Abstract
A specific multiplex PCR assay based on the amplification of parts of the 16S rRNA molecule was designed. Primers derived from variable regions of the 16S rRNA provided a means of easily differentiating the species Lactobacillus pontis and Lactobacillus panis. They could be clearly discriminated from the phylogenetically related species Lactobacillus vaginalis, Lactobacillus oris, and Lactobacillus reuteri and from other lactobacilli commonly known to be present in sourdough. Other strains isolated together with L. pontis from an industrial sourdough fermentation could be clearly separated from these species by comparative sequence analysis and construction of a specific PCR primer. For a fast identification a DNA isolation protocol based on the ultrasonic lysis of cells from single colonies was developed. To demonstrate the potential of such techniques for tracking these organisms in a laboratory-scale fermentation, we combined the specific PCR assay with direct DNA extraction from the organisms in the sourdough without previous cultivation.
The most prominent lactic acid bacteria (LAB) of the sourdough microflora belong to the genus Lactobacillus (9). Sourdough fermentations have been categorized by Böcker and coworkers (3) by taking into account the kind of propagation and the applied fermentation parameters resulting in typical microbial communities. Sourdoughs maintained by a continuous back-slopping over an extended period have been designated type I sourdoughs. They are characterized by a microflora which is mainly dominated by Lactobacillus sanfranciscensis (5). Type II sourdoughs are fermented over a longer period at elevated temperatures and higher water content. Typical lactobacilli isolated from this fermentations are Lactobacillus pontis and Lactobacillus panis, both endemic to cereal fermentations. Further species from type II fermentations have been recently itemized by Vogel et al. (14, 15).
L. pontis is close in the phylogenetic tree to Lactobacillus vaginalis, Lactobacillus oris, and Lactobacillus reuteri (13). Wiese and coworkers described L. panis as a new species which is phylogenetically related to L. vaginalis, L. oris, and L. reuteri (16).
Traditionally, physiological properties of bacteria, such as their capability to degrade carbohydrates and certain enzyme activities, have been used for identification purposes. Especially for LAB, reliable identification depending on these properties is almost impossible because of the similar nutritional requirements of different species due to adaption to a certain environment (1, 6). For this reason various approaches targeting the genotypes of bacterial cells, which reflect the natural relationship, have been described and steadily introduced into applied and research aspects of food microbiology. During the last decade rRNA emerged as a suitable target molecule for identification purposes (2, 4, 7).
In the framework of the description of L. pontis as a new species (14), a 16S rRNA-targeted oligonucleotide probe in the V1 region (11) of the 16S rRNA was designed. There were sufficient sequence variations to differentiate L. pontis from other sourdough lactobacilli. The 16S ribosomal DNA (rDNA) sequence of L. panis (16) revealed that this species is most closely related to L. oris, followed by L. vaginalis and L. pontis. The binding sites on the 16S rDNA of L. panis and L. pontis for the probe previously described for L. pontis are identical to each other. So far no evaluated system for the specific identification and differentiation of these two species has been available.
Therefore, we have developed 16S rDNA-targeted primers for a specific PCR to distinguish these two lactobacilli. Apart from the mentioned species, primers have been designed for a third sourdough lactobacillus (Lactobacillus sp.) originally isolated from an industrial type II rye fermentation. Comparative sequence analysis of 16S rDNA revealed the Lactobacillus sp. to be an intermediate between L. pontis and L. vaginalis, but no final systematic position has been established up to now. To demonstrate the applicability of the PCR system to trace these organisms during fermentation, we developed a laboratory-scale fermentation similar to an industrial sourdough process.
MATERIALS AND METHODS
Laboratory-scale fermentation.
The fermentation was performed in a 5-liter stirred reactor (Biostat; Braun, Melsungen, Germany) at 40°C. The substrate consisted of 900 g of rye bran mixed with 2,400 ml of preheated tap water. To start the fermentation, 90 g of 48-h-old sourdough of the corresponding industrial process was added.
Sampling.
Sourdough samples were collected aseptically. For the determination of the total cell count, expressed as CFU per milliliter of sourdough, samples were serially diluted 1:10 with NaCl (0.9% [wt/vol]) and plated on modified MRS (3) using a spiral plater (Spiralsystems, Inc., Cincinnati, Ohio). The plates were incubated under a modified atmosphere (90% N2, 10% CO2). To determine the total amount of aerobic bacteria in the flour without starter, plate count agar (Merck, Darmstadt, Germany) was used. Additionally, sourdough samples of 5 ml for the direct isolation of DNA from the organisms were taken and stored at −20°C.
Cultivation and storage of strains.
All reference organisms (see Table 2) were cultivated on mMRS (13). The incubation temperatures were 30°C for L. pontis, L. sanfranciscensis, and Lactobacillus farciminis and 37°C for L. panis, L. oris, L. vaginalis, and L. reuteri. Lactobacillus sp. strains TMW 1.655 and DSM 13145, isolated from an industrial fermentation process, and TMW 1.1104 and TMW 1.1098, isolated from previous laboratory-scale fermentations, were cultivated at 40°C. Stock cultures were stored at −80°C in 80% (wt/wt) glycerol.
TABLE 2.
Strains studied and results of specific PCR as indicated by visual bands on agarose gel
| Species | Strain | Other name | PCR signal with primera:
|
||
|---|---|---|---|---|---|
| LaponR | LapanR | LaspecR | |||
| L. pontis | TMW 1.84 | ATCC 51518 | + | − | − |
| L. pontis | TMW 1.85 | ATCC 51519 | + | − | − |
| L. pontis | TMW 1.56 | LTH 2587T | + | − | − |
| L. pontis | TMW 1.597 | LTH 3572 | + | − | − |
| L. pontis | TMW 1.1106 | LTH 2585 | + | − | − |
| L. pontis | TMW 1.1108 | LTH 3572 | + | − | − |
| L. panis | TMW 1.648 | DSM 6035 | − | + | − |
| L. panis | TMW 1.649 | DSM 6036 | − | + | − |
| L. speciesb | TMW 1.655 | − | − | + | |
| L. species | TMW 1.666 | − | − | + | |
| L. species | TMW 1.1104 | − | − | + | |
| L. species | TMW 1.1098 | − | − | + | |
| L. sanfranciscensis | TMW 1.53 | − | − | − | |
| L. farciminis | TMW 1.68 | − | − | − | |
| L. oris | TMW 1.16 | LMG 9848 | − | − | − |
| L. vaginalis | TMW 1.197 | LMG 12891 | − | − | − |
| L. reuteri | TMW 1.693 | DSM 20016T | − | − | − |
See Table 1.
L. species, Lactobacillus sp.
Selection of primers.
As the primer binding site, we selected helix 11 of the V2 region (11). The primer sequences obtained were subsequently checked with all small-subunit sequences in the Ribosomal Database Project database (10) using the check-probe function. To improve the sensitivity of the PCR and to include a positive control for DNA accessibility for the PCR, a multiplex PCR including two universal primers (616V and 609R) and a species-specific primer as the forward primer was developed. The sequences of the amplification primers are listed in Table 1. Reference strains are listed in Table 2.
TABLE 1.
Universal and specific primers applied in the multiplex PCR assay
| Sequencea | Specificity | Primer |
|---|---|---|
| AGAGTTTGATYMTGGCTCAG | Universal | 616V |
| ACTACYNGGGTATCTAAKCC | Universal | 609R |
| AGCCATCTTTGAAAT | L. pontis | LaponR |
| AACCATCTTTTATAC | L. panis | LapanR |
| AGCCTTCTTTTATAC | L. speciesb | LaspecR |
Sequences are given in the 5′-to-3′ direction. Mixed bases are given according to the International Union of Biochemistry code.
L. species, Lactobacillus sp.
DNA isolation protocols.
DNA for the development of the PCR system was isolated in accordance with the procedure of Lewington et al. (8).
For the DNA isolation of single colonies, an ultrasonic lysis protocol was developed. One colony (2- to 3-mm diameter) was suspended in 100 μl of lysis buffer (20 mM EDTA, 10 mM Tris [pH 7.9], 1% Triton X-100, 500 mM guanidine-HCl, 250 mM NaCl). Cells were lysed by 1 min of ultrasonication with the probe UP 50 H (Dr. Hielscher GmbH, Stahnsdorf, Germany). After the addition of 150 μl of cold (−20°C) ethanol the mixture was centrifuged over a spin column of the QIAamp tissue kit (Qiagen, Hilden, Germany) and finally eluted with 60 μl of buffer (10 mM Tris [pH 7.5]).
For isolation of bacterial DNA directly out of the sourdough a method based on enzymatic lysis was developed and evaluated. The sourdough sample of 5 ml was suspended in 10 ml of phosphate-buffered saline (PBS) (12) and centrifuged for 5 min at 1,500 × g. Ten milliliters of the supernatant was transferred into a new tube and centrifuged for 15 min at 5,000 × g. The pellet was resuspended in 4 ml of PBS, and 1.5 ml of this suspension was transferred in an Eppendorf tube and centrifuged for 5 min at 5,000 × g to collect the cell material. The supernatant was discarded. The pellet was resuspended in 180 μl of TES (50 mM Tris-HCl, 50 mM NaCl, 10 mM EDTA [pH 8.0]) containing 20 mg of lysozyme ml−1. For sufficient cell lysis it was incubated at 37°C for 40 min on a shaking platform (90 rpm). After the addition of 20 μl of proteinase K and 200 μl of lysis buffer AL (QIAamp tissue kit), the reaction mixture was incubated at 70°C for 30 min and for a further 30 min at 95°C. After the addition of 210 μl of cold ethanol (−20°C) it was centrifuged over a spin column (QIAamp tissue kit) and washed with 500 μl of the supplied buffers AW1 and AW2. The DNA was eluted with preheated (70°C) 10 mM Tris (pH 7.5) and stored at −20°C.
PCR conditions.
The amplification of the 16S rDNA was carried out in strips (Braun, Wertheim, Germany) on a Gradient Master thermocycler (Eppendorff, Hamburg, Germany) in a total volume of 25 μl. Primers were obtained from Interactiva (Ulm, Germany). In order to use the three sets of reaction mixtures (a set consisted of 609R, 616V, and the specific probe for L. pontis, L. panis, or the Lactobacillus sp.) in parallel, the optimum annealing temperature was determined by gradient PCR between 40 and 54°C with 1.1°C increments. The optimum annealing temperature was 46.5°C. The amplification conditions for the multiplex PCR assay with three primers were as follows: 0.5 μl of genomic DNA, 2.5 μl of 10× reaction buffer, 100 nM (each) deoxynucleoside triphosphate, 0.5 U of Taq polymerase (Amersham Pharmacia Biotech, Piscataway, N.J.), 10 pmol of the specific primer and 616V (universal; Table 1), 2.5 pmol of primer 609R (universal; Table 1), 1% dimethyl sulfoxide, and deionized H2O to a final volume of 25 μl. The amplification conditions were as follows: initial denaturation (94°C for 120 s) followed by 25 cycles of denaturation (94°C for 45 s), annealing (46.5°C for 60 s), and extension (72°C for 60 s). PCR products were electrophoretically separated on a 2% agarose gel and stained with ethidium bromide. As the size marker, a 100-bp ladder (Amersham Pharmacia Biotech) was used.
RESULTS AND DISCUSSION
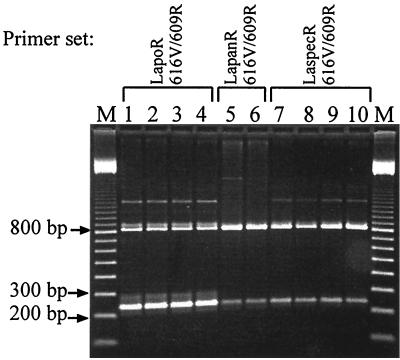
Figure 1 shows that it is possible to identify and discriminate between L. pontis, L. panis, and the closely related Lactobacillus sp. with the primer combinations deduced from the variable V2 region of the 16S rDNA. The primer combinations of 616V (universal) and the respective specific primers led to a 236-bp fragment for the three species. Primer 616V together with the universal reverse primer 609R allows the simultaneous amplification of an 800-bp fragment. In this way a combined specific identification together with a test of DNA accessibility was possible. So false-negative results could be avoided. The amplification of DNA isolated from strains of L. pontis and the Lactobacillus sp. led to an unspecific product of 1,300 bp. We accepted this as a compromise solution in order to have the possibility to run all three identification reactions in parallel. The specificity of the PCR assay was tested on several lactobacilli (Table 1). No cross-reactions appeared.
FIG. 1.
Multiplex PCR assay for the specific identification of L. pontis, L. panis, and the Lactobacillus sp. Primers indicated at the top are the specific primers for the identification of the following strains: L. pontis (LapoR) ATCC 51518 (lane 1), ATCC 51519 (lane 2), LTH 2587 (lane 3), and LTH 3572 (lane 4); L. panis (LapanR) DSM 6035 (lane 5) and DSM 6036 (lane 6); and Lactobacillus sp. strains (LaspecR) TMW 1.1098 (lane 7), TMW 1.1104 (lane 8), DSM 13145 (lane 9), and TMW 1.655 (lane 10). The specifically amplified fragment has a size of 236 bp (616V plus the specific reverse primer). The 800-bp fragment was amplified by the two universal primers 616V and 609R. Lane M, 100-bp ladder.
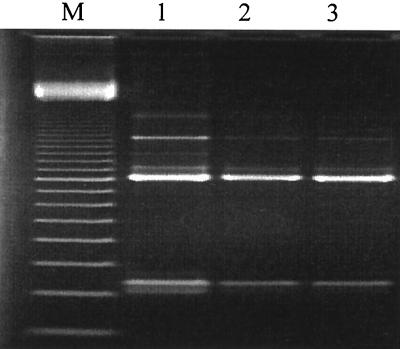
To have a tool for a fast and reliable identification of single pure cultures, the already-evaluated PCR assay was combined with a fast DNA isolation procedure. Time-consuming enzymatic lysis procedures and the variable susceptibility of bacteria to the lysozyme were overcome by ultrasonic treatment of the cells, with subsequent purification and concentration by binding DNA to a silica matrix. The cell material of a single colony was found to be sufficient for the PCR. Figure 2 illustrates a typical result for the amplification of DNA of representative strains of the investigated sourdough lactobacilli. The described PCR assay in combination with the fast-lysis procedure for even small amounts of cell material has major advantages over classical identification techniques, as physiological and biochemical identification is time-consuming and often not reliable. This is especially true for L. pontis, which exhibits a wide strain-dependent range of fermented carbohydrates, making a clear identification difficult. The fast ultrasonic lysis made it possible to identify this lactobacillus at the species level in 4 h. The application of this species-specific PCR technique for rapid identification provides an attractive alternative to conventional methods.
FIG. 2.
16S rDNA amplification of DNA isolated from a single colony of representative strains of the species L. pontis ATCC 51519 (lane 1), L. panis DSM 6035 (lane 2), and Lactobacillus sp. strain DSM 13145 (lane 3). The fragments were amplified with universal primer 616V and the respective species-specific reverse primers. The 800-bp fragment was amplified by the two universal primers 616V and 609R. Lane M, 100-bp ladder.
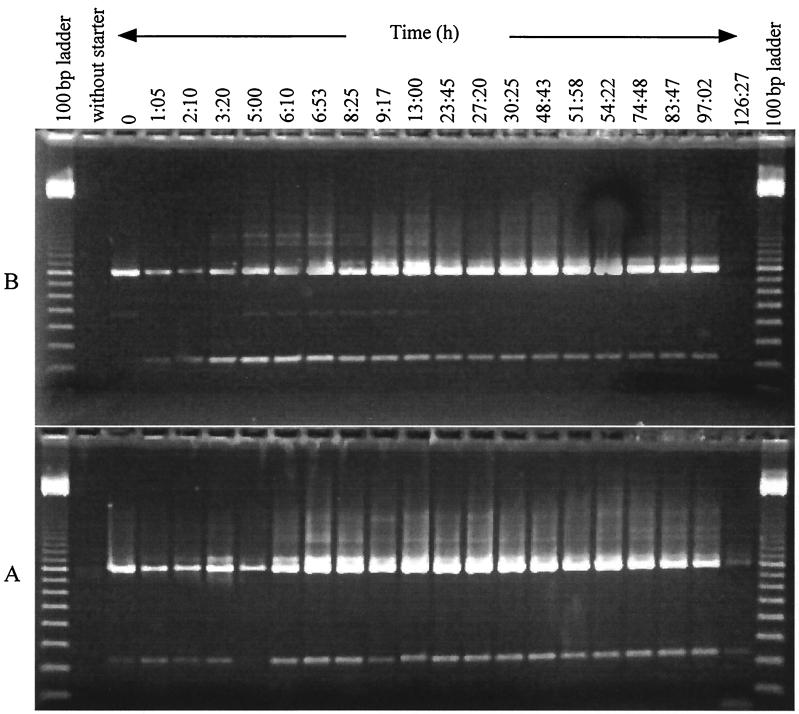
For the evaluation of the applicability of multiplex PCR for monitoring the lactobacilli from a mixed culture without prior cultivation, laboratory-scale sourdough fermentation was set up. The fermentation was based on rye bran and was started with sourdough from the corresponding industrial process which had been fermented for 48 h at similar conditions. Previous microbiological investigations of this as well as of laboratory-scale processes by randomly amplified polymorphic DNA typing and 16S rDNA sequence analysis showed that strains of the Lactobacillus sp., L. pontis, and Lactobacillus amylovorus predominated in the flora (unpublished results). As shown in Fig. 3, it was possible to specifically amplify DNA from L. pontis and the Lactobacillus sp. L. panis could not be identified in this fermentation, which resulted in the 800-bp fragment of the universal primers 616V and 609R only (a picture of the gel is not shown). Samples before starter addition showed no specific signal in both cases; even the universal primers targeting any bacterial DNA gave almost no signal (A) or no signal (B). This was confirmed by the classical microbiological investigation of plating on plate count agar, where a total cell count of 10 to 100 CFU was determined. On mMRS no LAB growth could be determined before starter was added. Directly after inoculation the total cell count was 9.3 × 106 CFU ml of sourdough−1, reaching its maximum of 3.2 × 109 CFU ml of sourdough−1 after 24 h and dropping to a final cell count of 5.5 × 107 CFU ml of sourdough−1 after 126 h. At this time for both primer combinations a signal with the two universal primers could be obtained, but only for L. pontis could a weak signal be recognized. During the fermentation the intensities of the signals for both the universally amplified product and the specific fragment increased, becoming lower after 54 h and disappearing completely for the Lactobacillus sp. after 126 h. The total cell count at the end of the fermentation seems to be a detection limit for directly extracted DNA from sourdough with this method. This limit seems to be high in comparison to those from other investigations in this field. For example, Zapparoli and Torriani (17) could amplify 102 CFU ml of diluted sourdough−1. Nevertheless the aim of this study was not to detect such small amounts but to have a fast and easy tool for detecting dominant lactobacilli from such fermentations. The direct isolation of total bacterial community DNA with a subsequent specific PCR can be considered a valuable tool for monitoring these lactobacilli in mixed microbial populations.
FIG. 3.
Specific amplification with primers for L. pontis (A) and the Lactobacillus sp. (B) of DNA extracted directly from the sourdough broth. The fermentation was monitored for over 126 h, starting from 0 h. At that time the starter was added. Without starter, sample before starter addition.
Moreover, no genotypic method to identify L. panis was available. Only a combined identification with L. pontis, not discrimination between these two species, was possible. With the presented specific PCR it could be demonstrated that it is possible to differentiate between these closely related species.
The Lactobacillus sp., a dominant element in the investigated fermentation, could be clearly discriminated from phylogenetically related species by genotypic identification techniques. To clarify the definitive phylogenetic position, further investigations are being prepared.
ACKNOWLEDGMENT
This work was supported by a grant of the European Union (FAIR project; CT 96 1126).
REFERENCES
- 1.Ampe F, Ben Omar N, Guyot J-P. Culture-independent quantification of physiologically-active microbial groups in fermented foods using rRNA-targeted oligonucleotide probes: application to pozol, a Mexican lactic acid fermented maize dough. J Appl Microbiol. 1999;87:131–140. doi: 10.1046/j.1365-2672.1999.00803.x. [DOI] [PubMed] [Google Scholar]
- 2.Betzl D, Ludwig W, Schleifer K H. Identification of lactococci and enterococci by colony hybridization with 23S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1990;56:2927–2929. doi: 10.1128/aem.56.9.2927-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böcker G, Stolz P, Hammes W P. Neue Erkenntnisse zum Ökosystem Sauerteig und zur Physiologie der sauerteigtypischen Stämme Lactobacillus sanfrancisco und Lactobacillus pontis. Getreide Mehl Brot. 1995;49:370–374. [Google Scholar]
- 4.Ehrmann M, Ludwig W, Schleifer K H. Reverse dot blot hybridization: a useful method for the direct identification of lactic acid bacteria in fermented food. FEMS Microbiol Lett. 1994;117:143–150. doi: 10.1111/j.1574-6968.1994.tb06756.x. [DOI] [PubMed] [Google Scholar]
- 5.Gänzle M G, Ehmann M, Hammes W P. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of sourdough fermentations. Appl Environ Microbiol. 1998;64:2616–2623. doi: 10.1128/aem.64.7.2616-2623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamad S H, Dieng M C, Ehrmann M A, Vogel R F. Characterization of the bacterial flora of Sudanese sorghum sourdough. J Appl Microbiol. 1997;83:764–770. doi: 10.1046/j.1365-2672.1997.00310.x. [DOI] [PubMed] [Google Scholar]
- 7.Klijn N, Weerkamp A H, De Vos W M. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl Environ Microbiol. 1991;57:3390–3393. doi: 10.1128/aem.57.11.3390-3393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewington J, Greenaway S D, Spillane B J. Rapid small scale preparations of bacterial genomic DNA, suitable for cloning and hybridization analysis. Lett Appl Microbiol. 1987;5:51–53. [Google Scholar]
- 9.Linko Y-Y, Javanainen P, Linko S. Biotechnology of bread baking. Trends Food Sci Technol. 1997;8:339–344. [Google Scholar]
- 10.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neefs J-M, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Vogel R F, Böcker G, Stolz P, Ehrmann M, Fanta D, Ludwig W, Pot B, Kersters K, Schleifer K H, Hammes W P. Identification of lactobacilli from sourdough and description of Lactobacillus pontis sp. nov. Int J Syst Bacteriol. 1994;44:223–229. doi: 10.1099/00207713-44-2-223. [DOI] [PubMed] [Google Scholar]
- 14.Vogel R F, Knorr R, Müller M R A, Steudel U, Gänzle M G, Ehrmann M A. Non-dairy lactic fermentations: the cereal world. Antonie Leeuwenhoek. 1999;76:403–411. [PubMed] [Google Scholar]
- 15.Vogel R F, Müller M, Stolz P, Ehrmann M. Ecology in sourdoughs produced by traditional and modern technologies. Adv Food Sci. 1996;18:152–159. [Google Scholar]
- 16.Wiese B G, Strohmar W, Rainey F A, Diekmann V. Lactobacillus panis sp. nov., from sourdough with a long fermentation period. Int J Syst Bacteriol. 1996;46:449–453. doi: 10.1099/00207713-46-2-449. [DOI] [PubMed] [Google Scholar]
- 17.Zapparoli G, Torriani S. Rapid identification and detection of Lactobacillus sanfrancisco in sourdough by species-specific PCR with 16S rRNA-targeted primers. System Appl Microbiol. 1997;20:640–644. [Google Scholar]