND METHODS
Strains and isolates.
Pseudomonas isolates from raw and pasteurized milk were obtained as part of Cornell University's Milk Quality Improvement Program shelf life testing program. In this program, bacterial numbers for pasteurized and raw milk samples from dairy plants in New York State are determined by various procedures including standard plate counts and psychrotrophic plate counts (21). Pasteurized milk samples are plated at the initial day, day 7, and day 14. For this study, putative Pseudomonas colonies were collected from representative plates and initially characterized by gram staining and testing for oxidase and catalase activities. All single colonies that were confirmed as putative Pseudomonas spp. (gram negative, oxidase positive, and catalase positive) were used for further characterization as described below.
Phenotypic characterization.
All isolates were initially characterized using API 20 NE strips (BioMerieux Vitek, Inc., Hazelwood, Mo.). Species identification (IDs) were obtained using the API database. Isolates were also phenotypically characterized by the Biolog system (Biolog, Hayward, Calif.), which is based on the differential utilization of 95 organic test substrates by the test organisms (3, 11). Biolog GN microplates were inoculated as described by the manufacturer and incubated at 30°C for 24 h. Formazan accumulation was measured at 4, 20, and 24 h using the Biolog microstation. Isolates were identified to the species level using the Biolog database. Biolog substrate utilization patterns were transformed from the octal code output of the Biolog workstation to a binomial code using Excel 5.0 (Microsoft, Seattle, Wash.). Binomial data were converted into a rich text format which was used as an input file for parsimony analysis using the Dollo parsimony method (DOLLOP in the software package PHYLIP version 3.57c) (8).
Enzyme production.
To determine production of proteases, lipases, and lecithinases, Pseudomonas isolates were plated on agar plates containing the appropriate substrates as described below. Production of proteolytic enzymes was determined on plate count agar (Difco, Detroit, Mich.) containing 10% skim milk powder (skim milk agar; Difco) (36). After incubation at 30°C for 72 h, plates were flooded with 1 N HCl to observe clearance zones. Lipase production was assessed using single-layer agar (36). Single-layer agar consists of 5% (wt/vol) clarified butterfat (15) and 1:7,500 Victoria blue B blended into tryptic soy agar (Difco). After incubation at 30°C for up to 5 days, plates were observed for the presence of colonies surrounded by dark blue zones. Lecithinase production was determined on plate count agar containing 10% egg yolk emulsion (egg yolk agar; Difco). After incubation at 30°C for up to 5 days, plates were observed for the presence of colonies surrounded by opaque zones.
Automated ribotyping.
All Pseudomonas isolates were characterized by automated ribotyping using the restriction enzyme EcoRI and the RiboPrinter (Qualicon Inc., Wilmington, Del.). All of the necessary operations are automated, and eight isolates can be typed within an 8-h time period (5). Briefly, bacterial isolates are grown overnight on a brain heart infusion agar (Difco) plate. A single colony from the plate is resuspended in lysis buffer, which is then added to the processing module. All subsequent steps are performed using standardized reagents (including prepoured agarose gels). Other restriction enzymes can be substituted for EcoRI, as desired. The template preparation, restriction enzyme digestion, gel electrophoresis, and blotting steps are completely automated. The blotted nucleic acids are hybridized with a sulfonated DNA probe. Hybridization is detected using alkaline phosphatase-labeled antibodies against sulfonated DNA. The presence of these antibodies is detected by capturing the emission of a chemiluminescent substrate with a charge-coupled device camera. The output is a densitometric scan depicting the restriction fragment distribution and molecular weight. This output is captured in the system's computer, which stores the ribotype pattern for each isolate. The ribotype patterns obtained are subsequently compared to patterns already in the RiboPrinter database (5). Relationships between ribotype patterns in the database can then be analyzed and bacterial identities can be predicted. Ribotype patterns for each isolate are compared against the patterns obtained for all other isolates, and similarity coefficients are calculated using the RiboPrinter's proprietary algorithm. All ribotype patterns with similarity values of >0.93 are initially grouped together to form a ribogroup. The suitability of ribotyping for differentiation of strains was determined using Simpson's numerical index as described by Hunter and Gaston (18).
16S rRNA sequencing.
One isolate from each cluster defined by Biolog results was further characterized by 16S rDNA sequencing. Lysozyme/proteinase K lysates of selected isolates were prepared as previously described (12). A 1-μl volume of 1:1,000 dilutions of the lysates was used for PCR; 1.5-kb fragments comprising the entire 16S rDNA open reading frame were amplified using primers 16S-P5SH (5′-TGA AGA GTT TGA TCA TGG CTC AG-3′) and 16S-DG74 (5′-AGG AGG TGA TCC AAC CGC A-3′) and Taq polymerase (PE Applied Biosystems, Foster City, Calif.). PCR conditions consisted of 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min, followed by a hold at 72°C for 5 min. PCR products were purified with the QIAquick PCR purification kit (Qiagen Inc., Chatsworth, Calif.) and were directly sequenced using the Perkin-Elmer cycle sequencing kit and an Applied Biosystems model 373A automated sequencer. Sequencing was performed using primers 16S-P5SH and P3-SH (5′-CTA CGG TTA CCT TGT TAC GAC TT-3′). BLASTN search analysis was used to compare the obtained sequence data with 16S rDNA sequence data deposited in GenBank (1). Alignment of 16S rRNA sequences was performed using the Clustal method in the software Megalign (DNAStar, Madison, Wis.). Phylogenetic analyses were performed using the parsimony method (DNAPARS in the software package PHYLIP version 3.57c) (8) as well as the programs SEQBOOT, CONSENSE, and DRAWTREE to perform bootstrap analysis.
Nucleotide sequence accession number.
The seven DNA sequences reported were deposited in GenBank under accession no. AF205133 through AF205138.
RESULTS
A total of 66 putative Pseudomonas isolates was obtained from raw milk and processed fluid milk products. To provide a benchmark for assessing homogeneity of characteristics among Pseudomonas isolates from dairy sources, we included an additional four isolates (B1-018 to B1-021) from vegetative sources (potato [2], mushroom, and apple) in our analyses. Isolates and their characteristics are summarized in Table 1.
TABLE 1.
Pseudomonas isolates and their characteristics
| Straina | Biolog ID (similarity) | Species ID by API 20 NEb | Enzyme activity for:
|
||
|---|---|---|---|---|---|
| Protease | Lipase | Lecithinase | |||
| Cluster A | |||||
| B1-054 | P. putida A (0.645) | P. putida | − | − | Brown |
| P. putida B (0.530) | |||||
| R1-057 | P. putida B1 (0.640) | P. putida | − | − | Brown |
| P. putida B1 (0.620) | |||||
| D1-034 | Deleya aesta (0.140) | P. fluorescens2 | + | + | − |
| Alcaligenes faecalis (0.351) | |||||
| D. marina (0.252) | |||||
| D. marina (0.611) | |||||
| B1-041 | P. putida A1 (0.815) | P. putida | − | − | − |
| B1-043 | P. putida A1 (0.811) | P. putida | − | − | − |
| R1-250 | P. putida A1 (0.835) | P. putida | − | − | − |
| P. putida A1 (0.834) | |||||
| D2-160 | P. putida A1 (0.891) | P. putida | − | − | − |
| D2-182 | P. putida A1 (0.501) | P. fluorescens | − | − | − |
| P. putida A1 (0.785) | |||||
| D1-082 | P. fulva (0.084) | P. putida | − | − | − |
| P. putida A1 (0.235) | |||||
| Cluster B1 | |||||
| D1-041 | P. corrugata (0.459) | P. fluorescens | + | +/− | + |
| P. tolasii (0.294) | |||||
| D1-043 | P. fluorescens A (0.374) | P. fluorescens10 | − | − | + |
| D1-077 | P. fluorescens A (0.635) | P. fluorescens | + | − | + |
| R1-146 | P. fluorescens A (0.796) | P. fluorescens | + | + | + |
| R1-051 | P. fluorescens A (0.632) | P. fluorescens1 | + | + | + |
| B1-069 | P. fluorescens A (0.793) | P. fluorescens1 | + | + | + |
| P. fluorescens A (0.866) | |||||
| R1-196 | P. fluorescens A (0.460) | P. fluorescens2 | + | + | + |
| R1-041 | P. fluorescens A (0.632) | P. fluorescens1 | + | + | + |
| R1-225 | P. fluorescens A (0.688) | P. fluorescens1 | + | + | + |
| R1-232 | P. fluorescens A (0.492) | P. fluorescens1 | + | + | + |
| R1-195 | P. tolasii (0.344) | P. fluorescens1 | + | + | + |
| R1-193 | P. tolasii (0.308) | P. fluorescens1 | + | + | + |
| P. tolasii (0.637) | |||||
| B1-052 | P. fluorescens A (0.977) | P. fluorescens1 | + | + | + |
| B1-045 | P. fluorescens A (0.552) | P. fluorescens1 | + | + | + |
| B1-048 | P. fluorescens A (0.552) | P. fluorescens1 | + | + | + |
| B1-047 | P. corrugata (0.301) | P. fluorescens1 | + | + | + |
| P. synxantha (0.653) | |||||
| B1-021c | P. fluorescens A (0.790) | P. fluorescens1 | + | + | + |
| D1-044 | P. corrugata (0.290) | P. fluorescens1 | + | +/− | + |
| P. corrugata (0.460) | |||||
| B1-044 | P. fluorescens A (0.760) | P. fluorescens1 | + | + | + |
| D2-059 | P. fluorescens A (0.748) | P. fluorescens | + | +/− | + |
| D1-019 | P. fluorescens A (0.875) | P. fluorescens1 | + | + | + |
| D1-025 | P. fluorescens A (0.647) | P. fluorescens7 | − | − | + |
| P. fluorescens A (0.526) | |||||
| D2-001 | P. fluorescens A (0.877) | P. fluorescens | + | +/− | + |
| P. fluorescens A (0.833) | |||||
| D2-004 | P. fluorescens A (0.661) | P. fluorescens | + | +/− | + |
| D1-040 | P. fluorescens A (0.806) | P. fluorescens | − | +/− | + |
| Cluster B2 | |||||
| B1-062 (B3) | P. fluorescens G (0.409) | P. fluorescens1 | + | + | − |
| P. fluorescens F (0.706) | |||||
| D1-028 | P. fluorescens A (0.801) | P. cepacia8 | + | + | + |
| D1-015 | P. fluorescens C (0.680) | P. pseudomallei6 | + | + | − |
| P. fluorescens C (0.794) | |||||
| D1-016 | P. fluorescens C (0.848) | P. pseudomallei6 | + | + | + |
| D1-048 | P. fluorescens C (0.760) | P. pseudomallei6 | + | + | + |
| P. fluorescens C (0.897) | (weak) | ||||
| D2-027 | P. fluorescens C (0.598) | P. fluorescens | − | − | − |
| P. fluorescens C (0.652) | |||||
| D2-048 | P. fluorescens C (0.729) | P. aeruginosa | + | +/− | + |
| B1-066 (B1) | P. fluorescens A (0.858) | P. fluorescens1 | + | + | + |
| B1-018d (B1) | P. fluorescens A (0.439) | P. fluorescens1 | − | − | − |
| P. tolasii (0.143) | |||||
| Cluster B3 | |||||
| B1-020d (B2) | P. fuscovaginae (0.702) | P. fluorescens | − | + | − |
| P. fuscovaginae (0.592) | |||||
| D1-045 | P. fluorescens B (0.467) | P. fluorescens1 | + | + | + |
| P. fluorescens G (0.609) | |||||
| B1-065 | P. fluorescens B (0.531) | P. fluorescens1 | + | + | + |
| P. fuscovaginae (0.610) | |||||
| B1-019e | P. marginalis (0.624) | P. fluorescens2 | + | + | − |
| P. corrugata (0.616) | |||||
| Cluster C | |||||
| B1-040 | P. fragi (0.794) | P. fluorescens3 | − | − | − |
| D1-081 | P. fragi (0.771) | P. putida | − | − | − |
| D1-018 | P. fragi (0.775) | P. putida | − | − | − |
| D1-014 | P. fragi (0.711) | P. fluorescens9 | − | + | − |
| P. fragi (0.635) | |||||
| D1-046 | P. fragi (0.833) | P. fluorescens9 | − | − | − |
| Cluster D | |||||
| D1-026 | P. pseudoalcaligenes (0.327) | P. putida | − | − | − |
| P. pseudoalcaligenes (0.329) | |||||
| D1-024 | P. fragi (0.450) | P. putida4 | − | − | − |
| P. cichorii (0.168) | |||||
| B1-033 | P. fragi (0.637) | P. putida4 | − | − | − |
| P. fulva (0.266) | |||||
| B1-060 | D. marina (0.513) | P. putida4 | − | − | − |
| D. marina (0.619) | |||||
| D1-027 | P. fulva (0.472) | P. putida4 | − | − | − |
| P. viridilivida (0.467) | |||||
| D2-329 | P. fragi (0.637) | P. putida | − | − | − |
| P. fragi (0.284) | |||||
| B1-032 | D. marina (0.415) | P. putida | − | − | − |
| P. fragi (0.168) | |||||
| B1-035 | D. marina (0.541) | P. putida | − | − | − |
| Cluster E | |||||
| B1-057 (A) | P. fragi (0.769) | P. putida | − | − | − |
| P. fragi (0.265) | |||||
| D2-017 | Acinetobacter genospecies 15 (0.417) | P. fluorescens | + | + | − |
| Acinetobacter genospecies 15 (0.438) | |||||
| D2-021 | Acinetobacter (0.445) | P. fluorescens | + | + | − |
| P. fragi (0.718) | |||||
| D2-036 | Psychrobacter immobilis (0.271) | P. fluorescens | + | + | − |
| Acinetobacter (0.556) | |||||
| D1-017 | P. fragi (0.418) | P. fluorescens | + | − | − |
| Acinetobacter genospecies 15 (0.389) | |||||
| D1-035 | Acinetobacter genospecies 15 (0.522) | P. fluorescens | + | + | − |
| P. mucidolens (0.346) | |||||
| D2-013 | P. fragi (0.799) | P. fluorescens | + | ++/− | − |
| B1-050 | Acinetobacter genospecies 15 (0.578) | P. putida5 | + | + | − |
| Acinetobacter genospecies 15 (0.580) | |||||
| D1-022 | Acinetobacter genospecies 15 (0.473) | P. putida5 | − | − | − |
| P. fulva (0.173) | |||||
| D1-021 | P. fragi (0.586) | P. putida5 | − | − | − |
| Acinetobacter genospecies 15 (0.602) | |||||
Each term in parentheses indicates the strain's resulting cluster designation when the phylogenetic analysis was repeated after eliminating substrate utilization data from the nine most variable Biolog substrates (see Results).
Species assigned by best likelihood; other possibilities indicated by superscript numbers as follows: 1, P. aureofaciens; 2, P. aureofaciens, P. chlororaphis; 3, P. putida; 4, P. fluorescens; 5, P. fluorescens, P. chlororaphis; 6, P. aeruginosa; 7, P. chlororaphis, P. aureofaciens; 8, P. aureofaciens, P. fluorescens; 9, P. putida, P. chlororaphis; 10, P. fluorescens, P. aureofaciens, P. cepacia.
Mushroom isolate.
Potato isolate.
Apple isolate.
Biochemical and phenotypic characterization.
Of the 66 dairy isolates tested, 38 (58%), 38 (58%), and 31 (47%) displayed protease, lipase, and lecithinase activity, respectively (Table 1). All 70 isolates were characterized using the Biolog system. Results after a 24-h incubation time were used for species identification and for parsimony analysis (16, 19). As preliminary work in our laboratory suggested the likelihood of variability of substrate utilization patterns for a given putative Pseudomonas isolate analyzed in duplicate with the Biolog system, we selected 38 isolates for duplicate analyses; 1 isolate was run in quadruplicate (Table 1). Table 2 summarizes the variability between duplicate analyses of substrate utilization patterns among the 95 Biolog substrates. Of the 95 substrates, 26 (27.4%) gave identical results between duplicate analyses; the remaining 69 substrates (72.6%) differed in utilization patterns between duplicates for at least one isolate examined. For example, glycogen utilization patterns differed between duplicate analyses for 55.3% of our isolates (Table 2).
TABLE 2.
Biolog substrates with differing oxidation patterns between duplicate isolate analysesa
| No. (%) of isolates that differed in oxidation patterns of the specified substrates between duplicate analyses (n = 38) | No. (%) of substrates that differed between duplicate samples | Substrates that differed between duplicate analyses |
|---|---|---|
| 1 (2.6) | 14 (14.7) | Tween 40, maltose, d-mannitol, sucrose, d-galacturonic acid, d-gluconic acid, d-glucosaminic acid, γ-hydroxybutyric acid, p-hydroxyphenylacetic acid, itaconic acid, α-ketobutyric acid, l-proline, γ-amino butyric acid, inosine |
| 2 (5.3) | 19 (20.0) | N-Acetyl-d-glucosamine, d-fructose, l-fucose, d-mannose, l-rhamnose, d-trehalose, citric acid, β-hydroxybutyric acid, α-ketoglutaric acid, α-ketovaleric acid, d-alanine, hydroxy-l-proline, l-ornithine, l-pyroglutamic acid, dl-carnitine, urocanic acid, uridine, 2,3-butanediol, glucose-6-phosphate |
| 3 (7.9) | 10 (10.5) | Tween 80, d-arabitol, m-inositol, d-raffinose, d-glucuronic acid, α-hydroxybutyric acid, l-alanine, l-histidine, d-serine, dl-α-glycerolphosphate |
| 4 (10.5) | 5 (5.3) | Acetic acid, dl-lactic acid, l-leucine, 2-aminoethanol, glycerol |
| 5 (13.1) | 6 (6.3) | Dextrin, d-melibiose, turanose, xylitol, propionic acid, putrescine |
| 6 (15.8) | 6 (6.3) | Methyl pyruvate, monomethyl succinate, l-alanyl-glycine, l-phenylalanine, l-serine, l-threonine |
| 7 (18.4) | 2 (2.1) | d-Galactose, formic acid |
| 8 (21) | 3 (3.2) | β-Methyl-d-glucoside, d-psicose, glucuronamide |
| 9 (23.7) | 1 (1.1) | Succinamic acid |
| 11 (28.9) | 1 (1.1) | Glycyl-l-glutamic acid |
| 14 (36.8) | 1 (1.1) | Alaninamide |
| 21 (55.3) | 1 (1.1) | Glycogen |
The 26 Biolog substrates (27.4% of total substrates) that are not listed did not differ in oxidation patterns between duplicate analyses of the same isolate.
Substrate utilization data for all strains, including the duplicate data, were used to construct a rooted tree using the Dollo parsimony method. The Dollo parsimony method is specifically suited for construction of the most parsimonious trees based on analyses of binomial data (e.g., ability or inability to utilize a specific substrate) with the assumption that loss of a characteristic is more likely than acquisition of a characteristic. Substrate utilization characteristics are complex traits that may involve multiple genetic elements; thus, the loss of ability to utilize one substrate is very unlikely to be evolutionarily equivalent to the loss of utilization of another substrate. Thus, no time scale is implied by the structure of a cladogram.
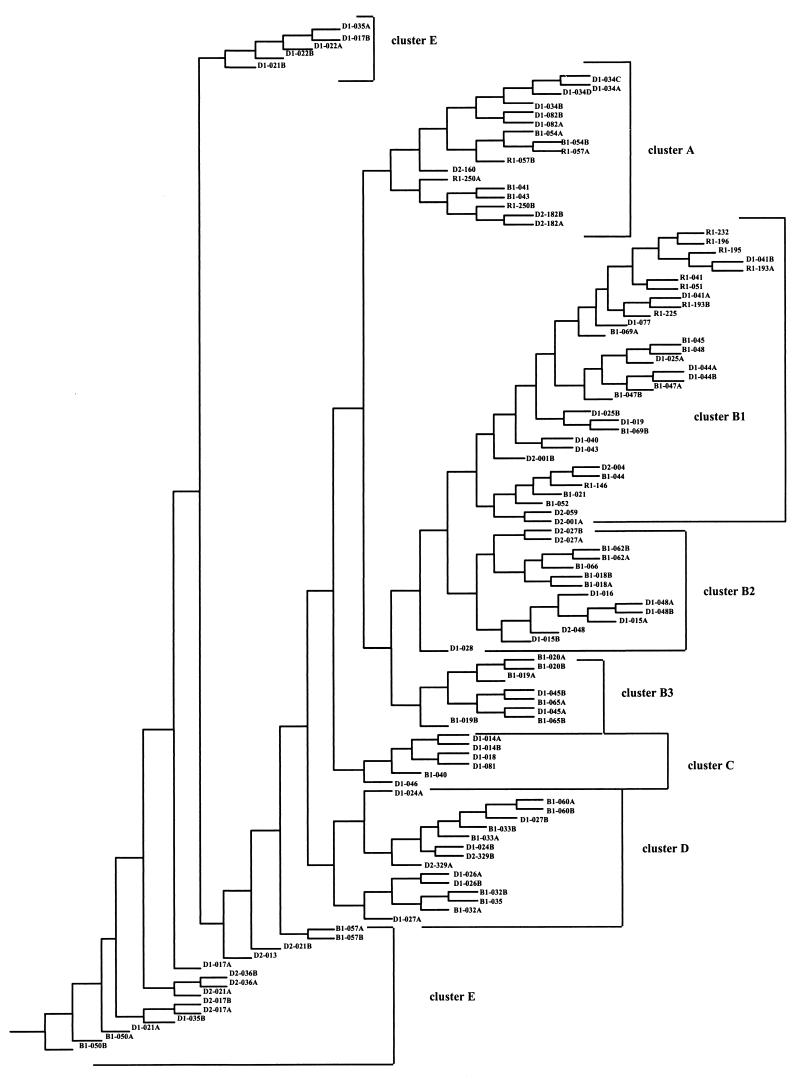
A total of 100 most parsimonious trees were obtained by Dollo parsimony analysis of substrate utilization patterns for the 95 Biolog substrates. These trees were used to calculate a consensus tree using CONSENSE in the software package PHYLIP (Fig. 1). To confirm the topology of this consensus tree, phylogenetic analysis was repeated after excluding the substrate utilization data from the nine most variable substrates (i.e., substrates that showed variability for >6 of the 38 isolates tested in duplicate [Table 2]), resulting again in the 100 most parsimonious trees. The use of 86 rather than 95 substrate utilization patterns for construction of the second consensus tree resulted in the following modifications: (i) B1-066 and B1-018 clustered in B2 rather than in B1; (ii) B1-062 clustered in B2 rather than in B3; (iii) B1-057 clustered in E rather than in A; and (iv) B1-020 clustered in B3 rather than in B2.
FIG. 1.
Simplified phenogram based on biochemical data obtained from oxidation patterns of 95 Biolog substrates for 70 putative Pseudomonas isolates. Thirty-eight isolates were run in duplicate, and one isolate was run in quadruplicate; these data were also included in the analyses. This phenogram, which is a consensus tree calculated using CONSENSE in the PHYLIP software package, was constructed using data from Dollo parsimony analyses of isolate substrate oxidation patterns.
Ribotyping.
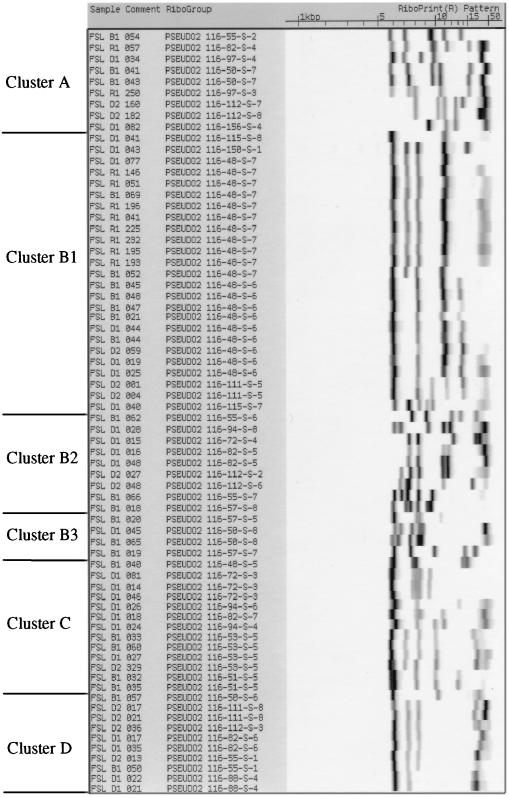
All isolates were characterized by automated ribotyping. The resulting ribogroup patterns are shown in Fig. 2. Initially, ribotype patterns with similarity coefficients of >0.93, as calculated based on band positions and intensities by the RiboPrinter's proprietary software, were considered identical and were grouped together as one ribogroup with the same designation (e.g., 11 isolates bear the ribogroup designation 116-48-S-7, as shown in Fig. 2). Further refinement of these groupings was performed by visual evaluation of closely related ribotype patterns. A total of 38 different ribogroups was found among the 70 isolates tested (Table 1). Thirteen ribogroups contained more than one isolate. With the exception of four ribogroups (116-48-S-6, 116-48-S-7, 116-72-S-3, and 116-82-S-6), isolates within a given ribogroup had the same activity profiles for protease, lipase, and lecithinase. Three of the four isolates from vegetative sources were represented by unique ribotype patterns (57-S-8, cluster B2; 57-S-5 and 57-S-7, cluster B3) that were not found among the 66 milk isolates.
FIG. 2.
EcoRI ribogroup patterns obtained in this study. Ribotypes were obtained using the automated RiboPrinter (Qualicon). Ribogroups are arranged in the same order as the clusters outlined in Table 1 and in Fig. 1. Strain designations are displayed on the left, and ribogroup patterns are shown on the right. For the ribotypes, the gel running direction is from right to left; i.e., the largest ribotype fragments are on the right side.
Simpson's index of discrimination was calculated to determine the discriminatory ability of ribotyping using EcoRI for the differentiation of dairy pseudomonads. The numerical value of this index (D) indicates the suitability of a given method for differentiating strains by estimating the probability that two unrelated strains are differentiated by a given typing method (18). As the numerical index approaches the maximum value of D = 1 (representing 100% discriminatory ability of a method), the higher the probability that a given method will be able to discriminate between two unrelated strains. Simpson's index of discrimination for automated ribotyping of dairy pseudomonads based on the 70 isolates characterized was 0.955, indicating that ribotyping with EcoRI provides good discriminatory capabilities between these strains.
16S rRNA sequencing.
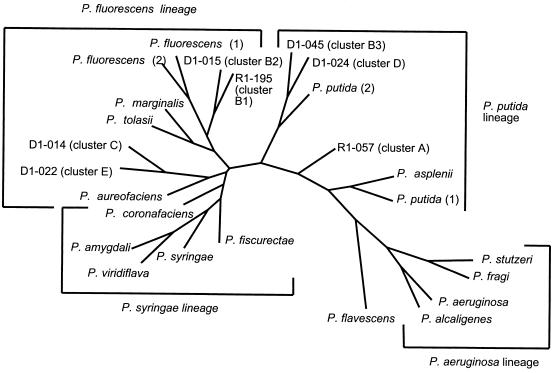
Partial DNA sequences of the 16S rRNA genes were obtained for seven Pseudomonas isolates. Isolates were selected for sequencing to provide one representative from each of the seven clusters (A, B1, B2, B3, C, D, and E). A total of 57 polymorphic nucleotides (i.e., nucleotides that are not identical in all seven isolates) were identified among a total of 1,362 nucleotides sequenced. These 16S rRNA sequences were used for phylogenetic analyses in combination with the previously described 16S rRNA sequences for the P. fluorescens intrageneric cluster and the P. aeruginosa lineage (22). A consensus tree of these 16S rRNA sequences constructed using the parsimony method is shown in Fig. 3. This tree shows that the 16S rRNA sequences obtained in this study cluster together with 16S rRNA sequences from the P. fluorescens intrageneric cluster. Specifically, isolates from clusters B1 (R1-195) and B2 (D1-015) cluster in the P. fluorescens lineage. The 16S rRNA sequences from one cluster E isolate (D1-022) and one cluster C representative (D1-014) cluster together with P. aureofaciens, which is also classified in the P. fluorescens lineages. The 16S rRNA sequences for the cluster D representative (D1-024, identified as P. putida by API 20 NE) and for a B3 cluster isolate (D1-045) cluster between the P. fluorescens and the P. putida lineage. Finally, the 16S rRNA sequence from one cluster A isolate (R1-057) groups closely to the P. putida lineage, as described below.
FIG. 3.
Unrooted bootstrap tree (100 replicates) for 16S rRNA sequences constructed by the parsimony method. The tree was constructed using SEQBOOT, DNAPARS, CONSENSUS, and DRAWTREE in the software package PHYLIP (8). The numbers at the nodes of the tree represent the bootstrap values for each node. Sequences used for this analysis were from isolates D1-014, D1-015, D1-022, D1-024, D1-045, R1-057, and R1-196 as well as from P. aeruginosa (GenBank accession no. Z76651), P. alcaligenes (Z76653), P. amygdali (Z76654), P. asplenii (Z76655), P. aureofaciens (Z76656), P. coronafaciens (Z76660), P. ficuserectae (Z76661), P. fluorescens (1, Z76662; 2, AF068010), P. fragi (D84014), P. marginalis (Z76663), P. putida (1, Z76667; 2, D86000), P. stutzeri (U26262), P. syringae (Z76669), P. tolasii (Z76670), and P. viridiflava (Z76671).
Description of clusters.
Based on our results, the 70 Pseudomonas isolates can be grouped into five main clusters, as described below.
Cluster A contains isolates that are predominantly characterized as P. putida by API 20 NE or Biolog. 16S rRNA sequence analyses also grouped this cluster close to P. putida, providing further confirmation that cluster A represents the species P. putida. Isolates in this cluster generally did not show protease, lipase, or lecithinase activity.
Cluster B contains isolates that are predominantly characterized as P. fluorescens by API 20 NE and by Biolog. 16S rRNA sequence analyses grouped clusters B1 and B2 with P. fluorescens and cluster B3 between the P. fluorescens and the P. putida lineages. These results suggest that cluster B represents the species P. fluorescens, although the taxonomic position of cluster B3 warrants further clarification. The majority (74%) of isolates in cluster B showed protease, lipase, or lecithinase activity.
Cluster C contains isolates that are predominantly characterized as P. fluorescens or P. putida by API 20 NE and as P. fragi by Biolog. By 16S rRNA sequence analyses, the cluster C representative grouped together with the P. fluorescens, whereas the one P. fragi 16S rRNA sequence available in GenBank (accession no. D84014 [2]) clustered into the P. aeruginosa lineage. Isolates in cluster C were generally negative for protease, lipase, or lecithinase activity.
Cluster D isolates were predominantly characterized as P. putida by API 20 NE. By Biolog, isolates in this cluster were classified as a variety of different species and genera, e.g., P. fragi, Deleya marina, or Acinetobacter spp. By 16S rRNA sequence analysis, the cluster D representative grouped with the P. putida lineage. Isolates in cluster D were generally negative for protease, lipase, or lecithinase activity.
Isolates in cluster E were identified as P. fluorescens or P. putida by API 20 NE. The Biolog system identification of isolates in this cluster as D. marina or as Acinetobacter spp. likely reflects a misclassification as both of these species are oxidase negative, whereas our isolates in this cluster were oxidase positive. All isolates in this cluster were negative for lecithinase activity but predominantly positive for protease and lipase activities. 16S rRNA sequence analyses grouped a representative (D1-022) from this cluster in the P. fluorescens lineage. Based on the cladogram constructed from Biolog data (Fig. 1), cluster E appears to represent a more diverse group than the other four clusters.
DISCUSSION
The goal of this project was to establish a taxonomic, molecular, and phenotypic framework to enable identification and, hence, tracking of Pseudomonas species found in dairy products. For this purpose, 70 putative Pseudomonas isolates obtained predominantly from processed milk samples were characterized by phenotypic methods, automated ribotyping, and 16S rRNA sequencing of representative isolates. Based both on phenotypic characterization by the Biolog system, which evaluates oxidation patterns of 95 different substrates by a given isolate, and on ribotyping, our isolates grouped into five main clusters. Despite the fact that the majority of Biolog substrates (72.6%) differed in utilization patterns between duplicate analyses of our isolates, only five isolates shifted cluster positions when the data from the nine most variable substrates were excluded from the analyses. All five clusters appear to represent saprophytic fluorescent pseudomonads, a subset of the genus Pseudomonas sensu stricto (25). Interestingly, none of our 66 dairy isolates grouped with the P. aeruginosa intrageneric cluster which includes the human pathogen P. aeruginosa or with the phytopathogenic fluorescent pseudomonads. Among the five clusters defined, clusters B and E contain a high frequency of isolates with protease, lipase, and lecithinase activities. Therefore, isolates in these groups represent spoilage organisms of particular concern to the dairy and food industries.
Species identification of dairy pseudomonads.
All dairy Pseudomonas isolates characterized in this study fall into groups within the rRNA homology group I of Pseudomonas (Pseudomonas sensu stricto), which is one of five rRNA-DNA homology groups within the genus Pseudomonas (25, 34). Based on rRNA-DNA hybridization studies, the rRNA homology group I can be further divided into three groups whose representative species are P. aeruginosa, P. fluorescens, and P. syringae (2, 25). rRNA homology group I can also be divided into two intrageneric clusters based on 16S rRNA sequencing data (22). These intrageneric clusters also differ significantly by biochemical criteria, as evaluated by the Biolog system (14) as well as by ribotyping (4). The P. aeruginosa intrageneric cluster contains lineages of P. aeruginosa, P. resinoverans, P. mendocina, and P. flavescens. The P. fluorescens intrageneric cluster includes the species P. fluorescens, P. marginalis, P. tolasii, P. chloraphis, P. aureofaciens, P. viridiflava, P. syringae, P. amygdali, P. coronafaciens, P. ficuserectae, P. cichorii, P. putida, P. asplenii, and P. agrici, which are grouped into five lineages. Three of these lineages represent saprophytic fluorescent pseudomonads (e.g., P. putida and P. fluorescens), while the two other lineages represent phytopathogenic species (e.g., P. syringae and P. asplenii). We show that all 66 Pseudomonas isolated from milk represent saprophytic fluorescent pseudomonads, which can be divided into five major clusters. Evidence that the five clusters defined in this study (Table 1; Fig. 1) represent saprophytic fluorescent pseudomonads include the following: (i) species ID by API 20 NE identified the majority of isolates (66 out of 70) as either P. putida or P. fluorescens; and (ii) isolates in clusters A and B were predominantly identified as P. putida or as P. fluorescens by Biolog, while clusters C, D, and E were predominantly identified as P. fragi, although Acinetobacter spp. were also a common identification in cluster D.
To further confirm the species identification of the five Pseudomonas clusters defined in this study, we obtained 16S rRNA gene sequences for one representative isolate from each cluster. These sequence data were used to perform a phylogenetic analysis in comparison with 16S rRNA gene sequences previously reported for representatives of the P. fluorescens, P. syringae, P. putida, P. flavescens, and P. aeruginosa lineages (22). The resulting phylogenetic tree showed a similar topology to the phylogenetic relationships previously derived by Moore et al. (22). All of our sequenced isolates clustered together with either the P. putida lineage or with the P. fluorescens lineage, both representing saprophytic fluorescent pseudomonads, while one P. fragi 16S rRNA sequence deposited in GenBank clustered in the P. aeruginosa lineage. This clustering is consistent with results obtained by Anzai et al. (2), who also found that P. fragi groups with the P. aeruginosa group based on 16S rRNA gene sequence analysis.
P. fluorescens can be divided into five biovars (I through V), while P. putida can be grouped into biovars A and B. Based on Biolog identification, we conclude that cluster A likely represents P. putida biovar A. Cluster D appears to also represent the species P. putida, possibly the P. putida biovar B, which has been shown by ribotyping to cluster separately from biovar A, possibly representing a species distinct from P. putida biovar A. Clusters B1 and B2 appear to represent the P. fluorescens biovars I (biovar A of Stanier et al. [31]) and III (biovar C of Stanier et al. [31]), respectively. Cluster B3 might represent the P. fluorescens biovar II (biovar B of Stanier et al. [31]), which also includes P. marginalis. Based on our current data, we cannot determine any clear correlation between clusters C and D and (a) specific biovar. In general, however, our results are consistent with a previous report by Johnson et al. (19), who showed that clusters defined by Biolog phenograms are generally in good agreement with biovar classifications.
Interestingly, the Biolog clusters defined in this study appear to be consistent with classification by EcoRI ribotyping, as the same EcoRI ribogroup was never present in two different Biolog clusters. Our results therefore suggest that ribogroups are generally consistent with respect to nomenspecies and biovars. This is in agreement with results by Brosch et al. (4), who found that 38 out of 41 ribogroups were homogeneous with respect to nomenspecies by SmaI and HincII ribotyping of a collection of 226 strains of Pseudomonas sensu lato. The general topology of the Biolog phenogram and the associated ribotypes shares important similarities with the SmaI and HincII ribotype clusters defined by Brosch et al. (4), including (i) P. putida biovars A and B form distinct clusters in both studies (our clusters A and D), (ii) isolates identified as P. fragi appear to cluster with P. putida biovar B (our cluster D), and (iii) P. fluorescens biovar I (our cluster B1) is distinct from other clusters.
Spoilage potential of Pseudomonas subsets.
Pseudomonas spp. are psychrotolerant organisms that can cause spoilage of milk and dairy products in two different ways. First, they can produce lipolytic and proteolytic enzymes which can be secreted into raw milk during preprocessing storage. Many of these enzymes survive pasteurization and can thus reduce the sensory quality and shelf life of processed fluid milk products (20). Second, Pseudomonas spp. are commonly present in milk as postprocessing contaminants and are therefore one of the major causes of bacterial spoilage in fluid milk products. Proteases and lipases (in particular lecithinases) produced by Pseudomonas spp. contribute to the spoilage of milk and dairy products as well as other foods (30). We have shown that among the five clusters of Pseudomonas spp. defined in this study, clusters B (P. fluorescens) and E (P. fluorescens or possibly P. fragi) contain a high frequency of isolates with protease, lipase, and lecithinase activities. Therefore, strains grouped in these clusters appear to represent spoilage organisms of particular concern to the dairy and food industries, particularly as P. fluorescens is reported as a common psychrotolerant spoilage organism in milk (7, 32, 33). Only 2 of 22 isolates in clusters A and D (P. putida) showed protease and/or lipase activity. This is in agreement with results by Swart et al. (32), who found that 43 out of 44 P. putida isolates from raw milk were negative for lipolysis and proteolysis. Therefore, we conclude that P. fluorescens strains are likely to represent the predominant cause of bacterial flavor defects in milk.
Our results with regard to Pseudomonas spp. isolated from fluid milk products are consistent with a variety of previous reports that also found that psychrotolerant milk spoilage flora can be classified predominantly as P. fluorescens, P. fragi, and P. putida (7, 33). Similarly, Swart et al. (32) reported that P. putida and P. fluorescens represent the most common gram-negative psychrotolerant species found in raw milk. The species P. fragi is not well defined, but isolates previously designated as P. fragi might be represented by strains classified in our clusters C, D, and E, as isolates in these clusters were commonly identified as P. fragi by Biolog.
We have also shown that isolates within a given ribogroup generally have the same enzyme activity profile. This agreement between clusters based on phenotypic and genetic characteristics is consistent with findings by Johnson et al. (19), who found good agreement between phenograms based on Biolog profiles and on repetitive extragenic palindromic PCR profiles for 41 phenanthrene-degrading fluorescent pseudomonads.
Our results also indicate that ribotypes are unique to different clusters of Pseudomonas spp. and that EcoRI ribotypes can be used to predict isolate classification into genetically and phenotypically coherent clusters of pseudomonads representing a Pseudomonas species or biovar. This interpretation is consistent with a previous report (4) that showed that Pseudomonas ribotypes carry taxonomic information in addition to typing information. Our results show that ribotyping offers a sensitive approach for typing Pseudomonas strains commonly isolated from raw and processed milk as determined by Simpson's index of discrimination. We therefore propose that ribotyping provides a suitable tool for tracking and characterizing Pseudomonas isolates from dairy and food systems. Ribotyping is a DNA subtyping method based on restriction polymorphisms adjacent to or within bacterial rRNA operons. Automated ribotyping as used in this study is based on the same principle as manual ribotyping; thus, the results and conclusions from our study also extend to manual ribotyping. Furthermore, we hypothesize that other genetic subtyping methods that rely on chromosomally encoded genetic differences would reveal similar correlations between genotypic and phenotypic groupings.
Conclusions.
We have assembled and characterized a Pseudomonas strain collection to evaluate the ability of biochemical and molecular methods to identify and characterize Pseudomonas isolates from dairy products and other foods. The five clusters of dairy Pseudomonas isolates identified in this study appear to represent the species P. putida (cluster A and D) and P. fluorescens (cluster B, C, and E). Clusters C, D, and E might also represent strains commonly designated as P. fragi, which has been classified into rRNA-DNA homology group I, but has not been otherwise well defined. Further studies will be necessary to allow clarification of its taxonomic relationship to other Pseudomonas spp. in the rRNA-DNA homology group I. Characterization of a variety of isolates allowed us to evaluate the ability of two different established identification systems (i.e., API 20 NE and Biolog) to identify putative pseudomonads. API 20 NE provided good identification of dairy Pseudomonas isolates to the species level. While the Biolog system differentiated among Pseudomonas isolates, the database did not provide reliable isolate species identification. Ribotyping allowed a high level of discrimination among dairy pseudomonads and thus presents a good tool for strain typing and fingerprinting of dairy spoilage pseudomonads. Ribotyping profiles appear to be unique to different genetically and phenotypically coherent clusters of Pseudomonas isolates, i.e., the same or similar ribotypes are not found in different clusters. This indicates that ribotypes could be used for characterization and identification of dairy Pseudomonas isolates, which will allow a specific ribotype to be used to predict the species and possibly the biovar of a given isolate. These conclusions are in agreement with a recent study by Brosch et al. (4), who showed that ribotyping with the restriction enzymes SmaI and HincII yielded taxonomic information and could be used to identify Pseudomonas strains to the species level.
Our results also confirm the long-term potential for molecular subtyping methods to complement and possibly replace phenotypic characterization methods. In many instances, molecular methods may be used not only for subtyping to facilitate tracking of pathogens and spoilage organisms but also to predict phenotypic characteristics and species identification. Molecular groupings may even replace many current taxonomic concepts in bacteriology. Cost comparisons between different subtyping methods show that commonly used molecular subtyping methods such as ribotyping and PFGE are still rather expensive ($60 to $100/isolate [Table 3]), although price differences between these different methods may not be significant (27).
TABLE 3.
Economic comparisons among DNA fingerprinting methods
| Fingerprinting method | Variable expense item
|
||||||
|---|---|---|---|---|---|---|---|
| Cost ($)
|
Technician expense
|
Cost ($)/isolate over 7 yr for:
|
|||||
| Equipment investment | Supplies/isolate | Time/8 isolates (h) | Cost ($)/isolate | 500 isolates/yr | 1,000 isolates/yr | 2,000 isolates/yr | |
| Automated ribotyping | 175,000a | 45.00b | 1 ($30) | 3.75 | 98.75 | 73.75 | 61.25 |
| Manual ribotyping | 11,800c | 10.00d | 16 ($480) | 60 | 73.37 | 71.70 | 70.84 |
| PFGE | 39,045e | 3.50f | 16 ($480) | 60 | 74.66 | 69.08 | 66.29 |
| Biolog | 32,236g | 6.47h | 2 ($60) | 7.50 | 23.18 | 18.57 | 16.27 |
Price for the RiboPrinter automated characterization system (Qualicon).
Price for ribotyping kit (Qualicon).
Includes power supply ($700), Submarine gel electrophoresis system ($600), Southern blotting system for electroblotting ($1,000), hybridization oven ($2,500), Gel Compare system ($5,000), and computer for Gel Compare ($2,000).
Expendable materials for EcoRI ribotyping as described by Bruce et al., including enzymes for DNA preparation, lysis, and restriction, membranes, agarose, and reagents for development of Southern blot and X-ray film.
Includes Bio-Rad CHEF Mapper system ($21,000), Video Gel Documentation system ($8,995), and Molecular Analyst/PC fingerprinting plus software ($9,950).
Expendable materials include a kit for the preparation of agarose plugs, restriction enzymes, agarose, and size markers.
Price for a complete Biolog system including Microstation with computer ($31,600), GN database ($616), and turbidity standard ($20).
Expendable materials include GN2 microplates, BUGM plates, and reservoirs.
ACKNOWLEDGMENTS
This publication was developed under the auspices of the Cornell University Center for Biotechnology, a New York State Center for Advanced Technology supported by the New York State Science and Technology Foundation and industrial partners. Part of this project was also supported by Dairy Management Inc.
We thank S. Kozlowski, B. Hammond, E. Witek, and S. Douglas for help with the isolation of Pseudomonas spp. and for providing Pseudomonas isolates. We also thank S. Murphy, M. Bodis, B. Miller, C. A. Batt, and all members of the Food Safety Laboratory for helpful discussions. We furthermore thank S. Beer and the members of his laboratory for access to their Biolog microstation and for help with Biolog analyses.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anzai Y, Kudo Y, Oyaizu H. The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int J Syst Bacteriol. 1997;47:249–251. doi: 10.1099/00207713-47-2-249. [DOI] [PubMed] [Google Scholar]
- 3.Bochner B R. “Breathprints” at the microbial level. ASM News. 1989;55:536–539. [Google Scholar]
- 4.Brosch R, Lefevre M, Grimont F, Grimont P A D. Taxonomic diversity of pseudomonads revealed by computer-interpretation of ribotyping data. Syst Appl Microbiol. 1996;19:541–555. [Google Scholar]
- 5.Bruce J. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 1996;50:77–81. [Google Scholar]
- 6.Bruce J L, Hubner R J, Cole E M, McDowell C I, Webster J A. Sets of EcoRI fragments containing ribosomal RNA sequences are conserved among different strains of Listeria monocytogenes. Proc Natl Acad Sci USA. 1995;92:5229–5233. doi: 10.1073/pnas.92.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cousin M A. Presence and activity of psychrotrophic microorganisms in milk and dairy products: a review. J Food Prot. 1982;45:172–207. doi: 10.4315/0362-028X-45.2.172. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 9.Foght J M, Westlake D W S, Johnson W M, Ridgway H F. Environmental gasoline-utilizing isolates and clinical isolates of Pseudomonas aeruginosa are taxonomically indistinguishable by chemotaxonomic and molecular techniques. Microbiology. 1996;142:2333–2340. doi: 10.1099/00221287-142-9-2333. [DOI] [PubMed] [Google Scholar]
- 10.Franck R. Quality counts. Dairy Herd Manage. 1997;34:24–27. [Google Scholar]
- 11.Frey P, Frey-Klett P, Garbaye J, Berge O, Heulin T. Metabolic and genotypic fingerprinting of fluorescent pseudomonads associated with the douglas fir-Laccaria bicolor mycorrhizosphere. Appl Environ Microbiol. 1997;63:1852–1860. doi: 10.1128/aem.63.5.1852-1860.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furrer B, Candrian U, Hoefelein C, Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin fragments. J Appl Bacteriol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths M W, Phillips J D, Muir D D. Post-pasteurization contamination—the major cause of failure of fresh dairy products. Hannah Res. 1984;1984:77–87. [Google Scholar]
- 14.Grimont P A D, Vancanneyt M, Lefevre M, Vandemeulebroecke K, Vauterin L, Brosch R, Kersters K, Grimont F. Ability of Biolog and Biotype-100 systems to reveal the taxonomic diversity of the pseudomonads. Syst Appl Microbiol. 1996;19:510–527. [Google Scholar]
- 15.Harris P L, Cuppett S L, Bullerman L B. A technique comparison of isolation of lipolytic bacteria. J Food Prot. 1990;53:176–177. doi: 10.4315/0362-028X-53.2.176. [DOI] [PubMed] [Google Scholar]
- 16.Holmes B, Costas M, Ganner M, On S L W, Stevens M. Evaluation of Biolog system for identification of some gram-negative bacteria of clinical importance. J Clin Microbiol. 1994;32:1970–1975. doi: 10.1128/jcm.32.8.1970-1975.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubner R J, Cole E M, Bruce J L, McDowell C I, Webster J A. Types of Listeria monocytogenes predicted by the positions of EcoRI cleavage sites relative to ribosomal RNA sequences. Proc Natl Acad Sci USA. 1995;92:5234–5238. doi: 10.1073/pnas.92.11.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter P, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson K, Andersen S, Jacobsen C S. Phenotypic and genotypic characterization of phenanthrene-degrading fluorescent Pseudomonas biovars. Appl Environ Microbiol. 1996;62:3818–3825. doi: 10.1128/aem.62.10.3818-3825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Fandino R, Olano A, Corzo N, Ramos M. Proteolysis during storage of UHT milk: differences between whole and skim milk. J Dairy Res. 1993;60:339–347. doi: 10.1017/s0022029900027680. [DOI] [PubMed] [Google Scholar]
- 21.Marshall R T, editor. Standard methods for the examination of dairy products. 16th ed. Washington, D.C.: American Public Health Association; 1993. [Google Scholar]
- 22.Moore E R B, Mau M, Arnscheidt A, Boettger E C, Hutson R A, Collins M D, van de Peer Y, de Wachter R, Timmis K N. The determination and comparison of 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst Appl Microbiol. 1996;19:478–492. [Google Scholar]
- 23.Morais P V, Mesquita C, Andrade J-L, daCosta M S. Investigation of persistent colonization by Pseudomonas aeruginosa-like strains in a spring water bottling plant. Appl Environ Microbiol. 1997;63:851–856. doi: 10.1128/aem.63.3.851-856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moseley W K. Pinpointing post-pasteurization contamination. J Food Prot. 1980;43:414. doi: 10.4315/0362-028X-43.5.414. [DOI] [PubMed] [Google Scholar]
- 25.Palleroni N J. Pseudomonadaceae. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Baltimore, Md: Williams and Wilkins; 1984. pp. 140–218. [Google Scholar]
- 26.Palleroni N J. Pseudomonas classification. Antonie Leeuwenhoek. 1993;64:231–251. doi: 10.1007/BF00873084. [DOI] [PubMed] [Google Scholar]
- 27.Pfaller M A, Wendt C, Hollis R J, Wenzel R P, Fritschel S J, Neubauer J J, Herwaldt L A. Comparative evaluation of an automated ribotyping system versus pulsed-field gel electrophoresis for epidemiological typing of clinical isolates of Escherichia coli and Pseudomonas aeruginosa from patients with recurrent Gram-negative bacteremia. Diagn Microbiol Infect Dis. 1996;25:5–8. doi: 10.1016/0732-8893(96)00082-x. [DOI] [PubMed] [Google Scholar]
- 28.Ralyea R D, Wiedmann M, Boor K J. Bacterial tracking in a dairy production system using phenotypic and ribotyping methods. J Food Prot. 1998;61:1336–1340. doi: 10.4315/0362-028x-61.10.1336. [DOI] [PubMed] [Google Scholar]
- 29.Schroder M J A. Origins and levels of post pasteurization contamination of milk in the dairy and their effects on keeping quality. J Dairy Res. 1984;51:59–67. doi: 10.1017/s0022029900023323. [DOI] [PubMed] [Google Scholar]
- 30.Shah N P. Psychrotrophs in milk: a review. Milchwissenschaft. 1994;49:432–437. [Google Scholar]
- 31.Stanier R Y, Palleroni N J, Doudorhoff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 32.Swart G J, Jooste P J, Mostert J F. Species identification and some physiological characteristics of Gram-negative psychrotrophic isolates from raw silo milk. S Afr Dairy Sci. 1990;22:31–36. [Google Scholar]
- 33.Ternström A, Lindberg A-M, Molin G. Classification of the spoilage flora of raw and pasteurized bovine milk, with special reference to Pseudomonas and Bacillus. J Appl Microbiol. 1993;75:25–34. doi: 10.1111/j.1365-2672.1993.tb03403.x. [DOI] [PubMed] [Google Scholar]
- 34.Tesar M, Hoch C, Moore E R B, Timmis K N. Westernprinting: development of a rapid immunochemical identification for species within the genus Pseudomonas sensu stricto. Syst Appl Microbiol. 1996;19:577–588. [Google Scholar]
- 35.Thomas S B, Druce R G. Psychrotrophic bacteria in refrigerated milk. Part III. Dairy Ind. 1969;34:501–505. [Google Scholar]
- 36.Vanderzant C, Splittstoesser D F. Compendium of methods for the microbiological examination of foods. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]