Abstract
Rhabdomyosarcoma (RMS) is a common soft tissue sarcoma in children that resembles developing skeletal muscle. Unlike normal muscle cells, RMS cells fail to differentiate despite expression of the myogenic determination protein MYOD. The TWIST2 transcription factor is frequently overexpressed in fusion-negative RMS (FN-RMS). TWIST2 blocks differentiation by inhibiting MYOD activity in myoblasts, but its role in FN-RMS pathogenesis is incompletely understood. Here, we show that knockdown of TWIST2 enables FN-RMS cells to exit the cell cycle and undergo terminal myogenesis. TWIST2 knockdown also substantially reduces tumor growth in a mouse xenograft model of FN-RMS. Mechanistically, TWIST2 controls H3K27 acetylation at distal enhancers by interacting with the chromatin remodelers SMARCA4 and CHD3 to activate growth-related target genes and repress myogenesis-related target genes. These findings provide insights into the role of TWIST2 in maintaining an undifferentiated and tumorigenic state of FN-RMS and highlight the potential of suppressing TWIST2-regulated pathways to treat FN-RMS.
Twist2 controls a gene regulatory program that promotes fusion-negative rhabdomyosarcoma.
INTRODUCTION
Cancer often arises because of the disruption of normal developmental programs and resulting unrestrained cell proliferation. Rhabdomyosarcoma (RMS), which accounts for approximately half of the soft tissue sarcomas in children, represents a classic example of the antagonism between oncogenesis and cell differentiation. Expression of the master muscle regulators myogenic differentiation 1 (MYOD) and myogenic factor 5 (MYF5) drives cell cycle exit and terminal differentiation of normal muscle lineage cells (1, 2). RMS cells, in contrast, express these transcription factors but fail to fully differentiate, resulting in continued cell divisions and tumor growth (3–5). There are two major RMS subtypes, fusion-positive RMS (FP-RMS) and the more prevalent fusion-negative RMS (FN-RMS) (6). While FP-RMS is mainly driven by a balanced chromosomal translocation generating oncogenic PAX3-FOXO1 or PAX7-FOXO1 fusion proteins, the molecular basis of FN-RMS is not understood (4, 7).
FN-RMS exhibits greater genomic instabilities than FP-RMS, however, mutations in oncogenes such as RAS or the TP53 tumor suppressor do not occur at a high incidence in FN-RMS tumors (8, 9). Analysis of genomic data from patient RMS revealed that frequent copy number alterations of oncogenes and tumor suppressors can drive FN-RMS pathogenesis (10). Strikingly, we showed that 72% (114 of 158 cases) of FN-RMS patient tumors contain copy number amplification events on the TWIST1 locus, TWIST2 locus, or both loci (11). These amplifications are consistent with increased expression of the TWIST genes in FN-RMS. In contrast, the amplification and expression of TWIST1 and TWIST2 are not substantially associated with FP-RMS cases (11). Therefore, the overrepresentation of TWIST amplification and expression in FN-RMS warrants characterizing its pathological role in this understudied pediatric sarcoma subtype.
The mammalian basic helix-loop-helix (bHLH) transcription factor Twist2 and its paralog Twist1 are important regulators of mesodermal cell identity and often antagonize the functions of lineage-determining transcription factors such as MyoD, Hand2, and Runx2 to spatiotemporally restrict cell fates (11–14). We and others have shown that prolonged expression of Twist1 and Twist2 in primary myoblasts, embryonic stem cells, or MyoD-expressing fibroblasts represses myogenic differentiation (11, 15, 16). Moreover, re-expression of Twist2 in differentiated primary myotubes reverted them to mononucleated myoblasts (11), and overexpression of Twist1 in uncommitted mesenchymal stem cells or muscle satellite cells in mice caused a marked reduction in muscle weight and myofiber diameter (17). These studies attest to the ability of the Twist proteins to inhibit muscle differentiation. Restoration of myogenic differentiation in tumor cells represents a potential therapeutic approach for FN-RMS cancers, but the implications of targeting TWIST in vivo have not been explored. Furthermore, the transcriptional and epigenetic regulation by TWIST in FN-RMS has not been systematically studied.
In this study, we explored the functions of TWIST2 in FN-RMS pathogenesis in vitro and in vivo. We show that knockdown (KD) of TWIST2 causes cell cycle arrest and promotes myogenic differentiation in FN-RMS cell lines and mouse tumor xenografts. By interrogating the TWIST2 interactome, we identified a druggable mechanism of TWIST2 function where TWIST2 associates with the SMARCA4 and CHD3 chromatin remodelers, leading to the blockade of muscle gene expression and persistent activation of growth-associated genes in FN-RMS. This bipotential role of TWIST2 is reflected in the deposition of H3 lysine 27 acetylation (H3K27ac) at enhancers of growth genes but not of myogenic genes in FN-RMS cells, and this is reversed upon TWIST2 KD. Together, these findings provide intriguing insights into the mechanistic basis of tumorigenesis in FN-RMS and a strong rationale for targeting of TWIST2-regulated pathways as a therapeutic option for FN-RMS.
RESULTS
TWIST2 KD inhibits growth and promotes myogenic differentiation of FN-RMS cells
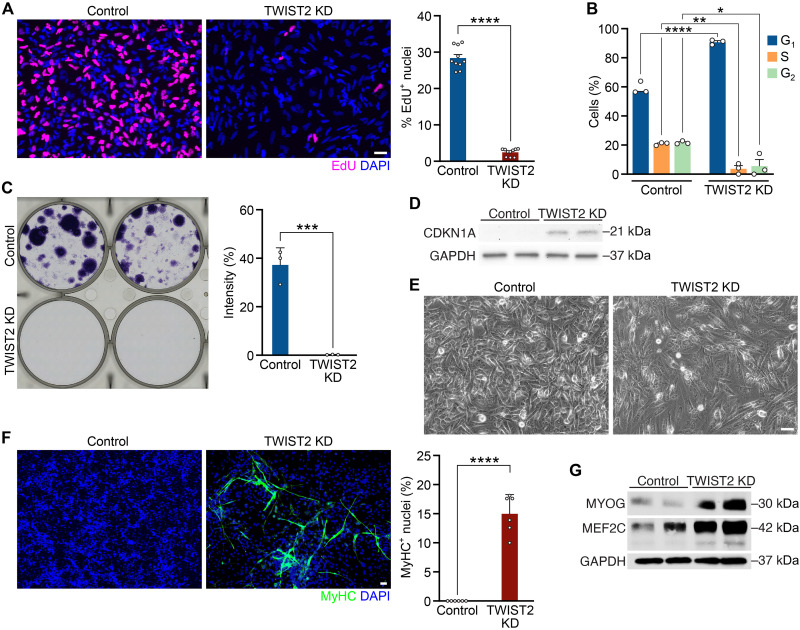
To explore the functions of TWIST in FN-RMS pathogenesis, we used an in vitro loss-of-function system to knockdown TWIST2 in FN-RMS cell lines. We generated stable FN-RMS cell lines expressing a doxycycline (Dox)–inducible lentiviral vector that harbors a short hairpin RNA (shRNA) targeting TWIST2. Treatment with Dox (10 μg/ml) for 4 days reduced the levels of TWIST2 protein (TWIST2 KD) compared to the untreated (Control) cells (fig. S1A). To survey the effect of TWIST2 KD, we first measured 5-ethynyl-20-deoxyuridine (EdU) incorporation as an indicator of proliferation of Control and TWIST2 KD rhabdomyosarcoma (RD) cells. RD is a commonly used patient tumor–derived FN-RMS cell line. Loss of TWIST2 resulted in a reduced number of EdU+ cells (Fig. 1A). A stable RD cell line expressing a Dox-inducible scrambled shRNA did not show changes in EdU incorporation upon treatment with Dox for 4 days, validating that Dox alone does not inhibit cell growth (fig. S1B).
Fig. 1. TWIST2 KD inhibits growth and promotes myogenic differentiation of FN-RMS cells.
(A) Left: EdU (magenta) immunocytochemistry of Control (untreated) and TWIST2 KD (treated with Dox) RD cells 4 days after ± Dox treatment. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Right: Quantification of the percentage of EdU+ nuclei. Scale bar, 50 μm. n = 10 biologically independent samples. (B) Quantification of the percentage of Control and TWIST2 KD RD cells in the G1, S, or G2 stages. n = 3 biologically independent samples. (C) Left: Single-cell colony formation assay for Control and TWIST2 KD RD cells 15 days after ± Dox treatment. Right: Quantification of the percent intensity of weighted colony area. n = 3 biologically independent samples. (D) Western blot showing an increase of CDKN1A protein level in TWIST2 KD RD cells compared to Control cells 4 days after ± Dox treatment. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control. (E) Phase contrast images of Control and TWIST2 KD RD cells 4 days after ± Dox treatment. Scale bar, 50 μm. (F) Left: Anti–myosin heavy chain (MyHC) (green) immunocytochemistry of Control and TWIST2 KD RD cells cultured for 5 days in differentiation media ± Dox. Nuclei were stained with DAPI (blue). Right: Quantification of percent of MyHC+ nuclei. Scale bar, 50 μm. n = 6 biologically independent samples. (G) Western blot showing increased protein levels of myogenin (MYOG) and myocyte enhancer factor 2C (MEF2C) in TWIST2 KD RD cells compared to Control cells 4 days after ± Dox treatment. GAPDH was used as the loading control. All statistical comparisons between groups were evaluated by unpaired Student’s t test, ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05. Data are presented as the mean ± SEM.
In a complementary approach, we performed cell cycle analysis on Control and TWIST2 KD RD cells. While more than 40% of the Control RD cells transitioned into the S and G2-M phases, only about 8% of the TWIST2 KD cells progressed past the G1-S phase, indicating cell cycle arrest (Fig. 1B). Using a colony formation assay to assess clonogenic capacity, we showed that TWIST2 KD caused a substantial reduction in colony coverage, calculated by the percent intensity of weighted colony area, compared to the Control experimental group (Fig. 1C). We performed EdU labeling in other human FN-RMS cell lines, including SMS-CTR, JR-1, RH18, and 381.T cell lines, all of which showed a decrease in EdU incorporation upon TWIST2 KD (fig. S1C). Moreover, fewer colonies were formed with TWIST2 KD in SMS-CTR and JR-1 cells (fig. S1D). Collectively, these results demonstrate that TWIST2 promotes oncogenic growth of multiple FN-RMS cell lines.
Consistent with a loss of proliferative and clonogenic potential, TWIST2 KD in RD cells resulted in the up-regulation of CDKN1A protein expression, a marker of cell cycle arrest that coincides with muscle-specific gene expression during myoblast differentiation (Fig. 1D) (18). Moreover, phase contrast images showed that FN-RMS cells became elongated upon TWIST2 KD (Fig. 1E and fig. S1E). This was accompanied by expression of myosin heavy chain 1 (MyHC), a marker of terminal muscle differentiation, in a subset of TWIST2 KD RD cells when cultured in differentiation media for 5 days (Fig. 1F), and increased protein expression of the myogenic regulatory transcription factors myogenin (MYOG) and myocyte enhancer factor 2C (MEF2C) (Fig. 1G and fig. S1A). These results show that TWIST2 actively maintains FN-RMS cells in an undifferentiated and tumorigenic state.
TWIST2 KD reduces FN-RMS tumor xenograft growth and induces terminal differentiation in vivo
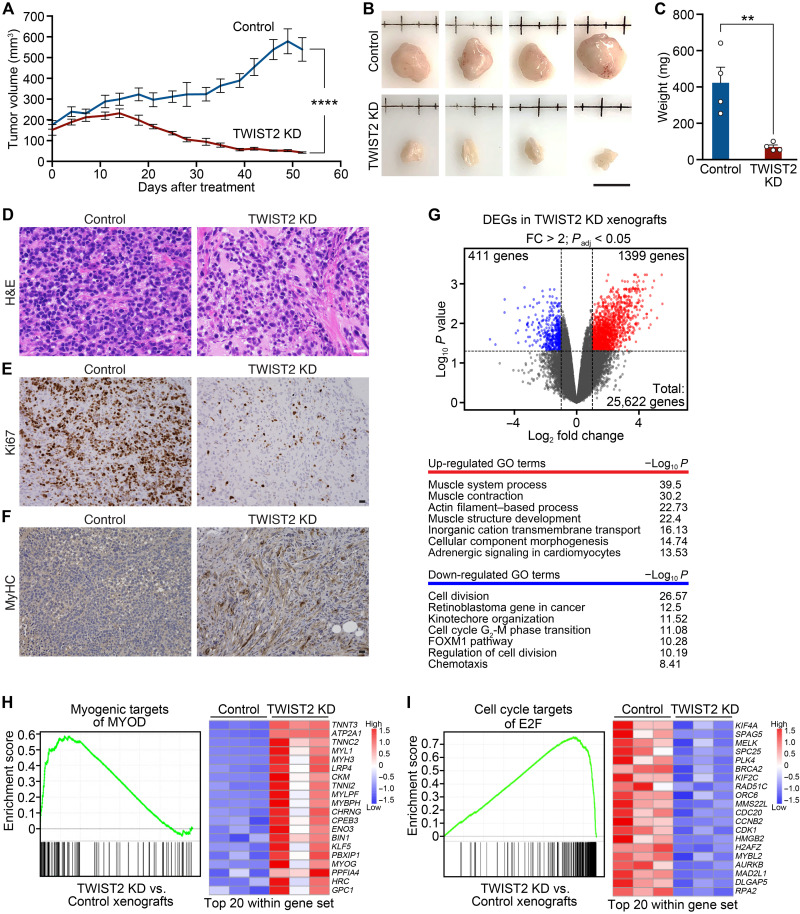
We assessed the effect of TWIST2 KD on tumor growth by injecting RD cells that stably expressed a Dox-inducible shTWIST2 into the flank of nonobese diabetic (NOD)–severe combined immunodeficient mice. Once the tumor xenografts grew to 100 to 200 mm3, we supplemented the drinking water of half the mice with Dox [2 mg/ml in 5% (w/v) sucrose] to initiate TWIST2 KD, while the remaining mice were left untreated as Control [5% (w/v) sucrose]. TWIST2 KD substantially reduced the size and weight of tumors at end point (52 days after treatment) (Fig. 2, A to C). Fewer cells were detected in TWIST2 KD tumors compared to Control tumors by hematoxylin and eosin (H&E) staining (Fig. 2D). Delayed growth in TWIST2 KD tumors was corroborated by a marked decrease in Ki67+ cycling nuclei (Fig. 2E). We also observed increased myogenic differentiation in TWIST2 KD tumors, shown by anti-MyHC staining (Fig. 2F). Together, these results imply that TWIST2 sustains FN-RMS tumor growth and prevents the tumor cells from terminally differentiating.
Fig. 2. TWIST2 KD reduces FN-RMS tumor xenograft growth and induces terminal differentiation in vivo.
(A) Growth curve of RD mouse tumor xenograft volume (mm3) plotted following the initiation of Dox treatment (Day 0) to TWIST2 KD. Control tumors were not treated with Dox. n = 8 mice for Control, 8 mice for TWIST2 KD. (B) Images of Control and TWIST2 KD tumors harvested 52 days after initiation of Dox treatment (the end point). Scale bar, 1 cm. (C) Weights (mg) of Control and TWIST2 KD tumors harvested at end point. n = 4 mice for Control, 4 mice for TWIST2 KD. (D) H&E of Control and TWIST2 KD tumors harvested at end point. Scale bar, 50 μm. (E) Immunohistochemistry for Ki67 on Control and TWIST2 KD tumors harvested at end point. Scale bar, 50 μm. (F) Immunohistochemistry of anti-MyHC (using the MF20 antibody) on Control and TWIST2 KD tumors harvested at end point. Scale bar, 50 μm. (G) Top: Volcano plot of the up-regulated genes (red) and down-regulated genes (blue) in TWIST2 KD RD tumor xenografts relative to the Control tumors identified by RNA-seq. A cutoff of fold change (FC) > 2 and Padj < 0.05 was used to identify differentially expressed genes (DEGs) by RNA-seq. Bottom: Top enriched Gene Ontology (GO) terms are shown. (H) Left: Gene set enrichment analysis (GSEA) plot showing positive enrichment for genes that are myogenic targets of MYOD in TWIST2 KD relative to Control tumors. Right: Heatmap plotting the gene expression of top 20 mRNA targets of MYOD. (I) Left: GSEA plot showing negative enrichment for genes that are cell cycle targets of E2 factor (E2F) in TWIST2 KD relative to Control tumors. Right: Heatmap plotting the gene expression of top 20 mRNA targets of E2F. All statistical comparisons between groups were evaluated by unpaired Student’s t test, ****P < 0.0001 and **P < 0.01. Data are presented as the mean ± SEM.
To reveal the transcriptional basis of TWIST2 function in FN-RMS, we profiled differences in gene expression between Control and TWIST2 KD tumor xenografts by RNA sequencing (RNA-seq). Differential enrichment gene analysis identified 1399 up-regulated genes and 411 down-regulated genes in TWIST2 KD tumors compared to Control (Fig. 2G). The up-regulated genes were enriched in Gene Ontology (GO) terms related to muscle contraction, muscle fiber development, and actin filament–based pathways (Fig. 2G). In contrast, genes that function in cell cycle processes were down-regulated in TWIST2 KD tumors. More specifically, gene set enrichment analysis revealed up-regulation of known myogenic targets of MYOD (Fig. 2H) (19) and down-regulation of cell cycle targets of E2 factor (E2F) transcription factors (Fig. 2I) (20). This global shift in the transcriptome away from proliferation and toward myogenesis upon TWIST2 KD explains the phenotypic changes in FN-RMS tumors lacking TWIST2.
We also performed RNA-seq on Control and TWIST2 KD samples in RD and SMS-CTR cells and evaluated the gene signatures that were shared by both cell lines. Myogenic differentiation was up-regulated, and a growth gene signature was down-regulated in both RD and SMS-CTR cell lines upon TWIST2 KD (fig. S2). These findings validate the phenotypic effects of TWIST2 described in tumor xenografts and in FN-RMS cells.
TWIST2 is a direct transcriptional activator of growth and inhibitor of myogenesis in FN-RMS
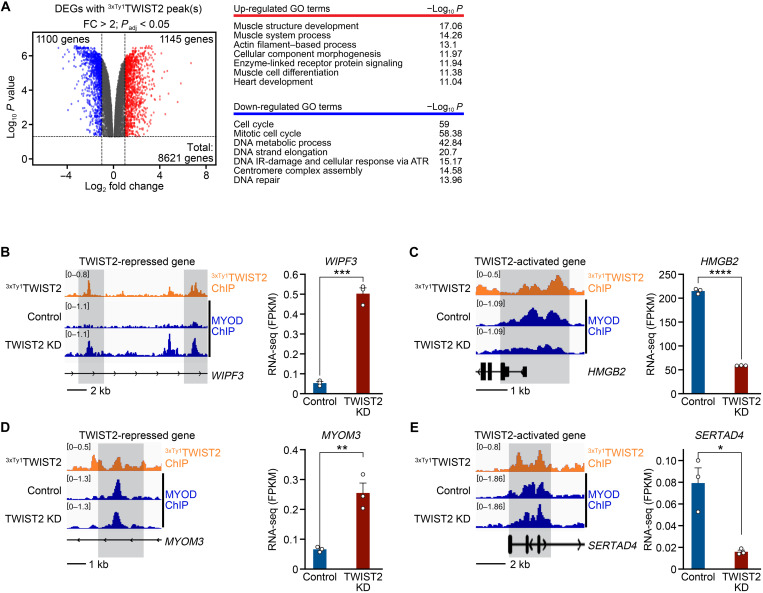
To examine the molecular function of TWIST2, we profiled genome-wide TWIST2 binding by infecting RD cells with lentiviruses expressing Ty1 epitope-tagged TWIST2 (3xTy1TWIST2) and performing chromatin immunoprecipitation and sequencing (ChIP-seq) for Ty1. A tagged version of TWIST2 was used because the available antibodies for TWIST2 are inadequate for ChIP. We confirmed that 3xTy1TWIST2 appropriately localized to the nucleus by immunostaining and did not affect RD cell proliferation (fig. S3A). ChIP-seq analysis identified TWIST2 binding sites (table S2) that were enriched mainly in intronic and intergenic regions, while only about 9% of the total peaks were located at promoters or transcription start sites (TSSs) (fig. S3B). These results are consistent with TWIST2 binding distribution in murine primary myoblasts (11). Furthermore, known motifs recovered from the TWIST2 peaks matched the consensus E-box motif, 5′-CANNTG-3′, with a preference for GC or TA at the third and fourth nucleotides (fig. S3C). E-box motifs with GC in their center dinucleotides can also be bound by myogenic bHLH transcription factors such as MYOD, MYF5, and MYOG and serve as sites of coregulation with TWIST2. Moreover, TWIST2 occupied a 5–base pair separated double E-box motif that has been described in TWIST target genes in Drosophila, murine myogenic cells, and human cell lines (11, 21). TWIST2-responsive elements were also enriched for the activator protein 1 (AP-1) motif, 5′-TGANTCA-3′, of basic leucine zipper (bZIP) transcription factors from the c-FOS, JUN, and ATF families, suggesting potential cooperation of these transcription factors with TWIST2.
To identify direct transcriptional targets of TWIST2 in FN-RMS, we annotated each TWIST2 peak to its nearest gene and integrated these data with RNA-seq data from Control and TWIST2 KD RD cells. We found 1145 genes that were up-regulated upon TWIST2 KD and harbor one or more TWIST2 peaks in their regulatory elements, indicating that TWIST2 directly represses these genes in RD cells (Fig. 3A and fig. S3E). The TWIST2-repressed direct target genes function in pathways regulating muscle formation and maturation (Fig. 3A). Moreover, a subset of these genes, whose expression in Control and TWIST2 KD RD cells is plotted in fig. S3D, was activated during the differentiation of human skeletal muscle myoblasts to myotubes in vitro (22), a process that requires down-regulation of TWIST2 expression. In contrast, 1100 genes were down-regulated upon TWIST2 KD and were bound by TWIST2 in their regulatory regions, suggesting that TWIST2 directly activates these genes in RD cells (Fig. 3A and fig. S3F). The TWIST2-activated genes function in pathways that maintain FN-RMS proliferation and tumorigenesis (Fig. 3A). Using quantitative reverse transcription polymerase chain reaction (qRT-PCR), we confirmed that these myogenic genes were up-regulated, and growth genes were down-regulated in SMS-CTR and 381.T cells upon TWIST2 KD (fig. S3, G to J). These direct target genes of TWIST2 are similarly regulated in human FN-RMS tumors, indicative of their relevance to FN-RMS pathophysiology. While the expression of TWIST2-activated target genes is increased in human FN-RMS tumors over control skeletal muscle, that of TWIST2-repressed target genes is decreased in patient FN-RMS over control skeletal muscle (fig. S3, K and L). Overall, these findings indicate that TWIST2 functions as a direct repressor of a myogenic gene program and as an activator of a growth gene program in FN-RMS cells.
Fig. 3. TWIST2 is a direct transcriptional activator of growth and inhibitor of myogenesis in FN-RMS.
(A) Left: Volcano plot of direct transcriptional TWIST2 target genes as identified by 3xTy1TWIST2 ChIP-seq and RNA-seq intersection in TWIST2 KD relative to Control RD cells. TWIST2 peaks whose nearest gene is up-regulated are shown in red, and TWIST2 peaks whose nearest gene is down-regulated are shown in blue. A cutoff of FC > 2 and false discovery rate (FDR) P value < 0.05 was used to identify DEGs by RNA-seq. Right: Top enriched GO terms are shown. (B to E) Left: ChIP-seq tracks where MYOD peaks in Control and TWIST2 KD RD cells are shown in blue, and 3xTy1TWIST2 peaks in RD cells are shown in orange. Right: Bar graph plotting FPKM (fragments per kilobase of exon per million mapped fragments) expression values from RNA-seq in Control and TWIST2 KD RD cells. (B) Left: Both TWIST2 and MYOD bound to the WIPF3 locus. In TWIST2 KD cells, MYOD binding was increased. Right: WIPF3 mRNA expression is increased in TWIST2 KD cells. (C) Left: Both TWIST2 and MYOD bound to the HMGB2 locus. In TWIST2 KD cells, MYOD binding was decreased. Right: HMGB2 mRNA expression is decreased in TWIST2 KD cells. (D) Left: Both TWIST2 and MYOD bound the MYOM3 locus. In TWIST2 KD cells, MYOD binding was not changed. Right: MYOM3 mRNA expression is increased in TWIST2 KD cells. (E) Left: Both TWIST2 and MYOD bound to the SERTAD4 locus. In TWIST2 KD cells, MYOD binding was not changed. Right: SERTAD4 mRNA expression is decreased in TWIST2 KD cells. All statistical comparisons between groups were evaluated by unpaired Student’s t test, ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05. Data are presented as the mean ± SEM.
TWIST2 regulates MYOD binding activity at a subset of gene loci in FN-RMS
We previously showed that Twist2 directly competes with MyoD binding at myogenic enhancers and redirects MyoD to growth and oncogenic enhancers in mouse primary myoblasts (11). To gain insights into the molecular interplay of MYOD and TWIST2 in RD cells, we examined MYOD binding profiles by performing ChIP-seq with an antibody for MYOD in Control and TWIST2 KD RD cells. We recovered a similar number of MYOD binding sites in both conditions (table S2). To interrogate changes in MYOD binding in response to TWIST2 KD, we annotated each MYOD peak to its nearest gene and clustered the differentially enriched MYOD peaks according to their neighboring gene expression. Then, by overlapping 3xTy1TWIST2 binding sites with the differential MYOD peaks, we showed that a change in MYOD binding occurred at very few direct targets of TWIST2, whereas at most loci, MYOD binding was maintained between Control and TWIST2 KD conditions (fig. S3, M and N). Among the 18,723 TWIST2 peaks mapped to TWIST2-repressed genes, only a small fraction (1193 peaks) overlapped with MYOD-gained peaks in TWIST2 KD cells (fig. S3M). These overlapping peaks represent myogenic loci at which TWIST2 directly competes with MYOD for binding. For example, expression of WIPF3 is inhibited by TWIST2 in Control RD cells (Fig. 3B). In TWIST2 KD cells, loss of TWIST2 binding enabled MYOD to bind to the WIPF3 locus, thus turning on WIPF3 gene expression. Similarly, Of 16,778 TWIST2 peaks mapped to TWIST2-activated genes, only 1164 peaks overlapped with MYOD lost peaks in TWIST2 KD cells (fig. S3N). These represent growth loci coactivated by TWIST2 and MYOD. For example, TWIST2 and MYOD both bound to the HMGB2 locus in Control cells, leading to activation of HMGB2 expression (Fig. 3C). TWIST2 KD led to decreased MYOD binding, resulting in decreased HMGB2 expression. Thus, in both these scenarios, TWIST2 alters the binding of MYOD to regulate gene expression.
In contrast, an overwhelming majority of the TWIST2-bound peaks were either not bound by MYOD or showed no change in MYOD binding between Control and TWIST2 KD RD cells (fig. S3, M and N). For example, in TWIST2 KD RD cells, expression of MYOM3 was activated, while SERTAD4 was inhibited, yet there was no change in MYOD binding at these loci (Fig. 3, D and E). These examples demonstrate that in contrast to myoblasts, TWIST2 does not modify MYOD binding activity to regulate global gene expression in FN-RMS cells.
TWIST2 controls enhancer activity and chromatin state in FN-RMS
Twist2 has been shown to mediate changes in chromatin accessibility, driving gene regulation in multiple tissues (11, 23). To determine its roles in FN-RMS, we compared the enhancer landscape in Control and TWIST2 KD RD cells by performing ChIP-seq for the active histone modification, H3K27ac (table S2), which is enriched in open accessible chromatin regions (24). We next annotated the differential H3K27ac peaks to their nearest genes and evaluated changes in gene expression. We found 3681 peaks with increased H3K27ac deposition in genes that were activated in TWIST2 KD cells, and 50,423 peaks with decreased H3K27ac in genes that were repressed upon TWIST2 KD (fig. S4A). Overall, the occurrence of H3K27ac deposition strongly correlated with gene expression.
The majority of the gained (98%) and lost (75%) H3K27ac peaks were in introns or intergenic regions, which highlights the role of distal enhancers in maintaining RD cell identity (fig. S4, B and C). Next, to identify enhancer regions that were directly regulated by TWIST2 binding, we intersected the above H3K27ac ChIP dataset with 3xTy1TWIST2 peaks in RD cells. We found 2321 TWIST2-bound peaks that gained the H3K27ac modification upon TWIST2 KD (Fig. 4A and fig. S4D). In contrast, 1845 TWIST2 peaks showed decreases in H3K27ac deposition upon TWIST2 KD (Fig. 4A and fig. S4E). The TWIST2-bound H3K27ac-gained peaks function in actin filament–based processes and platelet-derived growth factor receptor and Rho signaling pathways that regulate cell shape and cytoskeletal changes ahead of myogenic differentiation, while the TWIST2-bound HK27Ac lost peaks are involved in the cell cycle and the extracellular signal–regulated kinase and mitogen-activated protein kinase signaling pathways, consistent with the ability of TWIST2 to maintain FN-RMS cell proliferation (Fig. 4B). These results indicate that TWIST2 directly controls the activity of enhancers that dictate myogenic or growth gene expression.
Fig. 4. TWIST2 controls enhancer activity and chromatin state in FN-RMS.
(A) Heatmap of differentially enriched (DE) H3K27ac peaks in TWIST2 KD relative to Control RD cells and 3xTy1TWIST2 peaks at the same genomic loci. Only H3K27ac peaks whose nearest gene is similarly regulated by RNA-seq were plotted. Genomic regions ±10 kb from the peak center were plotted. A significance of FDR P value < 0.05 was used to identify DEGs and DE H3K27ac peaks, only DE peaks with a FC > 2 were plotted. IP, immunoprecipitation. (B) Top enriched Genomic Regions Enrichment of Annotations Tool (GREAT) terms are shown for the (top) 2321 TWIST2-bound H3K27ac-gained peaks in TWIST2 KD cells and the (bottom) 1845 TWIST2-bound H3K27ac lost peaks in TWIST2 KD cells. MAPK, mitogen-activated protein kinase; ERK, extracellular signal–regulated kinase. (C and D) Heatmaps showing transcription factor binding motifs uniquely enriched in enhancers that are (C) gained and (D) lost in TWIST2 KD RD cells. (E) Bar graph plotting FPKM expression values of E2F1, E2F2, and TFDP1 transcripts from RNA-seq in Control and TWIST2 KD RD cells. E2F1, E2F2, and TFDP1 mRNA expression is decreased in TWIST2 KD cells. n = 3 biologically independent samples. (F) qRT-PCR analysis of indicated genes in Control and TWIST2 KD RD cells infected with Empty3xFlag or a combination of E2F13xFlag, E2F23xFlag, and TFDP13xFlag for 3 days (relative to Empty3xFlag-infected Control cells, normalized to 18S rRNA). n = 3 biologically independent samples. All statistical comparisons between groups were evaluated by unpaired Student’s t test, ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05. Data are presented as the mean ± SEM.
Together, we found that TWIST2 maintains H3K27ac marks at enhancers that control growth gene expression while preventing H3K27ac deposition at enhancers that control muscle differentiation genes in RD cells. Loss of TWIST2, however, resulted in global shifts in enhancer activity.
TWIST2 and E2F transcription factors together activate a growth gene signature
Dysregulated gene expression in cancer depends on a circuitry of transcription factors that maintain cancer cell identity (25). It has been shown that bHLH transcription factors such as N-myc require the co-occupancy of other transcription factors to activate a cell type–specific gene program (26). To identify transcription factors that cooperate with TWIST2 to regulate enhancer activity, we searched for known transcription factor binding motifs that are differentially enriched at enhancers that are activated or repressed with TWIST2 KD. The gained H3K27ac peaks in TWIST2 KD were uniquely enriched in the cAMP responsive element (CRE) bound by certain bZIP transcription factors, e.g., ATF2, ATF4, ATF7, c-JUN, and CREB5, as well as the transcriptional enhanced associate (TEA) domain bound by TEAD transcription factors (Fig. 4C), suggesting that these transcription factors gain access to myogenic enhancers upon TWIST2 KD. Moreover, these transcription factors have been previously reported to facilitate terminal myogenesis (27, 28). DNA binding motifs of the E2F family of transcription factors were enriched in lost H3K27ac peaks in TWIST2 KD (Fig. 4D), which may contribute to maintaining RD cells in their proliferative state. We found that cell cycle target genes of E2F are down-regulated in TWIST2 KD tumor xenografts (Fig. 2I) and in RD, SMS-CTR, and 381.T cells (fig. S4, F to H), highlighting the importance of E2F function in FN-RMS.
E2F functions together with their dimerization partners, transcription factor Dp-1 (TFDP) proteins, to promote cell division (29). E2F1, E2F2, and their cofactor, TFDP1, were down-regulated in TWIST2 KD RD cells (Fig. 4E). To test whether the growth defect upon TWIST2 KD is due to down-regulation of these transcription factors, we overexpressed E2F1, E2F2, and TFDP1 together in TWIST2 KD RD cells (fig. S4I) and examined the expression of known E2F target genes that regulate cell cycle progression. qRT-PCR analysis revealed that the expression of these E2F target genes was decreased in TWIST2 KD cells, and overexpression of E2F1, E2F2, and TFDP1 partially rescued their expression (Fig. 4F). The E2F target genes analyzed are also direct transcriptional targets of TWIST2 (Fig. 3C). In addition, we observed a partial rescue of the proliferation defect in TWIST2 KD RD cells when E2F1, E2F2, and TFDP1 were overexpressed (fig. S4J). These results indicate that TWIST2 activates growth-related genes in FN-RMS in part via controlling E2F transcriptional activity.
BioID and mass spectrometry reveal that TWIST2 interacts with chromatin remodeling enzymes in FN-RMS cells
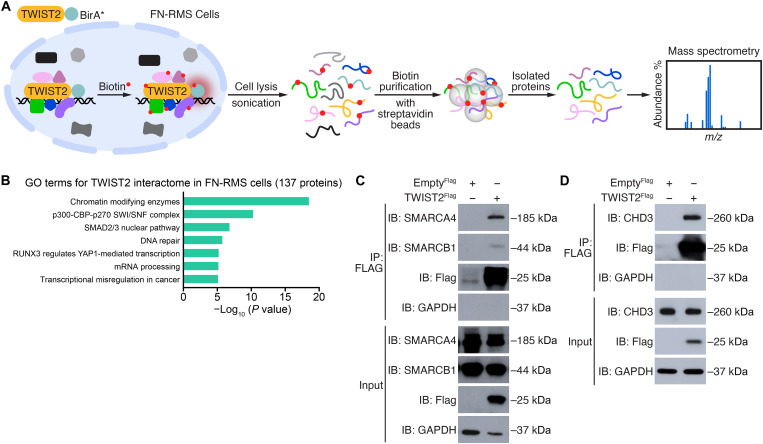
To explore the mechanism underlying TWIST2-mediated chromatin remodeling, we surveyed the TWIST2 interactome using proximity-dependent biotin identification (BioID) and mass spectrometry in RD and SMS-CTR cells (Fig. 5A). We first confirmed that the transiently expressed TWIST2miniTurbo construct used for BioID specifically localized to the nucleus (fig. S5A). We identified 137 putative TWIST2 interactors (fold change > 3; peptide-spectrum match > 2) that were enriched in the biotin-treated samples over the untreated samples in both cell lines (fig. S5B). GO analysis characterized these interactors as chromatin modifying enzymes, components of the switch/sucrose non-fermentable (SWI/SNF) complex, transcriptional and posttranscriptional regulators, and proteins in the DNA repair pathway (Fig. 5B). We investigated hits that regulate chromatin accessibility and are also expressed in FN-RMS patient tumors (30). Among these, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 4 (SMARCA4), an essential subunit of the SWI/SNF complex, and chromodomain helicase DNA binding protein 3 (CHD3), a core component of the nucleosome remodeling and deacetylase (NuRD) complex, were highly enriched in human FN-RMS tumors over control skeletal muscle (fig. S5, C and D).
Fig. 5. BioID and mass spectrometry reveal that TWIST2 interacts with chromatin remodeling enzymes in FN-RMS cells.
(A) Schematic of BioID performed in both RD and SMS-CTR cells independently to identify TWIST2-interacting proteins. A BirA* biotin ligase fused to TWIST2 (TWIST2-BirA*) labeled any proteins within 10 nm. After cell lysis and sonication, the biotin-labeled proteins were captured using streptavidin beads and identified by mass spectrometry. m/z, mass/charge ratio. (B) GO analysis of 137 high confidence TWIST2-interacting proteins that are common in RD and SMS-CTR cells. (C and D) Endogenous (C) SMARCA4 and SMARCB1 proteins and (D) CHD3 were pulled down after immunoprecipitation of Flag from lysates of RD cells infected with a lentivirus for Empty3xFlag or TWIST23xFlag. GAPDH was used as the loading control. IP, immunoprecipitate; IB, immunoblot.
We first confirmed the interactions of TWIST2 with SMARCA4 and SMARCB1 in RD cells by immunoprecipitating endogenous SMARCA4 and SMARCB1, respectively, with Flag-tagged TWIST2 protein (Fig. 5C). We also verified the observed interaction between SMARCA4 and TWIST2 in 381.T cells by using a SMARCA4 antibody to immunoprecipitate endogenous TWIST2 (fig. S5E). Moreover, we showed that TWIST2 interfaces with multiple SWI/SNF components, including the SMARCC1 and SMARCC2 core subunits and the SMARCD1 and SMARCD3 regulatory subunits (fig. S5F). The SWI/SNF complex recruits the p300 acetyl transferase to enable H3K27ac deposition and establish enhancer activity (31), raising the possibility that TWIST2 associates with SWI/SNF to similarly regulate enhancer function and gene expression in FN-RMS.
Next, we validated the interaction of TWIST2 with the endogenous CHD3 protein in RD cells (Fig. 5D). Reciprocally, we showed that endogenous CHD3 immunoprecipitated with TWIST23xFlag protein (fig. S5G). We also confirmed that TWIST2 interacts with additional components of the NurRD complex, such as methyl-CpG binding domain protein 3 (MBD3), GATA zinc finger domain containing 2A (GATAD2A), metastasis associated 1(MTA1), RB binding protein 4 (RBBP4), and histone deacetylase 1 (HDAC1), by overexpression of tagged constructs and co-immunoprecipitation (fig. S5H). In contrast to SWI/SNF, the NuRD complex plays an opposing role by removing H3K27ac from enhancers (32). This prompted us to ask whether TWIST2 acts as an activator and repressor of gene expression by interacting with functionally distinct chromatin remodeling complexes in FN-RMS. Toward this end, we interrogated the functions of SMARCA4 and CHD3 in FN-RMS cells.
Inhibition of SMARCA4 activity in FN-RMS cells represses TWIST2-activated growth genes
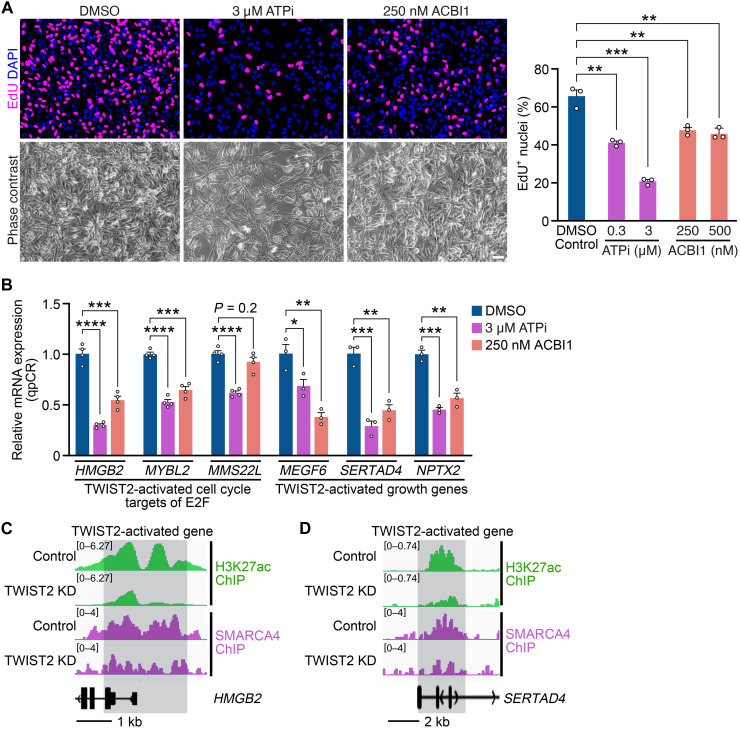
To test whether inhibiting SMARCA4 phenocopies TWIST2 KD in RD cells, we treated RD cells with two small-molecule inhibitors that target the adenosine triphosphatase (ATPase) subunit of SWI/SNF (33): an allosteric ATPase inhibitor (ATPi) (34) and a proteolysis targeting chimera (PROTAC) compound (ACBI1) (35). Increasing concentrations of both inhibitors led to reduced EdU incorporation in RD cells (Fig. 6A and fig. S6A). Therefore, we hypothesized that SMARCA4 cooperates with TWIST2 to activate growth gene expression by increasing their enhancer accessibility. To test this hypothesis, we analyzed the expression of TWIST2-activated genes in the presence of SMARCA4 inhibitors. Consistent with TWIST2 KD, ATPi and ACBI1 repressed the expression of these genes in RD cells (Fig. 6B and fig. S6B). In addition, shRNA-mediated KD of SMARCA4 (fig. S6C) recapitulated these results, demonstrating that SMARCA4 and TWIST2 regulate a similar growth gene signature in RD cells (fig. S6, D and F). We also showed that the inhibition of SMARCA4 with ATPi and SMARCA4 KD using shRNA led to decreased proliferation (fig. S6, E and F) and down-regulation of growth genes (fig. S6, G to J) in SMS-CTR and 381.T cells. These results extended the function of SMARCA4 to multiple FN-RMS cell lines. SMARCA4 inhibitors did not up-regulate the expression of myogenic genes (fig. S6K). Consistent with this finding, we did not observe morphological signs of myogenic differentiation in ATPi and ACBI1-treated cells (Fig. 6A), suggesting that the SWI/SNF complex is dispensable for inhibiting myogenesis in RD cells. Together, these findings confirmed that SWI/SNF chromatin remodelers activate the expression of growth-related target genes of TWIST2 in FN-RMS cells.
Fig. 6. Inhibition of SMARCA4 activity in FN-RMS cells represses TWIST2-activated growth genes.
(A) Left: EdU (magenta) immunocytochemistry of RD cells treated with DMSO, 3 μM ATPi, or 250 nM ACBI1 for 2 days followed by EdU incorporation for 4 hours. Nuclei were stained with DAPI (blue). Phase contrast images are shown below. Right: Quantification of the percentage of EdU+ nuclei using data from (A) and fig. S6A. Scale bar, 50 μm. n = 3 biologically independent samples. (B) qRT-PCR analysis of indicated genes in RD cells treated with DMSO, 3 μM ATPi, or 250 nM ACBI1 for 2 days (relative to DMSO, normalized to 18S rRNA). n = 3 biologically independent samples. (C and D) ChIP-seq tracks where H3K27ac peaks are shown in green, and SMARCA4 peaks are shown in purple. (C) ChIP-seq tracks of the MYOD-dependent HMGB2 locus in Control and TWIST2 KD RD cells. HMGB2 is a direct TWIST2-activated gene that showed decreases in SMARCA4 binding and H3K27ac deposition at its locus in TWIST2 KD cells. (D) ChIP-seq tracks of the MYOD-independent SERTAD4 locus in Control and TWIST2 KD RD cells. SERTAD4 is a direct TWIST2-activated gene that showed decreases in SMARCA4 binding and H3K27ac deposition at its locus in TWIST2 KD cells. All statistical comparisons between groups were evaluated by unpaired Student’s t test, ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05. Data are presented as the mean ± SEM.
To investigate SMARCA4 binding activity at TWIST2-regulated loci, we performed ChIP-seq for SMARCA4 in Control and TWIST2 KD RD cells. At TWIST2-activated loci such as HMGB2 (Fig. 6C) and SERTAD4 (Fig. 6D), SMARCA4 binding was decreased in TWIST2 KD cells, consistent with the decreased H3K27ac peaks at these loci. We previously showed that HMGB2 activation is dependent on MYOD binding (Fig. 3C), while activation of SERTAD4 is independent of MYOD in TWIST2 KD cells (Fig. 3E). This highlights that TWIST2 cooperates with SMARCA4 activity at loci of TWIST2-activated genes to promote gene expression, regardless of MYOD binding activity. Because these genes are involved in tumor-promoting pathways, the loss of SMARCA4 binding upon TWIST2 KD may explain the decrease in FN-RMS growth. At TWIST2-repressed loci such as WIPF3 (fig. S6L) and MYOM3 (fig. S6M), SMARCA4 binding was not changed upon TWIST2 KD, while H3K27ac peaks were increased. These results suggest that at TWIST2-repressed gene loci, SMARCA4 does not contribute to gene repression. Collectively, these data indicate that SMARCA4 activity determines the accessibility of TWIST2-activated growth loci.
CHD3 KD activates TWIST2-repressed myogenic genes in FN-RMS cells
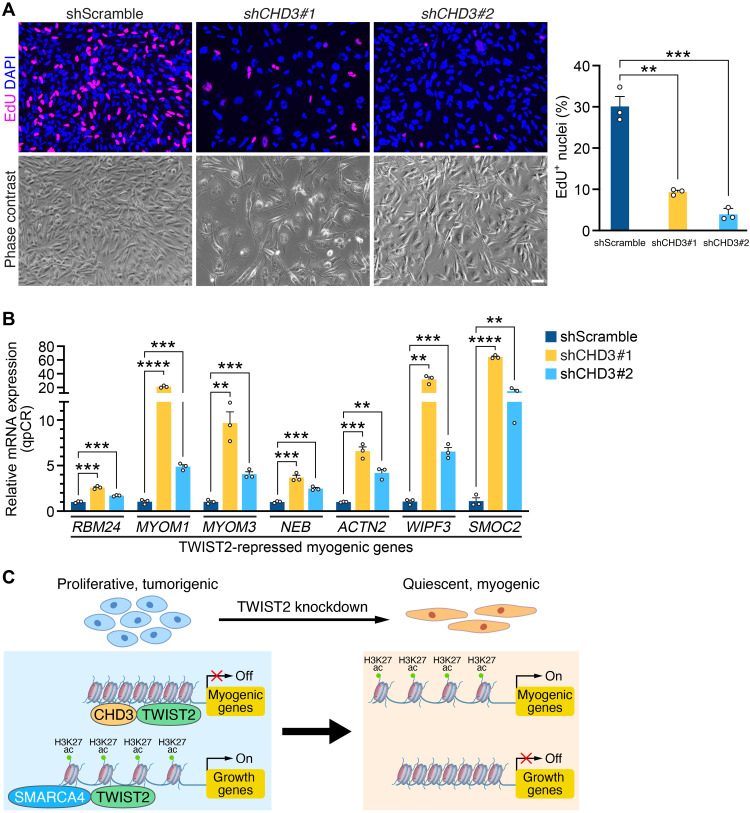
Last, to determine the functional consequences of the interaction between TWIST2 and CHD3, we performed shRNA-mediated CHD3 KD in RD cells (fig. S7A). Loss of CHD3 protein resulted in decreased proliferation and an elongated cell morphology, reflective of myogenic differentiation (Fig. 7A). Accordingly, qRT-PCR revealed that KD of CHD3 in RD cells resulted in the reactivation of TWIST2-repressed myogenic genes (Fig. 7B). We verified that CHD3 KD also decreases proliferation, leads to an elongated cell morphology, and up-regulates myogenic gene expression in SMS-CTR (fig. S7, B and D) and 381.T (fig. S7, C and E) cells. This suggests that TWIST2 recruits the deacetylase activity of the NuRD complex to suppress a myogenic gene program, which is activated similarly upon loss of TWIST2 or CHD3 in FN-RMS cells. Treating RD cells with entinostat, a class I–specific HDAC inhibitor, up-regulated the TWIST2-repressed gene signature (fig. S7F).
Fig. 7. CHD3 KD activates TWIST2-repressed myogenic genes in FN-RMS cells.
(A) Left: EdU (magenta) immunocytochemistry of RD cells infected with a shScramble control or shRNAs to knockdown CHD3 (shCHD3#1 or shCHD3#2) for 3 days followed by EdU incorporation for 4 hours. Nuclei were stained with DAPI (blue). Phase contrast images are shown below. Right: Quantification of the percentage of EdU+ nuclei. Scale bar, 50 μm. n = 3 biologically independent samples. (B) qRT-PCR analysis of indicated genes in RD cells infected with shScramble, shCHD3#1, or shCHD3#2 for 3 days (relative to shScramble, normalized to 18S rRNA). n = 3 biologically independent samples. (C) Model describing the mechanism of TWIST2 function in FN-RMS cells. In cycling cells, TWIST2 interacts with CHD3 at loci in myogenic genes to restrict H3K27ac deposition and turn off gene expression. TWIST2 also interacts with SMARCA4, and at growth gene loci, these proteins together enable H3K27ac deposition and activate gene expression. This is reversed in TWIST2 KD FN-RMS cells, where myogenic genes are activated fand growth genes are repressed, leading to cell cycle exit and terminal myogenic differentiation. All statistical comparisons between groups were evaluated by unpaired Student’s t test, ****P < 0.0001, ***P < 0.001, and **P < 0.01. Data are presented as the mean ± SEM.
In summary, analysis of the TWIST2 interactome revealed an elaborate mechanism for TWIST2-mediated activation or repression of gene expression. While the inhibition of SMARCA4 decreases expression of growth targets of TWIST2, inhibiting CHD3 increases the expression of myogenic targets of TWIST2 (Fig. 7C). We found that disrupting the activity of SMARCA4 or CHD3 chromatin remodelers reduced the oncogenic potential of FN-RMS cells by causing cell cycle arrest or pushing the tumor cells toward myogenic differentiation, together recapitulating the TWIST2 KD phenotype.
DISCUSSION
A major conclusion from this work is that the bHLH transcription factor, TWIST2, maintains FN-RMS cells in an undifferentiated, self-renewing, and tumorigenic state. We show that KD of TWIST2 in FN-RMS cells and tumor xenograft models causes cell cycle arrest and increases myogenic differentiation. Our study used ChIP-seq, RNA-seq, and proximal labeling proteomics to show that expression of TWIST2 in FN-RMS blocks the differentiation of proliferating tumor cells through its interaction with chromatin remodelers SMARCA4 and CHD3. We propose a model where TWIST2 binds to cis-regulatory loci of myogenic genes in FN-RMS cells to repress gene expression. CHD3 functions with TWIST2 to keep these loci repressed in FN-RMS cells. TWIST2 also binds to cis-regulatory loci of growth genes in FN-RMS to activate gene expression. SMARCA4 as well as E2F1 and E2F2 function with TWIST2 to keep these loci activated in FN-RMS cells.
Twist genes influence the differentiation status of sarcomas
To date, up-regulation of the TWIST protein has been reported in epithelial cancers, in which TWIST mediates cytoskeletal and cell shape changes that enable metastasis (36). TWIST proteins also promote the survival of oncogene-transformed cells by antagonizing p53 function (37, 38). In addition to FN-RMS, TWIST1 and TWIST2 are overexpressed in other mesenchymal tumors including osteosarcoma (39), synovial sarcoma (40), and leiomyosarcoma (41), yet their function in the pathogenesis of these sarcomas is not well understood. Our findings suggest that TWIST may function to maintain mesenchymal tumors in a lineage-restricted state by halting cellular differentiation. This phenomenon is akin to its role during normal development, where Twist expression is restricted to embryonic days 8.5 to 11.5 in mice, and failure to down-regulate Twist thereafter causes abnormal tissue patterning and cell fate misspecification (42).
Single-cell RNA-seq revealed a major population of undifferentiated cells that resemble the paraxial mesoderm in FN-RMS tumors but not in FP-RMS tumors (43). TWIST is responsible for specifying the paraxial mesoderm during embryogenesis (44). This may explain its abundance in FN-RMS and its underrepresentation in the more differentiated FP-RMS tumors. While this study focused on the role of TWIST2 in FN-RMS, its paralog, TWIST1, is also overexpressed specifically in FN-RMS tumors (11, 45). Because TWIST1 and TWIST2 exhibit a high degree of sequence similarity (14), it will be interesting to determine whether the two TWIST proteins act via similar or unique transcriptional mechanisms in FN-RMS.
The function of TWIST2 in FN-RMS extends beyond changes in MYOD binding at growth and myogenic loci
Prior studies in proliferating myoblasts have explored mechanisms that antagonize the induction of differentiation-specific gene programs in the presence of MyoD. A major conclusion from these studies is that Snai1, Snai2, and Twist2 bHLH transcription factors bind the canonical E-box binding motif (5′-CAGCTG-3′) in differentiation-associated genes, depriving MyoD access to these sites (46). While SNAI2 has been studied in FN-RMS, the authors did not comment on why MYOD binding activity changes at very few SNAI2-bound loci upon SNAI2 KD (46). Building on this observation, our work systematically shows that MYOD binding activity changes at a very small subset of all TWIST2-bound loci upon TWIST2 KD in FN-RMS cells. At most TWIST2-bound sites, MYOD does not bind or MYOD binding does not change between Control and TWIST2 KD conditions. This observation suggests that while MYOD binding is a prerequisite for myogenesis in normal muscle, it is not sufficient in FN-RMS, and changes in chromatin organization or DNA binding and transactivation by additional factors may be needed. These results support a previous studies’ conclusion that the overall pattern of MYOD binding and peak heights remains unchanged between RD cells and primary human myotubes (47, 48), strengthening our hypothesis that TWIST2 uses alternate mechanisms to activate or repress genes in the presence of MYOD.
To rationalize this, we analyzed TWIST2- and MYOD-bound loci in RD cells and revealed certain TWIST2-occupied motifs that are not highly enriched in MYOD peaks. TWIST2 binds to a double E-box motif and co-occupies AP-1 motifs that are typically bound by bZIP transcription factors. Prior ChIP-seq of an epethial to mesenchymal transition (EMT)-inducing bHLH transcription factor zinc finger E-box binding homeobox (ZEB1) also identified the AP-1 site among the most enriched motifs within ZEB1 peaks in a triple-negative breast cancer cell line (49). AP-1 proteins bind to their consensus motifs, allowing for ZEB1 recruitment and activation of the corresponding genes, demonstrating a context-specific function of bHLH transcription factors. This interdependence of the TWIST1 and AP-1 transcription factors has been investigated as a mechanism of EMT and tumor invasiveness (50), but a detailed understanding of their cooperation remains to be elucidated. Together, this suggests that TWIST2 regulates gene expression independent of MYOD activity, expanding the repertoire of TWIST2 function.
TWIST2-regulated pathways represent a therapeutic vulnerability in FN-RMS
Our study showed that TWIST2 acts closely with chromatin remodelers SMARCA4 and CHD3 to tightly control global gene expression. Inhibition of the ATPase subunits of the SWI/SNF complex has been described as a vulnerability in various cancers, including FP-RMS (33, 51). Small molecules directed against SMARCA4 enhanced myogenic differentiation in FP-RMS cells. However, up-regulation of differentiation genes is not observed upon inhibition of SWI/SNF activity in FN-RMS cells, suggesting that SMARCA4 mainly regulates growth in this RMS subtype. A study of Drosophila embryonic development demonstrated that Akirin, a transcription coregulator, activates Twist-regulated enhancers via the recruitment of the Bhrama-containing complex (the Drosophila SWI/SNF complex) to establish the somatic musculature (52). This supports our findings that SWI/SNF maintains the expression of a TWIST-dependent gene signature. Furthermore, TWIST1 has previously been shown to interact with components of the CHD4-containing NuRD complex to repress an epithelial gene signature, thus promoting lung metastasis of four T1 breast cancer cells (53). This bolsters our finding that TWIST2 cooperates with a CHD3-containing NuRD complex in FN-RMS to repress TWIST2-regulated enhancers and myogenic gene expression. It is likely that TWIST2- and NuRD-mediated gene repression occurs through HDACs, and previously, a class I–specific HDAC inhibitor, entinostat, was shown to have synergistic antitumor activity with vincristine in orthotopic allografts of FN-RMS (54). Overall, our results highlight that a tight control of epigenetic remodeling via TWIST2 is vital for the formation of FN-RMS, providing further rationale for targeting specific chromatin remodelers and TWIST2-regulated pathways for therapeutic benefit in this pediatric cancer.
MATERIALS AND METHODS
Experimental animals
Animal work described in this manuscript has been approved and conducted under the oversight of the UT Southwestern Institutional Animal Care and Use Committee (IACUC). Six- to 8-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (IMSR_JAX:00557) mice were used for the xenograft experiments. They were maintained at sterile conditions with five mice per cage. Animals were housed in a 12-hour light and 12-hour dark cycle in a temperature-controlled room in the Animal Research Center of UT Southwestern, with ad libitum access to water and food.
Cell lines and cell culture
RD, SMS-CTR, JR-1, RH18, 381.T, and Lenti-X 293 T cell lines, either provided by P. Houghton (St. Jude Children’s Research Hospital, Memphis, TN) or obtained from American Type Culture Collection (ATCC), were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich), supplemented with penicillin/streptomycin (100 U/ml), 2 mM l-glutamine, and 10% fetal bovine serum (FBS) (Gemini Bio Products) in 5% CO2 at 37°C. Cell lines were controlled for mycoplasma contamination using the Universal Mycoplasma Detection Kit (ATCC) and tested negative. All RMS cell lines were authenticated by short tandem repeat (STR) analysis profiling and positively matched.
Generation of stable and Dox-inducible shTWIST2 FN-RMS cells
To generate an inducible shTWIST2 (i-shTWIST2)–expressing plasmid, the stuffer DNA was removed from the pLKO-tet-On-puro backbone (Addgene #21915) (55, 56) by Age I/Eco RI digest and replaced with double-stranded oligos encoding shTWIST2 and Age I/ Eco RI sites (CCGGCGACGAGATGGACAATAAGATCTCGAGATCTTATTGTCCATCTCGTCGTTTTT, Sigma-Aldrich, TRCN0000020869).
Lentivirus was produced as mentioned in Supplementary Text. Forty-eight hours after lentivirus infection into FN-RMS cells of choice, they were replated. The next day, cells were selected with puromycin (1 μg/ml) added to DMEM with 10% tet system-approved FBS (Takara) and antibiotics (tet system growth medium) until the uninfected control cells detached. This technique established pooled puromycin-resistant stable cells, and cells are always maintained in puromycin-supplemented cell culture media. i-shTWIST2 cells (TWIST2-KD cells) were plated at a density of 6 × 104 cells per well of 12-well plates and were maintained in the tet system growth medium. The cells were treated with Dox (10 ng/ml; Sigma-Aldrich) for 4 days (TWIST2 KD) before analysis. Control cells were left untreated.
EdU labeling assay
Cells were treated with 10 μM EdU (Lumiprobe, 10540) for 4 hours. After that, cells were fixed with 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) at room temperature for 10 min, permeabilized with 0.3% Triton X-100 in PBS, followed by EdU staining by click chemistry (a label mix containing 8 μM sulfo-Cy5-azide, 2 mM CuSO4·5H2O, and 100 mM sodium ascorbate was applied to cells for 30 min). Next, nuclei were counter-stained with 4′,6-diamidino-2-phenylindole (DAPI) (1:2000; Invitrogen, D1306). Imaging was performed on a Keyence BZ-X700 microscope. Six fields at ×10 magnification were captured for each well. Quantification was carried out by counting the proportion of EdU+ nuclei/total DAPI-stained nuclei on the immunofluorescent staining images.
Inhibitor treatment and shRNA KD in RD cells
RD cells were plated at a density of 6 × 104 cells per well of 12-well plates and were maintained in DMEM with 10% FBS and antibiotics. Cells were treated with 0.3 or 3 μM BRM (Brahma)/BRG1 (Brahma-related gene-1) ATP inhibitor-1 (ATPi) (MedChemExpress, HY-119374), 250 or 500 nM PROTAC Baf degrader (ACBI1) (MedChemExpress, HY-128359), or 4 μM of entinostat (APExBIO, A8171) for 2 days before EdU incorporation and staining. For shRNA KD, cells were treated with shRNAs (predesigned pLKO.1-puro, Sigma-Aldrich) targeting CHD3 (shCHD3#1: CGCAAGCAAGTTAACTACAAT, Sigma-Aldrich, TRCN0000107974 and shCHD3#2: CCTCCCACACTGCCAAGTATA, Sigma-Aldrich, TRCN0000107970) or SMARCA4 (shSMARCA4#1: CGGCAGACACTGTGATCATT, Sigma-Aldrich, TRCN0000015552 and shSMARCA4#2: TGGAGCACAAACGCATCATG, Sigma-Aldrich, TRCN0000379829) for 3 days before EdU incorporation and staining.
Colony formation assay
i-shTWIST2 FN-RMS cells were plated at a density of 1 × 103 in six-well plates with 2 ml of tet system growth media. After 3 days, triplicate wells were treated with or without Dox (10 ng/ml). Medium (± Dox) was refreshed every 3 days. Then, 15 days later, cells were fixed in 4% PFA/PBS and stained with 0.5% crystal violet staining solution (0.125 g of crystal violet powder dissolved in 50 ml of 20% MeOH; Sigma-Aldrich) for 10 min at room temperature. Plates were rinsed six times with H2O and air-dried before scanning and quantification. Quantification was performed using the ColonyArea ImageJ plugin (57).
Immunofluorescence in cells
i-shTWIST2 RD cells were plated at a density of 1.3 × 105 cells per well of a six-well dish and treated with or without Dox (10 ng/ml) in tet approved growth media for 3 days. Cells were grown for 5 days in differentiation medium [DMEM with 2% horse serum, penicillin/streptomycin (100 U/ml), and 2 mM l-glutamine]. Cells were then fixed in 4% PFA/PBS for 10 min, permeabilized in 0.5% Triton X-100/PBS for 10 min, and incubated with monoclonal anti-MyHC (skeletal, Fast) antibody (clone MY-32) (1:400; Sigma-Aldrich, M4276) in 2% goat serum/PBS overnight at 4°C. Cells were next incubated with Alexa Fluor 488 goat anti-mouse (Invitrogen, 1:400) for 1 hour at room temperature. Cells were counterstained with DAPI and imaged using the Keyence BZ-X700 microscope. Images were processed in ImageJ, and quantification was carried out by counting the proportion of MyHC nuclei/total DAPI-stained nuclei.
Mouse RMS xenograft generation
Six- to 8-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (IMSR_JAX:00557) mice were used for the xenograft experiments. Experimental groups were randomly assigned. i-shTWIST2 stable RD cells were suspended in a 1:1 solution of PBS/Matrigel (Matrigel Matrix Phenol Red Free, LDEV-free; Corning, 356237). Then, 5 × 106 cells were injected subcutaneously. Water containing Dox (2 mg/ml) plus 5% (w/v) sucrose (TWIST2 KD) or 5% (w/v) sucrose (Control) was delivered immediately after tumors reach 100 to 200 mm3. Water with Dox was changed every 2 to 3 days. We used an electronic caliper to measure the length (the greatest longitudinal diameter) and width (the greatest transverse diameter) of tumors. We calculated tumor volume as tumor volume length * (width)2 * 0.52. The investigators were not blinded to the group allocation during the experiment or when assessing the outcome.
Histology and immunochemistry
Dissected tumors were fixed in 10% neutral-buffered formalin overnight, embedded in paraffin, and sectioned at 5-μm intervals. Hematoxylin counterstain was performed using standard procedures. All paraffin-embedded sections for immunohistochemistry were deparaffinized, heated in a microwave in 0.01 M sodium citrate buffer for antigen retrieval, treated with 3% H2O2 for 10 min, and rinsed in H2O and PBS. For the anti-Ki67 antibody (1:1000; Cell Signaling Technology, #9027) immunohistochemistry, sections were blocked in 10% goat serum in PBS followed by primary antibody incubation overnight at 4°C. For mouse monoclonal MF20 (1:10; Developmental Studies Hybridoma Bank (DSHB), supernatant), the M.O.M Immunodetection Kit (Vector Laboratories, BMK-2202) was used followed by primary antibody incubation overnight at 4°C. Detection of the primary antibody was performed by using the appropriate secondary biotinylated antibody (Vector laboratories, BA-1000), streptavidin peroxidase (SA-5004), and 3,3′-diaminobenzidine substrate (Vector Laboratories, SK-4100), with or without counterstaining with Gill’s hematoxylin (Dako, Carpinteria, USA). Negative controls were stained in parallel with either isotype nonspecific immunoglobulin G or only the primary antibody. Slides were imaged using the Keyence BZ-X700 microscope, 10× or 20× objective.
RNA extraction and qRT-PCR
To assess gene expression by qRT-PCR, total RNA was extracted from ventricles using TRIzol (Invitrogen, 15596018) according to the manufacturer’s protocol. cDNA was synthesize using iScript Reverse Transcription Supermix (Bio-Rad, 1708840). Gene expression was assessed using KAPA SYBR FAST kit (KAPA, KK4605) and quantified using the delta-delta Ct (ddCt) method. Primers are indicated in table S1.
Co-immunoprecipitation
Co-immunoprecipitation (CoIP) was performed as previously described (58), with modifications. Three days after infection, cells were scraped in ice-cold PBS and lysed in 800 μl of lysis buffer [50 mM tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% sodium deoxycholate, and 2× protease inhibitor tablet (Roche)] for 30 min on ice and cleared by centrifugation at 14,000g for 10 min at 4°C. Lysates (50 μl) were saved for “Input”. For each pull-down, 2 μg of Flag M2 antibody (Sigma-Aldrich) was bound to 20 μl of washed Dynabeads Protein G (Invitrogen). Lysates were then added to antibody-coupled beads. Reactions were incubated on a rotating platform for 3 hour at 4°C, and then beads were washed five times with 1 ml of wash buffer [50 mM tris-HCl at pH 7.5, 150 mM NaCl, 0.05% NP-40, and 2× protease inhibitor tablet (Roche)]. Immunoprecipitated proteins were eluted with 40 μl of elution buffer [36 μl of lysis buffer supplemented with 4 μl of 3× Flag peptide at 10 mg/ml (Sigma-Aldrich)] with shaking at 1000 rpm for 30 min at room temperature. Immunoblotting for Input and CoIP samples was performed as described earlier.
Statistics
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software Inc.) using a two-tailed unpaired t test, with P < 0.05 considered significant unless otherwise indicated. All data are displayed as means ± SEM unless otherwise indicated. False discovery rate correction for multiple comparisons followed the Benjamini-Hochberg procedure.
Acknowledgments
We thank S. Skapek’s and R. Gallindo’s laboratories for providing FN-RMS cell lines, J. Xu and Y. J. Kim from the Children’s Research Institute Sequencing Facility for Illumina sequencing, R. Kittler and V. Schmid from the McDermott Center Next Generation Sequencing (NGS) Core and P. Raj and C. Liang from the Genomics Core facilities for NGS, A. Lemoff from Proteomics Core for mass spectrometry, J. Shelton from the Molecular Histopathology Core for assisting with tissue processing and H&E, and L. Girard from the Sanger Sequencing Core for STR analysis of cell lines. We are grateful to J. Cabrera for graphics assistance and S. Li (Northwestern University), M. Cui (Boston Children’s Hospital, Harvard), M. Dos Santos, and Z. Wang (University of California San Diego) for the helpful discussion.
Funding: This work was supported by funds from the Cancer Prevention and Research Institute of Texas (CPRIT, RP200103 to E.N.O.) and NIH (AR071980 to N.L.). A.M.S. was supported by a Hamon Center of Regenerative Science and Medicine (CRSM) Fellowship.
Author contributions: A.M.S. and N.L. conceptualized and designed the study and wrote the manuscript. N.L., E.N.O., R.B.-D., and L.X. edited the manuscript. A.M.S., M.G.M., P.J., H.H., and K.C.H. performed the experiments. L.G. and K.C. performed the ChIP-seq and RNA-seq analysis and analyzed the human patient sequencing data. All authors discussed the results, vetted, and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE207170 and are publicly available as of the date of publication. A published GEO dataset on Gene Expression in Human Rhabdomyosarcoma was also used in this study (GSE108022; fig. S5, C and D) (30).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S7
Tables S1 and S2
References
REFERENCES AND NOTES
- 1.M. Buckingham, P. W. J. Rigby, Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225–238 (2014). [DOI] [PubMed] [Google Scholar]
- 2.C. F. Bentzinger, Y. X. Wang, M. A. Rudnicki, Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 4, a008342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. X. Skapek, A. Ferrari, A. A. Gupta, P. J. Lupo, E. Butler, J. Shipley, F. G. Barr, D. S. Hawkins, Rhabdomyosarcoma. Nat. Rev. Dis. Primers 5, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Saab, S. L. Spunt, S. X. Skapek, Myogenesis and rhabdomyosarcoma the Jekyll and Hyde of skeletal muscle. Curr. Top. Dev. Biol. 94, 197–234 (2011). [DOI] [PubMed] [Google Scholar]
- 5.S. J. Tapscott, M. J. Thayer, H. Weintraub, Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science 259, 1450–1453 (1993). [DOI] [PubMed] [Google Scholar]
- 6.M. Tsokos, The diagnosis and classification of childhood rhabdomyosarcoma. Semin. Diagn. Pathol. 11, 26–38 (1994). [PubMed] [Google Scholar]
- 7.B. E. Gryder, M. E. Yohe, H. C. Chou, X. Zhang, J. Marques, M. Wachtel, B. Schaefer, N. Sen, Y. Song, A. Gualtieri, S. Pomella, R. Rota, A. Cleveland, X. Wen, S. Sindiri, J. S. Wei, F. G. Barr, S. das, T. Andresson, R. Guha, M. Lal-Nag, M. Ferrer, J. F. Shern, K. Zhao, C. J. Thomas, J. Khan, PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 7, 884–899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Seki, R. Nishimura, K. Yoshida, T. Shimamura, Y. Shiraishi, Y. Sato, M. Kato, K. Chiba, H. Tanaka, N. Hoshino, G. Nagae, Y. Shiozawa, Y. Okuno, H. Hosoi, Y. Tanaka, H. Okita, M. Miyachi, R. Souzaki, T. Taguchi, K. Koh, R. Hanada, K. Kato, Y. Nomura, M. Akiyama, A. Oka, T. Igarashi, S. Miyano, H. Aburatani, Y. Hayashi, S. Ogawa, J. Takita, Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat. Commun. 6, 7557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. F. Shern, L. Chen, J. Chmielecki, J. S. Wei, R. Patidar, M. Rosenberg, L. Ambrogio, D. Auclair, J. Wang, Y. K. Song, C. Tolman, L. Hurd, H. Liao, S. Zhang, D. Bogen, A. S. Brohl, S. Sindiri, D. Catchpoole, T. Badgett, G. Getz, J. Mora, J. R. Anderson, S. X. Skapek, F. G. Barr, M. Meyerson, D. S. Hawkins, J. Khan, Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 4, 216–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.L. Xu, Y. Zheng, J. Liu, D. Rakheja, S. Singleterry, T. W. Laetsch, J. F. Shern, J. Khan, T. J. Triche, D. S. Hawkins, J. F. Amatruda, S. X. Skapek, Integrative Bayesian analysis identifies rhabdomyosarcoma disease genes. Cell Rep. 24, 238–251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. Li, K. Chen, Y. Zhang, S. D. Barnes, P. Jaichander, Y. Zheng, M. Hassan, V. S. Malladi, S. X. Skapek, L. Xu, R. Bassel-Duby, E. N. Olson, N. Liu, Twist2 amplification in rhabdomyosarcoma represses myogenesis and promotes oncogenesis by redirecting MyoD DNA binding. Genes Dev. 33, 626–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P. Bialek, B. Kern, X. Yang, M. Schrock, D. Sosic, N. Hong, H. Wu, K. Yu, D. M. Ornitz, E. N. Olson, M. J. Justice, G. Karsenty, A twist code determines the onset of osteoblast differentiation. Dev. Cell 6, 423–435 (2004). [DOI] [PubMed] [Google Scholar]
- 13.B. A. Firulli, D. Krawchuk, V. E. Centonze, N. Vargesson, D. M. Virshup, S. J. Conway, P. Cserjesi, E. Laufer, A. B. Firulli, Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat. Genet. 37, 373–381 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. L. Franco, J. Casasnovas, J. R. Rodriguez-Medina, C. L. Cadilla, Redundant or separate entities?—roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 39, 1177–1186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Hebrok, K. Wertz, E. M. Fuchtbauer, M-twist is an inhibitor of muscle differentiation. Dev. Biol. 165, 537–544 (1994). [DOI] [PubMed] [Google Scholar]
- 16.D. B. Spicer, J. Rhee, W. L. Cheung, A. B. Lassar, Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272, 1476–1480 (1996). [DOI] [PubMed] [Google Scholar]
- 17.P. Parajuli, S. Kumar, A. Loumaye, P. Singh, S. Eragamreddy, T. L. Nguyen, S. Ozkan, M. S. Razzaque, C. Prunier, J.-P. Thissen, A. Atfi, Twist1 activation in muscle progenitor cells causes muscle loss akin to cancer cachexia. Dev. Cell 45, 712–725.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.P. Zhang, C. Wong, D. Liu, M. Finegold, J. W. Harper, S. J. Elledge, p21CIP1 and p57KIP2 control muscle differentiation at the myogenin step. Genes Dev. 13, 213–224 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.I. L. de la Serna, Y. Ohkawa, C. A. Berkes, D. A. Bergstrom, C. S. Dacwag, S. J. Tapscott, A. N. Imbalzano, MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25, 3997–4009 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Liberzon, C. Birger, H. Thorvaldsdóttir, M. Ghandi, J. P. Mesirov, P. Tamayo, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. T. Chang, Y. Liu, K. Ayyanathan, C. Benner, Y. Jiang, J. W. Prokop, H. Paz, D. Wang, H. R. Li, X. D. Fu, F. J. Rauscher III, J. Yang, An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev. 29, 603–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Trapnell, D. Cacchiarelli, J. Grimsby, P. Pokharel, S. Li, M. Morse, N. J. Lennon, K. J. Livak, T. S. Mikkelsen, J. L. Rinn, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. Y. Kim, M. Park, J. Ohn, R. H. Seong, J. H. Chung, K. H. Kim, S. J. Jo, O. Kwon, Twist2-driven chromatin remodeling governs the postnatal maturation of dermal fibroblasts. Cell Rep. 39, 110821 (2022). [DOI] [PubMed] [Google Scholar]
- 24.M. P. Creyghton, A. W. Cheng, G. G. Welstead, T. Kooistra, B. W. Carey, E. J. Steine, J. Hanna, M. A. Lodato, G. M. Frampton, P. A. Sharp, L. A. Boyer, R. A. Young, R. Jaenisch, Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.B. E. Gryder, S. Pomella, C. Sayers, X. S. Wu, Y. Song, A. M. Chiarella, S. Bagchi, H. C. Chou, R. S. Sinniah, A. Walton, X. Wen, R. Rota, N. A. Hathaway, K. Zhao, J. Chen, C. R. Vakoc, J. F. Shern, B. Z. Stanton, J. Khan, Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat. Genet. 51, 1714–1722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. Zeid, M. A. Lawlor, E. Poon, J. M. Reyes, M. Fulciniti, M. A. Lopez, T. G. Scott, B. Nabet, M. A. Erb, G. E. Winter, Z. Jacobson, D. R. Polaski, K. L. Karlin, R. A. Hirsch, N. P. Munshi, T. F. Westbrook, L. Chesler, C. Y. Lin, J. E. Bradner, Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 50, 515–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. Joshi, G. Davidson, S. le Gras, S. Watanabe, T. Braun, G. Mengus, I. Davidson, TEAD transcription factors are required for normal primary myoblast differentiation in vitro and muscle regeneration in vivo. PLOS Genet. 13, e1006600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L. Daury, M. Busson, N. Tourkine, F. Casas, I. Cassar-Malek, C. Wrutniak-Cabello, M. Castellazzi, G. Cabello, Opposing functions of ATF2 and Fos-like transcription factors in c-Jun-mediated myogenin expression and terminal differentiation of avian myoblasts. Oncogene 20, 7998–8008 (2001). [DOI] [PubMed] [Google Scholar]
- 29.S. P. Chellappan, S. Hiebert, M. Mudryj, J. M. Horowitz, J. R. Nevins, The E2F transcription factor is a cellular target for the RB protein. Cell 65, 1053–1061 (1991). [DOI] [PubMed] [Google Scholar]
- 30.M. N. Hayes, K. McCarthy, A. Jin, M. L. Oliveira, S. Iyer, S. P. Garcia, S. Sindiri, B. Gryder, Z. Motala, G. P. Nielsen, J. P. Borg, M. van de Rijn, D. Malkin, J. Khan, M. S. Ignatius, D. M. Langenau, Vangl2/RhoA signaling pathway regulates stem cell self-renewal programs and growth in rhabdomyosarcoma. Cell Stem Cell 22, 414–427.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B. H. Alver, K. H. Kim, P. Lu, X. Wang, H. E. Manchester, W. Wang, J. R. Haswell, P. J. Park, C. W. M. Roberts, The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 8, 14648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A. Y. Lai, P. A. Wade, Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D. Laubscher, B. E. Gryder, B. D. Sunkel, T. Andresson, M. Wachtel, S. das, B. Roschitzki, W. Wolski, X. S. Wu, H. C. Chou, Y. K. Song, C. Wang, J. S. Wei, M. Wang, X. Wen, Q. A. Ngo, J. G. Marques, C. R. Vakoc, B. W. Schäfer, B. Z. Stanton, J. Khan, BAF complexes drive proliferation and block myogenic differentiation in fusion-positive rhabdomyosarcoma. Nat. Commun. 12, 6924 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. P. N. Papillon, K. Nakajima, C. D. Adair, J. Hempel, A. O. Jouk, R. G. Karki, S. Mathieu, H. Möbitz, R. Ntaganda, T. Smith, M. Visser, S. E. Hill, F. K. Hurtado, G. Chenail, H.-E. C. Bhang, A. Bric, K. Xiang, G. Bushold, T. Gilbert, A. Vattay, J. Dooley, E. A. Costa, I. Park, A. Li, D. Farley, E. Lounkine, Q. K. Yue, X. Xie, X. Zhu, R. Kulathila, D. King, T. Hu, K. Vulic, J. Cantwell, C. Luu, Z. Jagani, Discovery of orally active inhibitors of Brahma homolog (BRM)/SMARCA2 ATPase activity for the treatment of Brahma related gene 1 (BRG1)/SMARCA4-mutant cancers. J. Med. Chem. 61, 10155–10172 (2018). [DOI] [PubMed] [Google Scholar]
- 35.W. Farnaby, M. Koegl, M. J. Roy, C. Whitworth, E. Diers, N. Trainor, D. Zollman, S. Steurer, J. Karolyi-Oezguer, C. Riedmueller, T. Gmaschitz, J. Wachter, C. Dank, M. Galant, B. Sharps, K. Rumpel, E. Traxler, T. Gerstberger, R. Schnitzer, O. Petermann, P. Greb, H. Weinstabl, G. Bader, A. Zoephel, A. Weiss-Puxbaum, K. Ehrenhöfer-Wölfer, S. Wöhrle, G. Boehmelt, J. Rinnenthal, H. Arnhof, N. Wiechens, M.-Y. Wu, T. Owen-Hughes, P. Ettmayer, M. Pearson, D. B. McConnell, A. Ciulli, BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15, 672–680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Q. Qin, Y. Xu, T. He, C. Qin, J. Xu, Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 22, 90–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.S. Valsesia-Wittmann, M. Magdeleine, S. Dupasquier, E. Garin, A.-C. Jallas, V. Combaret, A. Krause, P. Leissner, A. Puisieux, Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell 6, 625–630 (2004). [DOI] [PubMed] [Google Scholar]
- 38.C. G. Pham, C. Bubici, F. Zazzeroni, J. R. Knabb, S. Papa, C. Kuntzen, G. Franzoso, Upregulation of Twist-1 by NF-κB blocks cytotoxicity induced by chemotherapeutic drugs. Mol. Cell. Biol. 27, 3920–3935 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. Entz-Werle, C. Stoetzel, P. Berard-Marec, C. Kalifa, L. Brugiere, H. Pacquement, C. Schmitt, M.-D. Tabone, J.-C. Gentet, R. Quillet, P. Oudet, P. Lutz, A. Babin-Boilletot, M.-P. Gaub, F. Perrin-Schmitt, Frequent genomic abnormalities at TWIST in human pediatric osteosarcomas. Int. J. Cancer 117, 349–355 (2005). [DOI] [PubMed] [Google Scholar]
- 40.K.-W. Lee, N. K. Lee, S. Ham, T.-Y. Roh, S.-H. Kim, Twist1 is essential in maintaining mesenchymal state and tumor-initiating properties in synovial sarcoma. Cancer Lett. 343, 62–73 (2014). [DOI] [PubMed] [Google Scholar]
- 41.S. Piccinin, E. Tonin, S. Sessa, S. Demontis, S. Rossi, L. Pecciarini, L. Zanatta, F. Pivetta, A. Grizzo, M. Sonego, C. Rosano, A. P. D. Tos, C. Doglioni, R. Maestro, A "twist box" code of p53 inactivation: Twist box: p53 interaction promotes p53 degradation. Cancer Cell 22, 404–415 (2012). [DOI] [PubMed] [Google Scholar]
- 42.M. P. O'Rourke, P. P. L. Tam, Twist functions in mouse development. Int. J. Dev. Biol. 46, 401–413 (2002). [PubMed] [Google Scholar]
- 43.A. G. Patel, X. Chen, X. Huang, M. R. Clay, N. L. Komarova, M. J. Krasin, A. Pappo, H. Tillman, B. A. Orr, J. McEvoy, B. Gordon, K. Blankenship, C. Reilly, X. Zhou, J. L. Norrie, A. Karlstrom, J. Yu, D. Wodarz, E. Stewart, M. A. Dyer, The myogenesis program drives clonal selection and drug resistance in rhabdomyosarcoma. Dev. Cell 57, 1226–1240.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Z. F. Chen, R. R. Behringer, twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9, 686–699 (1995). [DOI] [PubMed] [Google Scholar]
- 45.R. Maestro, A. P. D. Tos, Y. Hamamori, S. Krasnokutsky, V. Sartorelli, L. Kedes, C. Doglioni, D. H. Beach, G. J. Hannon, twist is a potential oncogene that inhibits apoptosis. Genes Dev. 13, 2207–2217 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.S. Pomella, P. Sreenivas, B. E. Gryder, L. Wang, D. Milewski, M. Cassandri, K. Baxi, N. R. Hensch, E. Carcarino, Y. Song, H. C. Chou, M. E. Yohe, B. Z. Stanton, B. Amadio, I. Caruana, C. de Stefanis, R. de Vito, F. Locatelli, Y. Chen, E. Y. Chen, P. Houghton, J. Khan, R. Rota, M. S. Ignatius, Interaction between SNAI2 and MYOD enhances oncogenesis and suppresses differentiation in Fusion Negative Rhabdomyosarcoma. Nat. Commun. 12, 192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.K. L. MacQuarrie, Z. Yao, A. P. Fong, S. J. Diede, E. R. Rudzinski, D. S. Hawkins, S. J. Tapscott, Comparison of genome-wide binding of MyoD in normal human myogenic cells and rhabdomyosarcomas identifies regional and local suppression of promyogenic transcription factors. Mol. Cell. Biol. 33, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. E. Yohe, B. E. Gryder, J. F. Shern, Y. K. Song, H.-C. Chou, S. Sindiri, A. Mendoza, R. Patidar, X. Zhang, R. Guha, D. Butcher, K. A. Isanogle, C. M. Robinson, X. Luo, J.-Q. Chen, A. Walton, P. Awasthi, E. F. Edmondson, S. Difilippantonio, J. S. Wei, K. Zhao, M. Ferrer, C. J. Thomas, J. Khan, MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci. Transl. Med. 10, eaan4470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.N. Feldker, F. Ferrazzi, H. Schuhwerk, S. A. Widholz, K. Guenther, I. Frisch, K. Jakob, J. Kleemann, D. Riegel, U. Bönisch, S. Lukassen, R. L. Eccles, C. Schmidl, M. P. Stemmler, T. Brabletz, S. Brabletz, Genome-wide cooperation of EMT transcription factor ZEB1 with YAP and AP-1 in breast cancer. EMBO J. 39, e103209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E.-H. Nam, Y. Lee, B. Moon, J. W. Lee, S. Kim, Twist1 and AP-1 cooperatively upregulate integrin α5 expression to induce invasion and the epithelial-mesenchymal transition. Carcinogenesis 36, 327–337 (2015). [DOI] [PubMed] [Google Scholar]
- 51.J. Cyrta, A. Augspach, M. R. de Filippo, D. Prandi, P. Thienger, M. Benelli, V. Cooley, R. Bareja, D. Wilkes, S. S. Chae, P. Cavaliere, N. Dephoure, A.-C. Uldry, S. B. Lagache, L. Roma, S. Cohen, M. Jaquet, L. P. Brandt, M. Alshalalfa, L. Puca, A. Sboner, F. Feng, S. Wang, H. Beltran, T. Lotan, M. Spahn, M. Kruithof-de Julio, Y. Chen, K. V. Ballman, F. Demichelis, S. Piscuoglio, M. A. Rubin, Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat. Commun. 11, 5549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. J. Nowak, H. Aihara, K. Gonzalez, Y. Nibu, M. K. Baylies, Akirin links twist-regulated transcription with the Brahma chromatin remodeling complex during embryogenesis. PLOS Genet. 8, e1002547 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.J. Fu, L. Qin, T. He, J. Qin, J. Hong, J. Wong, L. Liao, J. Xu, The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 21, 275–289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.N. Bharathy, N. E. Berlow, E. Wang, J. Abraham, T. P. Settelmeyer, J. E. Hooper, M. N. Svalina, Z. Bajwa, M. W. Goros, B. S. Hernandez, J. E. Wolff, R. Pal, A. M. Davies, A. Ashok, D. Bushby, M. Mancini, C. Noakes, N. C. Goodwin, P. Ordentlich, J. Keck, D. S. Hawkins, E. R. Rudzinski, A. Mansoor, T. J. Perkins, C. R. Vakoc, J. E. Michalek, C. Keller, Preclinical rationale for entinostat in embryonal rhabdomyosarcoma. Skelet. Muscle 9, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D. Wiederschain, W. Susan, L. Chen, A. Loo, G. Yang, A. Huang, Y. Chen, G. Caponigro, Y.-M. Yao, C. Lengauer, W. R. Sellers, J. D. Benson, Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009). [DOI] [PubMed] [Google Scholar]
- 56.S. Wee, D. Wiederschain, S. M. Maira, A. Loo, C. Miller, R. deBeaumont, F. Stegmeier, Y. M. Yao, C. Lengauer, PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. U.S.A. 105, 13057–13062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.C. Guzman, M. Bagga, A. Kaur, J. Westermarck, D. Abankwa, ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLOS ONE 9, e92444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. Thomas, F. Rehfeld, H. Zhang, T. C. Chang, M. Goodarzi, F. Gillet, J. T. Mendell, RBM33 directs the nuclear export of transcripts containing GC-rich elements. Genes Dev. 36, 550–565 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Z. Wang, M. Cui, A. M. Shah, W. Ye, W. Tan, Y.-L. Min, G. A. Botten, J. M. Shelton, N. Liu, R. Bassel-Duby, E. N. Olson, Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. Proc. Natl. Acad. Sci. U.S.A. 116, 18455–18465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.C. W. Law, Y. Chen, W. Shi, G. K. Smyth, voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.T. Wu, E. Hu, S. Xu, M. Chen, P. Guo, Z. Dai, T. Feng, L. Zhou, W. Tang, L. Zhan, X. Fu, S. Liu, X. Bo, G. Yu, clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. Lawrence, W. Huber, H. Pagès, P. Aboyoun, M. Carlson, R. Gentleman, M. T. Morgan, V. J. Carey, Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Y. Zhou, B. Zhou, L. Pache, M. Chang, A. H. Khodabakhshi, O. Tanaseichuk, C. Benner, S. K. Chanda, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.C. Y. McLean, D. Bristor, M. Hiller, S. L. Clarke, B. T. Schaar, C. B. Lowe, A. M. Wenger, G. Bejerano, GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.T. C. Branon, J. A. Bosch, A. D. Sanchez, N. D. Udeshi, T. Svinkina, S. A. Carr, J. L. Feldman, N. Perrimon, A. Y. Ting, Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S7
Tables S1 and S2
References