Abstract
Intestinal sulfate-reducing bacteria (SRB) growth and resultant hydrogen sulfide production may damage the gastrointestinal epithelium and thereby contribute to chronic intestinal disorders. However, the ecology and phylogenetic diversity of intestinal dissimilatory SRB populations are poorly understood, and endogenous or exogenous sources of available sulfate are not well defined. The succession of intestinal SRB was therefore compared in inbred C57BL/6J mice using a PCR-based metabolic molecular ecology (MME) approach that targets a conserved region of subunit A of the adenosine-5′-phosphosulfate (APS) reductase gene. The APS reductase-based MME strategy revealed intestinal SRB in the stomach and small intestine of 1-, 4-, and 7-day-old mice and throughout the gastrointestinal tract of 14-, 21-, 30-, 60-, and 90-day-old mice. Phylogenetic analysis of APS reductase amplicons obtained from the stomach, middle small intestine, and cecum of neonatal mice revealed that Desulfotomaculum spp. may be a predominant SRB group in the neonatal mouse intestine. Dot blot hybridizations with SRB-specific 16S ribosomal DNA (rDNA) probes demonstrated SRB colonization of the cecum and colon pre- and postweaning and colonization of the stomach and small intestine of mature mice only. The 16S rDNA hybridization data further demonstrated that SRB populations were most numerous in intestinal regions harboring sulfomucin-containing goblet cells, regardless of age. Reverse transcriptase PCR analysis demonstrated APS reductase mRNA expression in all intestinal segments of 30-day-old mice, including the stomach. These results demonstrate for the first time widespread colonization of the mouse intestine by dissimilatory SRB and evidence of spatial-specific SRB populations and sulfomucin patterns along the gastrointestinal tract.
The toxic gas hydrogen sulfide (H2S) is generated from sulfate during anaerobic respiration by sulfate-reducing Archaea and Bacteria (21, 58). A possible link between H2S and chronic intestinal disorders has been evoked by data indicating increased numbers of intestinal sulfate-reducing bacteria (SRB) and rates of sulfidogenesis in inflammatory bowel disease (IBD) patients compared to healthy humans (12, 37). Hydrogen sulfide selectively impairs the oxidation of n-butyrate by colonic epithelial cells (42). Because membrane lipid biosynthesis, ion absorption, mucin synthesis, and detoxification processes in colonocytes depend on the oxidation of n-butyrate, diminished n-butyrate metabolism is likely to compromise the epithelial cell barrier (42). Sulfide-induced damage of the epithelial barrier function would promote translocation of bacterial and food antigens, resulting in local inflammatory responses to normally benign antigens, an outcome consistent with histopathological features of IBD (16, 61). Chronic exposure to H2S might also perturb normal cycles of epithelial renewal in the intestine, thereby predisposing to proliferative disorders such as colon cancer.
Intestinal sulfate can be derived either from exogenous sources, namely sulfate in drinking water and dietary foodstuffs, or from endogenous sources such as sulfated mucins (sulfomucins), sulfate-conjugated bile, and chondroitin sulfate. Use of chemically bound, endogenous sulfate by SRB is facilitated through interactions with sulfatase-harboring bacteria (e.g., Bacteroides spp. [56]). Most goblet cells, a differentiated epithelial cell subtype that produces mucins, generate sulfomucins (22). The degree of sulfation, however, increases from proximal to distal segments of the intestine and is highest in those segments harboring dense bacterial populations, such as the cecum and colon (9, 20).
The ecology and taxonomy of intestinal SRB and their metabolic activities remain uncharacterized. Most studies of human intestinal SRB have relied on cultivation-based microbiological analyses of fecal samples (2, 3, 10, 11, 12, 38). Reports of laboratory mouse or rat intestinal SRB are also lacking, despite the common use of rodents as models of human IBD and colon cancer.
The present study defines the succession of intestinal SRB relative to the presence of sulfomucins in distinct anatomical segments of the mouse gastrointestinal tract using a culture-independent, molecular metabolic ecology (MME) approach that targets an enzyme essential for microbial sulfate reduction (Fig. 1 [35, 53]). The utility of such an approach for analysis and characterization of dissimilatory SRB populations was demonstrated recently by Schramm and coworkers (45), who screened aerated active sludge systems for sulfate-reducing organisms by targeting the dissimilatory sulfite reductase gene. Similarly, by targeting a conserved segment of the adenosine-5′-phosphosulfate (APS) reductase subunit A gene, we were able to detect the presence of organisms harboring that gene in distinct intestinal segments via PCR amplification from a community DNA sample and also to evaluate APS reductase gene expression via reverse transcription-PCR (RT-PCR) amplification from composite RNA samples. SRB-specific 16S ribosomal DNA (rDNA) probes were used for dot blot hybridization studies to substantiate results obtained by the MME technique.
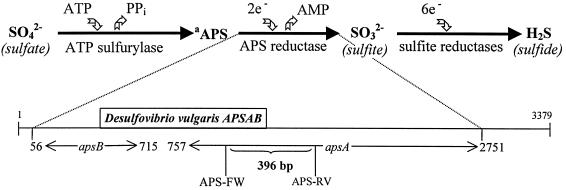
FIG. 1.
Biochemistry and genetics of dissimilatory sulfate reduction. The molecular ecology strategy outlined targets the dissimilatory sulfate reduction pathway and selectively amplifies the APS reductase A subunit gene or corresponding RNA transcripts from composite intestinal DNA or RNA samples, using the primer set APS-FW and APS-RV. The positions of nucleotides are according to the D. vulgaris APS reductase sequence (D. vulgaris APSAB, GenBank accession no. Z69372). The APSAB gene is 3,379 bp long and is compromised of subunit genes A and B, as indicated schematically above. The subunit genes A and B code for the enzyme APS reductase. aAPS, adenosine-5′-phosphosulfate.
MATERIALS AND METHODS
Animals and sample collections.
Animal procedures were approved by the Animal Care Committee of the University of Illinois and followed the Guide for the Care and Use of Laboratory Animals (32). DNA from intestinal samples of distinct segments of inbred C57BL/6J mice was isolated at distinct stages of development from birth to maturity. The C57BL/6J mice, a common and nonmutant laboratory strain, were offered daily laboratory rodent chow (Picolab mouse diet no. 20; PMI Nutrition International, Brantwood, Mo.) and autoclaved, reverse-osmosis water. At each sampling time (1, 4, 7, 14, 21, 30, 60, and 90 days after birth), five mice were sacrificed by CO2 asphyxiation followed by cervical translocation. Mucosal and luminal contents from stomach, proximal, middle, and distal segments of the small intestine (SI), cecum, and proximal and distal colon were collected from three mice of each age and stored at −80°C. For 1-, 4- and 7-day-old mice, tissues were homogenized to minimize sample loss; proximal and distal colon were not distinguished for these ages.
For histological analysis, 0.5- to 0.75-cm tissue sections were taken from two additional littermates at each sampling date. Tissue sections were fixed in Carnoy's for 2 h on ice (30). Tissues were then dehydrated in fresh 100% ethanol for 15 min, trimmed, placed in fresh 100% ethanol for 10 min, cleared in xylene for 10 min, and placed in fresh xylene for 10 min. After clearing, tissues were embedded in paraffin and cut in 2-μm-thick sections for mucin histochemistry.
Nucleic acid isolation.
For DNA isolation, intestinal samples were mixed with 10 ml of phosphate-buffered saline, vortexed thoroughly until homogenized, and then centrifuged at 30 × g for 2 min to remove interfering humic substances as described by Wilson and Blitchington (60). After 5 min of centrifugation at 12,000 × g, pellets were resuspended in 1 ml of lysis solution (lysozyme, 15 mg/ml) and incubated at 37°C for 30 min. After the addition of 1 ml of STS solution (0.15 M NaCl, 0.48 M Tris [pH 8], 10% sodium dodecyl sulfate) and additional incubation at 37°C for 30 min, samples were cooled to −80°C for three consecutive freeze-thaw steps. After the last thaw, 50 μg of proteinase K per ml was added, and the samples were incubated at 37°C for 30 min and then centrifuged at 6,000 × g for 20 min. Supernatants were then subjected to a phenol-based DNA extraction procedure as described previously (57). Total RNA was extracted from intestinal samples of three 30-day-old mice by a bead-beating, low-pH, hot-phenol extraction procedure (26, 51). Concentrations of DNA and RNA were determined spectrophotometrically, and the integrity of the nucleic acids was determined visually after electrophoresis in a 1% agarose gel containing ethidium bromide.
PCR and RT-PCR amplification.
PCR primers were based on sequence homology among the APS reductase genes from Desulfovibrio vulgaris (GenBank accession no. Z69372), Allochromatium vinosum (GenBank accession no. U84759), and Archaeoglobus fulgidus (GenBank accession no. X63435) (15). Forward (APS-FW; 5′-TGGCAGATMATGATYMACGGG-3′) and reverse (APS-RV; 5′-GGGCCGTAACCGTCCTTGAA-3′) primers were used to amplify a 396-bp fragment of the APS reductase subunit A gene (Fig. 1) with Y (T and C) and M (A and C) representing three degeneracies in the forward primer sequence. (The APS reductase primer set corresponds to conserved regions of bacterial and archaeal APS reductase gene sequences. While APS reductase-harboring archaea are also targeted with these primers, by convention, microbes contributing APS reductase PCR amplicons are termed SRB throughout this study.) DNA from APS reductase-positive (Desulfotomaculum ruminis, Desulfotomaculum thermobenzoicum, D. vulgaris, Desulfobacter curvatus, and Desulfovibrio salexigens) and APS reductase-negative (Escherichia coli, Enterococcus faecalis, and Bacteroides ovatus) bacterial species, grown in selective media (information can be found at the Deutsche Sammlung von Mikroorganismen und Zellkulturen website [http://www.dsmz.de]), was extracted as described above and screened via PCR as a positive control step. DNA obtained from the biofilm of a water sediment filter from which dissimilatory SRB were isolated and characterized was used as a positive environmental control. The biofilm was extracted from an in-line sediment filter in a rural well water distribution system and was characterized by a continuous film containing black precipitates and the emission of a H2S odor. SRB were isolated from the biofilm via a classical enrichment procedure (36), and DNA was subsequently extracted from the cultures as described above. A GC clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCG-3′) was added to the APS-RV primer for denaturing gradient gel electrophoresis (DGGE) analysis (31).
Hot-start PCR was performed with the Taq DNA polymerase kit from Takara Shuzo (Shiga, Japan). PCR mixtures of 48 μl contained 0.25 mM deoxynucleoside triphosphate (dNTP) mixture, 5 μl of 10× Ex Taq buffer (with MgCl2), and 200 ng of DNA. Samples were amplified in a GeneAmp PCR system 2400 (Perkin-Elmer, Norwalk, Conn.) using a hot-start PCR program: 95°C for 4.5 min, after which 2 μl of enzyme mixture containing 0.2 μl of 10× Ex Taq buffer (with MgCl2) and 2.5 U of Ex Taq DNA polymerase was added to each sample, followed by 35 cycles of 95°C for 30 s, 60°C for 55 s, and 72°C for 1 min, and then a cycle of 72°C for 7 min. Aliquots of 10 μl were analyzed by electrophoresis on a 2% (wt/vol) agarose gel containing ethidium bromide to verify amplicon sizes.
RT-PCR was performed with RNA isolated from intestinal samples of different segments of three 30-day-old mice with the GeneAmp Thermostable MuLV Reverse Transcriptase RNA PCR kit (Perkin-Elmer) to confirm the transcriptional activity of APS reductase in the mouse intestine. Reverse transcriptase mixtures of 20 μl contained 5 mM MgCl2 solution, 2 μl of 10× PCR Buffer II, 1 mM dNTP, 2.5 μM random hexamers, 20 U of RNase inhibitor, 50 U of murine leukemia virus reverse transcriptase, and 1,000 ng of RNA. The mixtures were incubated for 15 min at room temperature, for 20 min at 42°C, for 5 min at 95°C, and for 5 min at 5°C. After cDNA synthesis, 30 μl of the PCR mix was added. The PCR mix consisted of 3 μl of 10× PCR Buffer II, 25 pmol of the forward and reverse APS reductase primers, and 23 μl of H2O. The samples were amplified as described above, using 2.5 U of AmpliTaq DNA polymerase instead of the Ex Taq DNA polymerase. Replicate RNA samples from the stomach, distal SI, cecum, and distal colon were PCR amplified without the cDNA synthesis step to check for bacterial DNA contamination.
Dot blot hybridization with total DNA.
PCR of the APS reductase fragment reveals the presence of dissimilatory SRB in a particular environment. Genomic PCR, however, does not provide information on the number of organisms in a particular environment since one organism could theoretically provide sufficient amounts of DNA to yield a PCR signal. Thus, the presence of bacteria in a certain environment as detected by the MME approach does not necessarily reflect colonization of that environment. A signal in a dot blot hybridization assay only appears when the targeted organisms account for approximately 0.05 to 0.2% of total DNA (K. R. Hristova, R. I. Mackie, L. Raskin, and H. R. Gaskins, unpublished data). This technique allows detection only of the predominant organisms in a complex microbial ecosystem which would be expected to include colonizing organisms, while excluding transient or numerically insignificant populations. A dot blot hybridization assay was therefore performed to define colonization patterns in the mouse intestine.
DNA from distinct intestinal segments of 1-, 7-, 14-, 21-, and 60-day-old mice was used for dot blot hybridization, when the amount of DNA available from a particular segment was sufficient. The oligonucleotide probes used were as follows: for the domain Bacteria, S-D-Bact-0338-a-A-18 (Td = 54°C [1]) with E. coli as a positive control; for the Desulfobacter group, S-*-Dsb-0804-a-A-18 (Td = 46°C [7]) with D. curvatus as a positive control; for the Desulfovibrio group I, S-*-Dsv.sp-0698-a-A-18 (29) (Td = 55°C; Hristova et al., unpublished) with Desulfovibrio desulfuricans as a positive control; and for the genus Desulfotomaculum, S-G-Dtm-0229-a-A-18 (Td = 54°C [19]) with D. aeronauticum as a positive control. Synthetic oligonucleotide probes were 5′ end labeled with [γ-32P]ATP with polynucleotide kinase (Boehringer, Mannheim, Germany) as described previously (40). Immediately prior to membrane immobilization, DNA was denatured by adding of 3 volumes of 3% glutaraldehyde in 50 mM sodium phosphate (pH 7.0) to 1 volume of nucleic acid solution, incubated 10 min at room temperature, and diluted with double-distilled H2O containing 0.2 μl of bromophenol blue per ml. DNA isolated from pure cultures of positive control strains and mouse intestinal contents (100 ng in a total volume of 100 μl) were dot blotted onto Magna Charge nylon membranes (Micron Separation, Westboro, Mass.). Membranes were air dried and baked for 2 h at 80°C before hybridization. Prehybridizations, hybridizations, and washes were performed as described previously (40, 50). Final washes were performed at temperatures 6.5°C below the experimentally determined Td for DNA-RNA hybridization (listed above). The decreased Tw was necessary because DNA-DNA hybrids are less stable than DNA-RNA hybrids, and the experimentally determined difference in dissociation temperatures is approximately 6.5°C (33). Hybridization signals were captured using an Electronic Autoradiography Instant Imager (Packard Instruments, Meriden, Conn.). The hybridization signals were then used to determine the relative percentage of target rDNA in the samples. The abundance of Desulfobacter spp., members of Desulfovibrio group I, and Desulfotomaculum spp. in distinct intestinal segments over time was estimated by SRB-specific probes and expressed as a percentage of total bacterial rDNA.
DGGE analysis.
Parallel DGGE analysis was performed with the PCR samples from intestinal contents of a 30-day-old mouse. Controls were the APS reductase-positive strains (D. ruminis, D. curvatus, and D. desulfuricans). DGGE was performed as described previously (47) using a Bio-Rad D-Code System (Bio-Rad, Hercules, Calif.) to examine the relative diversity of mouse intestinal SRB. PCR fragments, obtained as described above except for the use of an APS reductase reverse primer with a GC clamp, were separated in 8% polyacrylamide gels in TAE buffer (20 mM Tris-acetate [pH 7.4], 10 mM sodium acetate, 0.5 mM sodium EDTA) containing 30 to 60% linear gradients of denaturant (100% denaturant corresponds to 7 M urea and 40% acrylamide-bisacrylamide stock solution, 37.5:1; Bio-Rad). Gradients were formed using a Bio-Rad Gradient Former Model 385, and gels were polymerized onto a gel support film (FMC, Rockland, Maine). PCR samples were applied to gels in aliquots of 3 μl per lane. A ladder, consisting of 200-bp 16S rDNA V3 amplicons amplified from DNA of Bacteroides thetaiotaomicron (VPI 5482), Bacteroides fragilis (VPI 2553), Ruminococcus albus strains AS7 and AS8 (laboratory collection), Streptococcus bovis (laboratory collection), E. coli K-12 NM522, Clostridium perfringens (laboratory collection), and Clostridium parvum (laboratory collection) with the primers 341F and 543R (31) was used to check for normal migration of the DGGE amplicons. Electrophoresis was performed at 60°C for 2 h at 150 V and subsequently for 2 h at 200 V. Gels were silver stained (28) and photographed using a Fotodyne FOTO/Analyst Investigator System (Fotodyne, Heartland, Wis.).
Cloning of PCR-amplified products, sequence, and phylogenetic analyses.
APS reductase amplicons were cloned, sequenced, and analyzed phylogenetically to verify the presence and examine the diversity of APS reductase sequences in the neonatal mouse intestine. APS reductase amplicons from the stomach, middle SI, and cecum of a 1-day-old mouse, from the stomach and middle SI of a 4-day-old mouse, from the biofilm of a water sediment filter (environmental control), and from D. desulfuricans and Desulfotomaculum aeronauticum were cloned in One Shot Competent E. coli INVαF′ using the Invitrogen TA Cloning Kit (Invitrogen, Carlsbad, Calif.). White colonies of ampicillin-resistant transformants were transferred to 5 ml of ampicillin-containing Luria-Bertani broth and grown overnight. Plasmid DNA was extracted by alkaline lysis as described previously (17). Plasmid DNA was digested by the restriction enzyme EcoRI and analyzed by electrophoresis in a 1.5% agarose gel to verify the insert size. For each intestinal segment, 2 of 10 inserts were randomly chosen for sequence analysis, except for only 1 insert from the middle SI of a 1-day-old mouse. Also, six clones from the filter biofilm and one clone for each APS reductase-positive strain were sequenced using the facilities of The Biotechnology Center of the University of Illinois. Sequences were aligned using the multiple sequence alignment program CLUSTAL W (54). Regions with gaps and ambiguities were excluded from the phylogenetic analysis. The two-parameter model of Kimura (23) was used for construction of neighbor-joining trees (43). The statistical significance of tree branches was evaluated by bootstrap analysis (8) involving the construction of 1,000 trees from resampled data.
Succession of sulfomucin-containing goblet cells.
Frosted microscope slides (Fisher Scientific, Pittsburgh, Pa.), supporting intestinal tissue sections from the mouse gastrointestinal tract, were deparaffinized, incubated in double-distilled H2O for 5 min, and stained for 16 h in a high iron diamine (HID) solution (48). After HID staining, tissues were washed in running tap water for 5 min and stained with alcian blue (pH 2.5) for 5 min (48). After being washed in the tap water for 2 to 3 min, tissues were dehydrated in 95% ethanol for 5 min, dehydrated in 100% ethanol for 5 min, cleared in xylene for 5 min, and mounted with Permount (Fisher Scientific, Pittsburgh, Pa.) on 1.5-mm-thick coverslips. Tissues were analyzed using a Nikon Optiphot-2 microscope (Nikon, Melville, N.Y.) and digitally captured using the Image-Pro Plus program, version 3.0 (Media Cybernetics, Silver Spring, Md.).
RESULTS
Validation of the MME strategy.
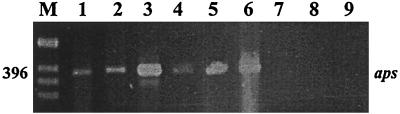
Positive control DNA samples (D. ruminis, D. thermobenzoicum, D. vulgaris, D. curvatus, and D. salexigens) yielded APS reductase PCR amplicons of the correct size (396 bp) in contrast to negative control DNA samples (E. coli, E. faecalis, and B. ovatus) for which amplicons were not detected (Fig. 2). A water sediment filter characterized by the presence of an H2S-producing biofilm from which dissimilatory SRB were isolated was screened for SRB using the MME strategy as an additional validation step. DNA extracted from the filter biofilm yielded APS reductase PCR amplicons of the correct size (Fig. 2). Mouse kidney DNA was used for PCR to test reactivity of the APS reductase primers with eukaryotic DNA; amplicons were not detected, confirming primer specificity.
FIG. 2.
PCR amplification of the 396-bp APS reductase fragment (aps) from positive control SRB strains (D. ruminis, lane 1; D. thermobenzoicum, lane 2; D. vulgaris, lane 3; D. curvatus, lane 4; and D. salexigens, lane 5) and the presence of APS reductase amplicons of the correct size (396 bp) from a sediment filter DNA sample (lane 6). PCR amplification of DNA from negative control bacteria strains (E. faecalis, lane 7; B. ovatus, lane 8; and E. coli, lane 9) did not yield APS reductase amplicons. M corresponds to a 1-kb ladder (Gibco BRL).
Succession of SRB in the mouse gastrointestinal tract.
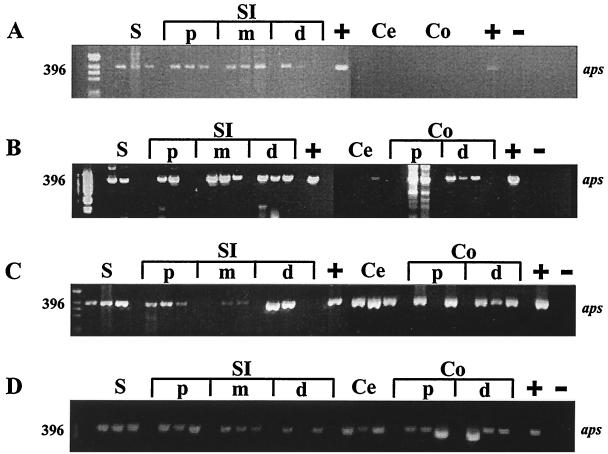
Amplicons of APS reductase of the correct size were detected from the stomach, and proximal, middle, and distal SI of 1-day-old mice (Fig. 3A) and of 4- and 7-day-old mice (data not shown). In the stomach and proximal and middle SI, 396-bp amplicons were consistently amplified from all mice, in contrast to the varied presence of APS reductase amplicons in the distal SI of 1-, 4-, and 7-day-old mice (2 of 3, 2 of 3, and 1 of 3 replicates, respectively). APS reductase amplicons were not detected from cecum or colon of 1-, 4-, or 7-day-old mice, except for a weak band from one of three 1-day-old replicate animals (Fig. 3A) and from cecal DNA from one of three 7-day-old mice (not shown).
FIG. 3.
Agarose gel showing presence or absence of intestinal SRB in distinct intestinal regions of C57BL/6J mice of different ages (A, 1 day after birth; B, 14 days; C, 21 days; D, 90 days) based on detection of APS reductase amplicons (aps) of the correct size (396 bp). D. vulgaris was used as a positive (+) control, and DNA from mouse kidney as a negative (−) control. Intestinal contents of three mice were analyzed at each sampling point. S, stomach; SI, small intestine; Ce, cecum; Co, colon; p, proximal; m, middle; d, distal.
Intestinal SRB were detected in the cecum and colon 14 days after birth as indicated by the presence of APS reductase amplicons in the distal colon of all mice assayed (3 of 3) and in cecum (1 of 3) and proximal colon (2 of 3) samples (Fig. 3B). APS reductase amplicons were detected in the stomach and proximal SI of two of three 14-day-old mice, while the middle and distal SI of 14-day-old mice consistently yielded APS reductase amplicons (3 of 3 mice; Fig. 3B).
The presence of SRB in the distal segments of the mouse intestine was more pronounced at weaning (21-day-old mice; Fig. 3C) than at 14 days after birth (Fig. 3B). APS reductase amplicons were detected in the cecum and distal colon of each of the 21-day-old replicate animals and in the proximal colon of two of three 21-day-old mice (Fig. 3C). SRB remained prominent in the stomach (3 of 3) and proximal SI (3 of 3) of 21-day-old mice, although APS reductase amplicons were not detected consistently at day 21 for the middle and distal SI (Fig. 3C). Similarly, APS reductase amplicons were detected in the stomach, SI, cecum, and colon of 30- and 60-day-old mice (not shown). Finally, APS reductase amplicons were detected from all intestinal segments of each replicate animal for 90-day-old mice (Fig. 3D).
Dot blot hybridization.
Intestinal DNA from 1-, 7-, 14-, 21-, and 60-day-old mice was used for dot blot hybridization with 16S rDNA probes as described in Materials and Methods. DNA samples from 1- and 7-day-old mice did not yield hybridization signals with dissimilatory SRB-specific probes; only two proximal colon samples yielded a signal with the Bacteria domain probe (not shown). Bacterial 16S rDNA signals were consistently detected in the cecum and colon of 14-, 21- and 60-day-old mice. Bacterial populations were detected in the middle and distal SI of one of three 14-day-old replicate animals. In 21-day-old mice, bacterial 16S rDNA signals were detected throughout the stomach and SI, except for the middle SI of two of three replicate animals. Bacterial populations were detected in the stomach of one of three 60-day-old replicate animals and in the proximal (3 of 3), middle (3 of 3), and distal (1 of 3) SI (not shown). The detection of signals with SRB-specific probes invariably coincided with the detection of signals with the Bacteria domain probe.
SRB colonization of the cecum and colon as detected by dot blot hybridization occurred by 14 days after birth, and SRB persisted in these segments of 21- and 60-day-old mice (Table 1). SRB 16S rDNA signals were not detected from proximal gastrointestinal samples of 14-day-old mice (Table 1). In 21-day-old mice, SRB belonging to the Desulfovibrio group I accounted for 2 and 1% of total bacterial 16S rDNA in the stomach and both the proximal and distal SI, respectively, and Desulfobacter spp. accounted for 2% of total bacterial 16S rDNA in the distal SI (Table 1). Otherwise, SRB signals were not detected among replicate 21-day-old mice for other proximal gastrointestinal segments (Table 1). SRB populations were detected throughout the gastrointestinal tract of 60-day-old mice, except in the distal SI (Table 1). The stomach and proximal and middle SI of 60-day-old mice were colonized by Desulfotomaculum spp., but their numbers were negligible in more distal intestinal segments in contrast to the Desulfobacter group and Desulfovibrio group I. Desulfobacter spp. were also more abundant than SRB belonging to the Desulfovibrio group I in distal intestinal segments, regardless of age.
TABLE 1.
Estimation of the relative abundance of distinct SRB groups derived from dot blot hybridization with 16S rDNA extracted from intestinal samples
| Segment | Probe | % Total bacterial rDNAa ± SE in animals:
|
||
|---|---|---|---|---|
| 14 days old | 21 days old | 60 days old | ||
| S | Dsb-0804 | ND | <dl | 4 ± 4b |
| Dsv-0698 | ND | 2 ± 1 | 5 ± 5b | |
| Dtm-0229 | ND | <dl | 2 ± 2b | |
| PS | Dsb-0804 | <dl | <dl | 10 ± 2 |
| Dsv-0698 | <dl | 1 ± 1 | 8 ± 3 | |
| Dtm-0229 | <dl | <dl | 4 ± 0 | |
| MS | Dsb-0804 | <dl | <dl | 6 ± 1 |
| Dsv-0698 | <dl | <dl | 3 ± 1 | |
| Dtm-0229 | <dl | <dl | 6 ± 0 | |
| DS | Dsb-0804 | <dl | 2 ± 1c | <dl |
| Dsv-0698 | <dl | 1 ± 1c | <dl | |
| Dtm-0229 | <dl | <dl | <dl | |
| C | Dsb-0804 | 14 ± 1 | 7 ± 1 | 14 ± 1 |
| Dsv-0698 | 12 ± 1 | 6 ± 1 | 6 ± 1 | |
| Dtm-0229 | 2 ± 0 | 1 ± 0 | <dl | |
| PK | Dsb-0804 | 21 ± 15c | 5 ± 3c | 21 ± 3 |
| Dsv-0698 | 2 ± 1c | 4 ± 1c | 2 ± 1 | |
| Dtm-0229 | 1 ± 1b | 0 ± 1b | <dl | |
| DK | Dsb-0804 | 14 ± 8 | 2 ± 2b | 15 ± 8c |
| Dsv-0698 | 3 ± 0 | 3 ± 1b | 2 ± 1c | |
| Dtm-0229 | <dl | <dl | <dl | |
The results are expressed as the percentage of total bacterial rDNA isolated from different intestinal segments (S, stomach; PS, proximal, MS, middle, and DS, distal, SI; C, cecum; PK, proximal, and DK, distal, colon) and represent the mean ± the standard error from three animal replicates. Oligonucleotide probes used were as follows: for domain Bacteria, S-D-Bact-0338-a-A-18 (1); for the Desulfobacter group, S-*-Dsb-0804-a-A-18 (7); for Desulfovibrio group I, S-*-Dsv.sp-0698-a-A-18 (29); and for the genus Desulfotomaculum, S-G-Dtm-0229-a-A-18 (19). ND, not determined. “<dl” indicates the value was below the detection limit (0.05 to 0.2%; see Materials and Methods).
SRB were detected in one of three animals.
SRB were detected in two of three animals.
SRB populations appeared to diminish at weaning as the percentage of SRB 16S rDNA decreased in all intestinal segments compared to 14-day-old mice (Table 1). SRB appeared to recover after weaning, as indicated by a general increase of the SRB contribution to total bacterial 16S rDNA in the intestines of 60-day-old mice compared to weaning age animals (21-day-old animals).
Variability in SRB presence in specific intestinal segments was detected among individual mice (Table 1). This outcome is somewhat surprising given that the mice were genetically identical, fed the same diet, and reared in the same environment. For example, Desulfobacter spp. DNA signals ranged from 0 to 53% of the total bacterial 16S rDNA in the proximal colon of the three replicate 14-day-old mice. Similarly, SRB colonized the stomach of only one of three replicate 60-day-old mice; SRB 16S rDNA accounted for 32% of total bacterial 16S rDNA in the stomach of this single mouse (Table 1).
DGGE-based SRB diversity.
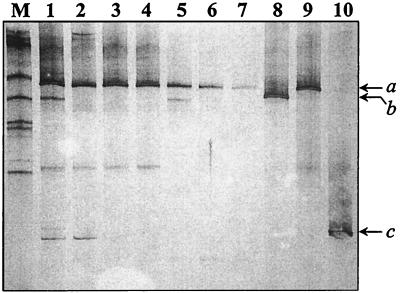
APS reductase amplicons from each intestinal segment of a 30-day-old mouse were analyzed by DGGE for initial determination of SRB diversity throughout the gastrointestinal tract in weaned, mature mice. DNA from the stomach and SI yielded a greater number of DGGE bands than DNA from the cecum and colon (Fig. 4). Comparison of the intestinal APS reductase DGGE bands with those from positive control strains revealed the presence of a SRB species closely related to D. ruminis in each intestinal segment, the presence of a SRB species closely related to D. desulfuricans in the stomach, cecum, and colon, and the presence of a SRB species closely related to D. curvatus in the stomach and proximal SI. Two additional APS reductase DGGE bands were observed in the stomach. One band exhibited a unique migration pattern, while the stomach APS reductase amplicon having the lower GC content corresponded to a DGGE band observed in the D. rumins and D. curvatus samples. This DGGE band was also present throughout the SI (Fig. 4).
FIG. 4.
DGGE analysis of APS reductase DNA amplicons comparing the banding patterns from distinct mouse intestinal regions (lane 1, stomach; lanes 2 to 4, proximal, middle, and distal SI; lane 5, cecum; and lanes 6 to 7, proximal and distal colon) with three positive control SRB strains (lane 8, D. desulfuricans [b]; lane 9, D. ruminis [a]; and lane 10, D. curvatus [c]). M corresponds to a synthetic marker comprised of known 16S rDNA sequences varying in GC content (see Materials and Methods).
Phylogenetic analysis of APS reductase sequences.
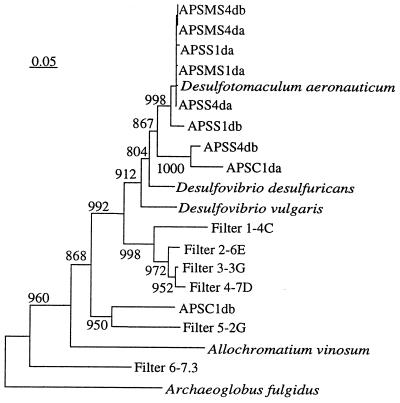
Phylogenetic analysis revealed that eight of the nine APS reductase sequences of intestinal origin formed a single tight cluster clearly separated from the APS reductase sequences of environmental origin (Fig. 5). The APS reductase fragment of the environmental isolate D. aeronauticum was also affiliated with this cluster. One of the intestinal sequences from the cecum of a 1-day-old mouse (APSC1db) was more closely related to sequences amplified from the water filter biofilm (Fig. 5). In general, APS reductase sequences amplified from the filter biofilm were more diverse with longer branch lengths than the intestinal sequences.
FIG. 5.
Phylogenetic placement of APS reductase sequences from intestinal and environmental samples. The archaeal sequence (Archaeoglobus fulgidus) was used as the outgroup for rooting the tree. Numbers above each node are confidence levels generated from 1,000 bootstrap trees (8). The scale bar is in fixed nucleotide substitutions per sequence position. Intestinal APS reductase sequences are amplicons from clones (a and b) from the stomach (APSS1da and APSS1db), middle SI (APSMS1da), and cecum (APSC1da and APSC1db) of a 1-day-old mouse and from the stomach (APSMS4da and APSS4db) and middle SI (APSMS4da and APSMS4db) of a 4-day-old mouse. Filters 1-4C, 2-6E, 3-3G, 4-7D, 5-2G, and 6-7.3 represent APS reductase amplicons from six clones from the biofilm of the water filter.
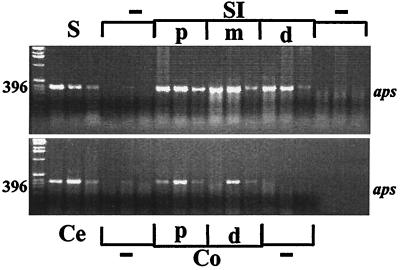
Intestinal APS reductase mRNA expression (RT-PCR).
APS reductase RT-PCR amplicons were detected in each intestinal segment of three replicate 30-day-old mice, including the stomach and proximal SI (Fig. 6). Most RT-negative control samples did not yield PCR amplicons, indicating general absence of contaminating bacterial DNA; weak signals were detected from one of three stomach and cecal samples.
FIG. 6.
Agarose gel comparing the presence and intensity of RT-PCR APS reductase amplicons (aps) in intestinal regions from three 30-day-old mice (stomach [S]; proximal [p], middle [m], and distal [d] SI; cecum [Ce]; and proximal [p] and distal [d] colon [Co]). Samples without reverse transcriptase served as a negative control (−) to screen for DNA contamination. APS reductase mRNA expression was observed in all intestinal regions.
Detection of sulfomucin-containing goblet cells.
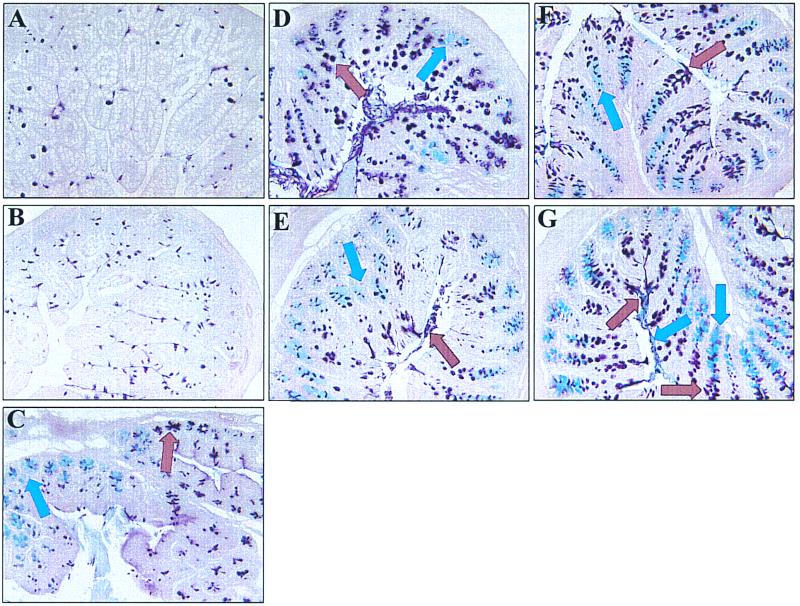
To compare the succession of intestinal SRB to the appearance of sulfomucins in the mouse intestine, tissue sections of different intestinal segments from mice at distinct stages of development were stained for sulfomucin (brown)- and sialomucin (blue)-containing goblet cells using conventional histochemical techniques as described in Materials and Methods (Fig. 7). Sulfomucin- and sialomucin-containing goblet cells were only sporadically detected in the stomach of mice of the ages examined (not shown). Sulfomucin-containing goblet cells were observed in the crypts and on villi in the SI and appeared to increase in number after weaning (Fig. 7A and B). Sialomucin-containing goblet cells were not detected in the SI at any of the ages examined (Fig. 7A and B). Mucin subtypes in the cecum underwent a gradual spatial shift from sialomucin in the proximal cecum to sulfomucin in the distal cecum (Fig. 7C). This spatial pattern of mucin distribution was established 4 days after birth and remained consistent at later ages. A mixture of sialomucin- and sulfomucin-containing goblet cells were detected in crypts and cuffs of the proximal and distal colon in 4-day-old mice. However, at this stage of development, sialomucins were not detected in the mucus layer covering the epithelium of proximal and distal colon (not shown). By 14 days after birth, a clear pattern of mucin distribution began to develop in the proximal and distal colon (Fig. 7D). In both colonic segments, sialomucin-containing goblet cells were detected only in colonic crypts, whereas sulfomucin-containing goblet cells were observed on colonic cuffs but not in crypts. The separation of sialomucin-containing (crypts) and sulfomucin-containing (cuffs) goblet cells became even more distinct in the distal colon after weaning (Fig. 7E to G). Although quantitative analyses were not performed, sialomucin-containing goblet cells appeared to become more dominant in the proximal colon after weaning, with only a few sulfomucin-containing goblet cells observed on top of the cuffs of the proximal colon in 90-day-old mice. The mucus layer in the proximal colon stained predominantly blue at this age, further indicating dominance of sialomucins over sulfomucins in this colonic segment (not shown). However, the mucus blanket in the distal colon of 90-day-old mice was comprised of distinct sulfo- and sialomucin layers (Fig. 7G).
FIG. 7.
Histological analysis of distinct intestinal regions of C57BL/6J mice by means of high iron diamine-alcian blue (pH 2.5) histology to differentiate sialated mucins (blue stain, blue arrow) from sulfated mucins (brown stain, brown arrow) (magnification, ×20). (A and B) Increase of sulfomucin-containing goblet cells in the distal SI before weaning (A, 14-day-old mouse) compared to after weaning (B, 30-day-old mouse). (C) Spatial mucin distribution in the cecum of a 90-day-old mouse. Sialated mucins (blue) predominate in the proximal cecum, while sulfated mucins (brown) predominate in the distal cecum. (D to G) Succession of mucin types in the mouse distal colon (D, 14 day old; E, 30 day old; F, 60 day old; and G, 90 day old).
DISCUSSION
In this report we describe for the first time dissimilatory SRB colonization of the mouse gastrointestinal tract including the stomach and SI. The present results indicate that SRB are major members of the mouse gastrointestinal microbiota since they accounted for 17 to 23% of the total bacterial 16S rDNA in the cecum and proximal colon of all postweaning mice examined. The detection of SRB in the mouse intestine complements earlier findings from cultivation-based studies of pig (3) and human (2, 11, 38) fecal SRB. However, Desulfovibrio spp. were found to be dominant in previous pig and human studies, while in our study Desulfobacter was a dominant SRB genus. The difference could reflect bias introduced by analyzing fecal versus intestinal samples or else the uniqueness or incomplete analysis of SRB populations among mammalian species examined to date.
Combined use of the dot blot hybridization and the MME techniques revealed a distinct colonization pattern among several SRB groups. Gram-positive Desulfotomaculum spp. resided preferentially in the stomach and SI both pre- and postweaning, while Desulfobacter spp. and, to a lesser extent, SRB related to D. desulfuricans were dominant in the distal segments. In general, SRB were found in greatest density in distal segments of the mouse intestine, which also contains large numbers of sulfomucin-containing goblet cells. We have obtained preliminary evidence that significant concentrations of endogenously secreted sulfate are present in the cecum and colon (B. Deplancke, K. Finster, V. J. McCracken, R. I. Mackie, and H. R. Gaskins, unpublished data), while others (24) have demonstrated that dietary sulfate is quantitatively absorbed in proximal segments of the mouse intestine. Differential sulfate concentrations in the colon compared to the small intestine may influence SRB population profiles based on the bioenergetic efficiency of community members. Liu and Peck (27) demonstrated that the growth of Desulfovibrio spp. is advantaged in sulfate-rich environments over Desulfotomaculum spp. because of the absence of significant electron-transfer-coupled phosphorylation in the latter species. However, the two SRB genera may be bioenergetically equivalent in environments containing low sulfate concentrations, where energy is derived predominantly from substrate phosphorylation. The initial demonstration of significant SRB populations in the mouse intestine as well as apparent differences in community profiles within distinct intestinal habitats justify further efforts to better characterize the physiological ecology of these bacteria.
While SRB density was greater in the large intestine, the stomach and SI appeared to harbor a somewhat more diverse SRB population in weaned mice. However, it must be noted that the MME approach has limited utility as a single tool to evaluate biodiversity because environmental APS reductase sequences may be differentially amplified or because multiple amplicons may migrate to similar positions in denaturant gels. Both outcomes are not uncommon for DGGE analysis of 16S rRNA products (13, 47), though we have limited observations on the potential for these problems to arise with APS reductase primers or amplicons. Moreover, conclusive taxonomic assignment based on APS reductase data is not possible at this stage due to the minimal nature of the APS reductase sequence database, which precludes knowledge of sequence variability and hence phylogenetic context. The general lack of intestinal isolates of various SRB genera also limits the utility of the APS reductase-based MME approach as a tool to analyze intestinal SRB diversity. All taxonomical designations based on the current MME approach are therefore presumptive. These findings emphasize the need for further isolation of intestinal SRB strains and further expansion of the APS reductase sequence databank.
Surprisingly, Desulfotomaculum spp. were detected in stomach and SI by 1 day after birth, as indicated by phylogenetic analysis of APS reductase sequences. The stomach and SI APS reductase sequences from 1- and 4-day-old mice generally formed one major cluster, related to D. aeronauticum, which groups with the intestinal isolate D. ruminis (49). Most Desulfotomaculum spp., such as D. ruminis, are able to use lactate as a carbon and energy source (4) and thus would be able to grow in the lactate-rich intestine of neonates (34, 55). Other predominantly lactate-utilizing SRB belong to the Desulfovibrio genus (39). Desulfotomaculum spp. are, however, spore formers, which might also have contributed to their presence but not overt colonization in newborn mice.
The mucus layer may be one mechanism by which SRB in more mature mice survive the acidic conditions of the stomach. The optimum pH range reported for dissimilatory SRB ranges from 7.5 to 8.0, and growth inhibition generally occurs at pH values lower than 5.5 or higher than 9 (41). While sulfate reduction has been observed in a peat bog and acid mine water at pH 3 to 4 (14), the growth of dissimilatory SRB isolated from those habitats was inhibited below a pH of 6. It was therefore concluded that SRB in acidic environments are present in microniches with higher and more favorable pH conditions. Similarly, SRB would not be expected to survive in the lumen of the stomach but may be protected from gastric acid in the mucus layer which functions as a H+ diffusion barrier (46). This hypothesis is supported by the present RT-PCR results from 30-day-old mice confirming SRB viability in the mouse stomach and by preliminary studies which have demonstrated microbial sulfide production in the stomach of 30-day-old mice and the presence of SRB also in gastric mucus of 7-day-old piglets (Deplancke et al., unpublished). Reduction of sulfate in the stomach by SRB is likely dependent on exogenous sulfate sources, since few sulfomucin-containing goblet cells were detected in the stomach mucosa. The present results demonstrate the importance of further defining ecological parameters mediating SRB colonization of the stomach given their potential to contribute to the development of gastric ulcers, as has been recognized previously for colonic ulcers (42).
Establishment of SRB populations in the cecum and colon did not occur until 14 days after birth, as indicated by dot blot hybridization results. This observation agrees with the earlier finding that anaerobic bacteria, such as Bacteroides species and a mixed group of anaerobic fusiform bacteria (the Bacillus-Clostridium group) do not appear in significant numbers in the mouse intestine until the animals first ingest solid food at 11 to 14 days after birth (25). Bacteroides spp. and other indigenous gut bacteria, including Clostridium and Ruminococcus spp., are able to degrade and utilize mucus as a carbon and energy source (5, 6, 18, 44, 52, 56). Interestingly, Willis et al. (59) demonstrated that B. fragilis and D. desulfuricans could be cocultured using sulfomucin as a single metabolizable substrate. B. fragilis released sulfate from sulfomucin and utilized remaining desulfated mucins as a carbon and energy source. As a consequence, short-chain fatty acids and sulfate were released into the medium, permitting the growth of D. desulfuricans. Further evidence on the role of mucin in influencing SRB populations comes from studies with a three-stage continuous culture model of the colon, which demonstrated that infusion of pig gastric mucin increased dissimilatory SRB numbers and activities in mixed cultures, although SRB in pure culture were unable to directly metabolize mucin (11). A similar syntrophic mechanism may have favored SRB colonization in the distal segments of 14-day-old mice based on histological analysis of goblet cells in the mouse cecum and colon. The simultaneous establishment of sulfate-cleaving organisms (e.g., Bacteroides spp.) in distal intestinal segments and the presence of numerous sulfomucin-containing goblet cells in the upper layers of the distal gut mucosa (Fig. 7D) may have promoted a bloom of dissimilatory SRB. These results indicate that SRB may be dependent on other intestinal bacteria and on endogenous sulfate sources secreted by their host to colonize the cecum and colon.
A link between IBD, particularly ulcerative colitis, and intestinal SRB would clearly be based upon the availability of sulfate in distal intestinal segments. However, the contribution of exogenous sulfate from sulfate-rich foods or sulfate in drinking water to the pool of available sulfate in distinct intestinal segments has not been clearly defined. Likewise, mechanisms of microbial depolymerization and desulfation of sulfomucins have not been adequately characterized in situ. For example, the relatively well defined relationship between B. fragilis and D. desulfuricans in vitro has not been examined in vivo, other similar relationships in the cecum and colon have not been reported, and the occurrence of syntrophic relationships between sulfate-cleaving bacteria and SRB in the SI has not been studied. Further investigation of both the role of dietary sulfate and sulfomucins will therefore be necessary to fully assess the impact of intestinal SRB on chronic intestinal diseases. The MME approach described here can be useful for detecting and identifying SRB in distinct intestinal environments and for studying the metabolic activity of SRB in situ. Identification of additional APS reductase sequences will complement such efforts and facilitate a better understanding of the phylogeny of intestinal APS reductase-harboring organisms.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant DK57940 (H.R.G.).
REFERENCES
- 1.Amann R I, Binder B J, Olsen R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beerens H, Romond C. Sulfate-reducing anaerobic bacteria in human feces. Am J Clin Nutr. 1977;30:1770–1776. doi: 10.1093/ajcn/30.11.1770. [DOI] [PubMed] [Google Scholar]
- 3.Butine T J, Leedle J A. Enumeration of selected anaerobic bacterial groups in cecal and colonic contents of growing-finishing pigs. Appl Environ Microbiol. 1989;55:1112–1116. doi: 10.1128/aem.55.5.1112-1116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell L L, Postgate J R. Classification of the spore forming sulfate-reducing bacteria. Bacteriol Rev. 1965;29:359–363. doi: 10.1128/br.29.3.359-363.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corfield A P, Wagner S A, Clamp J R, Kriaris M S, Hoskins L C. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase and glycosulfatase activities by strains of faecal bacteria. Infect Immun. 1992;66:3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corfield A P, Wagner S A, O'Donnell L J D, Durdey P, Mountford R A, Clamp J R. The roles of enteric bacterial sialidase, sialate O-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj J. 1993;10:72–81. doi: 10.1007/BF00731190. [DOI] [PubMed] [Google Scholar]
- 7.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. System Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 8.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 9.Forstner J F, Oliver M G, Sylvester F A. Production, structure, and biologic relevance of gastrointestinal mucins. In: Blauser M J, Smith P D, Ravdin J J, Greenberg H D, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 71–88. [Google Scholar]
- 10.Gibson G R, Macfarlene G T, Cummings J H. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J Appl Bacteriol. 1988;65:103–111. doi: 10.1111/j.1365-2672.1988.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 11.Gibson G R, Cummings J H, Macfarlane G T. Use of a three stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl Environ Microbiol. 1988;54:2750–2755. doi: 10.1128/aem.54.11.2750-2755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson G R, Cummings J H, Macfarlane G T. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Ecol. 1991;86:101–112. [Google Scholar]
- 13.Hansen C M, Tolker-Nielsen T, Givskov M, Mølin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 14.Hao O J, Chen J M, Huang L, Buglass R L. Sulfate-reducing bacteria. Crit Rev Environ Sci Technol. 1996;26:155–187. [Google Scholar]
- 15.Hipp W M, Pott A S, Thum-Schmitz N, Faath I, Dahl C, Truper H G. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology. 1997;143:2891–2902. doi: 10.1099/00221287-143-9-2891. [DOI] [PubMed] [Google Scholar]
- 16.Hollander D, Vadheim C, Brettholz E, Pettersen G M, Delahunty T, Rotter J I. Increased intestinal permeability in patients with Crohn's disease and their relatives. Ann Intern Med. 1986;105:883–885. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- 17.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 18.Hoskins L C, Boulding E T. Mucin degradation in human colon ecosystems. J Clin Investig. 1981;67:163–172. doi: 10.1172/JCI110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hristova, K. R., M. Mau, D. Zheng, R. I. Aminov, R. I. Mackie, H. R. Gaskins, and L. Raskin.Desulfotomaculum genus and group-specific small subunit rRNA hybridization probes for environmental studies. Environ. Microbiol., in press. [DOI] [PubMed]
- 20.Irimura T, Wynn D M, Hager L G, Cleary K R, Ota D M. Human colonic sulfomucin identified by a specific monoclonal antibody. Cancer Res. 1991;51:5728–5735. [PubMed] [Google Scholar]
- 21.Karkhoff-Schweizer R R, Huber D P W, Voordouw G. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl Environ Microbiol. 1995;61:290–296. doi: 10.1128/aem.61.1.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent P W, Marsden J C. A sulphated sialoprotein from sheep colonic mucin. Biochem J. 1963;87:38. [Google Scholar]
- 23.Kimura M. A simple model for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 24.Krijgsheld K R, Frankena H, Scholtens E, Zweens J, Mulder G J. Absorption serum levels and urinary excretion of inorganic sulfate after oral administration of sodium sulfate in the conscious rat. Biochim Biophys Acta. 1979;586:492–500. doi: 10.1016/0304-4165(79)90039-4. [DOI] [PubMed] [Google Scholar]
- 25.Lee A, Gemmel E. Changes in the mouse intestinal microflora during weaning: role of volatile fatty acids. Infect Immun. 1972;5:1–7. doi: 10.1128/iai.5.1.1-7.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C, Raskin L, Stahl D A. Microbial community structure in gastrointestinal tracts of domestic animals: comparative analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol Ecol. 1997;22:281–294. [Google Scholar]
- 27.Liu C-L, Peck H D. Comparative bioenergetics of sulfate reduction in Desulfovibrio and Desulfotomaculum spp. J Bacteriol. 1981;145:966–973. doi: 10.1128/jb.145.2.966-973.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Ritalahti K. Denaturing gradient gel electrophoresis (DGGE) protocol. ROME lab DGGE Workshop. East Lansing, Mich: Department of Microbiology and Center for Microbial Ecology, Michigan State University; 1996. [Google Scholar]
- 29.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 30.Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. 1997;40:782–789. doi: 10.1136/gut.40.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Research Council. Guide for the care and use of laboratory animals. NIH publication no. 85-23 (rev.). U.S. Washington, D.C.: Government Printing Office; 1985. [Google Scholar]
- 33.Oerther D B, Pernthaler J, Schramm A, Amann R, Raskin L. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge wastewater treatment systems. Appl Environ Microbiol. 2000;66:2154–2165. doi: 10.1128/aem.66.5.2154-2165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa K, Ben R A, Pons S, de Paolo M I L, Bustos Fernandes L. Volatile fatty acids, lactic acid and pH in the stools of breast-fed and bottle-fed infants. J Pediatr Gastroenterol Nutr. 1992;15:248–252. doi: 10.1097/00005176-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Peck H D. Comparative metabolism of inorganic sulphur compounds in microorganisms. Bacteriol Rev. 1962;26:67–94. doi: 10.1128/br.26.1.67-94.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfennig N, Widdel F, Trüper H G. The dissimilatory sulfate-reducing bacteria. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes. Vol. 1. Heidelberg, Germany: Springer-Verlag KG; 1981. pp. 926–940. [Google Scholar]
- 37.Pitcher M C L, Beatty E R, Cummings J H. The contribution of sulphate reducing bacteria and 5-aminosalicyclic acid to faecal sulphide in patients with ulcerative colitis. Gut. 2000;46:64–72. doi: 10.1136/gut.46.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pochart P, Dore J, Lemann F, Goderel I, Rambaud J C. Interrelations between populations of methanogenic archaea and sulfate-reducing bacteria in the human colon. FEMS Microbiol Lett. 1992;77:225–228. doi: 10.1016/0378-1097(92)90160-p. [DOI] [PubMed] [Google Scholar]
- 39.Postgate J R, Campbell L L. Classification of Desulfovibrio species, the non-sporulating sulfate-reducing bacteria. Bacteriol Rev. 1966;30:732–738. doi: 10.1128/br.30.4.732-738.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reis M A M, Almeida J S, Lemos P C, Carrondo M J T. Effect of hydrogen sulfide on growth of sulfate reducing bacteria. Biotechnol Bioeng. 1992;40:593–600. doi: 10.1002/bit.260400506. [DOI] [PubMed] [Google Scholar]
- 42.Roediger W E W, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci. 1997;42:1571–1579. doi: 10.1023/a:1018851723920. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Salyers A A. Energy sources of major intestinal fermentative anaerobes. Am J Clin Nutr. 1979;32:158–163. doi: 10.1093/ajcn/32.1.158. [DOI] [PubMed] [Google Scholar]
- 45.Schramm A, Santegoeds C M, Nielsen H K, Ploug H, Wagner M, Pribyl M, Wanner J, Amann R, De Beer D. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl Environ Microbiol. 1999;65:4189–4196. doi: 10.1128/aem.65.9.4189-4196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreiber S, Scheid P. Gastric mucus of the guinea pig: proton carrier and diffusion barrier. Am J Physiol. 1997;272:G63–G70. doi: 10.1152/ajpgi.1997.272.1.G63. [DOI] [PubMed] [Google Scholar]
- 47.Simpson J M, McCracken V J, White B A, Gaskins H R, Mackie R I. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J Microbiol Methods. 1999;36:167–179. doi: 10.1016/s0167-7012(99)00029-9. [DOI] [PubMed] [Google Scholar]
- 48.Spicer S S, Leppi T J, Stoward P J. Suggestions for a histochemical terminology of carbohydrate-rich tissue components. J Histochem Cytochem. 1965;13:599–603. doi: 10.1177/13.7.599. [DOI] [PubMed] [Google Scholar]
- 49.Stackebrandt E, Sproer C, Rainey F A, Burghardt J, Pauker O, Hippe H. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. int. J Bacteriol. 1997;47:1134–1139. doi: 10.1099/00207713-47-4-1134. [DOI] [PubMed] [Google Scholar]
- 50.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 205–248. [Google Scholar]
- 51.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanley R A, Ram S P, Wilkinson R K, Roberton A M. Degradation of pig gastric and colonic mucins by bacteria isolated from the pig colon. Appl Environ Microbiol. 1986;51:1104–1109. doi: 10.1128/aem.51.5.1104-1109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stille W, Truper H G. Adenylylsulfate reductase in some new sulfate-reducing bacteria. Arch Microbiol. 1984;41:1230–1237. [Google Scholar]
- 54.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomaszewski L, Hofman H, Brzozowska I. Lactic acid excretion with stools by premature babies, newborns, infants, and healthy children. Pediatr Pol. 1971;46:13–19. [PubMed] [Google Scholar]
- 56.Tsai H H, Sunderland D, Gibson G R, Hart C A, Rhodes J M. A novel mucin sulphatase from human feces: its identification, purification and characterization. Clin Sci (Colch) 1992;82:447–454. doi: 10.1042/cs0820447. [DOI] [PubMed] [Google Scholar]
- 57.Tsai Y L, Olsen B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Widdel F, Hansen T A. The dissimilatory sulfate- and sulfur-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 583–624. [Google Scholar]
- 59.Willis C L, Cummings J H, Neale G, Gibson G R. In vitro effects of mucin fermentation on the growth of human colonic sulphate-reducing bacteria. Anaerobe. 1996;2:117–122. [Google Scholar]
- 60.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyatt J, Vogelsang H, Hubl W, Waldhoer T, Lochs H. Intestinal permeability and the prediction of relapse in Crohn's disease. Lancet. 1992;341:1437–1439. doi: 10.1016/0140-6736(93)90882-h. [DOI] [PubMed] [Google Scholar]