Abstract
Recently, Cangelosi and Brabant used oligonucleotide probes targeting the precursor 16S rRNA of Escherichia coli to demonstrate that the levels of precursor rRNA were more sensitive to changes in growth phase than the levels of total rRNA (G. A. Cangelosi and W. H. Brabant, J. Bacteriol. 179:4457–4463, 1997). In order to measure changes in the levels of precursor rRNA in activated sludge systems, we designed oligonucleotide probes targeting the 3′ region of the precursor 16S rRNA of Acinetobacter spp. We used these probes to monitor changes in the level of precursor 16S rRNA during batch growth of Acinetobacter spp. in Luria-Bertani (LB) medium, filtered wastewater, and in lab- and full-scale wastewater treatment systems. Consistent with the previous reports for E. coli, results obtained with membrane hybridizations and fluorescence in situ hybridizations with Acinetobacter calcoaceticus grown in LB medium showed a more substantial and faster increase in precursor 16S rRNA levels compared to the increase in total 16S rRNA levels during exponential growth. Diluting an overnight culture of A. calcoaceticus grown in LB medium with filtered wastewater resulted in a pattern of precursor 16S rRNA levels that appeared to follow diauxic growth. In addition, fluorescence in situ hybridizations with oligonucleotide probes targeting total 16S rRNA and precursor 16S rRNA showed that individual cells of A. calcoaceticus expressed highly variable levels of precursor 16S rRNA when adapting from LB medium to filtered sewage. Precursor 16S rRNA levels of Acinetobacter spp. transiently increased when activated sludge was mixed with influent wastewater in lab- and full-scale wastewater treatment systems. These results suggest that Acinetobacter spp. experience a change in growth activity within wastewater treatment systems.
Oligonucleotide hybridization probes targeting the small subunit rRNA (16S and 16S-like rRNAs) and the larger rRNA of the large ribosomal subunit (23S and 23S-like rRNAs) have made it possible to determine the composition of microbial communities and to estimate the activity of microbial populations in numerous environments, including activated sludge wastewater treatment systems (for recent reviews, see references 5 and 26). For example, fluorescence in situ hybridization (FISH) has been used to quantify the biomass of specific filamentous microorganisms in activated sludge using a relationship between the number and length of individual target cells and biomass concentration (10). In addition, membrane hybridizations have been used to measure the activity of microbial populations in foaming activated sludge (11) and in activated sludge systems operated for enhanced biological phosphorus removal (20). Although these approaches may not work for all microbial populations—such as metabolically active microorganisms with low rRNA content (19) and metabolically inactive microorganisms with a high residual rRNA content (25, 32)—oligonucleotide hybridization probes targeting rRNA have been used successfully to study the microbial ecology of activated sludge (5).
To take full advantage of engineering efforts in rational design and optimization of activated sludge systems, results from oligonucleotide probe hybridizations remain to be interfaced with representations of microbial biomass and activity used in mathematical models of microbial growth and substrate utilization (21). This will only become possible when rapid and reliable methods for quantifying changes in the activity of microbial populations become available. One approach, initially suggested by DeLong and coworkers (12), is based upon determining the relationship between cellular ribosome content and growth rate of a target population. When this information is available, FISH signal intensity can be quantified with digital microscopy to determine the in situ cellular growth rate in environmental samples. This approach has been used to estimate the in situ cellular growth rates of a sulfate-reducing bacterium in biofilms (23), of Pseudomonas fluorescens in environmental mesocosms (6), of Pseudomonas strain B13(FR1) in marine microcosms (16), and of Pseudomonas putida in biofilms (18). Results of FISH targeting rRNA have been combined with a model of microbial growth and substrate utilization to study the zero-order degradation of toluene by P. putida (18, 22).
Recently, Cangelosi and Brabant (8) suggested an alternative approach for measuring in situ growth activity. Instead of using oligonucleotide hybridization probes to quantify the levels of rRNA, they designed oligonucleotide probes to monitor the expression of rRNA genes. In most bacteria, the 16S, 23S, and 5S rRNAs and a variety of tRNA genes are transcribed from polycistronic rrn operons (for a review, see reference 29). RNase III subsequently cleaves the full-length transcript into precursor RNA fragments. Each precursor RNA fragment contains a region of nucleotides at the 5′ end followed by a single mature RNA and an additional region of nucleotides at the 3′ end (15). For example, whereas the mature 16S rRNA has 1,542 nucleotides, the precursor 16S rRNA from rrnB in Escherichia coli contains 1,731 nucleotides, with 146 nucleotides at the 5′ terminus of the mature 16S rRNA and 43 nucleotides at the 3′ terminus (7, 33).
Cangelosi and Brabant (8) used oligonucleotide probes targeting the 5′ and 3′ regions of the precursor 16S rRNA of E. coli to show that the levels of precursor 16S rRNA vary significantly over growth phases. Their results indicate that, at least for some microbial populations, quantification of the expression of rrn genes can be used in place of direct measurement of cellular rRNA to monitor microbial growth in situ. By combining oligonucleotide probes targeting 16S rRNA and precursor 16S rRNA, the microbial community composition and growth activities of specific microbial populations may be measured in the same sample. In this study, we report on the design and characterization of oligonucleotide probes targeting precursor 16S rRNA sequences of Acinetobacter spp. as a model system for evaluating this approach in activated sludge. Subsequently, we used these probes to monitor changes in precursor 16S rRNA and total 16S rRNA levels during batch growth of a pure culture of Acinetobacter calcoaceticus and to monitor growth activity of Acinetobacter spp. within lab- and full-scale activated sludge wastewater treatment systems.
MATERIALS AND METHODS
Batch growth of microorganisms and sampling.
A. calcoaceticus ATCC 23055T and E. coli ATCC 11775T were cultured aerobically in Luria-Bertani (LB) medium (27) at 35°C on rotary shakers. Overnight cultures were diluted 20-fold into fresh LB medium or wastewater filtered through a 0.2-μm-pore-size filter (Nalgene, Rochester, N.Y.). Each batch growth study was conducted in duplicate. At sampling time points, the optical density of the culture was determined spectrophotometrically at a wavelength of 600 nm (OD600). Two samples (2 ml each) from each batch culture were removed and centrifuged at 14,000 × g, and the supernatant was decanted. One sample was immediately frozen in an ethanol-dry ice bath, and RNA was extracted by a bead-beating, low-pH, phenol-chloroform protocol (26, 30). The second sample was fixed in 4% paraformaldehyde for 1 h at room temperature and subsequently stored in 50% ethanol in phosphate-buffered saline (130 mM NaCl, 10 mM sodium phosphate buffer [pH 7.2]) at −20°C (9). To prepare pure cultures with a high fraction of precursor rRNA, overnight cultures of E. coli and A. calcoaceticus, diluted 20-fold into fresh LB medium, were treated with chloramphenicol (final concentration of 20 mg/liter) (Sigma, St. Louis, Mo.) after 1 h of growth at 35°C (17). The cultures remained at 35°C for 1 h, and samples were taken for extraction of RNA and fixation for FISH.
Probe synthesis and labeling.
For membrane hybridizations, oligonucleotide probes, synthesized by the University of Illinois Biotechnology Facility, were 5′ end labeled with [γ-32P]ATP (24). Oligonucleotides for FISH were conjugated with the cyanine dye Cy3 or Cy5 before purification with high-performance liquid chromatography (Interactive, Germany). Fluorescently labeled probes were diluted to 50 mg/liter with H2O and stored in 50-μl aliquots at −20°C in the dark. Probe and target sequences are listed in Table 1.
TABLE 1.
Oligonucleotide hybridization probes and target sequences
| Target organism or group and probea | Targetb (3′–5′) or probe (5′–3′) sequence | Source or reference |
|---|---|---|
| E. coli rrnA, -C, -D, -E, -F, -G, and -Hc | 3′ ACUCGUGACGUUUCAUGCGAAGAAAUUCCAUU 5′ | |
| E. coli rrnBc | 3′ ........U....................AUU 5′ | |
| S-S-E.coli-1543-a-A-24 | 5′ GCACTGCAAAGTACGCTTCTTTAA 3′ | This study |
| S-S-E.coli-1543-b-A-24 | 5′ GCACTACAAAGTACGCTTCTTTAA 3′ | This study |
| A. calcoaceticusd | 3′ CACCUAAGAAUGGUUAGCAGUUAGAAAGCAAUU 5′ | |
| A. baumanniie | 3′ ..............................AUU 5′ | |
| Acinetobacter sp.f | 3′ ..............................AUU 5′ | |
| Acinetobacter sp.g | 3′ ..............................AUU 5′ | |
| S-G-Acin-1543-a-A-24 | 5′ GATTCTTACCAATCGTCAATCTTT 3′ | This study |
| Acinetobacter spp., S-G-Acin-0659-a-A-24 | 5′ CTGGAATTCTACCATCCTCTCCCA 3′ | 20 |
| Domain Bacteria, S-D-Bact-0338-a-A-18 | 5′ GCTGCCTCCCGTAGGAGT 3′ | 3 |
| Almost all organisms, S-*-Univ-1390-a-A-18 | 5′ GACGGGCGGTGTGTACAA 3′ | 34 |
Named according to the nomenclature suggested in the Oligonucleotide Probe Database (1).
Periods in succeeding sequences indicate identical nucleotides.
Sequence from accession no. J01695; underlined sequence is mature 16S rRNA.
Sequence from accession no. U60278; underlined sequence is mature 16S rRNA.
Sequence from accession no. U60279; underlined sequence is mature 16S rRNA.
Sequence from Acinetobacter strain ATCC 19004, accession no. U60280; underlined sequence is mature 16S rRNA.
Sequence from Acinetobacter strain ATCC 17903, accession no. U60281; underlined sequence is mature 16S rRNA.
Preparation of recombinant rDNA plasmids.
Genomic DNA was extracted from overnight cultures of E. coli and A. calcoaceticus by a microwave lysis procedure (11). PCR amplification of near-complete 16S ribosomal DNA (rDNA) and the intergenic spacer region was conducted with primer pair S-D-Bact-0011-a-S-17 (5′-GTTTGATCCTGGCTCAG-3′) and L-Sc-gProt-1207-a-A-17 (5′-GCCTTCCCACATCGTTT-3′) using standard methods (11). The PCR products were subcloned with a TA Cloning Kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Recombinant plasmids were recovered with a lysozyme and sodium dodecyl sulfate (SDS) lysis and a phenol-chloroform extraction (27). Endonuclease restriction digests using EcoRI confirmed correct recombinants, and the recombinant rDNA plasmids were stored at −20°C in Tris-EDTA buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA [pH 8.0]).
Determination of optimal wash temperature.
For temperature of dissociation (Td) studies, total RNA extracted from chloramphenicol-treated cells and recombinant rDNA plasmid were denatured in 3 volumes of 2% glutaraldehyde, diluted to 0.5 mg/liter with dilution water (24) without poly(A), and 50 ng of each nucleic acid sample was applied by slot blotting to Magna Charge membranes (Micron Separation, Inc., Westboro, Mass.) (24). The membranes were baked for 2 h at 80°C, prehybridized for 2 h at 40°C, hybridized for 12 h at 30°C, and initially washed twice with 100 ml of wash buffer (1% SDS and 1× SSC [0.15 M NaCl plus 0.015 M sodium citrate]) for 1 h each at 30°C (34). The Td for each probe was experimentally determined by an elution method as previously described (34).
Quantitative membrane hybridizations.
For membrane hybridizations, RNA extracts were denatured as described above, 200 ng of RNA was applied in triplicate to Magna Charge membranes, and the membranes were baked for 2 h at 80°C (24). Prehybridizations were performed at 40°C for 2 to 6 h, and hybridizations took place at 30°C for 12 to 14 h. Membranes were initially washed twice for 1 h each in 100 ml of wash buffer at 30°C. Stringent washing was conducted for 30 min in 500 ml of fresh wash buffer at the experimentally determined wash temperature. The hybridization signal was quantified with an Electronic Autoradiography Instant Imager (Packard Instrument Company, Meriden, Conn.).
To calculate the average normalized hybridization signal, the following procedure was used. (i) Hybridization signals from triplicate slots of a single sample were averaged. (ii) The average hybridization signals were normalized per unit biomass by multiplying the average hybridization signal by the amount of RNA extracted from the sample and dividing this value by the amount of biomass per sample (biomass was expressed as OD600 or as the mass of suspended solids [SS]). (iii) Hybridization signals per unit biomass were normalized using the maximum hybridization signal per unit biomass measured in each replicate growth study. (iv) The normalized results from the replicate studies were averaged.
Standard deviations were calculated using the law of propagation of errors. Thus, the standard deviation (ςα/β) of α/β was ςα/β = (α/β) × [(ςα/α)2 + (ςβ/β)2]0.5, where ςα and ςβ are the standard deviations of the measurements of α and β, respectively.
The ratio of precursor 16S rRNA to total 16S rRNA (where total 16S rRNA is defined as the sum of precursor 16S rRNA and mature 16S rRNA) was calculated using a dilution series of RNA extracted from chloramphenicol-treated pure cultures of E. coli or A. calcoaceticus. The same dilution series was applied to the membranes hybridized with the precursor 16S rRNA and total 16S rRNA probe (probe S-*-Univ-1390-a-A-18 was used for the analysis of samples of pure cultures of E. coli and A. calcoaceticus, while probe S-G-Acin-0659-a-A-24 was used for the analysis of samples of activated sludge). Hybridization signals for samples hybridized with the precursor 16S rRNA probe or the total 16S rRNA probe were related to the dilution series located on the respective membranes. The use of an identical dilution series for both membranes permitted the calculation of the ratio of precursor 16S rRNA to total 16S rRNA.
Determination of optimal formamide stringency and FISH.
Chloramphenicol-treated cultures, fixed in 4% paraformaldehyde, were applied in a sample well on a Heavy-Teflon-Coated microscope slide (Cel-Line Associates, New Field, N.J.) and air dried. After dehydration with an increasing ethanol series (50, 80, and 95% [vol/vol] ethanol, 1 min each), each sample well was covered with 9 μl of hybridization buffer (0% [vol/vol] to 70% [vol/vol] formamide, 0.9 M NaCl, 100 mM Tris-HCl [pH 7.2], 0.1% SDS). A 1-μl portion of fluorescently labeled oligonucleotide probe (50 ng) was added to each well of the microscope slide. Hybridizations were conducted in a moisture chamber (4) for 1 to 2 h, in the dark, at 46°C. The slides were washed for 15 min at 48°C with 50 ml of prewarmed wash solution (500 to 20 mM NaCl, 20 mM Tris-HCl [pH 7.2], 0.1% SDS, 5 mM EDTA). Samples were counterstained with ice-cold, fresh 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) staining solution (1 mg of DAPI/liter) for 2 min, rinsed with ice-cold water, and rapidly air dried. Fixed, hybridized cells were mounted with a 1:4 (vol/vol) mixture of VectaShield (Vector Laboratories, Burlingame, Calif.) and Citifluor (Citifluor, Ltd., London, United Kingdom) immersion solutions and a coverslip. The optimal formamide stringency was determined with digital microscopy as previously described (9) with the modifications outlined below. Subsequent FISH experiments were performed using the optimal stringency.
Probe-conferred fluorescence was visualized with a Zeiss Axioplan 2 epifluorescence microscope (Carl Zeiss, Jena, Germany) equipped with a high-quality dichromatic filter set for green excitation (Chroma HQ 41007; Chroma Technology, Brattleboro, Vt.) using 50% intensity of the 100-W mercury lamp (AttoArk; Atto Instruments, Rockville, Md.). Between 10 and 30 digital images (12 bit, 4,096 possible gray values per pixel) were captured and stored on a personal computer with a Peltier-cooled slow-scan charge-coupled device camera (1317×1035 pixel array; SPOT; Diagnostic Instruments, Sterling Heights, Mich.) using the Windows-based image analysis software MetaMorph (Universal Imaging Corp., West Chester, Pa.). Image processing consisted of Unsharp Masking and subsequent median filtering (kernel size, 3×3 pixel) to reduce noise. The former process makes use of an edge detection algorithm that subtracts a downscaled (in our case, ×0.95 original brightness) low-pass filter (kernel size, 16×16 pixel) from the original image resulting in a “smoothed” image. Each pixel in the smoothed image is then multiplied by the reciprocal value of the relative difference between the downscaled image and the original image (in our case, 20-fold) in order to reestablish the original brightness range. Subsequently, images processed in this fashion were binarized by automatically setting a fixed threshold for each series. This binary mask was used to record the mean gray values of objects within the original images. Objects containing fewer than 100 pixels were discarded as dirt (e.g., salt crystals). Small cell aggregates that could not be split by edge detection were processed as single objects. A simple macro allowed the automatic evaluation of multiple image series without further user interference. Between 51 and 2,493 objects per series (mean, 408) were evaluated. Digital images of dual hybridizations were recorded with a laser scanning confocal microscope (LSM510; Carl Zeiss) and manipulated with LSM510 software version 2.0.2.
The average normalized mean gray value per object was calculated using the following procedure, and standard deviations were determined as described for quantitative membrane hybridizations above. (i) The average of gray values per object was determined for a single sample. (ii) Mean gray values were normalized using the maximum mean gray value measured in each replicate growth study. (iii) The normalized results from the replicate studies were averaged.
Lab-scale activated sludge wastewater treatment systems.
Three lab-scale activated sludge wastewater treatment systems, one completely mixed activated sludge (CMAS) system, and two sequencing batch reactors (SBR-1 and SBR-2) were operated with a hydraulic retention time (HRT) of 10 h and a mean cell residence time (MCRT) of 5 days. The CMAS system consisted of an aeration basin of 10 liters and a clarifier of 3 liters. This system was operated with an influent flow rate of 1.3 liter/h and a recycle ratio of 0.25. The mixed liquor volatile suspended solids (MLVSS) in the CMAS system was 987 mg/liter, and the food-to-microorganism ratio (F/M) was 0.11 mg of soluble chemical oxygen demand (S-COD) mg of volatile suspended solids (VSS)−1 day−1. SBR-1 was operated with an 8-h cycle time consisting of 20 min of filling with aeration, 340 min of aeration, 100 min of settling, and 20 min of decanting (4 liters of effluent were decanted). SBR-2 was operated with a 4-h cycle time consisting of 10 min of filling with aeration, 170 min of aeration, 50 min of settling, and 10 min of decanting (2 liters of effluent were decanted). The MLVSS in SRB-1 and SBR-2 were 673 mg/liter, and the F/M was 0.162 mg of S-COD mg of VSS−1 day−1. The systems were seeded with activated sludge from the Urbana-Champaign Sanitary District, Northeast Wastewater Treatment Plant (UCSD, NEWWTP), and were used to treat primary clarifier effluent collected from the UCSD, NEWWTP. After 20 days of operation, the systems were sampled over an 8-h period. The MLVSS levels were determined according to standard methods (12), and the S-COD levels of the influent and mixed liquor were determined with Hach Tests (Hach, Loveland, Colo.) according to the manufacturer's instructions.
Full-scale activated sludge wastewater treatment system.
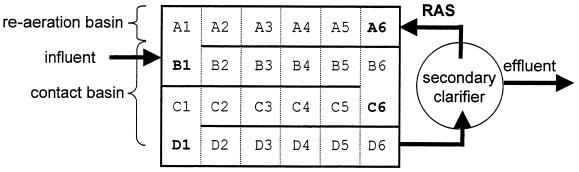
The UCSD, NEWWTP, treats on average 5.7 × 104 m3/day (15 million gallons/day) of municipal wastewater. The treatment plant reduces the average influent 5-day biochemical oxygen demand (BOD5) of 150 mg of BOD5/liter by 30% with primary clarification and by an additional 65% in a contact stabilization activated sludge system (T. Bachman, personal communication). The plant was operated with a target MCRT of 3.5 days and an F/M of approximately 0.2 g of BOD5 g of SS−1 day−1. The average mixed liquor suspended solids (MLSS) concentration in the contact basin was 1.1 g/liter. The typical return activated sludge (RAS) SS concentration was 5 g/liter, which resulted in an MLSS concentration of approximately 4.5 g/liter in the re-aeration basin. Figure 1 shows a schematic layout of the UCSD, NEWWTP. Samples were collected from across the contact basin (sites B1, C6, and D1, Fig. 1), from the re-aeration basin (site A6, Fig. 1), and from the RAS line (just before site A6, Fig. 1) at 9:00 h and 13:00 h on 24 September 1998 and at 8:00 h and 12:00 h on 10 October 1998.
FIG. 1.
Schematic of the UCSD, NEWWTP. Samples from the contact basin were removed from sites B1, C6, and D1. The re-aeration basin was sampled at site A6, while the RAS line was sampled immediately before the re-aeration basin.
RESULTS
Probe specificity and hybridization stringency.
Probes S-S-E.coli-1543-a-A-24 and S-S-E.coli-1543-b-A-24 were designed to target within the 3′ region of the precursor 16S rRNA of operons rrnA, -C, -D, -E, -F, -G, and -H and rrnB of E. coli, respectively. Probe S-G-Acin-1543-a-A-24 was designed to target within the 3′ region of the precursor 16S rRNA of the Acinetobacter genus. An advanced BLAST 2.0 (2) comparison of the oligonucleotide probe sequences, shown in Table 1, with the GenEMBL nonredundant nucleotide database was used to check the specificity of the probes. The BLAST analysis predicted that the oligonucleotide probes would hybridize with all intended targets. In addition, the analysis showed that the probes had at least two mismatches with nontarget sequences (with the exception of interoperon sequence similarity observed in E. coli).
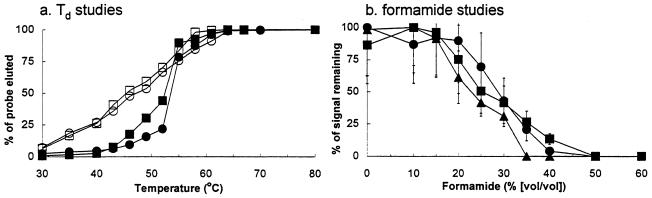
Results from the experimental determination of hybridization stringency for membrane hybridizations and FISH are shown in Fig. 2. The Td values, defined as the temperature at which 50% of the total amount of hybridized probe is washed off the membrane using an elution method (34), for probe S-S-E.coli-1543-a-A-24 were 45.5°C with rDNA and 52.4°C with RNA (Fig. 2a). The Td values for probe S-G-Acin-1543-a-A-24 were 47.2°C with rDNA and 53.5°C with RNA (Fig. 2a). The final wash temperature for subsequent hybridizations with these two probes was 53°C. The optimal formamide stringencies for oligonucleotide probes used for FISH were determined as the percentage of formamide that reduced maximum probe conferred fluorescent signal by 50%. Using pure cultures of E. coli and A. calcoaceticus, it was determined that the optimal formamide stringency was approximately 25% for the three probes examined (Fig. 2b).
FIG. 2.
Td studies for membrane hybridizations (a) and formamide stringency studies for FISH (b). The Td curves for probes S-S-E.coli-1543-a-A-24 (squares) and S-G-Acin-1543-a-A-24 (circles) were obtained using RNA extracted from chloramphenicol-treated E. coli and A. calcoaceticus (filled symbols) and recombinant rDNA plasmids (open symbols). Formamide stringency for probes S-S-E.coli-1543-a-A-24 (▴), S-S-E.coli-1543-b-A-24 (■), and S-G-Acin-1543-a-A-24 (●) was obtained using chloramphenicol-treated E. coli and A. calcoaceticus fixed in 4% paraformaldehyde.
Batch growth of E. coli in LB medium.
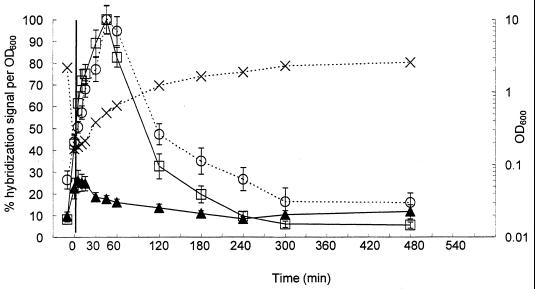
Figure 3 shows the changes in precursor 16S rRNA and total 16S rRNA during batch growth of E. coli in LB medium determined using membrane hybridizations. E. coli was diluted 20-fold into fresh LB medium from an overnight culture. The OD600 of the overnight culture was 2.19 (Fig. 3). After diluting the culture, the OD600 was determined to be 0.17. During the first 120 min of exponential growth, the OD600 increased from 0.17 to 1.24. As the cultures entered stationary phase, the OD600 increased from 1.24 to 2.59 after 480 min.
FIG. 3.
Results from membrane hybridizations of samples from batch growth of E. coli in LB medium. Probe S-S-E.coli-1543-a-A-24 was used to measure precursor 16S rRNA (□), and probe S-*-Univ-1390-a-A-18 was used to measure total 16S rRNA (○). The ratio of precursor 16S rRNA to total 16S rRNA (▴) was determined using a reference dilution series of RNA extracted from chloramphenicol-treated E. coli. Biomass was estimated by measuring the OD600 (×).
The average normalized hybridization signal obtained with the precursor 16S rRNA probe, S-S-E.coli-1543-a-A-24, reached a maximum 45 min after transfer of the culture to fresh LB medium (Fig. 3). At this time, the level of precursor 16S rRNA per unit biomass had increased approximately 12-fold since the transfer. Probe S-*-Univ-1390-a-A-18 was used to monitor the level of total 16S rRNA. A comparison of the normalized hybridization signal obtained with S-*-Univ-1390-a-A-18 for the overnight culture with the signal corresponding to the maximum normalized hybridization response (45 min after inoculation into fresh LB medium) demonstrated a fourfold increase in the level of total 16S rRNA per unit biomass.
The ratio of precursor 16S rRNA to total 16S rRNA for the overnight culture was <10% (Fig. 3). Immediately following inoculation into fresh LB medium, the ratio of precursor 16S rRNA to total 16S rRNA increased to 25%. This increase was concurrent with the more extensive increase in the level of precursor 16S rRNA compared to the modest increase in the level of total 16S rRNA during the early exponential growth phase. As the cultures entered stationary phase, after 120 min of growth, the ratio of precursor 16S rRNA to total 16S rRNA returned to approximately 10%.
Samples from the batch growth of E. coli in LB medium were also analyzed using FISH with oligonucleotide probes targeting precursor 16S rRNA. Although the signal intensity of FISH with the precursor 16S rRNA probes was detectable, the differences in signal intensity for the various samples were not statistically significant (data not shown).
Batch growth of A. calcoaceticus in LB medium.
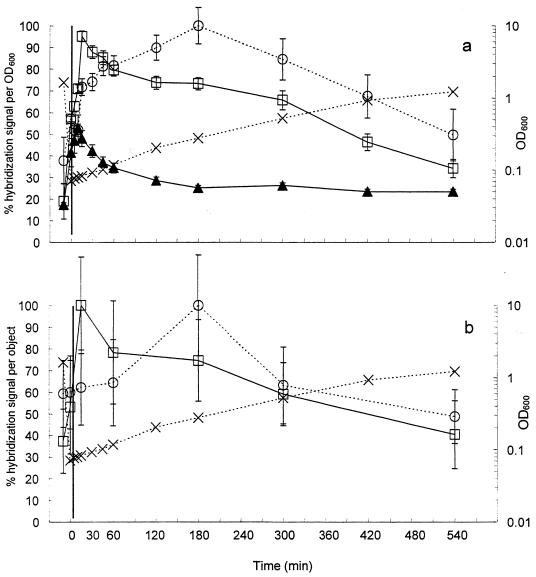
The results obtained with membrane hybridizations and FISH of batch cultures of A. calcoaceticus grown in LB medium are presented in Fig. 4a and 4b, respectively. The OD600 of the overnight culture, 1.64, was lowered to an OD600 of 0.07 after a 20-fold dilution into fresh medium (Fig. 4a). Throughout the study, the OD600 increased and reached a value of 1.22 after 540 min of growth.
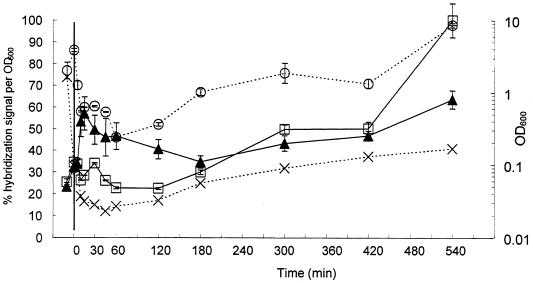
FIG. 4.
Results from membrane hybridizations (a) and FISH (b) of samples from batch growth of A. calcoaceticus in LB medium. Probe S-G-Acin-1543-a-A-24 was used to measure precursor 16S rRNA (□). For membrane hybridizations, probe S-*-Univ-1390-a-A-18 was used to measure total 16S rRNA (○), and for FISH, probe S-D-Bact-0338-a-A-18 was used to measure total 16S rRNA (○). The ratio of precursor 16S rRNA to total 16S rRNA (▴) was determined using a reference dilution series of RNA extracted from chloramphenicol-treated A. calcoaceticus. Biomass was estimated by measuring the OD600 (×).
Results from membrane hybridizations with the precursor 16S rRNA probe, S-G-Acin-1543-a-A-24, demonstrated an increase of approximately fivefold in the levels of precursor 16S rRNA after 15 min of incubation (Fig. 4a). In contrast, the use of probe S-*-Univ-1390-a-A-18, which targets total 16S rRNA, resulted in an increase of approximately 2.5-fold in the level of total 16S rRNA after 180 min of growth (Fig. 4a). The ratio of precursor 16S rRNA to total 16S rRNA rapidly increased from approximately 17% for the overnight culture, to a maximum of 53% 10 min after inoculation into fresh LB medium.
The results of the FISH show similar trends. The normalized hybridization signal with the precursor 16S rRNA probe, S-G-Acin-1543-a-A-24, increased approximately threefold during the first 15 min after transfer (Fig. 4b). The use of the total 16S rRNA probe, S-D-Bact-0338-a-A-18, resulted in less than a doubling in the level of total 16S rRNA after 180 min of growth (Fig. 4b).
Both membrane hybridization and FISH results indicated that the precursor 16S rRNA rapidly increased during the first 15 min of growth, while the total 16S rRNA reached a maximum only after 180 min of growth. In addition, as the OD600 returned to a level similar to the OD600 of the overnight culture, the levels of both precursor 16S rRNA and total 16S rRNA returned to levels similar to the overnight culture. The ratio of precursor 16S rRNA to total 16S rRNA calculated with the membrane hybridizations results decreased to 11% 540 min after transfer.
Batch growth of A. calcoaceticus in filtered wastewater.
An overnight culture of A. calcoaceticus, grown in LB medium, was diluted 20-fold with filtered (0.2-μm-pore-size filter) wastewater collected from the primary clarifier of the UCSD, NEWWTP. Results from measurements of OD600 and from membrane hybridizations with probes targeting precursor 16S rRNA and total 16S rRNA are presented in Fig. 5. The OD600 was 0.10 after the 20-fold dilution of the overnight culture into the filtered wastewater. During the first 45 min of growth, the OD600 decreased slightly to a minimum of 0.02 after 45 min. Subsequently, the OD600 increased to 0.17 540 min after the transfer, indicating an increase in the total biomass.
FIG. 5.
Results from membrane hybridizations of samples from batch growth of A. calcoaceticus in filtered wastewater. Probe S-G-Acin-1543-a-A-24 was used to measure precursor 16S rRNA (□), and probe S-*-Univ-1390-a-A-18 was used to measure total 16S rRNA (○). The ratio of precursor 16S rRNA to total 16S rRNA (▴) was determined using a reference dilution series of RNA extracted from chloramphenicol-treated A. calcoaceticus. Biomass was estimated by measuring the OD600 (×).
Changes in the levels of precursor 16S rRNA and total 16S rRNA were determined with membrane hybridizations using probes S-G-Acin-1543-a-A-24 and S-*-Univ-1390-a-A-18. Figure 5 shows that the normalized hybridization signal from precursor 16S rRNA increased slightly immediately following dilution with filtered wastewater and subsequently decreased to the lower levels observed in the overnight culture (with the exception of the data point obtained at 30 min). After 120 min of incubation, the levels of precursor 16S rRNA increased by a factor of 4 to reach a maximum after 540 min. Hybridization results with probe S-*-Univ-1390-a-A-18 showed that total 16S rRNA decreased from almost 90% to approximately 50% of the maximum signal from 0 to 60 min of batch growth (Fig. 5). Then, the levels of total 16S rRNA steadily increased, reaching a maximum after 540 min.
The ratio of precursor 16S rRNA to total 16S rRNA rapidly increased from 23 to >55% immediately following the transfer of the overnight culture to filtered wastewater. From 60 to 180 min, the ratio of precursor to total 16S rRNA decreased and subsequently increased to 64% after 540 min, concurrent with the more significant increase in precursor 16S rRNA compared to total 16S rRNA.
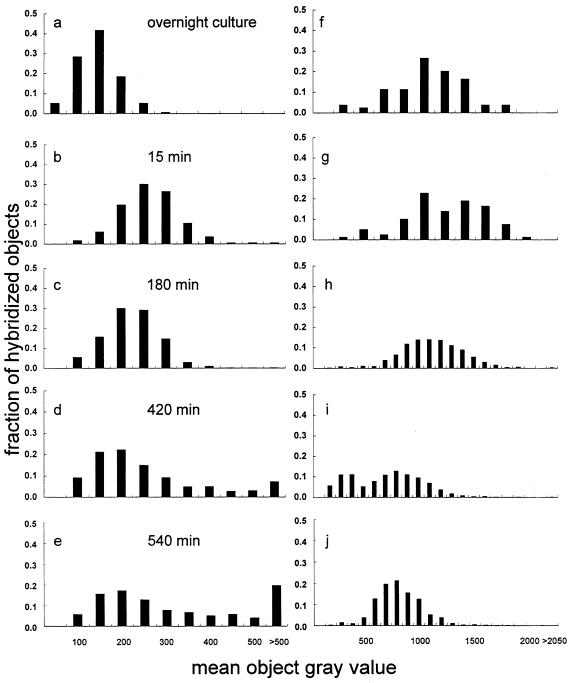
Figure 6 shows the results from FISH for growth of a culture of A. calcoaceticus in filtered wastewater. Results with the precursor 16S rRNA probe, S-G-Acin-1543-a-A-24, are shown in Fig. 6a to e, while results with the total 16S rRNA probe, S-D-Bact-0338-a-A-18, are shown in Fig. 6f to j. The precursor 16S rRNA results showed an upward shift in the distribution of the fraction of total hybridized objects. After 540 min of growth, approximately 20% of the total hybridized objects exhibited a hybridization signal with the precursor 16S rRNA probe greater than a mean object gray value of 500 (Fig. 6e). The total 16S rRNA results showed a slight downward shift in the distribution of the fraction of total hybridized objects from a mean object gray value of approximately 1,250 for the overnight culture (Fig. 6f) to a value of approximately 750 for the sample removed at 540 min (Fig. 6j).
FIG. 6.
Results from FISH of samples from the batch growth of A. calcoaceticus in filtered wastewater. The fraction of total hybridized objects with an average gray value between the value reported and the next lowest value are plotted in histogram format against mean object gray values. The fractions of objects hybridized with the precursor 16S rRNA probe, S-G-Acin-1543-a-A-24, are shown in panels a to e. Results with probe S-D-Bact-0338-a-A-18, which targets total 16S rRNA, are shown in panels f to j. Samples removed from the overnight culture are shown in panels a and f. Panels b and g, c and h, d and i, and e and j show samples removed after 15, 180, 420, and 540 min of growth, respectively.
Simultaneous FISH with oligonucleotide probes targeting precursor 16S rRNA and total 16S rRNA.
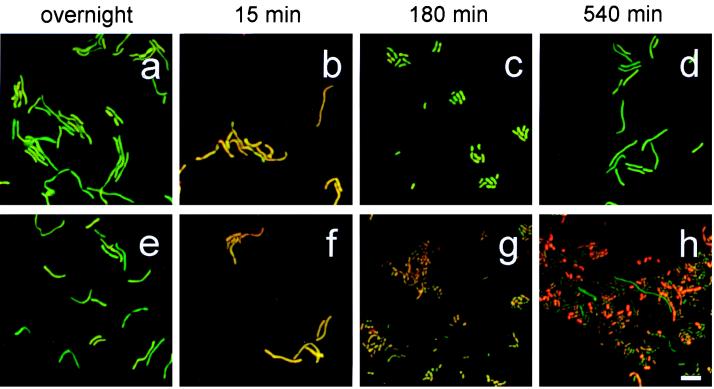
In Fig. 7, representative digital micrographs of samples of A. calcoaceticus transferred after overnight growth in LB medium to LB medium or to filtered wastewater are shown for various time points after transfer. The samples were simultaneously hybridized with probe S-G-Acin-1543-a-A-24, labeled with Cy5, and probe S-D-Bact-0338-a-A-18, labeled with Cy3. All digital micrographs were acquired using laser scanning confocal microscopy with constant settings (i.e., constant pinhole diameter, laser intensity, and electronic gain). Therefore, the hybridization signal from the precursor 16S rRNA probe, shown in red, and the hybridization signal from the total 16S rRNA probe, shown in green, can be compared for the images from the different samples. Figures 7a to d show images of A. calcoaceticus cultured in LB medium, while Fig. 7e to h are representative images of A. calcoaceticus cultured in filtered wastewater.
FIG. 7.
Representative digital micrographs from FISH with dual probes targeting precursor 16S rRNA (shown in red) and total 16S rRNA (shown in green). Results with samples from the batch growth of A. calcoaceticus in LB medium are shown in panels a to d. Panels e to h show results with samples from filtered wastewater. Samples removed from the overnight culture are shown in panels a and e. Panels b and f, c and g, and d and h show samples removed after 15, 180, and 540 min of growth, respectively. The bar is equal to 5 μm.
Dramatic morphologic plasticity was observed for A. calcoaceticus during batch growth in both LB medium and filtered wastewater. For instance, in overnight cultures (Fig. 7a and e), A. calcoaceticus exhibited a filamentous morphology with a characteristic length of approximately 10 μm per cell. In addition, some filamentous cells were arranged into longer filaments containing up to four cells. After 180 min, the dominant morphotype appeared to be diplococcoid cells with a characteristic length of approximately 1 to 2 μm per cell (Fig. 7c and g); A. calcoaceticus incubated in filtered wastewater was slightly smaller than A. calcoaceticus cultured in LB medium. After 540 min of growth, some A. calcoaceticus cells resumed growth in filamentous form (Fig. 7d and h). Filaments with a characteristic length of approximately 10 μm were the predominant morphotype in the culture grown in LB medium (Fig. 7d). In contrast, A. calcoaceticus cultured in filtered wastewater maintained a predominant diplococcoid morphology, although some filamentous cells were observed (Fig. 7h).
The simultaneous FISH with oligonucleotides targeting precursor 16S rRNA and total 16S rRNA allowed a direct comparison of the intracellular levels of precursor 16S rRNA and total rRNA. In Fig. 7, cells with a low ratio of precursor to total 16S rRNA have a characteristic green color. As the ratio of precursor to total 16S rRNA increased, individual cells appeared more orange. In agreement with the membrane hybridization (Fig. 4a and 5) and FISH (Fig. 4b and 6) results presented above, a comparison of cells from the overnight culture (Fig. 7a and e) with cells after 15 min of incubation (Fig. 7b and f) showed a dramatic increase in the ratio of precursor to total 16S rRNA. After 180 min of growth in LB medium, the ratio of precursor to total 16S rRNA decreased as cells changed from orange (Fig. 7b) to green (Fig. 7c). In contrast, A. calcoaceticus cultured in filtered wastewater maintained an elevated ratio of precursor to total 16S rRNA throughout batch growth (Fig. 5). Thus, most cells in Fig. 7g and h maintained an orange color.
Figure 7h supplies information in addition to the results obtained with membrane hybridizations (Fig. 5). As indicated in Fig. 6e and 7h, approximately 20% of the total cells exhibited a very strong hybridization signal with the precursor 16S rRNA probe, S-G-Acin-1543-a-A-24, after 540 min of growth. Close examination reveals that cells in Fig. 7h demonstrated a wide range of colors, from bright orange to green. These results support the observation that fluorescence intensity of the precursor 16S rRNA followed a bimodal distribution in Fig. 6e (i.e., in Fig. 6e, the mean object gray values of hybridization to precursor 16S rRNA for “dim” cells and “bright” cells were associated with mean object gray values of 200 and >500, respectively).
Precursor 16S rRNA and total 16S rRNA levels of Acinetobacter spp. in activated sludge wastewater treatment systems.
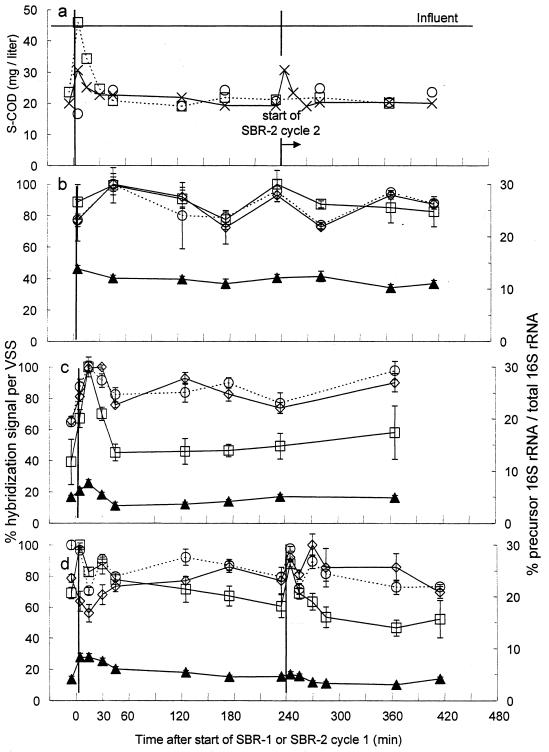
Figure 8a shows the results of S-COD analyses for the three lab-scale activated sludge systems. The S-COD concentrations in SBR-1 increased to the S-COD concentration of the influent when 4 liters of influent was pumped into the reactor at the start of the operating cycle and rapidly decreased within the first 45 min of operation. The S-COD concentrations in SBR-2 showed a similar pattern, with an increase in S-COD levels after the addition of 2 liters of influent, and a rapid consumption of S-COD within the first 45 min of aeration. As expected, the S-COD concentrations in the CMAS system were relatively constant.
FIG. 8.
S-COD concentrations in the influent wastewater and samples of mixed liquor from the lab-scale activated sludge wastewater treatment systems and results from membrane hybridizations with samples from the mixed liquor using oligonucleotide probes targeting precursor 16S rRNA and total 16S rRNA. The results of S-COD concentrations are shown in panel a: CMAS (○), SBR-1 (□), and SBR-2 (×). The results of membrane hybridizations with samples from the CMAS are shown in panel b, while the results with samples from SBR-1 and SBR-2 are shown in panels c and d, respectively. Precursor 16S rRNA was hybridized with probe S-G-Acin-1543-a-A-24 (□), while total 16S rRNA from Acinetobacter spp. and that from the total microbial community were measured with probes S-G-Acin-0659-a-A-24 (⧫) and S-*-Univ-1390-a-A-18 (○), respectively. The ratio of precursor 16S rRNA to total 16S rRNA (▴) was determined using a reference dilution series of RNA extracted from chloramphenicol-treated A. calcoaceticus.
Figure 8b, c, and d show the results of membrane hybridizations with samples from the lab-scale reactors. The levels of Acinetobacter precursor 16S rRNA in the CMAS system remained approximately constant during the 8 h of sampling (Fig. 8b). In contrast, the results for SBR-1 show a significant increase in the levels of Acinetobacter precursor 16S rRNA during the first 30 min of the operating cycle (Fig. 8c). After 30 min of aeration, the levels of precursor 16S rRNA declined sharply. The profile of Acinetobacter precursor 16S rRNA for SBR-2 was similar to that observed for SBR-1 (Fig. 8d). When influent was added to the reactor, the level of Acinetobacter precursor 16S rRNA rapidly increased for the first 30 min and then returned to lower levels. A second cycle exhibited the same results.
The ratio of precursor 16S rRNA to total 16S rRNA was calculated with results from membrane hybridizations with probes S-G-Acin-1543-a-A-24 and S-G-Acin-0659-a-A-24 (probe S-G-Acin-0659-a-A-24 was modified from probe ACA used by Wagner et al. [31; D. B. Oerther, J. Danalewich, E. Dulekgurgen, and L. Raskin, unpublished data]). In the CMAS system, the ratio of precursor to total 16S rRNA was relatively constant, with an average of approximately 10%. In contrast, the ratio of precursor to total 16S rRNA almost doubled, increasing from 5 to 8%, in SBR-1 upon the addition of influent. Similarly, the ratio of precursor to total 16S rRNA increased from 4 to 7% in SBR-2 after the first addition of influent, while the second addition of influent only resulted in a small increase.
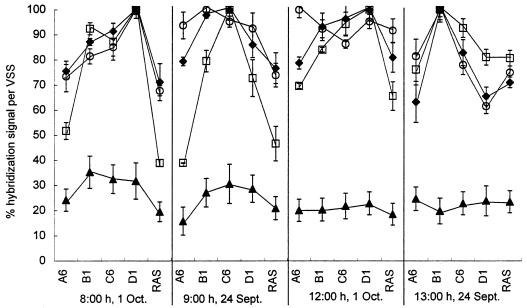
The results of membrane hybridization with samples from the UCSD, NEWWTP, are shown in Fig. 9. A comparison of the hybridization signals from the re-aeration basin (site A6, Fig. 1) and the contact basin (sites B1, C6, and D1, Fig. 1) indicates that the levels of Acinetobacter precursor 16S rRNA increased substantially with the addition of influent (in the contact basin). The levels of precursor 16S rRNA remained relatively high throughout the contact basin (from sites B1 to D1). The levels of precursor 16S rRNA in the RAS were significantly lower than in the contact basin. Early-morning samples (08:00 and 09:00 h) exhibited a larger increase in precursor 16S rRNA levels when the mixed liquor was moved from the re-aeration basin to the contact basin as compared to the mid-day samples (12:00 and 13:00 h). The ratio of precursor to total 16S rRNA increased for the early-morning samples (08:00 and 09:00 h) from approximately 23% in the reaeration basin to greater than 35% in the contact basin, but remained relatively constant for the mid-day samples (12:00 and 13:00 h).
FIG. 9.
Membrane hybridizations with samples from the UCSD, NEWWTP. Precursor 16S rRNA was measured using probe S-G-Acin-1543-a-A-24 (□), while total 16S rRNA from Acinetobacter spp. and from the total microbial community was measured with probes S-G-Acin-0659-a-A-24 (⧫) and S-*-Univ-1390-a-A-18 (○), respectively. The ratio of precursor 16S rRNA to total 16S rRNA (▴) was determined using a reference dilution series of RNA extracted from chloramphenicol-treated A. calcoaceticus.
DISCUSSION
Probe design and characterization.
A number of difficulties were encountered when designing precursor 16S rRNA probes. Searches of the GenEMBL database revealed few entries containing sufficient sequence information for the design of precursor 16S rRNA probes. Fortunately, four entries contained the sequences of the 3′ region of the precursor 16S rRNA of Acinetobacter spp. (Table 1). As previously discussed, each precursor 16S rRNA is excised from the full-length rrn transcript by RNase III (15). The RNase III cleavage site is a sequence-independent, secondary-structure-dependent stem produced by intrastrand hybridization between the 5′ and 3′ regions of each precursor 16S rRNA molecule. Since no information was available regarding the location of the RNase III cleavage site for precursor 16S rRNA in Acinetobacter spp., probe S-G-Acin-1543-a-A-24 was designed based upon the location of previously reported precursor 16S rRNA probes targeting E. coli (8). In addition, although one species of Acinetobacter is known to have seven copies of the rrn operon (13), no information was available regarding interoperon sequence divergence for Acinetobacter spp. Therefore, the specificity of S-G-Acin-1543-a-A-24 could not be fully examined in this study.
In order to use precursor 16S rRNA probes to follow changes in the growth activity of microorganisms in mixed cultures and environmental samples, stringent hybridization conditions are required. Therefore, we experimentally determined optimal hybridization stringencies using Td and formamide studies. Since the level of precursor 16S rRNA has been reported to be less than 10% of the level of mature 16S rRNA (8), we experimentally tested probe specificity against recombinant rDNA to rule out interference in quantifying precursor 16S rRNA due to hybridization to chromosomal rrn operons. The Td values obtained with rDNA were approximately 7°C lower than those obtained with rRNA (Fig. 2a). Thus, the final wash temperature of 53°C for the membrane hybridizations should have minimized hybridization signal from chromosomal rrn operons.
Precursor rRNA levels in E. coli: comparison with previous results.
Membrane hybridizations, using stringent experimental conditions, were used to monitor the levels of precursor 16S rRNA and total 16S rRNA during batch growth of E. coli in LB medium (Fig. 3). The rapid increase in the OD600 values suggested that E. coli underwent a rapid transition from stationary to exponential growth phase. Thus, the extensive lag phase observed in similar experiments by Cangelosi and Brabant (8) was not found with our experimental conditions. In agreement with previous reports (8, 17), we observed an immediate increase in the levels of precursor 16S rRNA after transferring E. coli to fresh medium (Fig. 3). However, Cangelosi and Brabant (8) reported a 50-fold increase in the levels of precursor 16S rRNA, while we observed only a 12-fold increase (Fig. 3). A number of hypotheses are offered for this apparent discrepancy. First, results from our hybridizations should reflect the stringent experimental conditions used in our assays. The hybridization signals reported by Cangelosi and Brabant (8) with oligonucleotide probes targeting six of the seven rrn operons in E. coli (rrnA, -C, -D, -E, -F, -G, and -H), rrnB alone, and all seven rrn operons were similar for all three probes, suggesting that hybridization conditions were not stringent and that the observed signals corresponded to the levels of all seven transcripts. Thus, the 50-fold increase reported by Cangelosi and Brabant probably reflects the increased transcription of all seven rrn operons. In contrast, our use of stringent hybridization conditions with probe S-S-E.coli-1543-a-A-24 should limit our hybridization results to a signal corresponding to the level of transcripts corresponding to only six of the seven rrn operons (rrnA, -C, -D, -E, -F, -G, and -H). Second, Cangelosi and Brabant reported that the level of precursor 16S rRNA for their overnight culture was below the detection limit of their slot blot hybridization assay (8). Thus, the 50-fold increase they observed was calculated as the increase from the detection limit of their assay to the maximum hybridization signal observed. In our experiment, the hybridization signal of the precursor 16S rRNA for the overnight culture of E. coli was above the detection limit of our slot blot assay. Therefore, it is likely that the overnight cultures were prepared differently and that those differences are responsible for the differences between the results of Cangelosi and Brabant and our results. If the initial level of precursor 16S rRNA was significantly higher in our overnight culture than in the overnight culture used by Cangelosi and Brabant, the difference in hybridization results as well as the difference between the lengths of the observed lag phases may be explained.
Precursor rRNA levels in Acinetobacter spp.
Acinetobacter spp. have been historically important in the study of activated sludge, especially in the area of enhanced biological phosphorus removal (20). Therefore, this genus was selected for further study. In agreement with the results for E. coli, precursor 16S rRNA levels increased rapidly at the onset of exponential growth and then declined as A. calcoaceticus entered the late-exponential-growth phase when cultured in LB medium (Fig. 4). The levels of total 16S rRNA reached a maximum during mid- to late-exponential-growth phase and declined throughout entry into stationary phase. These results suggest that the precursor 16S rRNA pool is much more sensitive to changes in growth phase than the total rRNA pool. Using membrane hybridizations, it was determined that the precursor 16S rRNA levels changed 5-fold versus a 2.5-fold change in total rRNA levels for A. calcoaceticus grown in LB medium. Although the results for the membrane hybridizations and the FISH showed similar trends, the increase in the levels of precursor 16S rRNA and total 16S rRNA was smaller with FISH (3- and <2-fold, respectively [Fig. 4b]) than with membrane hybridizations. A comparison of results obtained by Cangelosi and Brabant (8) with results reported by Licht et al. (17) suggests a similar difference between the results obtained with membrane hybridizations and FISH for measurements made with E. coli. As discussed above, Cangelosi and Brabant (8) used membrane hybridizations to detect a 50-fold increase in the levels of precursor 16S rRNA. In contrast, Licht et al. (17) only reported a ninefold increase in the levels of precursor 16S rRNA using FISH. This apparent discrepancy between the results obtained with membrane hybridizations and FISH may be due to the semiautomated image analysis procedures used to quantify the results of FISH. To quantify fluorescent objects with digital image analysis, a threshold signal intensity must be selected for each data series. This threshold signal intensity represents a balance between including nonspecific hybridization signals (e.g., edge effects) and excluding the hybridization signal of target objects. Therefore, selecting a conservative (e.g., higher) threshold signal intensity excludes some hybridization signal from target objects resulting in a reduction in the magnitude of the change in the levels of precursor 16S rRNA measured with FISH.
Although the levels of precursor 16S rRNA and total 16S rRNA appear to follow similar trends for E. coli and A. calcoaceticus cultured in batch in LB medium (Fig. 3 and 4), transferring A. calcoaceticus cultured in LB medium to filtered wastewater produced different results (Fig. 5). When transferred to filtered wastewater, the OD600 of A. calcoaceticus decreased, suggesting a lag phase or even a loss in biomass. In contrast, the levels of precursor 16S rRNA immediately increased, a result consistent with the results of A. calcoaceticus transferred to fresh LB medium.
A. calcoaceticus transferred to filtered wastewater developed subpopulations with different ratios of precursor 16S rRNA to total 16S rRNA (Fig. 7h). Corresponding subpopulations were, however, not observed in samples of A. calcoaceticus cultured in LB medium (Fig. 7d). FISH of the samples grown in batch in filtered wastewater with the Acinetobacter genus-specific total 16S rRNA probe S-G-Acin-0659-a-A-24 demonstrated that all of the DAPI-stained cells in the culture were Acinetobacter (data not shown). This result strongly suggests that it is unlikely that cells that did not hybridize strongly with the Acinetobacter genus-specific precursor 16S rRNA probe were contaminant microorganisms which were not removed by filtering the wastewater through the 0.2-μm-pore-size filter. Currently, we can only speculate as to why subpopulations of A. calcoaceticus may have developed upon transfer from LB medium to filtered wastewater. Differences in the physiological state of individuals within the population may have existed. For instance, some individual cells may have contained larger pools of storage compounds (e.g., polyhydroxalkanoates [28]), allowing them to express higher levels of precursor 16S rRNA after switching to a new mixture of substrates (i.e., reflecting a change in physiology suggested by diauxic growth conditions). Alternatively, individual cells may have developed asynchronous growth, producing elevated levels of precursor 16S rRNA during a specific stage in the production of daughter cells. Another hypothesis, based upon the work of Licht et al. (17), suggests that the processing of precursor 16S rRNA into mature 16S rRNA may have been inhibited in some individuals due to their susceptibility to unidentified rRNA processing inhibitors suspected to be present in fecal material. Nevertheless, we demonstrated that FISH with dual oligonucleotide probes targeting precursor 16S rRNA and total 16S rRNA can be used to visualize physiological differences between individual cells within a population.
Precursor rRNA levels in environmental samples.
To test the precursor 16S rRNA probes with environmental samples, we performed membrane hybridizations with RNA extracts from samples of lab- and full-scale activated sludge wastewater treatment systems. Due to the low abundance of Acinetobacter spp. in the examined samples, FISH was not performed. Hybridization results obtained with samples from the sequencing batch reactors appeared to correlate well with the results from the batch growth of A. calcoaceticus in LB medium and filtered wastewater. Since the aerated and unaerated HRTs of the CMAS system and SBR-1 were selected to be comparable, the microbial communities in these two systems experienced similar conditions, with the exception of differences due to continuous versus batch operation. Results suggest that each addition of influent to SBR-1 stimulated rapid growth of Acinetobacter spp. (Fig. 8c), a finding similar to the rapid growth observed after the transfer of the pure culture of A. calcoaceticus into fresh LB medium or filtered wastewater (Fig. 4 and 5). The levels of precursor 16S rRNA in SBR-2 showed similar trends (Fig. 8d).
The ratios of precursor 16S rRNA to total 16S rRNA were significantly lower for Acinetobacter in the sequencing batch reactors (3 to 6%) compared to the ratios of precursor to total 16S rRNA observed in LB medium (15 to 50%), in filtered wastewater (20 to 60%), and in the CMAS system (average of 10%). Since the only significant operational difference between the CMAS system and SBR-1 was continuous versus batch operation, Acinetobacter grown in a sequencing batch reactor appeared to maintain a smaller pool of precursor 16S rRNA.
Membrane hybridizations with precursor 16S rRNA- and total 16S rRNA-targeted probes were used to follow changes in the activity of the Acinetobacter population in a full-scale activated sludge wastewater treatment system. The levels of Acinetobacter precursor 16S rRNA were higher in the contact basin of the system than in the RAS and re-aeration basin (Fig. 9). The smaller difference in Acinetobacter precursor 16S rRNA levels in the mid-day samples was likely due to the lower influent BOD5 concentration experienced during this time of the day. Typically, the early-morning “first flush” results in a higher influent BOD5 level, while the mid-day influent BOD5 concentration is lower due to the dilution effect of lower- strength wastewater reaching the UCSD, NEWWTP (Bachmann, personal communication). The ratios of precursor 16S rRNA to total 16S rRNA for the full-scale system ranged from 15 to 35%. The ratio of precursor to total 16S rRNA was higher in the contact basin versus the RAS and re-aeration basin for the early-morning samples but was constant across the system during mid-day.
Conclusion.
Our results suggest that quantitative hybridizations with oligonucleotide probes targeting rRNA can be used to measure more than the composition of microbial communities. By combining hybridizations to both precursor 16S rRNA and total 16S rRNA, population size and in situ growth activity can be measured. However, care should be taken in the interpretation of precursor 16S rRNA levels. Already, unexpected effects such as the inhibition of E. coli precursor 16S rRNA processing by unidentified components of mouse intestinal contents have been reported (17). Moreover, our results point at remarkable heterogeneity in the levels of precursor 16S rRNA in Acinetobacter spp. transferred from LB medium to filtered wastewater. Finally, the observation that up to 64% of the total 16S rRNA in A. calcoaceticus existed in the precursor 16S rRNA form suggests that A. calcoaceticus may regulate the production of rRNA (and possibly ribosomes) using a strategy significantly different from that of E. coli. Although the methods presented in this study for measuring in situ growth activity appear promising, more data on rRNA processing and differences in precursor 16S rRNA pools of environmentally important microorganisms are required before this approach can be used widely to determine microbial growth parameters.
ACKNOWLEDGMENTS
We thank Bernhard Fuchs for technical assistance with quantification of whole-cell FISH signal intensity, and we would like to thank the UCSD for access to the NEWWTP.
Funding for this project was provided by the U.S. Department of Agriculture, Washington, D.C. (95-37500-1911), and the Max Planck Society, Munich, Germany. Daniel B. Oerther gratefully acknowledges additional financial support from the Richard S. and Mary A. Engelbrecht Fellowship, the Mavis Teaching Fellowship, and a Doctoral Dissertation Travel Grant from the University of Illinois.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R, Lemmer H, Wagner M. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol Ecol. 1998;25:205–215. [Google Scholar]
- 6.Boye M, Ahl T, Molin S. Application of a strain-specific rRNA oligonucleotide probe targeting Pseudomonas fluorescens Ag1 in a mesocosm study of bacterial release into the environment. Appl Environ Microbiol. 1995;61:1384–1390. doi: 10.1128/aem.61.4.1384-1390.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bram R J, Young R A, Steitz J A. The ribonuclease III site flanking 23S sequence in the 30S ribosomal precursor RNA of E. coli. Cell. 1980;19:393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- 8.Cangelosi G A, Brabant W H. Depletion of pre-16S rRNA in starved Escherichia coli cells. J Bacteriol. 1997;179:4457–4463. doi: 10.1128/jb.179.14.4457-4463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de los Reyes F L, Ritter W, Raskin L. Group-specific small subunit rRNA hybridization probes to characterize filamentous foaming in activated sludge systems. Appl Environ Microbiol. 1997;63:1107–1117. doi: 10.1128/aem.63.3.1107-1117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de los Reyes F L, Oerther D B, de los Reyes M F, Hernandez M, Raskin L. Characterization of filamentous foaming in activated sludge systems using oligonucleotide hybridization probes and antibody probes. Water Sci Technol. 1998;37:485–493. [Google Scholar]
- 11.de los Reyes M F, de los Reyes F L, Hernandez M, Raskin L. Quantification of Gordona amarae strains in foaming activated sludge and anaerobic digester systems using oligonucleotide hybridization probes. Appl Environ Microbiol. 1998;64:2503–2512. doi: 10.1128/aem.64.7.2503-2512.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 13.Gralton E M, Campbell A L, Neidle E L. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg A E, Clesceri L S, Eaton A D, editors. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]
- 15.Kindler P, Kiel T U, Hofschneider P H. Isolation and characterization of a ribonuclease III deficient mutant of Escherichia coli. J Biol Chem. 1973;251:53–69. doi: 10.1007/BF00333481. [DOI] [PubMed] [Google Scholar]
- 16.Leser T D, Boye M, Hendriksen N D. Survival and activity of Pseudomonas sp. strain B13(FR1) in a marine microcosm determined by quantitative PCR and an rRNA-targeting probe and its effects on the indigenous bacterioplankton. Appl Environ Microbiol. 1995;61:1201–1207. doi: 10.1128/aem.61.4.1201-1207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Licht T R, Tolker-Nielsen T, Holmstrøm K, Krogfel K A, Molin S. Inhibition of Escherichia coli precursor-16S rRNA processing by mouse intestinal contents. Environ Microbiol. 1999;1:23–32. doi: 10.1046/j.1462-2920.1999.00001.x. [DOI] [PubMed] [Google Scholar]
- 18.Møller S, Pedersen A R, Poulsen L K, Arvin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nold S C, Ward D M. Photosynthate partitioning and fermentation in hot spring microbial mat communities. Appl Environ Microbiol. 1996;62:4598–4607. doi: 10.1128/aem.62.12.4598-4607.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oerther D B, Danalewich J, Dulekurgen E, Leveque E, Freedman D L, Raskin L. Bioaugmentation of sequencing batch reactors for biological phosphorus removal: comparative rRNA sequence analysis and hybridization with oligonucleotide probes. Water Sci Technol. 1998;37:469–473. [Google Scholar]
- 21.Oerther D B, de los Reyes F L, Raskin L. Interfacing phylogenetic oligonucleotide probe hybridizations with representations of microbial populations and specific growth rates in mathematical models of activated sludge processes. Water Sci Technol. 1999;39:11–20. [Google Scholar]
- 22.Pedersen A R, Møller S, Molin S, Arvin E. Activity of toluene-degrading Pseudomonas putida in the early growth phase of a biofilm for waste gas treatment. Biotechnol Bioeng. 1997;54:131–141. doi: 10.1002/(SICI)1097-0290(19970420)54:2<131::AID-BIT5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescent in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raskin L, Rittmann B, Stahl D A. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raskin L, Capman W C, Sharp R, Poulsen L K, Stahl D A. Molecular ecology of gastrointestinal ecosystems. In: Mackie R I, White B A, Isaacson R E, editors. Gastrointestinal microbiology. 2. Gastrointestinal microbiology and host interactions. New York, N.Y: Chapman and Hall; 1997. pp. 243–298. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schembri M A, Bayly R C, Davies J K. Phosphate concentration regulates transcription of the Acinetobacter polyhydroxybutyrate acid biosynthetic genes. J Bacteriol. 1995;177:4501–4507. doi: 10.1128/jb.177.15.4501-4507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava A K, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 30.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner M, Amann R, Lemmer H, Manz W, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 33.Young R A, Steitz J A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci USA. 1978;75:3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]