Abstract
Genes showing differential expression related to the early G1 phase of the cell cycle during synchronized circadian growth of the toxic dinoflagellate Alexandrium fundyense were identified and characterized by differential display (DD). The determination in our previous work that toxin production in Alexandrium is relegated to a narrow time frame in early G1 led to the hypothesis that transcriptionally up- or downregulated genes during this subphase of the cell cycle might be related to toxin biosynthesis. Three genes, encoding S-adenosylhomocysteine hydrolase (Sahh), methionine aminopeptidase (Map), and a histone-like protein (HAf), were isolated. Sahh was downregulated, while Map and HAf were upregulated, during the early G1 phase of the cell cycle. Sahh and Map encoded amino acid sequences with about 90 and 70% similarity to those encoded by several eukaryotic and prokaryotic Sahh and Map genes, respectively. The partial Map sequence also contained three cobalt binding motifs characteristic of all Map genes. HAf encoded an amino acid sequence with 60% similarity to those of two histone-like proteins from the dinoflagellate Crypthecodinium cohnii Biecheler. This study documents the potential of applying DD to the identification of genes that are related to physiological processes or cell cycle events in phytoplankton under conditions where small sample volumes represent an experimental constraint. The identification of an additional 21 genes with various cell cycle-related DD patterns also provides evidence for the importance of pretranslational or transcriptional regulation in dinoflagellates, contrary to previous reports suggesting the possibility that translational mechanisms are the primary means of circadian regulation in this group of organisms.
Marine dinoflagellates represent a substantial fraction of bloom-forming phytoplankton in coastal areas of the world's oceans (4). In addition to the formation of red tides, some species of dinoflagellates produce a range of toxins that are poisonous to higher levels of the food web (49). One group of toxins synthesized by these harmful algal bloom organisms causes paralytic shellfish poisoning (PSP). The toxins included in this group are called saxitoxins (STX) and are all cyclic perhydropurine compounds differing in their specific toxicities (17, 49). Besides dinoflagellates, other organisms biosynthesize toxins causing PSP, such as, for example, bacteria (26, 51) and cyanobacteria (33, 50).
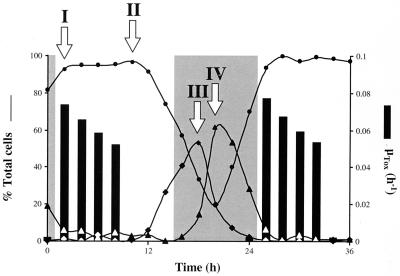
Classic mating experiments with toxic and nontoxic strains of Gymnodinium spp. and Alexandrium spp. have revealed that the inheritance of toxigenic potential and of toxin composition in dinoflagellates follows Mendelian rules (35, 43, 44). The nonrandom segregation patterns observed in the crossings also imply chromosomal localization of the genes involved in toxin biosynthesis and interconversions among derivatives (34, 44). A series of studies with synchronized cultures of Alexandrium fundyense has further shown that STX is accumulated during a discrete time period localized in the G1 phase of the cell cycle (Fig. 1) (52). The combination of these two factors, nuclear genes and discontinuous toxigenesis, gave rise to the hypothesis that STX biosynthesis could be regulated at the transcriptional level, with the genes responsible for toxin production being activated in a cyclic pattern. Furthermore, comparative analysis of cells harvested when toxin accumulation is on or off would allow the identification of genes that are expressed while STX is produced and that are therefore potentially related to toxigenesis. (Note that we use the term toxigenesis to mean toxin accumulated in the cell, ignoring losses due to leakage or catabolism.)
FIG. 1.
STX biosynthesis and the cell cycle in synchronously growing A. fundyense. Lines show relative distributions of cells in the different cell cycle stages (G1 [●], S [⧫], and G2 plus M [▴]). Positive toxin production rates (bars; μTOX) were limited to the first 8 to 10 h of the G1 cell cycle phase. No toxin production was detected for the remainder of the cell cycle. Arrows denote sampling points for the DD analysis protocol; two were during G1, the first (I) when toxin was accumulated and the second (II) after it stopped, one was during the S phase (III), and the last one was during G2 (IV). Shaded areas, dark periods of the cycle of 14 h of light and 10 h of darkness under which A. fundyense was synchronized.
Due to the harmful effects of STX production on humans and the environment, much work has been done on the ecology and physiology of STX biosynthesis in A. fundyense as well as on monitoring and predicting outbreaks of blooms caused by this organism. One of the key elements in studying bloom dynamics is the determination of in situ growth rates for Alexandrium. Current approaches rely on the analysis of the relative distributions of cells in the different cell cycle stages (9, 10). Alternative methods in which cell cycle-related genes exhibiting cyclic expression patterns are used as indicators of population growth rates have yet to be developed for dinoflagellates (25, 32). Dinoflagellates have a typical eukaryotic cell cycle with regulatory mechanisms resembling those of other eukaryotes (42, 56). The identification of genes whose expression is cell cycle dependent would be the first step toward the rational development of new methods for measuring cell growth rates in Alexandrium.
Recent advances in the development of molecular tools aimed at analyzing differences between complex genomes at the level of gene expression show great promise in this regard. One such tool is differential display (DD), a genetic approach that allows the analysis of differentially expressed genes in eukaryotes (28, 57). This technique has been used successfully to identify developmentally, environmentally, or hormonally regulated genes in protozoa, plants, animals, and humans (2, 21, 48, 54, 55). In this procedure, the two or more sets of differentially expressed mRNAs to be compared are used as templates to generate the corresponding cDNAs. Subsequently, the cDNA fragments are amplified by means of PCR using a combination of an mRNA-specific oligo(dT) “anchor” primer together with an arbitrary decamer primer in the presence of radioactively labeled deoxynucleoside triphosphates (dNTPs). After separation by denaturing polyacrylamide gel electrophoresis, differentially amplified cDNAs are identified by autoradiography. These cDNAs are further cloned and used as probes to screen cDNA libraries of the organism of interest.
The goal of this study was to identify genes for which expression was correlated to cell cycle progression and possibly toxin production in Alexandrium. Differential display was used to analyze mRNA expression patterns among cells differing in their cell cycle stage and toxigenic status. A first approach consisted of comparing phylogenetically closely related toxic and nontoxic strains of Alexandrium tamarense and A. fundyense. A second approach consisted of the identification of genes for which expression cycles were correlated to progression through the cell cycle or to the superimposed, discontinuous pattern of STX biosynthesis in A. fundyense. Several genes were identified, and their possible roles in cell cycle regulation and STX biosynthesis were explored.
MATERIALS AND METHODS
Organisms.
Batch cultures of the nontoxic strains A. tamarense PGt183 (Plymouth, United Kingdom), Pe1V (Galicia, Spain), and WKS-1 (Tanabe Bay, Japan) and the toxic strains A. tamarense GtPP03 (Perch Pond, Mass.), AtSL12 (St. Lawrence estuary, Canada), and GtLI21 (Moriches Bay, N.Y.) and A. fundyense GtCA28 (Cape Ann, Mass.) and Gt7 (Bay of Fundy, Canada) were used in this study. Cultures were maintained at 20°C in f/2 medium (14) modified by the addition of H2SeO3 (10−8 M) and by reducing the concentration of CuSO4 to 10−8 M. Vineyard Sound seawater (filtered through a 0.2-μm-pore-size filter; 31°/oo salinity) was used as the medium base. Throughout the experiment, irradiance of ca. 250 μmol of photons m−2 s−1 was provided by cool white fluorescent bulbs on a cycle of 14 h of light and 10 h of darkness. Cultures were harvested in mid-exponential phase.
Synchronization of A. fundyense.
Cultures of A. fundyense GtCA28 (see above for details) were synchronized by a dark-induced block/release method (52). Low-density (approximately 500 cells ml−1), exponentially growing cultures (on a cycle of 14 h of light and 10 h of darkness) of A. fundyense strain GtCA28 were left in continuous darkness for 82 h and then reexposed to the original light-darkness cycle. At the end of the dark blocking period, all the cells had divided once more and were arrested in the G1 phase of their cell cycle. Cell size and density were monitored with a Multisizer apparatus (Coulter Electronics, Hialeah, Fla.) every 2 h for a period of 2 days. Samples for DNA analysis (50,000 to 100,000 cells) were taken with the same periodicity and were preserved in 5% formaldehyde at −20°C. The samples were treated with the DNA-specific stain propidium iodide (PI) (4 μg of PI ml−1; 250 U of RNase A ml−1) and analyzed on a FACScan flow cytometer (Becton Dickinson).
RNA purification.
Cells (106 to 3 × 106) were harvested through centrifugation at 4,400 × g for 5 min at 4°C. The pellet was deep-frozen in liquid N2 and then resuspended in 1 ml of RNA STAT-60 (Teltest “B”, Inc., Friendswood, Tex.). The suspensions were placed in a N2 cell disruption bomb, and the cells were ruptured in three consecutive cycles of nitrogen decompression at 2,000 lb/in2 (Parr Instrument Company, Moline, Ill.). Homogenates were processed by following the manufacturer's protocol (RNA precipitation for 1 h), with an additional RNA precipitation in the presence of sodium acetate (pH 5.2; final concentration, 0.3 M) and 2 volumes of ethanol for 1 h at −70°C. RNA was quantified and stored at −70°C.
DD.
DD of mRNA was performed as previously described by Liang et al. (27, 28). The reverse transcription and PCR were modified as follows. Total RNA (0.2 μg) was reverse transcribed with a mixture containing 200 U of Moloney murine leukemia virus reverse transcriptase (RTase; Superscript, RNase H−; GIBCO-BRL), 40 μM dNTPs, and a 1 μM concentration of one of the four different degenerate oligo(dT) primers (NBI, Plymouth, Minn.) T12VN (where V stands for dA, dC, or dG and N stands for dA, dC, dG, or dT). Control reactions were performed with total RNA in the absence of RTase or with RNase-treated RNA in the presence of RTase. The resulting cDNA was then amplified by PCR with a combination of the respective degenerate oligo(dT) primer and one random decamer primer (10-mer kit A; Operon Technologies) in the presence of [α-33P]dATP on a Perkin-Elmer 2400 thermocycler. The reactions were performed in duplicate as a control for replicability of band patterns. All components of the PCR remained the same as originally described except for a 10-fold increase in the concentration of the random decamer primer (2 μM). The samples were denatured at 94°C for 15 s directly preceding PCR. Parameters for the 40-cycle PCR were as follows: denaturation at 94°C for 15 s, annealing at 40°C for 1 min, and extension at 72°C for 1 min. At the end an additional extension at 72°C for 10 min was performed. The radiolabeled PCR products were analyzed on 6% denaturing polyacrylamide gels.
Cloning and sequencing.
Bands identified as differentially expressed on the DD gels (details in Results) were excised, extracted from the acrylamide by boiling in H2O, and precipitated in the presence of 0.3 M sodium acetate–0.5 mg of glycogen ml−1–4 volumes of ethanol. The DNA was then reamplified using the same primer and the PCR conditions described above, except that the dNTP concentration was increased 10-fold. The PCR product was cloned using the pCR-TRAP cloning system (GenHunter Corp., Brookline, Mass.). Cloned fragments were sequenced on a LiCor automated sequencing system using pCR-TRAP-specific primers (Aidseq; GenHunter Corp.).
cDNA library construction and screening.
Approximately 108 cells of exponentially growing A. fundyense were harvested, and their RNA was processed as described above. One milligram of fresh total RNA was further purified for poly(A)+ mRNA with an oligo(dT) column (5Prime→3Prime, Inc., Boulder, Colo.). Five micrograms of poly(A)+ mRNA was used for construction of a lambda phage cDNA library (ZAP cDNA cloning kit; Stratagene, La Jolla, Calif.). The library was further amplified (2 × 105 PFU/μl), and approximately 106 plaques were screened on each round.
DD probes for screening the library were obtained by labeling the cloned cDNA of interest with digoxigenin (DIG)-11-dUTP (Genius nonradioactive nucleic acid labeling and detection system, Roche, Indianapolis, Ind.) in a 30-cycle PCR with the following parameters: denaturation at 94°C for 30 s, annealing at 52°C for 40 s, and extension at 72°C for 1 min. At the end an additional extension at 72°C for 5 min was performed. The final concentrations of dNTPs in the reaction mixture were optimized to the following values: 20 μM dATP, dCTP, and dGTP, 13 μM dTTP, and 7 μM DIG-dUTP. The yield of DIG-labeled PCR product was estimated as indicated by the manufacturer, and hybridizations were conducted overnight in a standard hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2% [wt/vol] blocking reagent, 0.1% N-lauroylsarcosine, 0.2% sodium dodecyl sulfate [SDS]), with a probe concentration of 10 ng ml−1 at a temperature of 65°C. Positive plaques were detected using the chemiluminescent substrate CSPD (Roche). Plaques were further purified in a second and if necessary third screening until single plaques could be isolated. The phagemid representing the cDNA of interest was then excised in vivo, cloned, and sequenced following the manufacturer's protocols (ZAP cDNA cloning kit; Stratagene).
RNA blot analysis.
Ten micrograms of total DNase-treated RNA representing each of the four cell cycle stages described below was loaded on positively charged nylon membranes (Boehringer Mannheim, Indianapolis, Ind.). The DD probes were labeled as described above. Hybridizations were performed overnight in high-SDS concentration buffer, as recommended by the manufacturer, at 42°C (Genius nonradioactive nucleic acid labeling and detection system; Roche). Detection of positive dots was performed as detailed above for the cDNA library screenings.
Sequence analysis.
The nucleotide sequences obtained were compared to sequences in the nonreductant GenBank, EMBL, DDBJ, and PDB sequence databases using the Basic Local Alignment Search Tool program (BLAST) in its versions for nucleotides (BLASTN) and amino acids (BLASTX) (3). The corresponding amino acid sequence was predicted using the universal genetic code. The deduced protein sequences were further characterized with the sequence analysis software program MacVector (Oxford Molecular Limited). Percent similarities were determined based on the following similarity groupings for amino acids: (i) H, Q, E, D, R, K, N, G; (ii) V, L, I, M; (iii) C, S, A, T; (iv) F, Y; (v) P; and (vi) W (amino acids were counted as similar when more than half of the amino acids at a particular position on the aligned sequences were included in one of the six subgroupings).
Nucleotide sequence accession numbers.
The DNA sequences determined in this study have been assigned GenBank accession no. AF105295 (Sahh), AF105293 (Map), and AF105294 (HAf).
RESULTS
DD of mRNA from toxic and nontoxic Alexandrium.
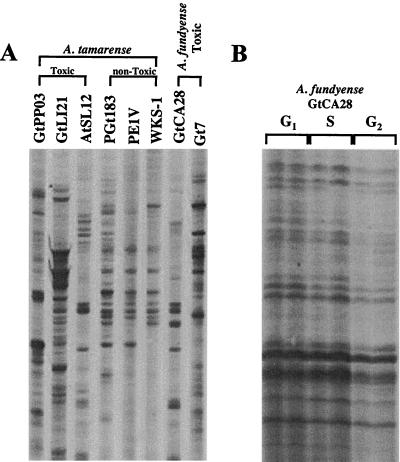
DD of several toxic and nontoxic strains of A. tamarense and two toxic A. fundyense strains (see Materials and Methods for specific strain information) was done using T12VC as the degenerate oligo(dT) primer and OP-03 as the random decamer. High variability was observed among the DD patterns of the different strains of A. tamarense analyzed (Fig. 2A). Interspecific variations between A. tamarense and A. fundyense were also high (Fig. 2A).
FIG. 2.
Comparison of inter- and intraspecific DD analysis of dinoflagellates. (A) Three toxic strains of A. tamarense, three nontoxic strains of A. tamarense, and two toxic strains of A. fundyense were compared. The inter- and intrageneric variabilities were high. (The image is a composite of lanes from one gel rearranged for clarity.) (B) Three different cell cycle stages in synchronized cultures of A. fundyense (GtCA28) were analyzed. In contrast to what was seen in panel A, the patterns were identical and hence allowed the identification of differentially expressed bands (Fig. 4).
DD of synchronized cultures of A. fundyense.
DD of mRNA was applied to synchronized cells of A. fundyense in different cell cycle stages (Fig. 2B) and while toxin production was on or off (52). Two samples were drawn during G1, one while toxin production was active (Fig. 1, I) and a second one once toxigenesis was over (II); a third sample comprised cells in S phase (III), and the fourth one represented cells that were predominantly in G2 (IV). A total of 80 different anchor-plus-random-primer combinations were used to ensure total representation of the mRNA populations (27). All samples were run in duplicate, and, in order to avoid false positives, only those bands present in both reactions were included in the analysis. The average identity between band patterns for every primer combination was approximately 97%.
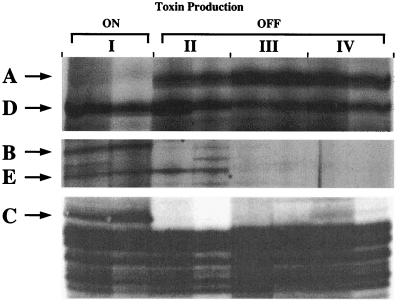
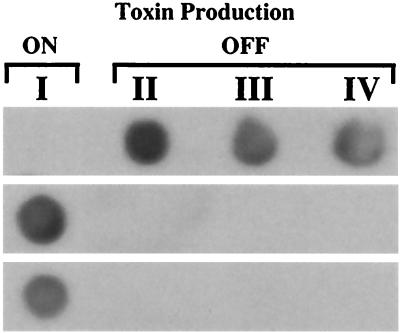
Twenty-four bands were identified as differentially expressed at the four time points selected. The various expression patterns observed are summarized in Table 1. Several different expression patterns were observed. Most bands were present throughout the cell cycle (i.e., not differentially expressed), as exemplified by band D in Fig. 3. Others showed cell cycle stage-specific expression, as was the case for band E in the same figure, which was induced throughout G1 but which was not detectable during S and G2. Only those bands corresponding to genes showing suppression (band A) or induction (bands B and C) during toxigenesis at the beginning of G1 were isolated for further characterization (Fig. 3). The three bands were singled out by these criteria, excised, cloned, and subsequently used as probes to recover the corresponding cDNAs from an A. fundyense cDNA library. The DD expression patterns were also confirmed by RNA dot blot analysis using total RNA from the equivalent cell cycle stages (I through IV) on the 24-h cycle immediately following the cycle analyzed by DD (Fig. 4). As described below, the three genes were similar to those encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein (HAf).
TABLE 1.
Cell cycle-related differential expression patterns observed in this study
| No. of DD bands showing specific pattern | Presence (+) or absence (−) of band on DD sequencing gel in:
|
|||
|---|---|---|---|---|
| G1 (early) | G1 (late) | S | G2 | |
| 8 | − | − | + | + |
| 4 | − | + | + | − |
| 3 | + | + | + | − |
| 3 | + | + | − | + |
| 2 | + | − | − | − |
| 2 | − | + | − | − |
| 1 | − | + | + | + |
| 1 | + | + | − | − |
FIG. 3.
DD comparing cells of A. fundyense in three different cell cycle stages (G1, S, and G2) and while STX production was turned on or off (I through IV refer to the four sampling points in Fig. 1; double lanes represent duplicate reverse transcription-PCRs). Band A is downregulated, while bands B and C are upregulated, relative to toxin production. Band D is constitutively expressed, while band E is G1 specific (expressed in early and late G1).
FIG. 4.
RNA dot blot analysis confirming the induction of DD bands A, B, and C (shown from top to bottom) in A. fundyense. Total RNA (10 μg per dot) corresponding to four equivalent sampling points (I through IV; Fig. 1) on the 24-hour cycle immediately following the cycle analyzed by DD was hybridized with cDNA probes generated by PCR of cloned cDNA fragments obtained from the DD gels (see Results).
S-Adenosylhomocysteine hydrolase.
The 131-bp band A was downregulated during toxin biosynthesis (Fig. 3 and 4). Its sequence showed that only the random decamer had been applied during the PCR amplification step of DD (data not shown). It has been shown previously that a significant proportion of DD bands can result from the amplification of cDNA with only one primer, either the anchored, mRNA-specific primer or the random decamer (15, 27). These bands are equally reproducible and represent potentially transcriptionally regulated genes. Such single-primer bands are the result of the random decamer sequence and its inverse complement being present on the same cDNA. Generation of these bands resembles arbitrarily primed PCR fingerprinting of RNA, a technique similar to DD, which relies on the use of only one primer for the generation of cDNAs representative of expressed sequences in eukaryotic and prokaryotic cells (58, 59).
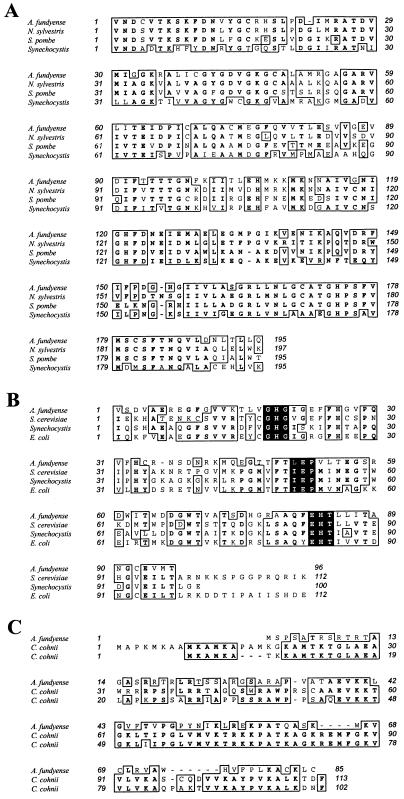
The alignment of the partial protein, as deduced from the rescued cDNA fragment, showed a 90% similarity with S-adenosylhomocysteine hydrolases from eukaryotes and prokaryotes. Some specific values are as follows (GenBank accession numbers are in parentheses): tobacco (Nicotiana sylvestris; BAA03709), 85% similarity and 75% identity; yeast (Schizosaccharomyces pombe; CAA17833), 75% similarity and 62% identity; and a cyanobacterium (Synechocystis sp.; P74008), 63% similarity and 44% identity (Fig. 5A).
FIG. 5.
Partial alignments of the A. fundyense amino acid sequences determined in this study. (A) S-Adenosylhomocysteine hydrolase aligned with tobacco (N. sylvestris), yeast (S. pombe), and a cyanobacterium (Synechocystis sp.). (B) Methionine aminopeptidase aligned with yeast (S. cerevisiae), a cyanobacterium (Synechocystis sp.), and E. coli. The shaded areas correspond to three of the five highly conserved copper binding regions that have been described for this enzyme (see text). (C) Histone-like protein aligned with two different histone-like protein sequences from C. cohnii. Boxes denote higher than 50% identity (boldface) or similarity (lightface) among organisms.
Methionine aminopeptidase.
The 115-bp band B was upregulated during toxin biosynthesis in G1 (Fig. 3 and 4). The fragment rescued from the cDNA library corresponded to the 3′ end of the mRNA (data not shown). The alignment of the deduced protein with the carboxy ends of three other methionine aminopeptidases showed a similarity of 70%. Some specific values are as follows (GenBank accession numbers are in parentheses): yeast (Saccharomyces cerevisiae; AAB67398), 61% similarity and 44% identity; a cyanobacterium (Synechocystis sp.; BAA10466), 61% similarity and 42% identity; and Escherichia coli (PO7906), 63% similarity and 46% identity (Fig. 5B). In addition, three of the five highly conserved copper binding regions that have been described for this enzyme were identified in the A. fundyense sequence (Fig. 5B; shaded areas). The other two metal-binding domains fall within the missing section of the sequence.
Histone-like protein.
The 318-bp band C was upregulated during toxigenesis, coinciding with the beginning of the G1 phase of the cell cycle (Fig. 3 and 4). A 577-bp cDNA with a 255-bp open reading frame (ORF) was recovered (data not shown). The database search aligned this sequence to the sequences encoding two histone-like proteins from Crypthecodinium cohnii Biecheler, GenBank accession no. Q01239 and Q01238 (Fig. 5C). The combined similarity was 60%, with each of the two different histone-like proteins showing a similarity of 52% and an identity of 35%. The ORF obtained from the cDNA library encodes a protein with 85 amino acids and a molecular mass of approximately 9.3 kDa.
DISCUSSION
In this study we report the identification of several transcriptionally regulated genes that are differentially expressed throughout the cell cycle of A. fundyense and are either up- or downregulated during toxigenesis. The ability to obtain cells at different times from a single culture of A. tamarense while the cells were progressing synchronously through the cell cycle and toxin production was turned on or off was essential to this study and provided a robust system for identifying molecular factors related to the cell cycle and possibly toxigenesis. The advantages of using DD over other techniques, such as subtractive hybridization, to identify differentially expressed genes was that it only required small amounts of RNA for the analysis of relative mRNA levels and resolved up and downregulation simultaneously.
Synchronized growth and DD.
The application of DD to the identification of genes that are cell cycle regulated or expressed during periods of toxin accumulation in Alexandrium spp., and hence potentially related to toxin biosynthesis, was contingent on the design of a system that would allow the comparison of cells in different cell cycle stages. Cells of a single, synchronously growing isolate were collected at different cell cycle stages and while toxin production was turned on or off. In addition, a second experimental setup for the identification of toxigenesis-related genes was possible: phylogenetically close toxic and nontoxic strains of Alexandrium spp. were compared by DD analysis. As discussed above, mutant selection was not a realistic option.
Extensive work has been done on the phylogeny of the Alexandrium group using rRNA gene sequences (46). Based on the phylogenetic distance information we ran DD gels comparing toxic and nontoxic strains of A. tamarense that were closely related, but their genetic variation was still large enough to make it impossible to identify differentially expressed genes over the general variation between DD patterns due to interstrain dissimilarities (Fig. 2). Variations in DD patterns arise from differences in the nucleotide sequences of the mRNAs. Changes in just one nucleotide in the region binding to the DD primers can result in no annealing of a specific primer, and hence no amplification of the mRNA. This same mRNA might be amplified by a different set of primers. Therefore, the absence of a specific band does not mean that the corresponding gene is not expressed but rather indicates that it is not amplified by this particular set of primers. The similarity among the DD patterns has to be at least 90 to 95% to make the comparison meaningful. Variabilities of the magnitude observed in Fig. 2A result in an impossibility to make any prediction about the actual regulation of the genes identified as differentially expressed because of the uncertainty about the identity between similar bands. Interspecific comparisons between A. tamarense and A. fundyense were equally uninformative because of variations (Fig. 2A). The sensitivity of DD to changes in sequences was such that the cells to be compared had to be identical in their genetic makeup to avoid a noisy background.
The alternative was to take advantage of the discontinuity in toxin accumulation over the cell cycle in synchronously growing cells of A. fundyense that we had previously reported (52). When we performed DD on samples drawn at different times throughout the cell cycle of A. fundyense, the patterns of the reverse transcribed and amplified mRNA components analyzed were largely identical, resulting in a uniform background over which specific, differentially expressed genes could be identified (Fig. 2B). One potential problem with our experimental setup was that despite the remarkable degree of synchrony achieved, a significant overlap of cell cycle stages was still present (Fig. 1). Samples III and IV (Fig. 1) represent a mixture of cells in different cell cycle stages with only a predominance (55 and 62%, respectively) of cells in either S phase or G2. This could result in a compromised analysis of true S or G2 gene expression and could potentially mask the expression patterns of those genes that might be turned off only in S or only in G2. The focus of our research was on the identification of genes expressed in early G1 and absent during the rest of the cell cycle or, conversely, suppressed in early G1 and present throughout the rest of the cell cycle.
S-Adenosylhomocysteine hydrolase.
The enzyme S-adenosylhomocysteine hydrolase (encoded by Sahh) has a key role in the regulation of the S-adenosylmethionine (SAM) pathway, one of the main elements of one-carbon metabolism in the cell. It catalyzes the reversible cleavage of S-adenosylhomocysteine, the side product of SAM-mediated methylation reactions, into adenosine and homocysteine. Similar to our observation for A. fundyense is the finding that this enzyme is transcriptionally regulated in other organisms. Transcription of Sahh is induced by pathogenic attack in parsley (23), and the gene is differentially expressed over the progression of developmental stages in lucerne (1).
The connection Sahh downregulation in A. fundyense might bear to STX biosynthesis lies in the postulated presence of one SAM-dependent methylation step in the early phases of the synthesis of the STX backbone (49). By regulating the supply of SAM, this pathway could limit or enhance the activity of the toxigenic enzymes. Such a correlation, though, would most probably be nonspecific for toxin biosynthesis, since SAM is involved in a wide spectrum of anabolic and catabolic processes in the cell (e.g., methionine degradation).
Methionine aminopeptidase.
Methionine aminopeptidase (encoded by Map) is responsible for cleaving the amino-terminal methionine during the processing of most nascent proteins. It is cobalt dependent, and its active site contains two zinc ion binding domains (6). The partial Alexandrium sequence reported here contains three of the five highly conserved domains responsible for its metal-binding character. The function of this enzyme in other organisms clearly relates to regulation of protein synthesis and enzyme activities in general and is essential for the viability of the organism. Map genes are present and active throughout the entire life cycle of a cell. Depending on the organism, the number of different Map genes in a cell can vary depending on the developmental status or cell cycle stage of the cell (6). Regulation further happens at both the transcriptional and posttranslational levels in different organisms (6).
Protein biosynthesis regulation at this primary level might have a direct influence on toxin biosynthesis via complex interactions between different components of the general cellular metabolism, but no straight correlation to STX biosynthesis can be inferred. Other scenarios cannot be completely excluded, as recent reports have shown that in addition to its protein-editing function, methionine aminopeptidase also acts as an inhibitor of eukaryotic initiation factor 2α phosphorylation and is involved in the regulation of angiogenesis in higher eukaryotes (13), implying that not all the functions of Map are known yet.
Histone-like protein.
Dinoflagellate nuclei are unique among eukaryotes in that their chromosomes remain condensed throughout the entire cell cycle, in contrast to the unwinding and relaxation of the chromatin observed in all other eukaryotic organisms (reviewed in reference 40). In addition to this, dinoflagellates also lack the typical histone proteins and nucleosome structures that define nuclei of other eukaryotic organisms (18, 38). Nonetheless, dinoflagellate chromosomes have been shown to contain an alternative suite of basic proteins, the so-called histone-like proteins (39, 41). No clear function has been ascribed to these proteins, but the evidence gathered so far suggests that rather than having an intrinsic structural or stabilizing function they might be involved in transcriptional regulation or, perhaps, initiation (45).
Band C of the DD analysis rescued a full-length cDNA encoding an amino acid sequence with a high similarity to those of histone-like proteins from the dinoflagellate Crypthecodinium cohnii. Despite a difference in their sizes, these proteins exhibit remarkably similar hydropathy and antigenicity profiles (data not shown). The core structure consists of one small and highly hydrophobic region flanked by two larger and very hydrophilic domains. This overlap in molecular characteristics might relate to the function of these proteins. A differential or cyclic de novo synthesis of histone-like proteins similar to the one seen here has been suggested for C. cohnii based on immunocytochemical studies (12). Cell cycle-dependent expression has, for example, been shown for histones in plants (8). No information is available on histone-like proteins in A. fundyense.
Cell cycle-related genes in A. fundyense.
The isolation of a histone-like protein illustrates the fact that DD analysis of any population of synchronized cells is, by definition, an ideal system to identify genes related to cell cycle progression. As mentioned above, histone-like proteins are closely associated with DNA and its structure or some of its functions, which means that its biosynthesis is probably regulated in a DNA- or cell cycle-dependent manner.
As mentioned in the introduction, one of the challenges in studying bloom dynamics is the determination of in situ growth rates for Alexandrium. Current approaches rely on the analysis of the relative distributions of cells in the different cell cycle stages (9, 10). These analyses can be done at a species-specific level by combining the flow-cytometric determination of relative DNA distributions with the precise detection of organisms using rRNA-based species-specific probes (47). An alternative approach consists of using cell cycle-related genes as indicators of population growth rates (24, 25, 32, 37). The species-specific detection of a cell cycle-related gene and its concomitant quantification will, potentially, allow the determination of precise in situ growth rates of Alexandrium and other harmful algal bloom species in the future.
DD and STX biosynthesis in A. fundyense.
Biosynthetic pathways can be regulated at many different levels. Expression of a specific enzyme can be regulated at the transcriptional level, through the induction or suppression of gene transcription itself, or at the pretranslational level, which includes mRNA editing, regulation of translation, and posttranslational mechanisms such as protein maturation and enzyme inhibition. Little is known about the regulation of STX biosynthesis in dinoflagellates. Enhancement and inhibition of toxigenesis have been shown under various environmental conditions (5, 7, 16). Toxin content and variations in generation time or the length of the cell cycle have been suggested to be correlated (5). More recently, STX biosynthesis has also been shown to be coupled to the G1 phase of the cell cycle (52). All this evidence led us to the hypothesis that STX biosynthesis might be regulated in a cell cycle-dependent manner.
Many cell cycle-dependent genes are known to be transcriptionally regulated in eukaryotes (8, 36). Based on this and our ability to synchronize cultures of A. fundyense, we set out to identify genes regulated in patterns that correlated with episodes of toxin accumulation during the early part of the G1 phase of the cell cycle. Out of the three genes we isolated, only one might bear some correlation to STX biosynthesis. The enzyme S-adenosylhomocysteine hydrolase regulates the supply of methylating units, SAM, in the cell and hence affects not just the methylation step in STX biosynthesis but also other methyl-dependent pathways in the cells. No genes with unknown functions were isolated in this study.
In the future, alternative approaches to the identification of genes related to toxigenesis could include the comparative analysis by DD of cells growing under different physiological conditions, such as phosphorus limitation or increased arginine supply, that are known to induce toxin accumulation in Alexandrium (5).
Genetic regulation in A. fundyense.
Few data on genetic regulation in dinoflagellates exist. The best-studied system is the regulation of luciferin binding protein (LBP) in Gonyaulax polyedra (29, 31). The synthesis of LBP is under circadian control, and its corresponding mRNA is present throughout the cell cycle. Translation is repressed in the presence of a protein that binds specifically to a 3′-untranslated region of the mRNA. The regulation of two other families of proteins, the peridinin chlorophyll a binding proteins and the chlorophyll a to c binding proteins, has been studied at a physiological level similar to that at which STX production has been studied (20, 22, 53). The activation pattern of the cell cycle regulatory protein CDC2 kinase has been characterized in Gambierdicus toxicus (56). CDC2 kinase is constitutively expressed throughout the cell cycle and is activated only in mitotic cells. Evidence for the presence of this ubiquitous regulator of cell cycle progression in eukaryotes has also been gathered with C. cohnii (42).
The common denominator to all the previous systems is translational regulation. In this study we show for the first time that a large component of transcriptional or pretranslational regulation does indeed occur in dinoflagellates. Previous reports had suggested the possibility that translational mechanisms could be the primary means of circadian regulation in these organisms (30). We have not only shown the induction and suppression of the three genes described here, but also observed transcriptional or pretranslational regulation of about 20 other genes with various DD patterns (Table 1). A complete picture of the importance of transcriptional regulation will only be achieved by the introduction of such global observation technologies as DNA chip expression profiling to the study of dinoflagellate regulation.
DD and dinoflagellates.
This study illustrates the potential of applying DD to the identification of transcriptionally regulated genes in dinoflagellates and other phytoplankton. Many of these organisms cannot be cultivated or, when finally cultivated, do not produce enough material for traditional molecular studies. This is not a problem with DD due to the amplification step inherent to the method. DD is also a useful way to identify gene function in organisms where mutant isolation is not feasible. In this study, we adapted DD to the analysis of different cell cycle stages in Alexandrium. Other applications include the study of different life cycle stages of phytoplankton and the isolation of specific genes induced by environmental stresses. Examples for these applications on higher eukaryotes already exist in the literature. Sexual-stage-specific mRNAs have been isolated in Eimeria bovis, a parasitic protozoan, and genes involved in the different steps of hematopoiesis have been identified in mice (2, 15). Tissue-specific markers have been observed in the rhodophyte Porphyra perforata (19). Genes induced by ozone stress in Arabidopsis thaliana and genes induced by phosphate depletion in rat kidney have been isolated (11, 48).
In conclusion, transcriptional or pretranslational regulation of gene expression in dinoflagellates can no longer be ignored, as demonstrated by the identification of over 20 different gene products in A. fundyense that are differentially expressed over the cell cycle of the organism. We have further shown the potential to use DD to look for mechanisms that could underlie cell cycle control or the regulation of STX biosynthesis. Finally, this study has also documented the potential of applying DD to the identification of genes that are related to certain physiological or developmental processes in phytoplankton.
ACKNOWLEDGMENT
This work was supported by a grant from the National Science Foundation (OCE 94-15536).
Footnotes
Contribution no. 9757 of the Woods Hole Oceanographic Institution.
REFERENCES
- 1.Abrahams S, Hayes C M, Watson J M. Expression patterns of three genes in the stem of lucerne (Medicago sativa) Plant Mol Biol. 1995;27:513–528. doi: 10.1007/BF00019318. [DOI] [PubMed] [Google Scholar]
- 2.Abrahamsen M S, Johnson R R, Hathaway M, White M W. Identification of Eimeria bovis merozoite cDNAs using differential mRNA display. Mol Biochem Parasitol. 1995;71:183–191. doi: 10.1016/0166-6851(94)00052-o. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Anderson D M. Toxic algal blooms and red tides: a global perspective. In: Okaichi T, Anderson D M, Nemoto T, editors. Red tides: biology, environmental science, and toxicology. New York, N.Y: Elsevier Science Publishing; 1989. pp. 11–16. [Google Scholar]
- 5.Anderson D M, Kulis D M, Sullivan J J, Hall S, Lee C. Dynamics and physiology of saxitoxin production by the dinoflagellate Alexandrium spp. Mar Biol. 1990;104:511–524. [Google Scholar]
- 6.Arfin S M, Kendall R L, Hall L, Weaver L H, Stewart A E, Matthews B W, Bradshaw R A. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc Natl Acad Sci USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer G L, Sullivan J J, Andersen R J, Harrison P J, Taylor F J R. Effects of nutrient limitation on toxin production and composition in the marine dinoflagellate Protogonyaulax tamarensis. Mar Biol. 1987;96:123–128. [Google Scholar]
- 8.Callard D, Mazzolini L. Identification of proliferation-induced genes in Arabidopsis thaliana. Characterization of a new member of the highly evolutionarily conserved histone H2A.F/Z variant subfamily. Plant Physiol. 1997;115:1385–1395. doi: 10.1104/pp.115.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter E J, Chang J. Species-specific phytoplankton growth rates via diel DNA synthesis cycles. 1. Concept of the method. Mar Ecol Prog Ser. 1988;43:105–111. [Google Scholar]
- 10.Chang J, Carpenter E J. Species-specific phytoplankton growth rates via diel DNA synthesis cycles. 5. Application to natural populations in Long Island Sound. Mar Ecol Prog Ser. 1991;78:115–122. [Google Scholar]
- 11.Custer M, Spindler B, Verrey F, Murer H, Biber J. Identification of a new gene product (diphor-1) regulated by dietary phosphate. Am J Physiol. 1997;273:F801–F806. doi: 10.1152/ajprenal.1997.273.5.F801. [DOI] [PubMed] [Google Scholar]
- 12.Géraud M-L, Herzog M, Soyer-Gobillard M O. Nucleolar localization of rRNA coding sequences in Prorocentrum micans Ehr. (Dinomastigote, kingdom protoctista) by in situ hybridization. BioSystems. 1991;26:61–74. doi: 10.1016/0303-2647(91)90038-m. [DOI] [PubMed] [Google Scholar]
- 13.Griffith E C, Su Z, Turk B E, Chen S, Chang Y H, Wu Z, Biemann K, Liu J O. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem Biol. 1997;4:461–471. doi: 10.1016/s1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 14.Guillard R R L, Ryther J H. Studies of marine phytoplanktonic diatoms. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 15.Guimarães M J, McClanahan T, Bazán J F. Application of differential display in studying developmental processes. In: Liang P, Pardee A B, editors. Differential display methods and protocols. Totowa, N.J: Humana Press Inc.; 1997. pp. 175–194. [DOI] [PubMed] [Google Scholar]
- 16.Hall S. Toxins and toxicity of Protogonyaulax from the northeast Pacific. Ph.D. thesis. Fairbanks: University of Alaska; 1982. [Google Scholar]
- 17.Hall S, Strichartz G, Moczydlowski E, Ravindran A, Reichardt P B. The saxitoxins: sources, chemistry and pharmacology. In: Hall S, Strichartz G, editors. Marine toxins: origin, structure and molecular pharmacology. Washington, D.C.: American Chemical Society; 1990. pp. 29–65. [Google Scholar]
- 18.Herzog M, Soyer M O. Distinctive features of dinoflagellate chromatin. Absence of nucleosomes in a primitive species, Prorocentrum micans E. Eur J Cell Biol. 1981;23:295–302. [PubMed] [Google Scholar]
- 19.Hong Y-K, Sohn C H, Polne-Fuller M, Gibor A. Differential display of tissue-specific messenger RNAs in Porphyra perforata (Rhodophyta) thallus. J Phycol. 1995;31:640–643. [Google Scholar]
- 20.Johnsen G, Nelson N B, Jovine R V M, Prézelin B B. Chromoprotein- and pigment-dependent modeling of spectral light absorption in two dinoflagellates. Mar Ecol Prog Ser. 1994;114:245–258. [Google Scholar]
- 21.Joseph R, Dou D, Tsang W. Molecular cloning of a novel mRNA (neuronatin) that is highly expressed in neonatal mammalian brain. Biochem Biophys Res Commun. 1994;201:1227–1234. doi: 10.1006/bbrc.1994.1836. [DOI] [PubMed] [Google Scholar]
- 22.Jovine R V M, Triplett E L, Nelson N B, Prézelin B B. Quantification of chromophore pigments, apoprotein abundance and isoelectric variants of peridinin-chlorophyll a-protein complexes (PCPs) in the dinoflagellate Heterocapsa pygmea grown under variable light conditions. Plant Cell Physiol. 1992;33:733–741. [Google Scholar]
- 23.Kawalleck P, Plesch G, Hahlbrock K, Somssich I E. Induction by fungal elicitor of S-adenosyl-L-methionine synthetase and S-adenosyl-L-homocysteine hydrolase mRNAs in cultured cells and leaves of Petroselinum crispum. Proc Natl Acad Sci USA. 1992;89:4713–4717. doi: 10.1073/pnas.89.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemp P F, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerkhof L, Ward B B. Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl Environ Microbiol. 1993;59:1303–1309. doi: 10.1128/aem.59.5.1303-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kodama M, Ogata T, Sato S. Bacterial production of saxitoxin. Agric Biol Chem. 1988;52:1075–1077. [Google Scholar]
- 27.Liang P, Averboukh L, Pardee A B. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 29.Machabée S, Wall L, Morse D. Expression and genomic organization of a dinoflagellate gene family. Plant Mol Biol. 1994;25:23–31. doi: 10.1007/BF00024195. [DOI] [PubMed] [Google Scholar]
- 30.Milos P, Morse D S, Hastings J W. Circadian control over synthesis of many Gonyaulax proteins is at a translational level. Naturwissenschaften. 1990;77:87–89. doi: 10.1007/BF01131782. [DOI] [PubMed] [Google Scholar]
- 31.Mittag M, Lee D-H, Hastings J W. Circadian expression of the luciferin-binding protein correlates with the binding of a protein to the 3′ untranslated region of its mRNA. Proc Natl Acad Sci USA. 1994;91:5257–5261. doi: 10.1073/pnas.91.12.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mountain H A, Sudbery P E. The relationship of growth rate and catabolite repression with WHI2 expression and cell size in Saccharomyces cerevisiae. J Gen Microbiol. 1990;136:733–737. doi: 10.1099/00221287-136-4-733. [DOI] [PubMed] [Google Scholar]
- 33.Negri A P, Jones G J, Blackburn S I, Oshima Y, Onodera H. Effect of culture and bloom development and of sample storage on paralytic shellfish poisons in the cyanobacterium Anabaena circinalis. J Phycol. 1997;33:26–35. [Google Scholar]
- 34.Oshima Y. Chemical and enzymatic transformation of paralytic shellfish toxins in marine organisms. In: Lassus P, Arzul G, Erard E, Gentien P, Marcaillou C, editors. Harmful marine algal blooms. Paris, France: Lavoisier; 1995. pp. 475–480. [Google Scholar]
- 35.Oshima Y, Blackburn S I, Hallegraeff G M. Comparative study on paralytic shellfish toxin profile of the dinoflagellate Gymnodinium catenatum from three different countries. Mar Biol. 1993;116:471–476. [Google Scholar]
- 36.Price C, Nasmyth K, Schuster T. A general approach to the isolation of cell cycle-regulated genes in the budding yeast, Saccharomyces cerevisiae. J Mol Biol. 1991;218:543–556. doi: 10.1016/0022-2836(91)90700-g. [DOI] [PubMed] [Google Scholar]
- 37.Ralling G, Bodrug S, Linn T. Growth rate-dependent regulation of RNA polymerase synthesis in Escherichia coli. Mol Gen Genet. 1985;201:379–386. doi: 10.1007/BF00331327. [DOI] [PubMed] [Google Scholar]
- 38.Rizzo P J. Comparative aspects of basic chromatin proteins in dinoflagellates. BioSystems. 1981;14:433–443. doi: 10.1016/0303-2647(81)90048-4. [DOI] [PubMed] [Google Scholar]
- 39.Rizzo P J. Biochemistry of the dinoflagellate nucleus. In: Taylor F J R, editor. The biology of dinoflagellates. Palo Alto, Calif: Blackwell Scientific Publications; 1987. pp. 143–173. [Google Scholar]
- 40.Rizzo P J. The enigma of the dinoflagellate chromosome. J Protozool. 1991;38:246–252. [Google Scholar]
- 41.Rizzo P J, Morris R L. Some properties of the histone-like protein from Chrypthecodinium cohnii (HCc) BioSystems. 1984;16:211–216. doi: 10.1016/0303-2647(83)90005-9. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez M, Cho J W, Sauer H W, Rizzo P J. Evidence for the presence of cdc2-like protein kinase in the dinoflagellate Cryptecodinium cohnii. J Eukaryot Microbiol. 1994;40:91–96. [Google Scholar]
- 43.Sako Y, Kim C, Ishida Y. Mendelian inheritance of paralytic shellfish poisoning toxin in the marine dinoflagellate Alexandrium catenella. Biosci Biotechnol Biochem. 1992;56:692–694. doi: 10.1271/bbb.56.692. [DOI] [PubMed] [Google Scholar]
- 44.Sako Y, Naya N, Yoshida T, Kim C-H, Uchida A, Ishida Y. Studies on stability and heredity of PSP toxin composition in the toxic dinoflagellate Alexandrium. In: Lassus P, Arzul G, Erard E, Gentien P, Marcaillou C, editors. Harmful marine algal blooms. Paris, France: Lavoisier; 1995. pp. 345–350. [Google Scholar]
- 45.Sala-Rovira M, Geraud M L, Caput D, Jacques F, Soyer-Gobillard M O, Vernet G, Herzog M. Molecular cloning and immunolocalization of two variants of the major basic nuclear protein (HCc) from the histone-less eukaryote Crypthecodinium cohnii (Pyrrophyta) Chromosoma (Berl) 1991;100:510–518. doi: 10.1007/BF00352201. [DOI] [PubMed] [Google Scholar]
- 46.Scholin C A, Hallegraeff G M, Anderson D M. Molecular evolution of the Alexandrium tamarense ‘species complex’ (Dinophyceae): dispersal in the North American and west Pacific regions. Phycologia. 1995;34:472–485. [Google Scholar]
- 47.Scholin C A, Roman M L, Miller P E. DNA probe-based assays for rapid detection of toxic algal species in environmental samples. J Phycol. 1996;32(3):43. [Google Scholar]
- 48.Sharma Y K, Davis K R. Isolation of a novel Arabidopsis ozone-induced cDNA by differential display. Plant Mol Biol. 1995;29:91–98. doi: 10.1007/BF00019121. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu Y. Microalgal metabolites. Chem Rev. 1993;93:1685–1698. [Google Scholar]
- 50.Shimizu Y, Norte M, Hori A, Genenah A, Kobayashi M. Biosynthesis of saxitoxin analogues: the unexpected pathway. J Am Chem Soc. 1984;106:6433–6434. [Google Scholar]
- 51.Silva E S, Sousa I. Experimental work on the dinoflagellate toxin production. Arq Inst Nacion Saude (Portugal) 1981;VI:381–387. [Google Scholar]
- 52.Taroncher-Oldenburg G, Kulis D M, Anderson D M. Toxin variability during the cell cycle of the dinoflagellate Alexandrium fundyense. Limnol Oceanogr. 1997;42:1178–1188. [Google Scholar]
- 53.Triplett E L, Jovine R V M, Govind N S, Roman S J, Chang S S, Prézelin B B. Characterization of two full-length cDNA sequences encoding apoproteins of peridinin-chlorophyll a-protein (PCP) complexes. Mol Mar Biol Biotechnol. 1993;2:246–254. [PubMed] [Google Scholar]
- 54.Utans U, Liang P, Wyne L R, Karnovsky M J, Russell M E. Chronic cardiac rejection: identification of five upregulated genes in transplanted hearts by differential mRNA display. Proc Natl Acad Sci USA. 1994;91:6463–6467. doi: 10.1073/pnas.91.14.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Knaap E, Kende H. Identification of a gibberellin-induced gene in deepwater rice using differential display of mRNA. Plant Mol Biol. 1995;28:589–592. doi: 10.1007/BF00020405. [DOI] [PubMed] [Google Scholar]
- 56.Van Dolah F M, Leighfield T A, Sandel H D, Hsu C K. Cell division in the dinoflagellate Gambierdiscus toxicus is phased to the diurnal cycle and accompanied by activation of the cell cycle regulatory protein cdc2 kinase. J Phycol. 1995;31:395–400. [Google Scholar]
- 57.Wan J S, Sharp S J, Poirier G M-C, Wagaman P C, Chambers J, Pyati J, Hom Y-L, Galindo J E, Huvar A, Peterson P A, Jackson M R, Erlander M G. Cloning differentially expressed mRNAs. Nat Biotechnol. 1996;14:1685–1691. doi: 10.1038/nbt1296-1685. [DOI] [PubMed] [Google Scholar]
- 58.Welsh J, Chada K, Dalal S S, Cheng R, Ralph D, McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992;20:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong K K, McClelland M. Stress-inducible gene of Salmonella typhimurium identified by arbitrarily primed PCR of RNA. Proc Natl Acad Sci USA. 1994;91:639–643. doi: 10.1073/pnas.91.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]