Abstract
An oxidative pathway for the mineralization of 2,4-dinitrotoluene (2,4-DNT) by Burkholderia sp. strain DNT has been reported previously. We report here the isolation of additional strains with the ability to mineralize 2,4-DNT by the same pathway and the isolation and characterization of bacterial strains that mineralize 2,6-dinitrotoluene (2,6-DNT) by a different pathway. Burkholderia cepacia strain JS850 and Hydrogenophaga palleronii strain JS863 grew on 2,6-DNT as the sole source of carbon and nitrogen. The initial steps in the pathway for degradation of 2,6-DNT were determined by simultaneous induction, enzyme assays, and identification of metabolites through mass spectroscopy and nuclear magnetic resonance. 2,6-DNT was converted to 3-methyl-4-nitrocatechol by a dioxygenation reaction accompanied by the release of nitrite. 3-Methyl-4-nitrocatechol was the substrate for extradiol ring cleavage yielding 2-hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid, which was converted to 2-hydroxy-5-nitropenta-2,4-dienoic acid. 2,4-DNT-degrading strains also converted 2,6-DNT to 3-methyl-4-nitrocatechol but did not metabolize the 3-methyl-4-nitrocatechol. Although 2,6-DNT prevented the degradation of 2,4-DNT by 2,4-DNT-degrading strains, the effect was not the result of inhibition of 2,4-DNT dioxygenase by 2,6-DNT or of 4-methyl-5-nitrocatechol monooxygenase by 3-methyl-4-nitrocatechol.
2,6-Dinitrotoluene (2,6-DNT) and 2,4-dinitrotoluene (2,4-DNT) occur as soil and groundwater contaminants at former 2,4,6-trinitrotoluene (TNT) production sites and in the wastewater from the commercial production of feedstocks for polyurethane foam (23). Twenty years after the cessation of TNT production in the United States, the manufacturing sites are still heavily contaminated with both 2,4- and 2,6-DNT even though 2,4-DNT-mineralizing bacteria can be readily isolated from the contaminated material (26). Commercial manufacture of DNT results in the release of DNT to industrial and municipal waste treatment systems (information found at the Environmental Health Center website [http://safety.webfirst.com/ehc/ew/chemical.htm] and in the TOXNET Toxics Release Inventory [http://six.nlm.nih.gov/sis1]). The unpredictable presence of DNT in the waste streams sent to the treatment plants can cause upsets in the ability of activated sludges to effectively remove the organic components in the waste streams (11). 2,4- and 2,6-DNT are priority pollutants (13), and industrial waste streams from DNT-manufacturing facilities are specifically regulated by the U.S. Environmental Protection Agency (40 CFR 261.32).
Contaminated munitions manufacturing sites are ready sources of bacteria able to mineralize 2,4-DNT, but bacteria able to grow on 2,6-DNT have been more elusive. The bacterial pathway for degradation of 2,4-DNT (8, 28) is initiated by dioxygenation of 2,4-DNT, which results in the formation of 4-methyl-5-nitrocatechol (4M5NC) and the release of nitrite; monooxygenation of 4M5NC then yields 2-hydroxy-5-methylquinone, which is subsequently reduced to 2,4,5-trihydroxytoluene prior to ring cleavage. The initial goal of the work was to get a sense of the distribution of bacteria able to degrade DNT. During that study, we discovered strains able to degrade 2,6-DNT and focused the rest of the work on the pathway. In order to allow rational design of bioremediation systems for 2,4-DNT-contaminated sites, it is necessary to understand the degradation of 2,6-DNT and how the degradation pathways interact.
We have examined the effects of 2,6-DNT on the degradation of 2,4-DNT by several 2,4-DNT-degrading strains. We also report here the isolation of bacteria able to use 2,6-DNT as the sole source of carbon, nitrogen, and energy and the initial steps in the 2,6-DNT degradative pathway.
(Preliminary accounts of this work have been presented previously [S. F. Nishino and J. C. Spain, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. Q-380, p. 452, 1996; S. F. Nishino and J. C. Spain, Abstr. 2nd SETAC World Congr., abstr. PW253, p. 277, 1995; S. F. Nishino and J. C. Spain, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. Q-348, p. 513, 1997].)
MATERIALS AND METHODS
Isolation and growth of bacteria.
Soil and groundwater samples were obtained from a number of sites contaminated by DNT (10 sites), and activated sludges were obtained from industrial waste treatment systems (9 sites) that receive DNT-containing waste streams. One milliliter of water or activated sludge or 1 g of soil was inoculated into 100 ml of nitrogen-free minimal medium (3) (BLK) containing 2,4-DNT (100 μM) or 2,6-DNT (50 μM) as the sole source of carbon and nitrogen. Cultures were incubated at 30°C with shaking (250 rpm). DNT concentrations were monitored by high-performance liquid chromatography (HPLC) (see below). Transfers to fresh BLK were made when concentrations of DNT in the culture fluid decreased. After several transfers (2 to 14 months), samples were spread on dilute (one-fourth strength) tryptic soy agar or on DNT plates (see below) and incubated for 1 to 6 weeks. Freshly grown isolates were inoculated into 96-well microtiter plates containing BLK (100 μl/well) with either 2,4-DNT (100 μM), 2,6-DNT (50 μM), or a mixture of 2,4- and 2,6-DNT (100 and 50 μM, respectively) and incubated at 30°C. After 3 to 5 days, nitrite and ammonia were measured in the culture fluids.
Strains were characterized by standard procedures (24) and by GN and GP Microplates (Biolog, Inc., Hayward, Calif.). 16S ribosomal DNA (rDNA) analysis was provided by Fred Rainey of the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH.
Amberlite XAD-7 resin was added to some cultures to provide a gradual but continuous release of DNT. DNT was added to an empty flask to give an amount equal to a final concentration of 1 to 4 mM. The DNT was dissolved in a small amount of acetone which was evaporated under a stream of air to leave a coat of fine DNT crystals in the bottom of the flask. Appropriate amounts of BLK and XAD-7 resin (washed three times with methanol, 10 g [hydrated weight]/liter or 3.5 g [dry weight]/liter) were added to the DNT-coated flask prior to autoclaving. The procedure resulted in a final dissolved DNT concentration of 20 to 200 μM after autoclaving and cooling. Agar plates containing DNT and XAD-7 resin were prepared in a similar manner except that the resin was ground to a paste in a mortar and pestle and added at a concentration of 7 g (hydrated weight)/liter with a final DNT concentration of 3 mM and 1.8% agar.
Pure cultures were routinely maintained on BLK agar containing DNT and XAD-7 resin. For experiments with induced cells, cultures were incubated at 30°C with shaking at 250 rpm for 2 to 4 weeks in BLK containing XAD-7 resin and 2,4- or 2,6-DNT (3 mM). When cultures became dense and DNT in the aqueous phase disappeared, cells were harvested by centrifugation after filtration through glass wool to remove the resin. Cells were washed twice with phosphate buffer (0.02 M, pH 7.0) before use in subsequent experiments. Uninduced cells were grown overnight in tryptic soy broth.
Escherichia coli JM109(pDTG603) (29, 30) containing todE (encoding 3-methylcatechol 2,3-dioxygenase) and several other genes encoding enzymes involved in toluene degradation was provided by D. T. Gibson of the University of Iowa. The tod operon was induced in glucose-grown cells with IPTG (isopropyl-β-d-thiogalactopyranoside) as previously described (29). Cells were harvested and washed as described above, and the pellet was stored frozen until needed.
Preparation of cell extracts.
2,6-DNT-grown cultures were incubated (2 to 4 h) with penicillin G (100 U/ml) prior to harvest. Washed cells were broken by three passages through a French pressure cell at 32,000 lb/in2. The exudate was centrifuged at 34,000 × g for 60 min at 4°C for most purposes or at 100,000 × g for preparation of soluble fractions. The pellet was discarded, and the supernatant was stored on ice or frozen until used.
The ring fission enzyme was partially purified from extracts of 2,6-DNT-grown cells. Cell extracts were fractionated by anion-exchange chromatography on a MonoQ HR 10/10 column (Pharmacia Biotech, Uppsala, Sweden). The protein was loaded on the column in potassium phosphate buffer (10 mM, pH 7.0) and eluted with a linear NaCl gradient (0 to 1.0 M) in phosphate buffer. Fractions that produced a yellow product from 3-methylcatechol were combined and used in subsequent experiments.
Chemicals.
3-Methyl-4-nitrocatechol (3M4NC) was prepared biologically by the action of 2,4-DNT-grown Burkholderia cepacia strain R34 on 2,6-DNT. 3M4NC and 2,6-DNT were extracted from cell-free culture fluid with a C18, 35-cm3 solid-phase extraction cartridge (Millipore, Milford, Mass.). 3M4NC was eluted from the cartridge with methanol-water (30:70). The methanol was removed by flash evaporation, the aqueous phase was acidified, and the 3M4NC was extracted into ethyl acetate. The ethyl acetate was removed by flash evaporation, and the residue was dissolved in a small amount of acetonitrile and purified by semipreparative HPLC. For some preparations, E. coli HB101(pJS332), which contained a 5.9-kb NsiI-EcoRV fragment encoding the initial 2,4-DNT dioxygenase of B. cepacia R34 in plasmid pGEM7(+) (Promega, Madison, Wis.) (R. Jain, unpublished results), was used to transform 2,6-DNT to 3M4NC in the presence of 10 mM glucose.
2,3,6-Trihydroxytoluene and 2-hydroxy-3-methylquinone were synthesized as previously described (7). 4M5NC and 2,4,5-trihydroxytoluene were generously supplied by Ronald Spanggord (SRI International, Menlo Park, Calif.). Amberlite XAD-7 was from Sigma (St. Louis, Mo.). All other chemicals were of the highest grade commercially available.
Synthesis and purification of nitroaliphatic pathway intermediates.
The partially purified ring cleavage enzyme was used to convert 3M4NC (200 μM) to the ring cleavage product (compound X) in potassium phosphate buffer (1 mM, pH 7.0). After complete conversion of the 3M4NC, protein was removed from the reaction mixture by passing the solution through a 5,000-molecular-weight-cutoff membrane (Amicon, Beverly, Mass.). Water was removed by lyophilization. The dried preparation was stored under nitrogen gas at −80°C. Samples used for nuclear magnetic resonance (NMR) analysis were dissolved in D2O just prior to analysis.
The metabolite (compound Y) produced from the ring fission product was synthesized by incubation of 3M4NC (200 μM) with crude cell extract in potassium phosphate buffer (1 mM, pH 7.0). Protein was removed as described above, and the filtrate was lyophilized to concentrate the compound. After acidification to pH 2.5, the compound was extracted into ethyl acetate, and the ethyl acetate was evaporated under vacuum. The compound was dissolved in deuterated ether for NMR analysis.
Enzyme assays.
3-Methylcatechol-2,3-dioxygenase (14) and 4M5NC monooxygenase activity (8) were measured as previously described. 3M4NC dioxygenase activity was measured by monitoring the increase in the A375 of the 3M4NC ring cleavage product. Reaction mixtures for 3M4NC dioxygenase contained 0.1 μmol of 3M4NC, 9.8 μmol of sodium phosphate (pH 7.0), and cell extract (0.1 to 0.5 mg of protein) in a final volume of 1 ml. The molar extinction coefficients of compounds X and Y (E375 = 16.8 and 16.1 mM−1 cm−1, respectively) were estimated by assuming complete conversion of 3M4NC to the respective products under conditions of excess enzyme. Some cell extracts were dialyzed for 4 h against two changes of phosphate buffer (0.02 M, pH 7.0) before use. Some cell extracts were preincubated with ferrous or ferric sulfate (50 μM) for 5 min prior to the assay.
Respirometry.
Oxygen uptake was measured polarographically with a Clark-type oxygen electrode connected to a YSI Model 5300 biological oxygen monitor.
Analytical methods.
Nitrite and ammonia concentrations were measured by standard methods (24). Protein was measured as previously described (25). HPLC analyses for DNT and methylnitrocatechols were performed as previously described (21) or on two Zorbax Reliance CN cartridge columns connected in series (4 mm inside diameter [i.d.] by 8 cm; 5 μm). The mobile phase consisted of a 75:25 ratio of part A (13.5 mM trifluoroacetic acid in water) to part B (6.75 mM trifluoroacetic acid in acetonitrile), delivered at a flow rate of 2 ml/min. Semipreparative HPLC was performed with an Adsorbosphere C18 column (10 mm [i.d.] by 25 cm; 10 μm), with a mobile phase of 85:15 part A-part B, respectively, delivered at a flow rate of 4 ml/min. The ring cleavage product and subsequent metabolites were analyzed using a Synchropak SCD (4.6 mm [i.d.] by 25 cm; Micra Scientific, Northbrook, Ill.) reversed-phase column with a mobile phase of potassium phosphate buffer (100 mM, pH 7.0) at a flow rate of 0.5 ml/min.
Spectrophotometric analyses were performed on a Cary 3E UV-visible light (UV-VIS) spectrophotometer (Varian Associates, Sunnyvale, Calif.). Gas chromatography-mass spectrometry (GC-MS) analyses were performed on an HP5890 gas chromatograph equipped with a 30-m DB-5 fused-silica capillary column and an HP5971 mass selective detector. Liquid chromatography (LC)-MS-MS analyses were conducted by J. V. Johnson of the Department of Chemistry, University of Florida. NMR analyses were performed by T. Gedris of the NMR Laboratory, Chemistry Department, Florida State University.
RESULTS
Isolation and identification of 2,4- and 2,6-DNT-degrading bacteria.
DNT disappearance accompanied by accumulation of nitrite in enrichment cultures began after several days to several weeks of incubation with 2,4-DNT provided as the sole source of carbon, nitrogen, and energy and after several weeks to several months of incubation with 2,6-DNT. Most contaminated soil and groundwater samples yielded both 2,4- and 2,6-DNT-degrading strains. Enrichments prepared using activated sludge as initial inocula were not as successful. Many of the activated sludges transformed 2,4- and 2,6-DNT to 2-amino-4-nitrotoluene or 2-amino-6-nitrotoluene, probably through the action of nonspecific nitroreductases. The aminonitrotoluenes often accumulated without further transformation, and efforts to isolate DNT-degrading strains from such cultures were discontinued. Pure cultures isolated from other enrichments were tested for release of ammonia and nitrite from DNT. None of the strains released ammonia when provided with DNT as the sole carbon and nitrogen source. Strains that released substantial amounts of nitrite were transferred to fresh medium. Strains that removed DNT from culture fluids during a 7-day incubation, accompanied by stoichiometric release of nitrite and without the accumulation of aminonitrotoluenes, were considered presumptive DNT-degrading bacteria.
Approximately 30 strains that degraded 2,4-DNT were isolated from soil and surface water collected at Radford Army Ammunition Plant (Table 1). Two of the 30 strains grew notably faster on 2,4-DNT than did the only previously described 2,4-DNT-degrading isolate, Burkholderia sp. strain DNT (28). The two new isolates were determined to be B. cepacia by partial 16S rDNA analysis and designated strains R34 and PR7. Strain R34 accumulated substantially less 4M5NC during induction than did Burkholderia sp. strain DNT or strain PR7. Samples collected from Volunteer Army Ammunition Plant yielded three different types of 2,4-DNT-degrading strains. Two clusters that appeared distinct by Biolog GN microplate reactions gave identical partial 16S rDNA sequences and have been designated Alcaligenes sp. They are represented by strain JS867 (Alcaligenes denitrificans Biolog cluster) and strain JS871 (Alcaligenes xylosoxidans Biolog cluster). The third cluster, represented by B. cepacia strain JS872 (21), exhibited particularly rapid growth on 2,4-DNT. Depletion of 2,4-DNT (1 mM) by JS872 took place in 24 h compared to 3 to 5 days for strain DNT and 2 to 3 days for strains PR7 and R34. All 2,4-DNT-degrading strains examined to date use the same pathway as Burkholderia sp. strain DNT for degradation of 2,4-DNT. None of the 2,4-DNT-degrading isolates was capable of growth on 2,6-DNT.
TABLE 1.
Identification of selected DNT-degrading strains
| Strain | Identification
|
Sourcea; yr isolated | DNT isomer degraded | Gram stain reaction | Activity
|
||
|---|---|---|---|---|---|---|---|
| 16S rDNA | Biolog | Oxb | Catc | ||||
| R34 | Burkholderia cepacia | Pseudomonas sp. | 1; 1992 | 2,4 | − | + | + |
| PR7 | Burkholderia cepacia | Pseudomonas sp. | 1; 1992 | 2,4 | − | + | + |
| JS850 | Burkholderia cepacia | None | 3; 1995 | 2,6 | − | − | + |
| JS863 | Hydrogenophaga palleronii | None | 2; 1995 | 2,6 | − | − | + |
| JS867 | Alcaligenes sp. | Alcaligenes denitrificans | 2; 1995 | 2,4 | − | + | + |
| JS871 | Alcaligenes sp. | Alcaligenes xylosoxidans | 2; 1995 | 2,4 | − | + | + |
| JS872 | Burkholderia cepacia | Pseudomonas cepacia | 2; 1995 | 2,4 | − | + | + |
| JS881 | Pseudomonas putida | Pseudomonas sp. | 1; 1992 | 2,6 | − | − | + |
Source: 1, Radford Army Ammunition Plant, soil and surface water; 2, Volunteer Army Ammunition Plant, soil and surface water; 3, West Virginia, activated sludge.
Ox, oxidase.
Cat, catalase.
The first 2,6-DNT-degrading strains (Table 1) were enriched from soil samples from the Volunteer Army Ammunition Plant. The partial 16S rDNA sequence of the isolate that grew most rapidly on 2,6-DNT, strain JS863, was 99% identical to the 16S rDNA of Hydrogenophaga palleronii. B. cepacia strain JS850, also identified by partial 16S rDNA analysis, was isolated from activated sludge from an industrial waste treatment system of a DNT-manufacturing facility. None of the 2,6-DNT-degrading isolates could grow with 2,4-DNT provided as the sole growth substrate.
Inhibition of 2,4-DNT degradation by 2,6-DNT.
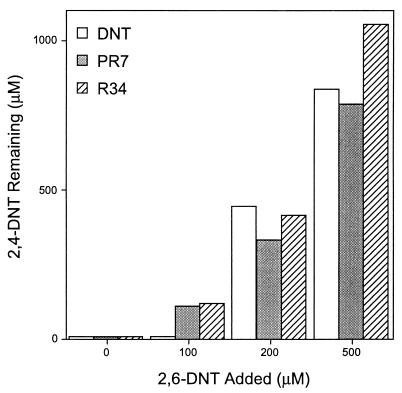
2,4-DNT-grown cultures of strains DNT, PR7, and R34 were provided with mixtures of 2,4- and 2,6-DNT. After 3 days (Fig. 1), 2,4-DNT remained in all cultures provided with 200 μM or more 2,6-DNT while cultures without added 2,6-DNT completely removed 2,4-DNT within 1 to 2 days. When the strains were grown on succinate with NH4Cl added as nitrogen source, addition of 2,4- or 2,6-DNT at concentrations of ≥100 μM reduced the cell densities achieved during a 24-h incubation, whereas 10 and 25 μM DNT had little effect on the final cell densities. The results indicate that high concentrations of either isomer of DNT inhibit growth of DNT-degrading strains on simple substrates.
FIG. 1.
Inhibition of 2,4-DNT degradation by 2,6-DNT. 2,4-DNT-degrading isolates were grown on 2,4-DNT, washed, and then suspended in BLK containing 2,4-DNT (1,000 μM) plus 2,6-DNT. Bars depict 2,4-DNT remaining after a 3-day incubation.
Production and identification of 3M4NC.
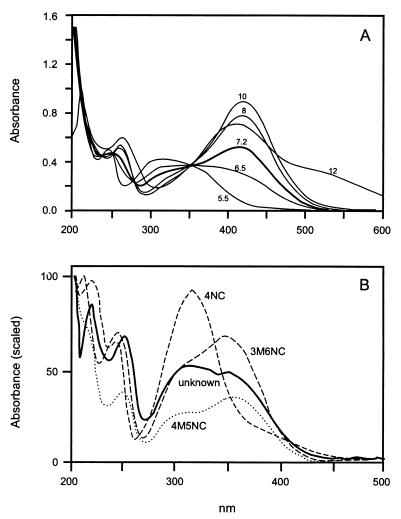
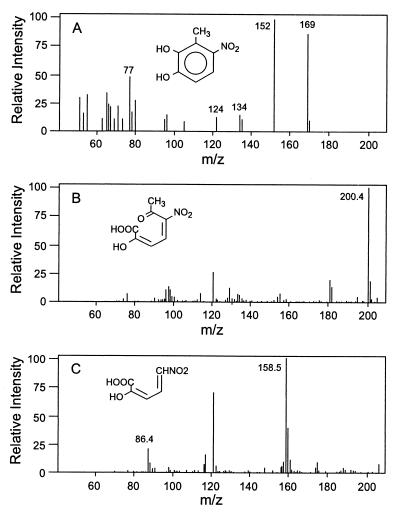
HPLC analysis revealed the accumulation of a yellow metabolite in culture fluids after growth of strains PR7 and R34, but not in culture fluids of strain DNT, when 2,6-DNT was present with 2,4-DNT. The metabolite had a different HPLC retention time from that of 4M5NC, but the pH-dependent UV-VIS spectrum was similar to that of 4M5NC (Fig. 2). The metabolite was purified from 12-liter cultures that were grown on 2,4-DNT and then incubated with 2,6-DNT. The mass spectrum of the purified yellow metabolite (Fig. 3A) was very similar to that previously described for 4M5NC (28). As with 4M5NC, there was an apparent molecular ion at 169 and a base peak at 152, which indicates the loss of a hydroxyl group from an aromatic compound with a nitro group and a methyl group in an ortho configuration. Smaller peaks were also very similar. The NMR spectrum (Table 2) revealed a pair of interacting protons on the aromatic ring, one ortho to a hydroxyl group and therefore more upfield, and the other ortho to a nitro group and therefore more downfield. Based upon the UV-VIS, mass, and NMR spectra, the metabolite was conclusively identified as 3M4NC. The maximum concentration of 3M4NC (70 μM) accumulated in cultures incubated with 100 μM 2,6-DNT. Cultures provided with higher concentrations of 2,6-DNT accumulated lower concentrations of 3M4NC as more of the 2,6-DNT remained untransformed.
FIG. 2.
UV-VIS spectra of yellow metabolite that accumulated when 2,4-DNT-degrading strains were provided with 2,6-DNT. (A) Effect of pH. (B) Spectrum of yellow metabolite compared with other nitrocatechols (pH 7.0).
FIG. 3.
Mass spectra of 2,6-DNT metabolites. (A) 3M4NC analyzed by GC-MS. (B) 2-Hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid analyzed by LC-MS. (C) 2-Hydroxy-5-nitropenta-2,4-dienoic acid analyzed by LC-MS.
TABLE 2.
13C and 1H NMR data for 2,6-DNT metabolitesa
| Compound | Position | δ (ppm)
|
J (Hz) | |
|---|---|---|---|---|
| 13C | 1H | |||
| MNC | 1 | ND | ||
| 2 | ||||
| 3 | ||||
| 4 | ||||
| 5 | 7.5 (d) | 3J (5-H,6-H) = 8.8 | ||
| 6 | 6.9 (d) | |||
| 7 (CH3) | 2.4 (s) | |||
| Compound X | 1 (COOH) | 199.3 | ||
| 2 | 175.9 | |||
| 3 | 117.3 | 6.8 (d) | 3J (3-H,4-H) = 15.9 | |
| 4 | 144.4 | 7.9 (d) | ||
| 5 | 130.1 | |||
| 6 | 201.1 | |||
| 7 (CH3) | 33.0 | 2.5 (s) | ||
| Compound Y | 1 (COOH) | 167.6 | —b | |
| 2 | 153.5 | —b | ||
| 3 | 105.9 | 6.1 (d) | 3J (3-H,4-H) = 11.9 | |
| 4 | 134.9 | 7.8 (dd) | 3J (4-H,5-H) = 13.3 | |
| 5 | 142.7 | 7.2 (d) | 4J (3-H,5-H) = 1.5 | |
MNC, methylnitrocatechol; ND, not determined.
Signal too weak to detect.
Toxicity of 3M4NC to 2,4-DNT-degrading bacteria.
4M5NC is toxic to strain DNT at concentrations above 2 μM (9). Accumulation of 3M4NC in culture fluids of 2,4-DNT-grown strains provided with 2,6-DNT suggested that 3M4NC might be toxic to 2,4-DNT-degrading strains. The purified 4M5NC monooxygenase from strain DNT (9) was assayed for its ability to oxidize 3M4NC. Concentrations of 1 to 10 μM 3M4NC neither served as a substrate nor inhibited the oxidation of 4M5NC by the monooxygenase. When 2,4-DNT-grown cells were incubated with 3M4NC or 2,6-DNT, oxidation of 4M5NC and 2,4-DNT was not markedly inhibited (Table 3). The results suggest that the inability of 2,4-DNT-grown cells to metabolize 2,4-DNT in the presence of 2,6-DNT was not due to the effect of 2,6-DNT or 3M4NC on the 2,4-DNT dioxygenase or the 4M5NC monooxygenase. In addition, neither 3M4NC nor 4M5NC, each of which is very stable at room temperature under aqueous conditions, has ever been detected in DNT-contaminated soil. We conclude that, if a metabolite is involved in inhibition of DNT degradation, it is not one of the methylnitrocatechols.
TABLE 3.
Effect of 2,6-DNT and 3M4NC on 2,4-DNT-degrading strains
| Substratea | Oxygen uptake (nmol/min/mg of protein) by strain:
|
||||
|---|---|---|---|---|---|
| DNT | PR7 | JS867 | JS871 | JS872 | |
| 2,4-DNT | 3.3 | 2.6 | 13.2 | 9.8 | 10.9 |
| 2,6-DNT | <0.1 | <0.1 | 3.0 | 0.8 | <0.1 |
| 2,4-DNT + 2,6-DNT | 1.8 | 2.9 | 17.3 | 9.8 | 8.3 |
| 4M5NC | 12.7 | 5.9 | 12.1 | 9.8 | 11.7 |
| 3M4NC | 4.8 | <0.1 | 0.8 | <0.1 | 0.3 |
| 4M5NC + 3M4NC | 12.0 | 4.9 | 8.3 | 8.8 | 9.1 |
Substrates were provided at 50 μM.
Growth of 2,6-DNT-degrading strains.
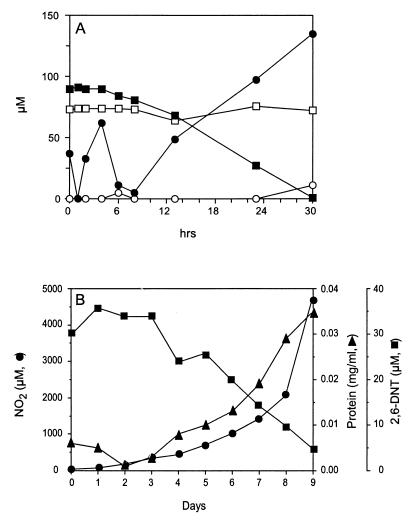
Growth on 2,6-DNT by strains JS863 and JS850 was accompanied by the accumulation of 1.6 to 1.8 mol of nitrite per mol of 2,6-DNT (Fig. 4A). Growth was slow and followed a 3- to 10-day lag period. At 2,6-DNT concentrations of 100 μM or below, DNT removal was complete. At 250 μM 2,6-DNT, the rate of degradation was reduced, and at 500 μM, 2,6-DNT degradation was completely inhibited. When 2,6-DNT (1 to 3 mM) was provided as being adsorbed to XAD-7 resin, both nitrite and protein increased as the concentration of DNT in the aqueous phase decreased (Fig. 4B). Growth on 2,6-DNT was inhibited in the presence of other carbon sources tested (glucose, succinate, acetate, glycerol, yeast extract, aspartate, glutamate, alanine, and proline), and 2,6-DNT was not used as a nitrogen source if an additional carbon source but no other nitrogen source was provided (data not shown). The protein content of cultures after growth on 2,6-DNT was comparable to the protein content of cultures grown on succinate. JS850 lost the ability to degrade 2,6-DNT following multiple passages on nonselective medium, which suggests that at least one of the genes encoding enzymes for the 2,6-DNT degradative pathway is plasmid encoded. 2,6-DNT degradation was not inhibited by nitrite concentrations up to 100 mM.
FIG. 4.
Growth of strain JS850 in BLK with 2,6-DNT. (A) 2,6-DNT (squares) and nitrite (circles) inoculated with 2,6-DNT-grown cells (solid symbols) and in an uninoculated control (open symbols). (B) Aqueous-phase concentrations of 2,6-DNT, nitrite, and soluble protein were measured in a culture of strain JS850 grown in BLK medium containing 3 mM 2,6-DNT adsorbed to XAD-7 resin.
Production of 3M4NC by 2,6-DNT-degrading strains.
During the lag period before growth on 2,6-DNT, many of the 2,6-DNT-degrading isolates transiently accumulated a yellow metabolite in the culture medium. The transient accumulation and subsequent metabolism of this yellow compound by 2,6-DNT-degrading strains suggested that the yellow compound was an intermediate in the 2,6-DNT degradative pathway. The metabolite was purified and identified as 3M4NC by HPLC, UV-VIS, and mass spectral analyses. A typical culture of JS863 grown with 2,6-DNT (65 μM) accumulated a maximum of 3 μM 3M4NC.
Respirometry.
Simultaneous adaptation studies with resting cells of strains JS850 and JS863 grown on 2,6-DNT indicated that 2,6-DNT, 3M4NC, and catechol, but not 2,4-DNT, TNT, 4M5NC, 2,4,5-trihydroxytoluene, 2-methyl-3-nitrophenol, or 2-amino-6-nitrotoluene, stimulated oxygen uptake (Table 4). 2,3,6-Trihydroxytoluene appeared to stimulate oxygen uptake in JS850 but not in JS863. The results suggested that the 2,6-DNT-degrading strains have a narrow substrate specificity. Lack of increased oxygen uptake after incubation with 2-methyl-3-nitrophenol and 2-amino-6-nitrotoluene also suggested that neither a monooxygenase nor a nitroreductase was involved in the initial attack.
TABLE 4.
Oxygen consumption by washed cells after growth on 2,6-DNT
| Substratea | O2 uptake (nmol/min/mg of protein)b by strain:
|
|
|---|---|---|
| JS850 | JS863 | |
| 2,6-DNT | 7.5 | 1.6 (2.5)c |
| 3M4NC | 11.5 | 3.3 (1.4) |
| 2,3,6-Trihydroxytoluene | 14.4 | 0.6 |
| 4M5NC | 0.6 | 0.9 |
| 3-Methyl-6-nitrocatechol | 0.2 | <0.1 |
| 4-Nitrocatechol | 1.0 | 0.1 |
| Catechol | 5.5 | 1.8 |
| Toluene | <0.1 | <0.1 |
| TNT | 1.6 | 0.3 |
| 2,4-DNT | <0.1 | <0.1 |
| 2,6-Dinitrophenol | <0.1 | <0.1 |
| 2-Methyl-3-nitrophenol | <0.1 | <0.1 |
| 2-Amino-6-nitrotoluene | <0.1 | <0.1 |
| 2,4,5-Trihydroxytoluene | 0.3 | 3.0 |
Substrates were provided at 50 μM.
Yeast extract- and tryptic soy broth-grown cells showed no increased uptake with 2,6-DNT and 3M4NC. Other substrates were not tested.
Number in parentheses is stoichiometry of O2 consumption.
Enzyme studies.
Crude cell extracts prepared from 2,6-DNT-grown strains JS863 and JS850 converted 3M4NC to a yellow compound (compound Y) that only slowly disappeared upon overnight incubation. Typical reaction rates for conversion of 3M4NC to compound Y were 35 ± 6 nmol/min/mg of protein. Compound Y had an absorbance maximum at 375 nm. The A375 decreased upon acidification and returned when the mixture was returned to a neutral pH or made basic. The behavior of compound Y was consistent with that of a meta-ring cleavage product. Repetitive scans during the reaction revealed an isosbestic point at 435 nm (Fig. 5). Addition of ferrous or ferric iron, NAD+, or NADP+ did not affect the reaction. Preincubation of cell extract (100 μl) with H2O2 (5 μl of 30%) greatly reduced the enzyme activity. The activity was stable in frozen extracts but was reduced by 90% in extracts heated to 50°C for 10 min and abolished in extracts that were heated to 55°C for 15 min. No nitrite release was detected upon conversion of 3M4NC to compound Y. The enzyme was not active with 4M5NC, 3-methyl-6-nitrocatechol, 4-nitrocatechol, 2-hydroxy-3-methylquinone, or 2,3,6-trihydroxytoluene; however, 3-methylcatechol was slowly converted (at a rate 32 to 35% of that observed with 3M4NC) to a yellow compound (compound Z) with absorbance maxima at 385 and 320 nm, which are identical to those of the product of meta-ring cleavage of 3-methylcatechol by toluene-grown Pseudomonas putida (14). Neither 3M4NC nor 3-methylcatechol was transformed by cell extracts prepared from strain JS850 or JS863 grown on tryptic soy broth.
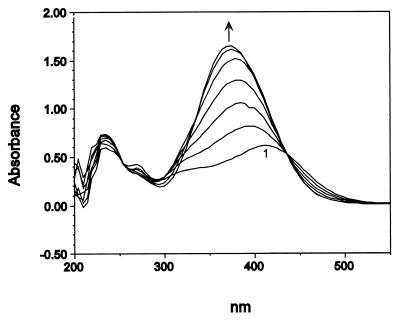
FIG. 5.
Conversion of 3M4NC to compound Y by crude cell extracts of JS863. An extract from a 2,6-DNT-grown culture was incubated in phosphate buffer (20 mM, pH 7.0) with 100 μM 3M4NC. The reaction was initiated by the addition of the catechol (scan 1). Scans were recorded at 2-min intervals. Compound Y had a single absorbance maximum at 375 nm.
When 3M4NC was incubated with cell extracts heated to 50°C for 10 min, 3M4NC was converted to a yellow product (compound X) whose absorbance spectrum differed from that of compound Y. Compound X had absorbance maxima at 394 and 326 nm at pH 7.0. Subsequent addition of an unheated extract of JS850 or JS863 to the heat-treated assay mixture resulted in the conversion of compound X to compound Y. Unheated extract of JS850 or JS863 converted 3M4NC to compound Y in solutions buffered between pH 7 and pH 10. The reaction was inhibited at pH 4 and pH 11. At pH 5, compound X was produced (initial rate, 32 ± 8 nmol/min/mg of protein). At pH 6, compound X was produced and only slowly transformed to compound Y. The results suggest that the crude cell extracts convert 3M4NC to compound Y via compound X in at least two enzymatic steps.
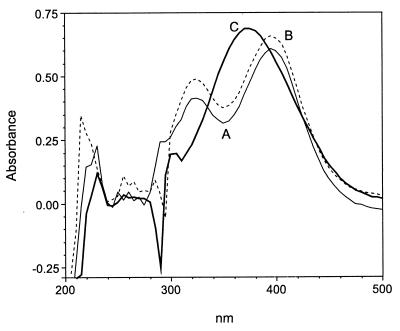
Cell extracts prepared from E. coli JM109 containing pDTG603 (carrying the cloned todE gene encoding 3-methylcatechol-2,3-dioxygenase) converted 3M4NC to a compound with an absorbance spectrum identical to that of compound X. Addition of unheated cell extracts of JS850 or JS863 converted the compound to one with an absorbance spectrum identical to that of compound Y (Fig. 6) (initial rate, 36 ± 1 nmol/min/mg of protein). Cell extracts containing pDTG603 converted 3-methylcatechol to 2-hydroxy-6-oxohepta-2,4-dienoic acid (30), which had an absorbance spectrum and an HPLC retention time identical to those of compound Z. Addition of cell extracts of JS850 or JS863 had no effect on the spectrum of 2-hydroxy-6-oxohepta-2,4-dienoic acid. 3-Methylcatechol-2,3-dioxygenase catalyzed the complete conversion of 3-methylcatechol 15 times faster than the complete conversion of 3M4NC. The rate of conversion of 3M4NC declined rapidly after the reaction was initiated by addition of substrate. Conversion of the last 40% of the substrate required 2.5 times as much time as the conversion of the first 60% of the substrate. We did not distinguish between loss of enzyme activity and product inhibition. Extracts from JS850 and JS863 maintained the same rate for the entire reaction period. The reaction rates given above are consistent with oxygen uptake rates. The results indicate that the enzyme that produced compound X from 3M4NC is a catechol-2,3-dioxygenase with a different substrate specificity from that of 3-methylcatechol-2,3-dioxygenase.
FIG. 6.
(A and B) Reaction of heat-treated extract from cells of 2,6-DNT-grown JS850 with 3M4NC (A) and heat-treated extract from cells of E. coli JM109(pDTG603) (B) which expresses a cloned 3-methylcatechol-2,3-dioxygenase gene. (C) Addition of fresh (not heat-treated) cell extract from cells of 2,6-DNT-grown JS850 to the reaction product shown by line B.
Identification of compounds X and Y.
Compounds X and Y were accumulated and purified as described in Materials and Methods. Compound X was unstable at low pH and did not partition into organic solvents at neutral pH, necessitating concentration of the compound by lyophilization without further purification. LC-MS of compound X revealed a base peak at m/z 200 (Fig. 3B) which corresponded to the [M—H]− ion. The fragment with a m/z of 120 is an artifact of a gas-phase reaction involving acetate-acetic acid from the LC mobile phase and, as demonstrated by LC-MS-MS of the 200 mass ion (data not shown), is unrelated to compound X. A [M—H]− mass of 200 is consistent with that expected for a dioxygenolytic product of 3M4NC. LC-MS-MS of the [M—H]− ion resulted in a fragment with a m/z of 129 ([H3C—C(⩵O)—C(—NO2)⩵CH2—CH2]−), and MS analysis of the fragment of m/z 129 revealed fragment ions of m/z 86 ([O2N—CH⩵CH—CH2]−) and m/z 59 ([O2N—CH:]−). The observed fragmentation is consistent with the product of proximal meta-ring cleavage of 3M4NC but not with the product of distal meta-ring cleavage or ortho-ring cleavage of 3M4NC.
The 13C NMR of compound X revealed seven carbon resonances for which putative assignments have been made, and 1H NMR showed a set of coupled protons and a singlet peak at 2.5 ppm arising from protons of a methyl group (Table 2). The results are consistent with the structure of the 3M4NC proximal meta-ring cleavage product, 2-hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid.
Compound Y was stable (several minutes) under acidic conditions but decomposed upon overnight storage at pH 3.5. The LC-MS of compound Y (Fig. 3C) revealed a base peak at m/z 158 and, as further demonstrated by LC-MS-MS of the ([M—H]−) ion, a major fragment at m/z 86 ([O2N—CH⩵CH—CH2]−). As described above, the peak at m/z 120 is an artifact. The 13C and 1H NMR spectra (Table 2) revealed five carbon resonances and three olefinic proton signals that were nearly identical to those previously reported for 2-hydroxy-5-nitropenta-2,4-dienoic acid (6). The mass spectrum and NMR analyses unequivocally identified compound Y as 2-hydroxy-5-nitropenta-2,4-dienoic acid. The structure of 2-hydroxy-5-nitropenta-2,4-dienoic acid is consistent with the postulated product of the hydrolytic loss of acetate from 2-hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid.
DISCUSSION
DNT-contaminated sites around TNT-manufacturing plants have been the source of many 2,4-DNT-degrading bacterial strains and, as reported here, 2,6-DNT-degrading bacteria. Soil slurry reactors (21) and fluid bed reactors (15) inoculated with mixtures of specific DNT-degrading strains have demonstrated degradation and mineralization (21) of mixtures of the 2,4- and 2,6-DNT isomers. Preliminary evidence indicates, however, that in mixed cultures, high concentrations of 2,6-DNT inhibit the degradation of 2,4-DNT and high concentrations of 2,4-DNT inhibit the degradation of 2,6-DNT. We determined the initial steps in the 2,6-DNT catabolic pathway in order to gain an understanding of how the DNT isomers affect overall DNT degradation when both 2,4- and 2,6-DNT are present. To our knowledge, this is the first description of the degradative pathway for 2,6-DNT and the first reported bacterial production of 3M4NC and 2-hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid.
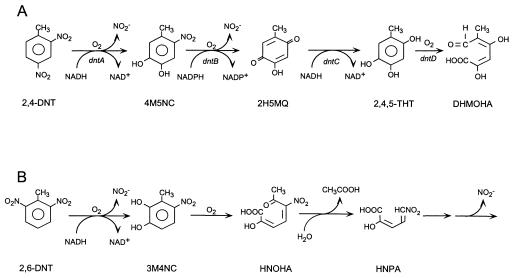
The transient accumulation of 3M4NC during induction of the 2,6-DNT degradation pathway suggested that 3M4NC is a pathway intermediate. Simultaneous induction studies indicated that 3M4NC is the product of an initial dioxygenation as in the 2,4-DNT degradation pathway, rather than sequential monooxygenation of 2,6-DNT to 2-methyl-3-nitrophenol and then 3M4NC. The results of the simultaneous induction studies and the release of both nitro groups of 2,6-DNT as nitrite suggested that the 2,6-DNT pathway might be analogous to the 2,4-DNT pathway (Fig. 7A). However, neither simultaneous induction nor enzyme assays indicated that 2,3,6-trihydroxytoluene, the 2,6-DNT analog of 2,4,5-trihydroxytoluene of the 2,4-DNT pathway, was involved in the 2,6-DNT catabolic pathway. Studies with cell extracts conclusively demonstrated that, after the initial dioxygenation, the two pathways diverged.
FIG. 7.
Comparison of the 2,4-DNT (A) and 2,6-DNT (B) catabolic pathways. The 2,4-DNT pathway intermediates are 2,4-DNT, 4M5NC, 2H5MQ (2-hydroxy-5-methylquinone), 2,4,5-THT (2,4,5-trihydroxytoluene), and DHMOHA (2,4-dihydroxy-5-methyl-6-oxohexa-2,4-dienoic acid). The 2,6-DNT pathway intermediates are 2,6-DNT, 3M4NC, HNOHA (2-hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid), and HNPA (2-hydroxy-5-nitropenta-2,4-dienoic acid).
Enzyme assays with crude and partially purified cell extracts revealed the presence of an extradiol ring cleavage dioxygenase and a hydrolase that catalyzed reactions subsequent to the initial dioxygenation of 2,6-DNT. The nitro group of 3M4NC is eliminated by unknown reactions subsequent to ring fission. In contrast, the nitro group of 4M5NC is eliminated prior to ring cleavage in the 2,4-DNT degradative pathway (8, 28). Elimination of the nitro group prior to ring fission is also the case in pathways for degradation of 4-nitrophenol (12, 27), nitrobenzene (20), 2-nitrotoluene (10), 3-nitrotoluene (1), and 3-nitrobenzoate (4, 19). Only a few other pathways have been reported in which a nitroaliphatic compound is the result of ring cleavage. Rhodococcus erythropolis HL 24-1 and HL 24-2 produced the dead-end metabolite 4,6-dinitrohexanoate during growth on 2,4-dinitrophenol (17). Later studies showed the reductive production of analogous dead-end metabolites when the strains were grown on substituted 2,4-dinitrophenols (16). Rhodococcus sp. strain RB1 grew on 2,4-dinitrophenol via 3-nitroadipate (2). The authors could not eliminate the possibility that 4,6-dinitrohexanoate was produced by cleavage of a hypothetical Meisenheimer intermediate and was then oxidized to 3-nitroadipate. Alcaligenes eutrophus JMP 134 and A. eutrophus JMP 222 grew on 2,6-dinitrophenol, with the stoichiometric release of nitrite (6). The key intermediate in the pathway is 4-nitropyrogallol, which serves as the ring fission substrate. The ring of 4-nitropyrogallol was opened between the 2- and 3-hydroxy positions to yield 2-hydroxy-5-nitromuconic acid. The steps leading to the subsequent elimination of the second nitro group have not been determined for the Alcaligenes strains. 2-Hydroxy-5-nitropenta-2,4-dienoic acid is thought to be a dead-end product of spontaneous decarboxylation of the nitromuconic acid. In contrast to the above pathways, 2-hydroxy-5-nitropenta-2,4-dienoic acid synthesis is enzyme catalyzed in the 2,6-DNT pathway. The fact that 2-hydroxy-5-nitropenta-2,4-dienoic acid has not been detected in the culture fluids during growth on 2,6-DNT suggests that it is subject to further productive metabolism by 2,6-DNT-degrading bacteria.
Catechol undergoes meta-cleavage to 2-hydroxy-6-oxohexa-2,4-dienoic acid (hydroxymuconic semialdehyde), which can be converted to 2-oxopent-4-enoic acid in two ways (18). An NAD+-dependent dehydrogenase can oxidize the hydroxymuconic semialdehyde to the enol form of 4-oxalocrotonate which is converted to the keto form by the action of a tautomerase. The keto compound is enzymatically decarboxylated to 2-oxopent-4-enoic acid. Alternatively, the hydroxymuconic semialdehyde may be directly converted to 2-oxopent-4-enoic acid by the action of a hydrolase. In contrast, in the 3-methylcatechol and the 2,6-DNT degradative pathways, the hydroxymuconic semialdehyde analogs have methyl group substituents on the 6-carbon so that the oxo groups exist as ketones rather than aldehydes and cannot be acted upon by a dehydrogenase. Thus, direct enzymatic hydrolysis of 2-hydroxy-4-nitro-6-oxohepta-2,4-dienoic acid with loss of acetate is the only plausible route to 2-hydroxy-5-nitropenta-2,4-dienoic acid. The additional observation that cell extracts converted 3M4NC only to 2-hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid at pH 5.0 is consistent with inhibition of a hydrolase. The aromatic ring cleavage product hydrolases are serine hydrolases, members of the αβ hydrolase fold family (5). Enzymes of this family require the deprotonation of a serine to generate a nucleophilic residue in the active site (22). At acidic pH, the active-site serine would remain protonated, reducing the catalytic efficiency of the hydrolase.
Crude cell extracts from induced cultures of JS850 and JS863 eliminate the nitro group from nitroalkanes (nitroethane and nitromethane) without any additional cofactors (data not presented), but the activity remains to be linked to the release of nitrite from a nitroaliphatic intermediate in the 2,6-DNT catabolic pathway. If the activity is indeed part of the 2,6-DNT pathway, then nitrite release is at least two enzyme reactions down the pathway from 2-hydroxy-5-nitropenta-2,4-dienoic acid production. That is, transformation of 2-hydroxy-5-nitropenta-2,4-dienoic acid which has not been detected in cell extracts would require at least one reaction to yield a product that the nitro group-eliminating activity could then act upon.
Based on the above results, we propose the following pathway for 2,6-DNT degradation (Fig. 7B). Dioxygenase attack at either of the nitro groups converts 2,6-DNT to 3M4NC with the elimination of nitrite. The aromatic ring is opened by an extradiol ring cleavage dioxygenase resulting in 2-hydroxy-5-nitro-6-oxohepta-2,4-dienoic acid. By analogy to the 3-methylcatechol meta-ring cleavage pathway, hydrolytic attack would produce 2-hydroxy-5-nitropenta-2,4-dienoic acid accompanied by the loss of acetate.
Previous work demonstrated that the highly specific 4M5NC monooxygenase from the 2,4-DNT pathway does not attack 3M4NC (9). The 3M4NC-2,3-dioxygenase from the 2,6-DNT pathway also appears to be highly specific for 3M4NC, with the only other substantial activity being against 3-methylcatechol. Because there was no detectable activity with 4-nitrocatechol, it seems that the methyl group in the 3-position is the determining factor in substrate recognition by the enzyme. In addition, 3-methyl-6-nitrocatechol was not attacked, which suggests that compounds that are substituted at the 6-position are not recognized by the enzyme.
3M4NC was initially identified in 2,4-DNT-degrading cultures that were incubated with 2,6-DNT. This result was not surprising, as we had previously reported the limited ability of the 2,4-DNT-degrading strain DNT to transform other nitrotoluenes (28). Additionally, the genes that encode the α subunit of the nitroarene dioxygenases have been shown to share a striking degree of nucleotide sequence similarity (G. R. Johnson and J. C. Spain, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. Q-344, p. 512, 1997). We were able to use a cloned 2,4-DNT dioxygenase gene to synthesize substantial quantities of 3M4NC from 2,6-DNT, but 2,4-DNT was clearly the preferred substrate. Although the cloned genes were overexpressed, the yields were low, generally around 30%, perhaps indicating that only subtle changes in amino acid sequence are required to affect substrate specificity or perhaps indicating that some crucial component of the regulatory system was missing in the clone.
The above observation and the puzzling question of why DNT persists in environments known to harbor effective DNT-degrading organisms highlight our lack of understanding of the induction and regulation of the DNT degradative pathways. A related question concerns the evolutionary origin and distribution of the genes involved in DNT degradation. The genes for the initial dioxygenases involved in 2,4- and 2,6-DNT degradation are all closely related, even though the distribution of the organisms harboring the genes is quite discontinuous. And while the initial dioxygenases are closely related, the remainder of the pathways are clearly different. The difference raises the question of why divergent pathways evolved to degrade the two isomers. We are currently working to determine the mechanisms involved in regulating the degradation of mixtures of DNT.
ACKNOWLEDGMENTS
We thank Joe Hughes and Chaun Yue Wang for helpful discussions; Sol Resnick, Joe Wander, and Ronald Spanggord for insight into interpretation of NMR and LC-MS spectra; and Ronald Spanggord for many helpful suggestions for synthesis of metabolite standards.
This work was supported in part by the Air Force Office of Scientific Research and the Strategic Environmental Research and Development Program Federal Integrated Biotreatment Research Consortium. G.C.P. acknowledges the support of a National Research Council Postdoctoral Research Associateship.
REFERENCES
- 1.Ali-Sadat S, Mohan K S, Walia S K. A novel pathway for the biodegradation of 3-nitrotoluene in Pseudomonas putida. FEMS Microbiol Ecol. 1995;17:169–176. [Google Scholar]
- 2.Blasco R, Moore E, Wray V, Pieper D, Timmis K, Castillo F. 3-Nitroadipate, a metabolic intermediate for mineralization of 2,4-dinitrophenol by a new strain of a Rhodococcus species. J Bacteriol. 1999;181:149–152. doi: 10.1128/jb.181.1.149-152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruhn C, Lenke H, Knackmuss H-J. Nitrosubstituted aromatic compounds as nitrogen source for bacteria. Appl Environ Microbiol. 1987;53:208–210. doi: 10.1128/aem.53.1.208-210.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartwright N J, Cain R B. Bacterial degradation of nitrobenzoic acids. Biochem J. 1959;71:248–261. doi: 10.1042/bj0710248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Díaz E, Timmis K N. Identification of functional residues in a 2-hydroxymuconic semialdehyde hydrolase. J Biol Chem. 1995;11:6403–6411. doi: 10.1074/jbc.270.11.6403. [DOI] [PubMed] [Google Scholar]
- 6.Ecker S, Widmann T, Lenke H, Dickel O, Fischer P, Bruhn C, Knackmuss H-J. Catabolism of 2,6-dinitrophenol by Alcaligenes eutrophus JMP134 and JMP222. Arch Microbiol. 1992;158:149–154. [Google Scholar]
- 7.Flaig V W, Salfeld J-C. UV-Spektren und Konstitution von p-Benzochinonen. Justus Liebigs Ann Chem. 1958;618:117–139. [Google Scholar]
- 8.Haigler B E, Nishino S F, Spain J C. Biodegradation of 4-methyl-5-nitrocatechol by Pseudomonas sp. strain DNT. J Bacteriol. 1994;176:3433–3437. doi: 10.1128/jb.176.11.3433-3437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haigler B E, Suen W-C, Spain J C. Purification and sequence analysis of 4-methyl-5-nitrocatechol oxygenase from Burkholderia sp. strain DNT. J Bacteriol. 1996;178:6019–6024. doi: 10.1128/jb.178.20.6019-6024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigler B E, Wallace W H, Spain J C. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl Environ Microbiol. 1994;60:3466–3469. doi: 10.1128/aem.60.9.3466-3469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horan N J. Biological wastewater treatment systems: theory and operation. Chichester, United Kingdom: John Wiley & Sons; 1990. [Google Scholar]
- 12.Jain R K, Dreisbach J H, Spain J C. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl Environ Microbiol. 1994;60:3030–3032. doi: 10.1128/aem.60.8.3030-3032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keith L H, Telliard W A. Priority pollutants I—a perspective view. Environ Sci Technol. 1979;13:416–423. [Google Scholar]
- 14.Klecka G M, Gibson D T. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl Environ Microbiol. 1981;41:1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lendenmann U, Spain J C, Smets B F. Simultaneous biodegradation of 2,4-dinitrotoluene and 2,6-dinitrotoluene in an aerobic fluidized-bed biofilm reactor. Environ Sci Technol. 1998;32:82–87. doi: 10.1002/(sici)1097-0290(19990620)63:6<642::aid-bit2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 16.Lenke H, Knackmuss H-J. Initial hydrogenation and extensive reduction of substituted 2,4-dinitrophenols. Appl Environ Microbiol. 1996;62:784–790. doi: 10.1128/aem.62.3.784-790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenke H, Pieper D H, Bruhn C, Knackmuss H-J. Degradation of 2,4-dinitrophenol by two Rhodococcus erythropolis strains, HL 24-1 and HL 24-2. Appl Environ Microbiol. 1992;58:2928–2932. doi: 10.1128/aem.58.9.2928-2932.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray K, Duggleby C J, Sala-Trepat J M, Williams P A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972;28:301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- 19.Nadeau L J, Spain J C. Bacterial degradation of m-nitrobenzoic acid. Appl Environ Microbiol. 1995;61:840–843. doi: 10.1128/aem.61.2.840-843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino S F, Spain J C. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl Environ Microbiol. 1995;61:2308–2313. doi: 10.1128/aem.61.6.2308-2313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino S F, Spain J C, Lenke H, Knackmuss H-J. Mineralization of 2,4- and 2,6-dinitrotoluene in soil slurries. Environ Sci Technol. 1999;33:1060–1064. [Google Scholar]
- 22.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, Sussman J L, Verschueren K H G, Goldman A. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 23.Popp J A, Leonard T B. The hepatocarcinogenicity of dinitrotoluenes. In: Rickert D E, editor. Toxicity of nitroaromatic compounds. Washington, D.C.: Hemisphere Publishing Corporation; 1985. pp. 53–60. [Google Scholar]
- 24.Smibert R M, Krieg N R. Phenotypic characterization. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 607–654. [Google Scholar]
- 25.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 26.Spain J C. Bacterial degradation of nitroaromatic compounds under aerobic conditions. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Publishing Corp.; 1995. pp. 19–35. [Google Scholar]
- 27.Spain J C, Gibson D T. Pathway for biodegradation of p-nitrophenol in a Moraxella sp. Appl Environ Microbiol. 1991;57:812–819. doi: 10.1128/aem.57.3.812-819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1: nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 30.Zylstra G J, McCombie W R, Gibson D T, Finette B A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988;54:1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]