Abstract
Genetic heterogeneity of denitrifying bacteria in sediment samples from Puget Sound and two sites on the Washington continental margin was studied by PCR approaches amplifying nirK and nirS genes. These structurally different but functionally equivalent single-copy genes coding for nitrite reductases, a key enzyme of the denitrification process, were used as a molecular marker for denitrifying bacteria. nirS sequences could be amplified from samples of both sampling sites, whereas nirK sequences were detected only in samples from the Washington margin. To assess the underlying nir gene structure, PCR products of both genes were cloned and screened by restriction fragment length polymorphism (RFLP). Rarefraction analysis revealed a high level of diversity especially for nirS clones from Puget Sound and a slightly lower level of diversity for nirK and nirS clones from the Washington margin. One group dominated within nirK clones, but no dominance and only a few redundant clones were seen between sediment samples for nirS clones in both habitats. Hybridization and sequencing confirmed that all but one of the 228 putative nirS clones were nirS with levels of nucleotide identities as low as 45.3%. Phylogenetic analysis grouped nirS clones into three distinct subclusters within the nirS gene tree which corresponded to the two habitats from which they were obtained. These sequences had little relationship to any strain with known nirS sequences or to isolates (mostly close relatives of Pseudomonas stutzeri) from the Washington margin sediment samples. nirK clones were more closely related to each other than were the nirS clones, with 78.6% and higher nucleotide identities; clones showing only weak hybridization signals were not related to known nirK sequences. All nirK clones were also grouped into a distinct cluster which could not be placed with any strain with known nirK sequences. These findings show a very high diversity of nir sequences within small samples and that these novel nir clusters, some very divergent from known sequences, are not known in cultivated denitrifiers.
Denitrification is a dissimilatory process where oxidized nitrogen compounds (NO3− and NO2−) are used as alternative electron acceptors for energy production. Nitrogen oxides are reduced stepwise to gaseous end products (NO, N2O, and N2) which are concomitantly released, leading to a loss of fixed nitrogen to the environment. In marine coastal sediments, 40 to 50% of external inputs of dissolved inorganic nitrogen are removed by denitrification (23), resulting in an unbalanced nitrogen budget in the ocean (5, 7). Accumulation of the intermediates NO and N2O contributes to global warming and the destruction of the ozone layer (6). Thus, the underlying communities of denitrifying bacteria and their responses to environmental conditions need to be understood. Commonly, 16S rDNA molecules are tools to investigate microbial communities which avoid limitations of culturability (9, 11, 16). Lately, several functional genes were shown to be useful for the same purpose (17, 19, 28) because their phylogeny is congruent or very similar to phylogenetic relationships based on 16S rRNA gene (rDNA) analyses. In many cases, functional genes provide a resolution below species level because of higher evolutionary rates of the less conserved functional molecules. Additionally, this approach may indicate functional diversity in the environment.
An approach involving 16S rDNA does not appear to be suitable to investigate communities of denitrifying bacteria, as denitrification is widespread among phylogenetically unrelated groups (33). Common metabolic functions have been used for detection and community analysis of denitrifying bacteria such as nitrite and nitrous oxide reduction (3, 12, 21, 22). These key steps of the denitrification process are catalyzed by nitrite and nitrous oxide reductase, respectively. Two structurally different but functionally equivalent enzymes catalyze nitrite reduction: a copper and a cytochrome cd1-containing nitrite reductase encoded by the genes nirK and nirS, respectively. The nirK and nirS genes were useful targets for PCR primers to detect communities of denitrifying bacteria in samples from freshwater environments and activated sludge (3, 12). In the present study, we successfully applied these PCR methods and subsequent clone analysis to detect denitrifying bacteria in sections of marine sediment cores from Puget Sound and from the Washington margin. A cultivation study was set up concomitantly with sediment samples from the Washington margin to isolate representative denitrifying bacteria from the same environment. Cultivation of denitrifiers with nir genes closely related to sequences from the cloning experiment would allow details of function of novel gene sequences to be explored.
MATERIALS AND METHODS
Sediment sampling.
Sediment cores investigated came from offshore Washington coast and Puget Sound, Washington. Sampling stations and their locations are described in Table 1. The Washington coast sediment cores were separated each into 10 sections of 0.5 or 1.0 cm. Sections were collected (November 1997) into sterile polypropylene bags and stored at 4°C until isolation of denitrifying bacteria and DNA extraction in Michigan. The Puget Sound sediment core was collected in March 1998, divided into three sections (1.0 to 1.5 cm, 1.5 to 2.0 cm, and 6.0 to 6.5 cm), and placed in sterile polypropylene bags. Samples were stored on ice, shipped to Michigan on dry ice, and stored at −20°C until DNA extraction.
TABLE 1.
Characteristics of sampling stations
| Station | Latitude, longitude | Water depth (m) | Penetration depth into sediment (cm)
|
Carbon (%) | Bottom water O2 concn (μM) | Isolate(s) | Clones
|
||
|---|---|---|---|---|---|---|---|---|---|
| O2 | NO3− | nirK | nirS | ||||||
| Washington margin | |||||||||
| 301 (A) | 46°48′60"N, 124°37′20"W | 119 | 0.4 | 0.4 | 1.69 | 151 | A3-5 | ||
| 303 (B) | 46°47′42"N, 125°21′80"W | 1,936 | 0.9 | 0.9 | 2.34 | 75 | B9-12 | wB | wA-C |
| 304 (C) | 46°45′05"N, 126°00′58"W | 2,530 | 2.2 | 6.0 | 1.62 | 93 | C10-1 | wC | wD-F |
| 305 (D) | 46°44′97"N, 127°59′96"W | 2,664 | 3.4 | 21.0 | 1.18 | 112 | D4-14, D7-6, D9-1 | ||
| 306 (E) | 46°29′60"N, 124°43′22"W | 630 | 0.6 | 0.6 | 2.52 | 27 | E4-2 | ||
| 307 (F) | 46°26′61"N, 124°47′22"W | 997 | 0.7 | 0.8 | 3.05 | 23 | F8-5, F9-2 | ||
| Puget Sound | |||||||||
| Carkeek | 47°43′50"N, 122°23′90"W | 182 | 0.4 | 0.4 | ∼2 | 202 | pA/pB/pC | ||
Isolation of denitrifying bacteria and growth conditions.
To isolate denitrifying bacteria from the Washington margin (Table 1), samples from 10 discrete depth intervals from six stations were diluted in 10-fold series, and each dilution was incubated anaerobically at room temperature in nutrient broth (Difco Laboratories, Detroit, Mich.) supplemented with artificial seawater (3% salt [14]) and KNO3 (5 mM). After 2 weeks, denitrifying activity was monitored by nitrate disappearance using the Szechrome reagent (Polysciences, Inc., Warrington, Pa.). The highest dilution from each sample showing denitrifying activity was used for bacterial isolation on nutrient agar. At least four bacteria differing in colony morphology, color, and size were picked from each sample. Prior to sequencing, unique isolates were differentiated by repetitive extragenic palindromic-PCR fingerprint analysis (18). The 16S rDNA gene sequencing was performed as described previously (32), and the general taxonomic position was determined with the BLAST program from the Genetics Computer Group program package (10).
Denitrifying bacteria were also enriched from the Puget Sound core in 10-fold serial dilutions but in marine medium (Gibco BRL, Gaithersburg, Md.) supplemented with 5 mM KNO3 and incubated anaerobically. Additionally, these enrichments were supplemented with either yeast extract (0.5 mg ml−1) or Casitone (0.5 mg ml−1). After incubation for 8 months at 22°C, the enrichments were tested microscopically for growth and qualitatively for the removal of nitrate and nitrite, using test sticks (Merck, Darmstadt, Germany).
Extraction and purification of DNA.
DNA was prepared from pure cultures, environmental samples, and enrichments. (i) Genomic DNA from pure cultures was obtained by lysozyme-proteinase K-sodium dodecyl sulfate (SDS) treatment. DNA was purified by precipitation with hexatrimethylammonium bromide (Sigma Chemical Co., St. Louis, Mo.), phenol-chloroform extraction, and subsequent precipitation with ethanol (1). (ii) Total DNA was extracted from sediment sections of Washington margin sediment cores 303 and 304 (sections 0.5 to 1.0 cm [1,936 m] and 0.0 to 0.5 cm [2,530 m]) and from the three sections from Puget Sound (Table 1). DNA from 5 g of sediment each was extracted by the method of van Elsas and Smalla (29), using the freeze-thaw procedure. The method was modified by using an additional proteinase K treatment (50 μl of a 20-mg ml−1 solution) after incubation with SDS. (iii) Total DNA was extracted from 5 ml of liquid enrichment culture of the 10−2 dilution. Cells were harvested by sequential centrifugation (13,600 × g, 15 min, 22°C) in the same 2-ml tube, resuspended in 1.5 ml of 120 mM sodium phosphate buffer (pH 8.0). DNA was extracted as described for step ii. DNA was quantified and analyzed spectrophotometrically at 230, 260, and 280 nm.
PCR amplification of nir fragments.
Fragments of the nirK and nirS gene were amplified using primer pairs nirK1F-nirK5R for nirK and nirS1F-nirS6R for nirS, developed by Braker et al. (3). Touchdown PCR was done in a model 9600 thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) using conditions as described elsewhere (3) except that the annealing temperature during the first 10 cycles was shifted to 56 to 51 and to 54°C during the following 25 cycles. Additionally, the amount of primer was doubled to 1 μM each primer, and to increase specificity of the reactions, bovine serum albumin (400 ng μl−1; Boehringer Mannheim) was added. Aliquots of 10 μl of the reactions were analyzed by electrophoresis on 2% (wt/vol) agarose gels (Gibco BRL) followed by 15 min of staining with ethidium bromide (0.5 mg liter−1). Bands were visualized by UV excitation.
Cloning of nirK and nirS PCR products from sediment samples and enrichments.
For nirK, PCR products from sediment samples were purified by eluting the bands from an agarose gel (2% [wt/vol]) using the QIAquick gel extraction kit (Qiagen, Chatsworth, Calif.) as specified by the manufacturer. Eluted nirK PCR (3 μl) products or amplified nirS PCR products (3 μl) were cloned using the TA cloning kit (Invitrogen, San Diego, Calif.). One hundred clones for each sediment sample were randomly selected for further analysis. nirK or nirS PCR products were cloned from enrichments, and eight clones each were analyzed. A small amount of cell material picked up with a toothpick was resuspended in a preprepared 25-μl PCR mix, and inserts were amplified as described elsewhere (31). Aliquots (2.5 μl) were analyzed on 2% (wt/vol) agarose gels. PCR product (0.2 μl) from clones containing inserts of the estimated size were further used for a 25-μl nested PCR amplification. In this step, the nirK and nirS primer pairs and conditions were used as described for the initial PCR. Aliquots of the PCR were analyzed on 2% (wt/vol) agarose gels.
RFLP screening of nirK and nirS clone libraries.
PCR products were first screened by restriction fragment length polymorphism (RFLP). For nirK clones, PCR products were digested in two separate reactions with 0.3 U of restriction enzymes HaeIII and MspI (Gibco BRL) at 37°C overnight. For nirS clones, PCR products were digested in a single reaction with 0.3 U of each restriction enzyme HhaI and MspI (Gibco BRL) at 37°C overnight. The digested products were separated by gel electrophoresis in 3.5% (wt/vol) Metaphor agarose (FMC Bioproducts, Rockland, Maine) in freshly prepared 1× Tris-borate-EDTA buffer at 4°C with 7 V cm−1 for 4 h. The RFLP patterns were analyzed and clustered with the GelCompar software (Applied Maths, Kortrijk, Belgium) applying the unweighted pair group method using arithmetic averages (UPGMA) and the Jaccard algorithm. The resulting clusters were additionally compared by eye.
Dot blot hybridization.
One microliter, or 1 μl of a dilution of up to 1:3, of each nested PCR product from the clones was spotted onto a positively charged nylon membrane (Zeta-Probe; BioRad, Hercules, Calif.) and fixed by UV cross-linking for 45 s at 302 nm. Four different probes for each gene were used to hybridize with cloned PCR products: for nirK, PCR products obtained with primer pair nirK1F-nirK5R from Pseudomonas sp. strain G-179 (30), Rhodobacter sphaeroides f. sp. denitrificans (20), Blastobacter denitrificans DSM 1113, and Alcaligenes sp. strain DSM 30128. For nirS, PCR products were generated with primer pair nirS1F-nirS6R from Pseudomonas stutzeri JM300, Pseudomonas aeruginosa NCTC 6750, Roseobacter denitrificans ATCC 33942, and two marine denitrifying isolates, B9-12 and D4-14 (this study). Probes generated from P. stutzeri JM300 and P. aeruginosa were used in a mixture of equal amounts (25). PCR products were purified by elution from an agarose gel (2% [wt/vol]) by using the QIAquick gel extraction kit (Qiagen) as specified by the manufacturer. The probes were randomly labeled by using the digoxigenin DNA labeling and detection kit (Boehringer Mannheim) as specified by the manufacturer. After prehybridization of the membranes at 42°C for 6 h in DIG Easy Hyb solution (Boehringer Mannheim), hybridization was performed at 42°C overnight in a DIG Easy Hyb solution containing the specific probe (25 ng ml−1). After hybridization, the membrane was washed twice for 5 min at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) SDS and twice at 56°C for nirK and at 58°C for nirS for 15 min in 0.5× SSC–0.1% (wt/vol) SDS. Subsequently, the hybridization of the digoxigenin-labeled probe was detected by an enzyme-linked immunoassay with nitroblue tetrazolium–X-phosphate as the substrate as specified by the manufacturer (Boehringer Mannheim). Strength of hybridization signals was assigned visually as 0 (no hybridization signal) to 6 (strong hybridization signal).
Southern blot hybridization.
Aliquots of nirK PCR products (10 μl) from Puget Sound DNA extracts and from one Washington coast (0.5 to 1.0 cm, 1,936 m) DNA extract and a nirK PCR product from Alcaligenes sp. strain DSM 30128 were separated in an 2% (wt/vol) agarose gel. The DNA was transferred onto a positively charged nylon membrane (Bio-Rad) by capillary transfer (26) and UV cross-linked to the membrane with UV light (45 s at 302 nm). Hybridization with the probes for nirK was performed as described above.
Sequencing of nir products and phylogenetic analysis.
For DNA sequencing, amplified products were purified with the QIAquick PCR purification kit (Qiagen) as specified. DNA sequences were determined by direct sequencing with a model 373A DNA sequencer (Applied Biosystems Inc., Foster City, Calif.) by using dye terminator chemistry. The primers used for sequencing were nirK5R for nirK and nirS1F and nirS6R for nirS.
Nucleotide sequences and deduced amino acid sequences were aligned with sequences from the EMBL database, using the CLUSTAL program from the Genetics Computer Group program package (10), and dendrograms were constructed by using the programs SEQBOOT, DNADIST, DNAPARS, PROTDIST, PROTPARS, and CONSENSUS from the PHYLIP version 3.5c package (8).
Nucleotide sequence accession numbers.
nirK and nirS gene sequences from marine sediments and enrichment cultures have been deposited in the EMBL nucleotide sequence database under accession no. AJ248392 through AJ248448.
RESULTS
Nitrite reductase genes of marine denitrifying isolates.
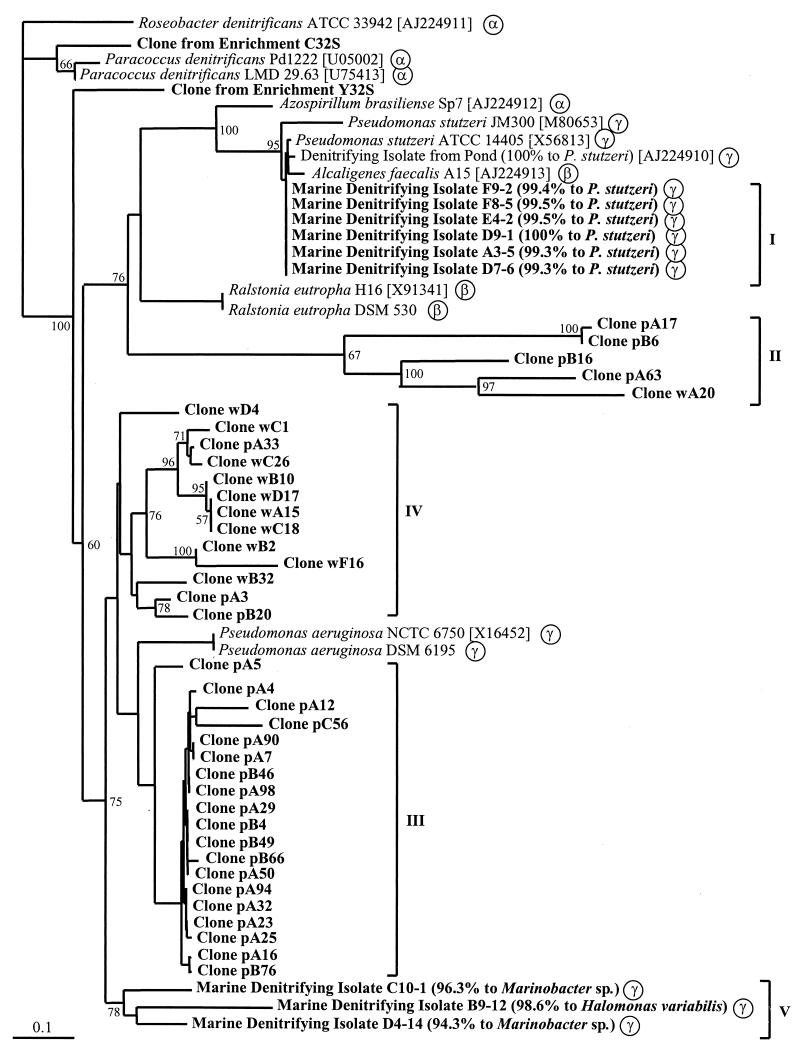
Nine denitrifying isolates shown to be distinct by REP-PCR (data not shown) were investigated for the presence of the nirK or the nirS gene. All of these isolates showed amplification only with the nirS-specific primer combination resulting in one PCR product of the expected size (890 bp), and none reacted with the nirK-specific primer pair. Identity of the nirS products was confirmed by sequencing. The general taxonomic position of the isolates was determined by sequencing of the 16S rDNA genes and by comparison to the EMBL database. All isolates were members of the γ-Proteobacteria. Six of them were close relatives of P. stutzeri, two were related to Marinobacter sp., and one was related to Halomonas variabilis. The branching within the nirS gene tree and the general taxonomic position are shown in Fig. 1.
FIG. 1.
Phylogenetic relationship of nirS gene products (partial, 112 amino acids; accession numbers in brackets). The dendrogram was generated by phylogenetic distance analysis with a neighbor-joining algorithm with Roseobacter denitrificans [AJ224911] as the outgroup. Values indicate the percentage of 100 replicate trees supporting the branching order. Bootstrap values below 50 are omitted. Scale bar, 10 mutations per 100 sequence positions. Clones obtained from Puget Sound and Washington coast samples are labeled p and w, respectively. Phylogenetic positions of pure cultures based on 16S rDNA genes are indicated by α, β, and γ for the subgroups of the Proteobacteria. General taxonomic positions of marine isolates based on 16S rDNA genes in percent similarity determined by BLAST search is given in parenthesis after isolate designations. Roman numbers indicate the clusters of nirS sequences from isolates and environmental clones.
Amplification of nir genes from sediment samples and enrichments.
DNA of high molecular mass was extracted from all sediment samples, with 1 g of wet sediment yielding up to 12.5 μg of purified DNA. To confirm the suitability of DNA extracts for PCR amplification, 16S rDNA genes were successfully amplified from total DNA extracts from all sediments. Amplification using the nirS-specific primer pair was successful for sediment samples from Puget Sound and the Washington coast, resulting in a single PCR product of the predicted size (approximately 890 bp). PCR products of the copper-containing nitrite reductase using the nirK-specific primer pair were obtained only from the Washington coast sediment DNA extracts, not from the Puget Sound sediments. Failure to amplify nirK fragments from these sediment samples also occurred when the DNA extraction and PCR procedure were repeated. PCR products of the predicted size (514 bp) were weak and showed one additional unspecific band for the DNA extract from the 2,530-m Washington coast sample. Therefore, bands of the predicted size were eluted from an agarose gel and concentrated before cloning. A Southern blot confirmed that no amplification was achieved with the nirK-specific primers from Puget Sound samples, while nirK products were detected from a Washington coast sample (1,936 m) as a positive control (data not shown).
nirK PCR products were obtained only from the enrichment supplemented with yeast extract, but the nirS-specific primers amplified fragments from both enrichments supplemented with yeast extract or Casitone. The PCR-amplified regions of the nirK and nirS genes were cloned successfully from both the Washington coast and Puget Sound sediment samples and from the enrichments.
RFLP analysis of nirK and nirS clones.
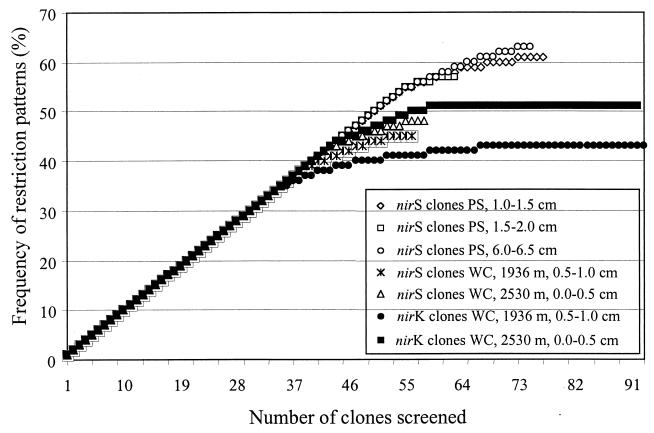
A total of 185 nirK clones (93 from the Washington coast 1,936 m and 92 from 2,530 m) and 229 nirS clones (77 from Puget Sound 1.0 to 1.5 cm, 63 from 1.5 to 2.0 cm, 75 from 6.0 to 6.5 cm; 56 from the Washington coast 1,936 m and 58 from 2,530 m) were screened by RFLP. For nirK clones, the analysis resulted in one dominant group of clones occurring in both samples from the Washington coast comprising 29 and 37% of all nirK clones. Several minor groups were also detected, but 34 and 40% of nirK clones were unique to one sediment sample. For nirS clones, no dominant group was found. Groups consisted of no more than five redundant clones, and the amount of clones unique to one sediment sample ranged from 55 to 70% (Puget Sound 1.0 to 1.5 cm, 55%; 1.5 to 2.0 cm, 61%; 6.0 to 6.5 cm, 59%; Washington coast 1,936 m, 70%; Washington coast 2,530 m, 67%). Rarefraction analysis showed a similar level of diversity for nirK and nirS sequences from the Washington coast samples, whereas it indicated a higher level of diversity of nirS sequences from the Puget Sound sediment samples (Fig. 2). Comparison of RFLP patterns from nirS clones did not indicate any congruence to those from marine denitrifying isolates and other denitrifying strains (Fig. 1).
FIG. 2.
Rarefraction curves indicating the diversity of denitrifying bacteria as detected by RFLP analysis of cloned nitrite reductase genes (nirK and nirS) from Puget Sound and Washington coast samples. nirK clones were digested with the tetrameric restriction enzymes MspI and HaeIII, and nirS clones were digested with MspI and HhaI in a single reaction. PS and WC stand for Puget Sound and Washington coast, respectively.
Analysis of clones from enrichments showed one dominant group of five nirS clones and three unique RFLP patterns from the enrichment supplemented with Casitone and one single pattern each for nirK and nirS clones from the enrichment supplemented with yeast extract. None of these patterns matched those of clones from the sediment samples or from marine denitrifying isolates or denitrifying strains.
Dot blot hybridization of nirK and nirS clones.
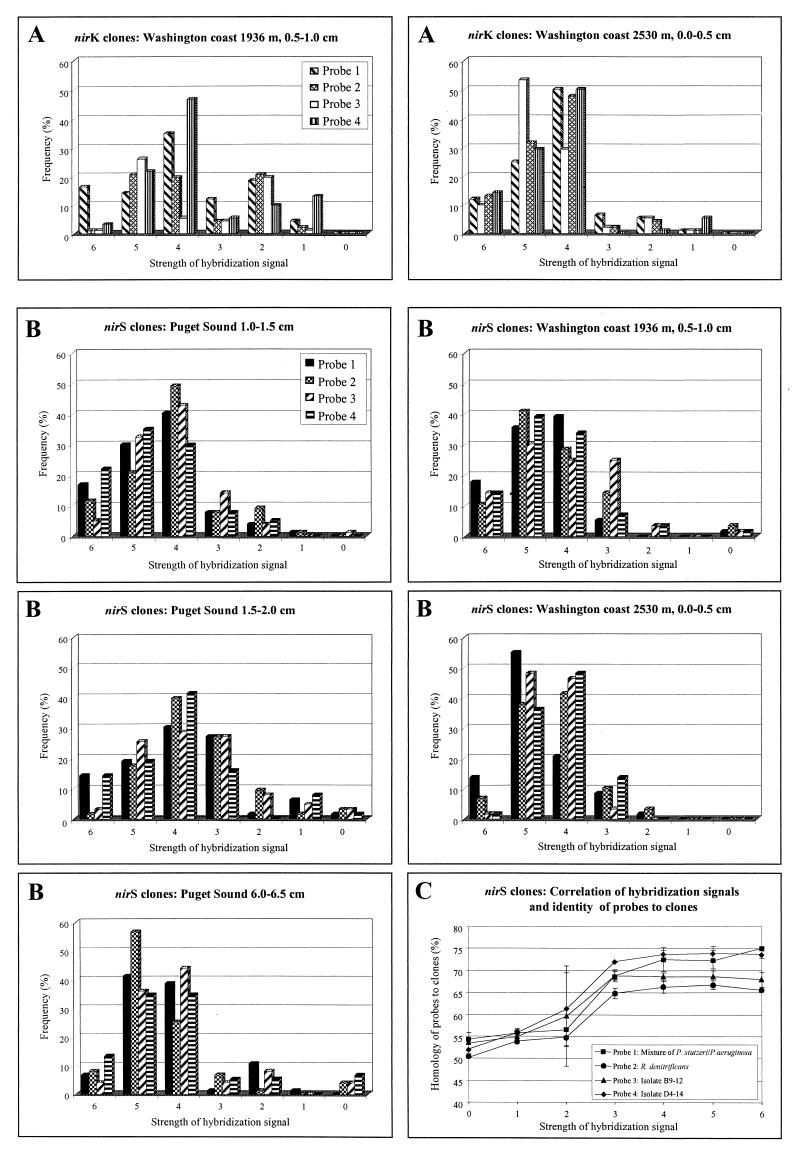
To determine the specificity of cloned nirK and nirS PCR products, all clones were hybridized to four probes for each gene. The probes were selected to show identity levels of 68.2 to 74.6% for nirK and 61.0 to 76.6% for nirS to each other. The stringency conditions were set to detect >70% identity to achieve a broad reactivity to unrelated clones. With a few exceptions, hybridization signals of all four probes for a specific clone did not vary more than three strengths. The same differences were found for clones with the same RFLP pattern due to uneven amounts of the DNA spotted. For nirK clones, a visible hybridization signal (signal strength 1 or stronger) to all probes was detected. nirK clones most frequently showed hybridization signals of 4 and stronger, with a predominance of signal strength 4 according to the dominant group of nirK clones in these sediment samples (Fig. 3A).
FIG. 3.
Dot blot hybridization of nirS and nirK clones obtained from Puget Sound and Washington coast sediment samples. (A) Cloned nirK fragments were hybridized to four different probes: nirK PCR products from Pseudomonas sp. strain G-179 (1), from B. denitrificans (2), from R. sphaeroides f. sp. denitrificans (3), and from Alcaligenes sp. (4). Strength of hybridization signals was assigned as 0 (no hybridization signal) to 6 (strong hybridization signal). Frequency (%) expresses the total number of clones hybridizing with each probe. (B) Cloned nirS fragments were hybridized to four different probes: nirS PCR products from P. stutzeri/P. aeruginosa (1), from P. denitrificans (2), from marine isolate B9-12 (3), and from marine isolate D4-14 (4). Strength of hybridization signals was assigned as for panel A. (C) Strength of nirS hybridization signals was correlated to homology values of the probes to 37 sequenced nirS clones. Bars show standard deviation.
Except for two nirS clones which did not hybridize to any of the probes, all clones hybridized to at least one of the probes. nirS clones showing signal strength of 4 and stronger (Fig. 3B) were dominant in all sediment samples investigated. Thus, nirS clones in these sediments show dominance of conserved sequences in the amplified sample based on hybridization signals of 3 and stronger. This signal strength was correlated to a sequence homology of 65% and higher (Fig. 3C). None of the probes generally showed stronger or weaker signals, indicating no nir subfamily dominance in any samples.
Sequence analysis of nirK and nirS clones.
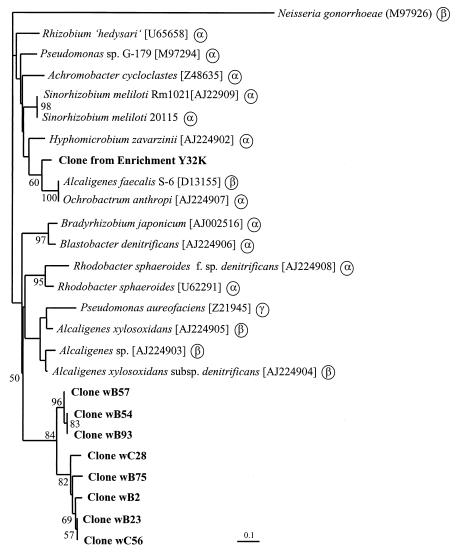
Partial nirK sequences (336 bp) were obtained from eight clones representing the dominant RFLP groups and showing hybridization signals of 3 or stronger; they were confirmed as nirK genes. Sequences from nirK clones showing weaker hybridization signals could not be defined as nirK sequences by alignment or comparison to the EMBL database using FASTA or BLAST search. Levels of nucleotide identity of the nirK clones were from 78.6 to 100.0%, and levels of identity of the deduced amino acids were from 89.3 to 100% within the sequenced region (data not shown). A similar level of conservation over the gene (63.1 to 74.7% for the full sequence, in comparison to 65.6 to 79.9% for the sequenced 336 bp at the C terminus for the published sequences) supported the usefulness of this region for phylogenetic purposes. The aniA gene from Neisseria gonorrhoeae was not considered in this analysis but was used in the generation of the dendrogram. This gene encodes a putative gonococcal nitrite reductase with significant homologies to copper-containing nitrite reductases from denitrifiers (15). However, the level of conservation of this sequence in comparison to all nirK sequences is significantly lower (37.4 to 44.7% and 29.1 to 34.6% for nucleotides and amino acids, respectively, within the sequenced region). Dendrograms of nucleotide sequences and deduced amino acid sequences generated by neighbor joining or parsimony were consistent for the major clustering except for minor differences within the clusters derived of sequences from known denitrifying strains. Regions of insertion or deletion were omitted from the analysis because of uncertain alignment. Trees (Fig. 4) showed two major clusters with the sequence from the enrichment clone branching with nirK-containing denitrifiers belonging to the α-Proteobacteria except for Alcaligenes faecalis S-6 (β-Proteobacteria). Sequences from sediment clones clustered with none of these sequences. The second cluster consisted of known denitrifiers belonging to the α-, β-, or γ-Proteobacteria and clones from marine sediment samples. The latter form a distinct subcluster of nirK sequences unknown in cultured organisms, suggesting that these genes are derived from novel denitrifiers.
FIG. 4.
Phylogenetic relationship of nirK gene products (partial, 111 amino acids; accession numbers in brackets). The dendrogram was generated by phylogenetic distance analysis with a neighbor-joining algorithm with B. denitrificans [AJ224906] as the outgroup. Values indicate the percentage of 100 replicate trees supporting the branching order. Bootstrap values below 50 are omitted. Scale bar, 10 mutations per 100 sequence positions. Phylogenetic positions of pure cultures based on 16S rDNA genes are indicated by α, β, and γ for the subgroups of the Proteobacteria.
As rarefraction analysis showed a higher level of diversity of nirS than nirK clones (Fig. 2) in marine sediment samples, 35 clones were chosen for partial sequencing (336 to 348 bp). Clones selected were from groups with distinct restriction patterns or were nonredundant clones without (clones wA20 and pB95) or with weak (clones pA17, pA63, pB6, and pB16) hybridization signals. Sequencing confirmed that all nirS clones from marine sediment samples and enrichments were nirS sequences except clone pB95, for which a matching sequence could not be found by FASTA and BLAST search. Nucleotide identity ranged from 45.3 to 100% and 40.0 to 100.0% for identity of deduced amino acids (data not shown). Similar levels of conservation of the entire gene (72.2 to 94.2%) in comparison to 69.5 to 95.0% for the sequenced region from published sequences suggests the usefulness of the sequenced region for phylogenetic purposes. Dendrograms were identical with both the distance and parsimony methods except for minor differences in the positions of clones wD4, pA5, and wB32. Regions of insertion or deletion were omitted from the analysis because of uncertain alignment. Trees showed three major clusters of nirS sequences with clones from marine sediment samples clustering in three distinct subclusters (Fig. 1). None of the clones was closely related to the nir gene of any isolate, whereas clone C32S from the enrichment supplemented with Casitone was related to the nirS gene of Paracoccus denitrificans within the first major cluster. Clone Y32S from the enrichment supplemented with yeast extract was located on a distinct branch. One major cluster consisted of a subcluster of closely related nirS sequences of all P. stutzeri strains and related isolates (Fig. 1, cluster I). A second subcluster within this branch consisted of very distantly related nirS sequences from both environments, Puget Sound and the Washington coast (cluster II). Within the other major cluster, the remaining nirS clones formed subclusters mostly according to the environment from which they were obtained. None of the clones from offshore Washington coast branched with a group of sequences from Puget Sound clones (cluster III) which clustered together with nirS sequences of P. aeruginosa. Clones from the Washington coast formed a separate subcluster (cluster IV), although three clones from Puget Sound branched within this cluster. A third branch within this cluster was formed by three marine denitrifying isolates (cluster V). The distant relatedness and separate clustering of the nirS clones from marine sediments suggest that groups of novel denitrifying bacteria inhabit these sediments.
DISCUSSION
PCR approaches amplifying 514- and 890-bp fragments from nirK and nirS genes from natural denitrifying populations, respectively, were applied to investigate the community structure of denitrifiers in marine sediment samples from two habitats in the Pacific Northwest. With the nirS primer pair, amplification was possible with all DNA extracts from all sediment samples tested. With the nirK primer pair, amplification was not possible with DNA extracts from Puget Sound. Even Southern blot hybridization with four different nirK probes did not detect any amplification from the Puget Sound sediment samples. The nirK primers seemed to amplify nirK genes closely related to those from which the primers were designed (3). Thus, they may not have detected whether more distantly related nirK genes were present in Puget Sound sediments. There was a small group of clones which could not be specified as nirK sequences although these hybridized weakly to the probes (Fig. 3A). Even though some of these sequences were partial open reading frames, BLAST search or alignments did not show close relationships to known nirK sequences and to each other. Additionally, analysis of the deduced amino acid sequences within the sequenced region showed that histidine B255 (Achromobacter cycloclastes numbering), which is not a copper ligand but close to the active site (2), was not conserved among these sequences but was conserved among all nirK sequences and the aniA sequence of N. gonorrhoeae. Comparison of deduced amino acids among nirK sequences revealed only two amino acids within this region which were specific for the marine clones, thus suggesting that the nirK primers indeed showed bias toward well-conserved nirK sequences. On the other hand, within the nirK gene tree the marine nirK sequences clustered together in a distinct subcluster of marine sequences, indicating that the marine nirK containing denitrifier genes were different from known nirK genes. Any suggestion about the phylogenetic affiliation of these denitrifiers was not possible. However, their grouping within nirK sequences from Proteobacteria suggests that they might be members of the Proteobacteria (Fig. 4).
For nirS, all except one clone could be identified as nirS by hybridization and sequencing. BLAST search and alignments did not show any homology to nirS sequences for this clone. A second clone (wA20) not hybridizing to any of the nirS probes was confirmed as a very distantly related nirS sequence. Levels of nucleotide identity were from 45.3 to 58.7% to other nirS sequences but analysis of deduced amino acids revealed conservation of key amino acids among all partial sequences. Histidine 352 (P. aeruginosa numbering) was conserved among all sequences whereas His 302 was not conserved among the distantly related clones (clone pA17, pA63, pB6, pB16, and wA20) and His 314 was not conserved among Azospirillum brasiliense Sp7 and clone pB16 but was conserved in all other sequences (data not shown). This suggests only His 352 as a heme d1 ligand (24). Despite weak hybridization signals, all nirS clones hybridizing to any of the probes were specific nirS sequences amplified by the nirS primer pair used in this study. Furthermore, despite a dominance of clones with hybridization signals of 4 and stronger, these nirS primers also amplified fragments from less conserved nirS genes, reflected by the clones with hybridization signals of 3 and weaker (Fig. 3B). Strength of hybridization signals was correlated to the level of sequence conservation of nirS clones. For signal strength of 3 or weaker, there was a linear correlation of increasing signal strength to the increasing level of identity between probes and clones. For signal strength of 4 and stronger, it was assumed that an increase in strength was due to more conserved regions which strongly hybridized to the probes, although the overall level of identity for the sequenced clones to the probes did not exceed 65 to 75% (Fig. 3C).
RFLP analysis of clones and denitrifying strains revealed that clones from the marine sediment samples did not match any nir sequence from the database. Sequence analysis of selected clones confirmed these findings and grouped the nirS clones into three distinct subclusters of novel marine sequences within the nirS gene tree, suggesting that there were completely different nir genes if not groups of denitrifiers in this habitat. Interestingly, most of the clones grouped according to the environment from which they were obtained, suggesting that there are distinct populations of denitrifying bacteria within Puget Sound sediments and offshore Washington coast sediments.
The nirS trees show generally the same clustering as the 16S rDNA trees with the exception of the nirS genes of Alcaligenes faecalis A15 (β-Proteobacteria) and A. brasiliense Sp7 (α-Proteobacteria) grouping with the nirS genes of P. stutzeri (γ subgroup). Therefore, it is likely that at least the more closely related clones from Puget Sound and the Washington coast belonged to Proteobacteria. More obscure remains the phylogenetic affiliation of the very unrelated cluster (cluster II) of nirS clones. Unfortunately, but not unexpectedly, clones from enrichments and nirS sequences from the marine denitrifying isolates did not group closely with the nirS environmental clones (Fig. 1). Further cultivation strategies are desirable for recovering the organisms with these more novel sequences so that any unique functional diversity can be identified.
With the caveat of primer preference for more conserved nirS genes and possible bias during PCR (27), it was assumed that these results reflect the community structure of denitrifying bacteria in these sediments, at least more so than isolation studies. Rarefraction analysis detected a high level of diversity within nirS genes in the sediment samples, and analysis of RFLP patterns showed that there were distinct populations present even in individual sections of the Puget Sound core although the first two sections were adjacent (1.0 to 1.5 cm and 1.5 to 2.0 cm). Clones were not redundant among Puget Sound sediment samples and those from the Washington coast. Only a few clones were redundant among the sections of the Puget Sound core. This suggested an independent development of cytochrome cd1-dependent nitrite reductase genes, especially in these two environments which are spatially distinct on a bacterial size scale and differ in quality and quantity of carbon input. Higher input of less degraded organic matter into Puget Sound sediments than into Washington coast sediments (13) together with long time and distance scales may have led to the development of distinct bacterial populations and hence nirS genes.
Comparison of the identity levels of nucleotides and of amino acids revealed that in many cases, the amino acids were less conserved than the nucleic acids which indicated that most mutations were not neutral and thus did not occur at the third base but within the first two bases of the codons. High rates of amino acid exchange indicating low selection pressure may be more likely under environmental conditions in which genes are not necessary for survival. For denitrification genes, this can readily occur where either aerobic growth or nitrate-free anaerobic conditions often predominate. Bioturbation in marine sediments disturbs chemical gradients periodically exposing denitrification genes to nonselective conditions. The enhanced bioturbation rates in shallow sediments than in deeper sediments (4) suggest lower selection pressure on Puget Sound versus Washington coast nirS gene populations, perhaps enhancing evolutionary rates of this functional gene.
ACKNOWLEDGMENTS
We are grateful to Stephen C. Nold for setting up the enrichments and for providing these to us and to Peter Hirsch for critical reading of the manuscript.
This work was supported by DOE grant DE-FG02-98ER62535.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 2.Berks B C, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 3.Braker G, Fesefeldt A, Witzel K-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpenter R, Peterson M L, Bennett J T. 210Pb-derived sediment accumulation and mixing rates for the Washington continental slope. Mar Chem. 1982;48:135–164. [Google Scholar]
- 5.Codispoti L A. Is the ocean losing nitrate? Nature. 1995;376:724. [Google Scholar]
- 6.Conrad R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) Microbiol Rev. 1996;60:609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devol A H. Direct measurements of nitrogen gas fluxes from continental sediments. Nature. 1991;349:319–321. [Google Scholar]
- 8.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genetics Computer Group. GCG program manual. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 11.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallin S, Lindgren P-E. PCR detection of genes encoding nitrite reductases in denitrifying bacteria. Appl Environ Microbiol. 1999;65:1652–1657. doi: 10.1128/aem.65.4.1652-1657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedges, J. I., F. S. Hu, A. H. Devol, H. E. Hartnett, E. Tsamakis, and R. G. Keil. Sedimentary organic matter preservation: a test for selective degradation under oxic conditions. Am. J. Sci., in press.
- 14.Lyman J, Fleming R H. Composition of sea water. J Mar Res. 1940;3:134–167. [Google Scholar]
- 15.Mellies J, Jose J, Meyer T F. The Neisseria gonorrhoeae gene aniA encodes an inducible nitrite reductase. Mol Gen Genet. 1997;256:525–532. doi: 10.1007/s004380050597. [DOI] [PubMed] [Google Scholar]
- 16.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. Diversity of deep-sea hydrothermal vent Archaea from Loihi Seamount, Hawaii. Deep-Sea Res. 1998;45:303–317. [Google Scholar]
- 17.Pichard S L, Campbell L, Paul J H. Diversity of the ribulose bisphosphate carboxylase/oxygenase form I gene (rbcL) in natural phytoplankton communities. Appl Environ Microbiol. 1997;63:3600–3606. doi: 10.1128/aem.63.9.3600-3606.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rademaker J L W, de Bruijn F J. Characterization and classification of microbes by REP-PCR genomic fingerprinting and computer-assisted pattern analysis. In: Caetano-Anollés G, Greshoff P M, editors. DNA markers: protocols, applications and overviews. J. New York, N.Y: Wiley & Sons, Inc.; 1997. pp. 151–171. [Google Scholar]
- 19.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satoh T, Hoshino Y, Kitamura H. Rhodopseudomonas sphaeroides forma sp. denitrificans, a denitrifying strain as a subspecies of Rhodopseudomonas sphaeroides. Arch Microbiol. 1976;108:265–269. doi: 10.1007/BF00454851. [DOI] [PubMed] [Google Scholar]
- 21.Scala D J, Kerkhof L J. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett. 1998;162:61–68. doi: 10.1111/j.1574-6968.1998.tb12979.x. [DOI] [PubMed] [Google Scholar]
- 22.Scala D J, Kerkhof L J. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl Environ Microbiol. 1999;65:1681–1687. doi: 10.1128/aem.65.4.1681-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seitzinger S P. Denitrification in aquatic sediments. In: Revsbech N P, Sørensen J, editors. Denitrification in soil and sediment. New York, N.Y: Plenum Press; 1990. pp. 301–322. [Google Scholar]
- 24.Silvestrini M C, Falcinelli S, Ciabatti I, Cutruzzolà F, Brunori M. Pseudomonas aeruginosa nitrite reductase (or cytochrome oxidase): an overview. Biochimie. 1994;76:641–654. doi: 10.1016/0300-9084(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 25.Smith G B, Tiedje J M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992;58:376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:624–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Elsas J D, Smalla K. Extraction of microbial community DNA from soils. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–11. [Google Scholar]
- 30.Ye R W, Fries M R, Bezborodnikov S G, Averill B A, Tiedje J M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl Environ Microbiol. 1993;59:250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Palumbo A V, Tiedje J M. Sensitive detection of a novel class of toluene-degrading denitrifiers, Azoarcus tolulyticus, with small-subunit rRNA primers and probes. Appl Environ Microbiol. 1997;63:2384–2390. doi: 10.1128/aem.63.6.2384-2390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zumft W G. The denitrifying prokaryotes. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 554–582. [Google Scholar]