Abstract
Phylogeny and the distribution of symbiotic bacteria in the mixed segment of the wood-eating termite Nasutitermes takasagoensis (Shiraki) were studied. Bacterial 16S rRNA genes (rDNA) were amplified from the mixed segment of the gut by PCR, and two kinds of sequences were identified. The phylogenetic tree was constructed by neighbor-joining and maximum parsimony methods to identify symbionts harbored in the mixed segment. They are classified as low-G+C-content gram-positive bacteria and are most closely related to the genus Clostridium. The distribution of these bacteria throughout the whole gut was examined by PCR using specific primers, which suggested that they are confined to the mixed segment despite the presence of bacteria throughout the gut. In situ hybridization indicated that the symbiotic bacteria were localized to the ectoperitrophic space between the midgut wall and the peritrophic membrane in the mixed segment. Electron microscopy revealed the close association between these bacteria and the mesenteric epithelium, suggesting that they have some interactions with the gut tissue of termites.
It is widely known that termites cannot survive without intestinal microorganisms. The physiological functions of symbiotic prokaryotes in termites are extremely diverse and include cellulose digestion (21, 40), hemicellulose digestion (43, 47), acetogenesis (5, 29, 34), hydrogenesis (46, 47), methanogenesis (5, 35), sulfate reduction (28), and nitrogen fixation (39, 48). Moreover, intestinal bacteria contribute to creating suitable conditions for symbiotic flagellates through the production of nutrients and the maintenance of the pH and anaerobic conditions in “lower termites” (7). Such microorganisms are usually distributed in the enlarged part of the hindgut, called “the paunch”; metabolites from the microorganisms are considered to be absorbed across the hindgut wall (23). It has also been reported that termites belonging to the Termitidae, which do not have symbiotic protozoa and are known as “higher termites,” possess bacteria not only in the hindgut but also in the mixed segment (1, 12).
The mixed segment is a part of the gut present only in higher termites, where it is situated between the midgut and the first proctodeal segment (2). The basic structure of the mixed segment is such that the mesenteric epithelium occupies half of the gut wall and the proctodeal epithelium covers the remaining area. Though the role of the mixed segment is still ambiguous, some physiological characteristics have recently been clarified. One of the most significant features in the mixed segment is an elevated pH (3, 7, 8). In the case of the wood-eating higher termite Nasutitermes nigriceps, a neutral pH value in the midgut increases in the mixed segment and reaches 10.23 ± 0.46 in the first proctodeal segment (7). In contrast, oxygen concentration decreases in the mixed segment and becomes zero in the first proctodeal segment (7). Bacteria have been observed under such conditions in the ectoperitrophic space of the mixed segment, which is between the gut wall and the peritrophic membrane (1). Cocci and rods have been reported there in the wood-eating termite N. exitiosus (12), while spirochetes and actinomycete-like bacteria have been reported in the soil-feeding termites Cubitermes severus and Procubitermes aburiensis (1). However, the classes and roles of the mixed-segment bacteria are still unknown. In the present study, we report the phylogenetic relationship and distribution of symbiotic bacteria in the mixed segment of the wood-eating higher termite N. takasagoensis.
MATERIALS AND METHODS
Termites.
N. takasagoensis termites were collected at Iriomote island in the Okinawa prefecture (Japan) and kept at room temperature with their nest materials. Mature worker insects were used throughout the investigations unless otherwise indicated.
Amplification and cloning of rDNA.
A gut was dissected from a termite in STE (0.15 M NaCl, 10 mM EDTA, 25 mM Tris-HCl; pH 8.0), and the mixed segment was collected in a microtube with 10 μl of STE containing 2 mg of proteinase K (Sigma Chemical Co., St. Louis, Mo.) per ml. The mixed segment was homogenized and incubated at 37°C for 1 h. The homogenate was boiled for 5 min, diluted 10 times with sterilized distilled water, and used as a PCR template. PCR was performed using primers fD1 and rP2, which are designed for most eubacterial 16S ribosomal DNA (rDNA) (53). The reaction mixture (50 μl) consisted of 0.15 mM deoxynucleotides (Takara, Tokyo, Japan), 0.2 μM forward primer, 0.2 μM reverse primer, 2 μl of the PCR template, and 0.05 U of recombinant Taq DNA polymerase (Takara) per μl in PCR buffer (Takara). The temperature regimen for 30 cycles of PCR was 94°C for 30 s, 55°C for 1 min, 72°C for 2 min, and 72°C for 5 min as a final extension after the last cycle.
We designed the specific primers rSymA (5′-GCACCGAAACTCTCATCCCG-3′, positions 806 to 787) and rSymB (5′-GGCACCGAAGGTTTCCCTCC-3′, positions 805 to 786) for 16S rDNA of the symbiotic bacteria NT-1 and NT-2, respectively, and checked for the presence of the bacteria in various gut regions. The gut from a termite was dissected in STE and divided into the foregut, midgut, mixed segment, first proctodeal segment, enteric valve plus paunch, and colon plus rectum. Each gut segment was collected in a microtube with 100 μl of STE containing 2 mg of proteinase K (Sigma) per ml. The gut segments were homogenized and incubated at 50°C for 3 h. The homogenate was boiled for 5 min and diluted 100 times with sterilized distilled water, and 0.5 μl of the diluted sample was used as a PCR template. PCR was performed using primers fD1 and rP2, primers fD1 and rSymA, or primers fD1 and rSymB. The composition of the reaction mixture (10 μl) and the PCR regimen were the same as described above.
T-vector plasmid was prepared according to the method of Marchuk et al. (31) using pBluescript II KS(+) (Stratagene, La Jolla, Calif.). The amplified DNA fragments and the T vectors were ligated using a DNA Ligation Kit (version 2; Takara). The ligated mixture was introduced into Escherichia coli JM 109 (Toyobo, Tokyo, Japan), and transformants were selected on Luria-Bertani plates containing 100 μg of ampicillin per ml. The inserted DNA fragments were amplified directly from the positive white colonies by PCR (44), and the fragment sizes were confirmed by 1% agarose gel electrophoresis.
Nucleotide sequencing and phylogenetic analysis.
A DNA fragment inserted into plasmid was amplified by PCR by using M13-20 and reverse primers, and the DNA was used for sequencing templates after purification with Sephacryl S-300 HR spin columns (Pharmacia Biotech, Uppsala, Sweden). The sequences of all positive clones were determined in both opposite orientations by using ABI PRISM Dye Primer Cycle Sequencing Kits (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.) with a DNA Sequence System (model 373S; Perkin-Elmer). The homology of the sequences obtained was examined with the MPsrch program at the DNA Information and Stock Center at the National Institute of Agrobiological Resources, Tsukuba, Japan. Related sequences were aligned with CLUSTAL W 1.7 software (50) for multiple sequence alignment, and the nucleotides of ambiguous alignments were corrected or removed manually. Tree topology was built using the maximum parsimony method with PAUP 4.0 and the neighbor-joining method of Kimura's two-parameter model with PHYLIP 3.5. Relative support for the different nodes was assessed by using 1,000 bootstrap replicates with seven random additional replicates per bootstrap replicate.
In situ hybridization.
Oligonucleotide probes (5′-CCCATTCCCCATGCGGTGTTAAGG-3′) and (5′-TCCCTGTATGATGCCATACTGTGA-3′) were designed using complementary sequences in positions of 16S rRNA from 184 to 161 for NT-1 and 182 to 159 for NT-2 and were labeled with digoxigenin (DIG)-11-ddUTP using a DIG Oligonucleotide 3′-End Labeling Kit (Boehringer Mannheim GmbH Biochemica, Mannheim, Germany). The gut was removed from a termite and fixed with 70% ethanol at 4°C for 1 h. The gut was dehydrated in an ordinary ethanol series, xylen, and then embedded in paraffin. The gut was sectioned at 6 μm and rehydrated in the ethanol series in reverse order. The sections were rinsed briefly in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS containing 0.2% Tween 20 (PBST). The sections were then rinsed with PBST and treated with 0.1% lysozyme in a 0.1 M Tris-HCl buffer (pH 8.0) containing 50 mM EDTA at 4°C for 20 min. Prehybridization was performed at room temperature for 1 h, and hybridizations were carried out at 37°C overnight in the hybridization buffer described by Hirota et al. (22), including 10 pmol of a labeled probe per ml. A mixture of 10 labeled and 100 pmol of nonlabeled probes per ml was applied to the sections as negative controls. After hybridization, the specimens were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature twice for 30 min each and in 1× SSC at room temperature for 10 min. The specimens were incubated with 10% normal goat serum in PBST for 1 h and with alkaline phosphatase-conjugated anti-DIG antibody (Boehringer Mannheim) in PBST containing 2 mM levamisole at 37°C for 2 h. The specimens were rinsed with PBST containing 2 mM levamisole and stained using a DIG Detection Kit (Boehringer Mannheim).
Whole-cell hybridization of symbiotic bacteria.
The mixed segments and hindguts (paunches) were collected from five termites, and the gut contents were washed out in 200 μl of 0.2-μm (pore-size) filtered Solution U (52). Paraformaldehyde was added to be 4% and fixed at 4°C for 1.5 h. The gut contents were briefly rinsed with PBST for three times and suspended with 20 μl of 50% ethanol. The suspension was spread on coverslips and then air dried for 2 h. The aforementioned probes were labeled with rhodamine using the Label IT Rhodamine Labeling Kit (PanVera Co., Madison, Wis.) and, to strengthen the hybridization signals, the primers rSymA and rSymB were also labeled with rhodamine and used as supplementary probes. Hybridization was performed in a culture dish using the mixture of the probes (at 50 pmol/ml) in the hybridization buffer at 37°C overnight. After hybridization, the coverslips were briefly washed in 4× SSC at room temperature and then washed in 2× SSC followed by 1× SSC for 10 min each at room temperature. The gut contents on the coverslips were counterstained with 1 μg of DAPI (4′,6′-diamidino-2-phenylindole) per ml (in 0.2-μm-filtered, distilled water), and the morphologies of the bacteria were observed with an epifluorescence microscope.
Electron microscopy.
The midguts and mixed segments from mature workers were fixed with 2.5% glutaraldehyde in 0.05 M phosphate buffer (pH 7.4) for 2 h at 4°C. The specimens were rinsed three times in the same buffer, for 10 min each time, at 4°C. The specimens were then postfixed with 1% osmium tetraoxide in 0.05 M phosphate buffer for 1 h at 4°C, dehydrated in an ordinary ethanol series, and embedded in Spurr's epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope (JEM-100C; JEOL, Ltd., Tokyo, Japan).
Nucleotide sequence accession numbers.
The sequence data determined in this study are in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession numbers AB031327 and AB031328 for NT-1 and NT-2, respectively.
RESULTS
Composition of intestinal bacteria in the mixed segment.
Bacterial 16S rRNA genes were amplified using primers fD1 and rP2, which resulted in an approximately ∼1.5-kb band in an agarose gel. Amplified fragments of prokaryotic rDNA were cloned into E. coli JM109, and 22 clones were partially sequenced. The same procedure was repeated using the mixed segment from another individual, and 20 clones were partially sequenced. The cloned rDNA sequences were composed of 11 NT-1 clones, 7 NT-2 clones, and 4 other clones in the first investigation. On the other hand, there were 13 NT-2 clones, 4 NT-1 clones, and 3 other clones in the second examination. The same procedure was repeated using another individual from a different colony, resulting in 8 clones that were NT-1, 5 clones that were NT-2, and 3 other clones, making a total of 16. These data indicate that intestinal bacteria primarily consist of two species in the mixed segment of N. takasagoensis.
Phylogeny of symbiotic bacteria in the mixed segment.
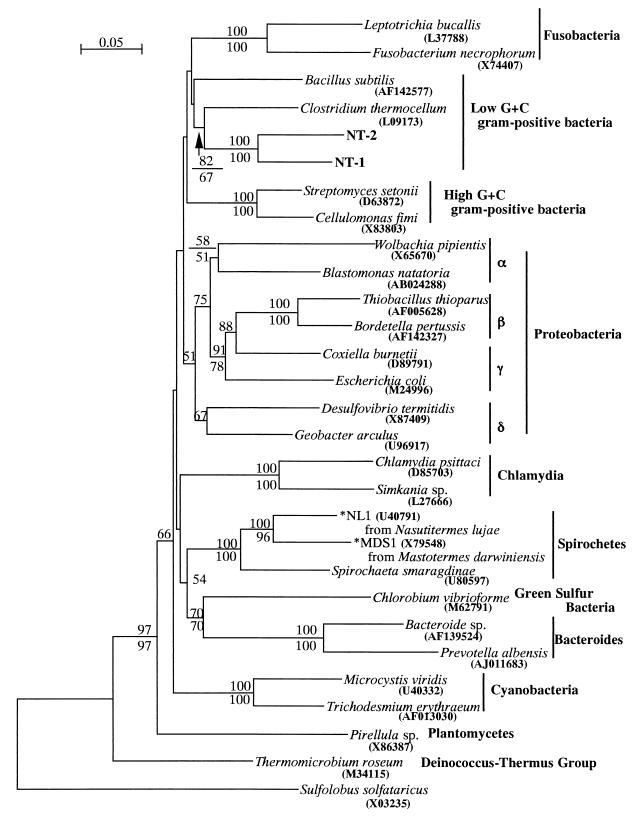
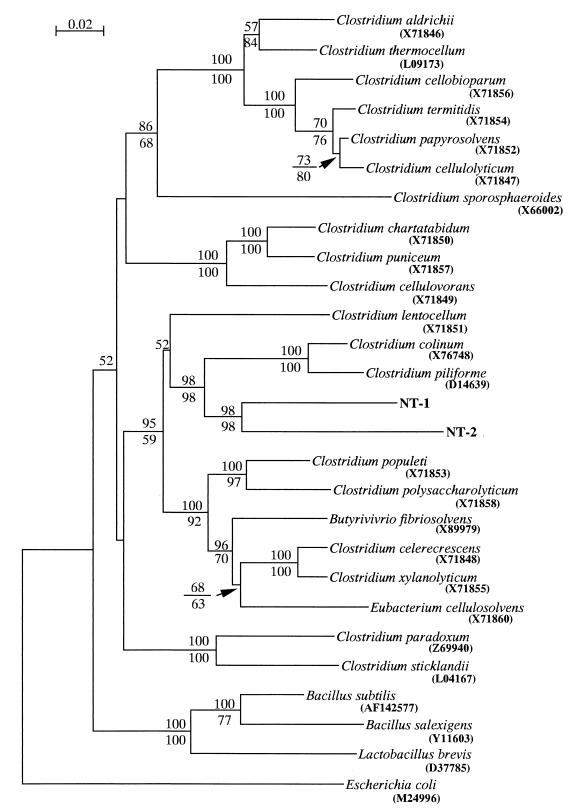
Totals of 1,442 and 1,440 bp of 16S rDNA were determined for NT-1 and NT-2, respectively; the sequence of NT-1 was 85.6% identical to that of NT-2. The sequences of both NT-1 and NT-2 clones were subjected to a homology search with the MPsrch program. No sequence in the database showed more than 90% identity to these sequences. The highest identity to NT-1 and NT-2 was obtained from Tizzer's organism (45), Clostridium piliforme (formerly known as Bacillus piliformis), which showed 85.8 and 83.7% identities to NT-1 and NT-2, respectively. The phylogenetic tree was constructed by neighbor-joining and maximum-parsimony methods, and both methods resulted in essentially the same topology. The symbiotic bacteria were placed into the low-G+C-content gram-positive group of bacteria (Fig. 1). Further phylogenetic analysis was performed within the low-G+C-content gram-positive bacterial group. Among them, both NT-1 and NT-2 were most likely to be assigned to the genus Clostridium, and they showed a monophyletic relationship with C. piliforme and C. colinum (Fig. 2), with 98% bootstrap supports for neighbor-joining and maximum-parsimony methods.
FIG. 1.
Eubacterial phylogenetic tree constructed by the neighbor-joining method on the basis of 1,189 unambiguously aligned bases. The archaebacterial 16S rDNA sequence of Sulfolobus solfataricus is used as an outgroup. The scale bar represents a 0.05 substitution per nucleotide position. Numbers above and below the branches indicate the percentage bootstrap support out of 1,000 resampling data from neighbor-joining and maximum parsimony methods, respectively (values of <50% are not shown). Accession numbers are indicated in parentheses. Spirochetes marked with an asterisk have been affiliated with the genus Treponema (30, 38).
FIG. 2.
Phylogenetic tree of low-G+C-gram positive group constructed by the neighbor-joining method on the basis of 1,245 unambiguously aligned bases. The scale bar represents a 0.02 substitution per nucleotide position. Numbers above and below the branches indicate the percentage bootstrap support out of 1,000 resampling data from neighbor-joining and maximum parsimony methods, respectively (values of <50% are not shown). The sequence from E. coli is used as an outgroup. Accession numbers are indicated in parentheses.
Distribution of the symbiotic bacteria in the gut.
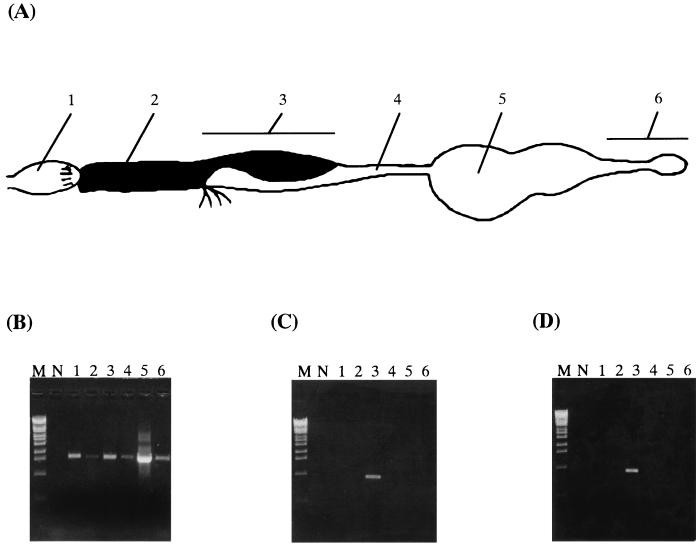
Based on these sequences, we designed specific primers rSymA and rSymB to study the distribution of the symbiotic bacteria in the gut. A gut from a termite was dissected and divided into six segments: the foregut, the midgut, the mixed segment, the first proctodeal segment, the second plus the third proctodeal segments (the enteric valve and paunch), and the forth plus the fifth proctodeal segments (the colon and rectum) (Fig. 3A). PCR was performed to study general distribution of bacteria using primers fD1 and rP2 to each gut segment. This PCR revealed an expected fragment in all gut segments (Fig. 3B), suggesting that bacteria are present throughout the gut. We then performed PCR using the primer set rSymA-fD1 or rSymB-fD1, which is specific to NT-1 or NT-2, respectively. The expected bands were detected only in the mixed segment for both NT-1 and NT-2 (Fig. 3C and D), suggesting that they are confined to the mixed segment.
FIG. 3.
Distribution of bacteria in the gut of N. takasagoensis. (A) A schematic drawing of the gut showing the following: 1, foregut; 2, midgut; 3, mixed segment; 4, first proctodeal segment; 5, enteric valve and paunch; and 6, colon and rectum. (B) PCR using general eubacterial primers. (C) PCR using specific primers to NT-1. (D) PCR using specific primers to NT-2. M, molecular marker (λ-Eco T14I digest); N, negative control without template. The number of each lane corresponds to the numbers in panel A.
Infection of the symbiotic bacteria in colony members of N. takasagoensis.
It is difficult to properly select each stage of the termites since caste systems are not well studied in N. takasagoensis. Therefore, we randomly selected 10 soldiers and 10 workers of various sizes and collected the mixed segments from all selected termites. PCR was performed using the specific primers for NT-1 and NT-2 to determine whether these bacteria infected various stages of the termites. The expected bands for NT-1 and NT-2 were detected in all worker and soldier termites (data not shown). This result suggests that NT-1 and NT-2 infect most termites in a colony.
In situ hybridization.
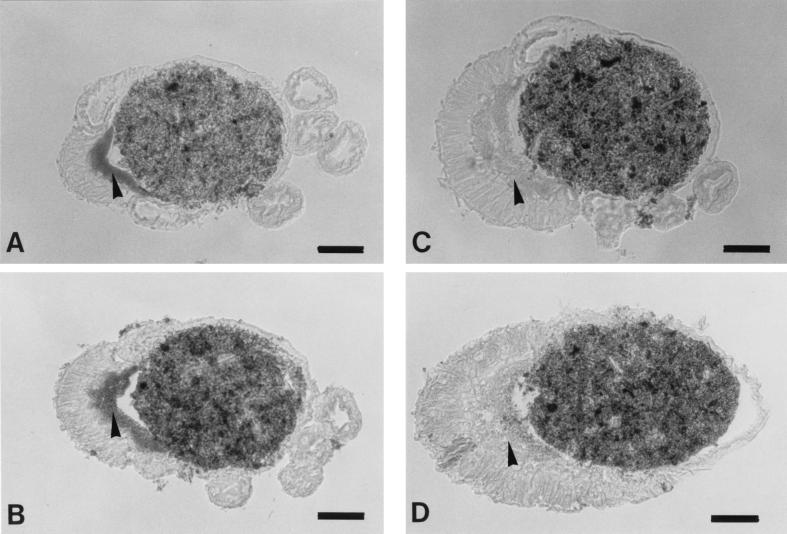
An in situ hybridization technique was applied to paraffin sections of the gut with short oligonucleotide probes (24 bp) specific to the 16S rRNAs of NT-1 and NT-2. The hybridization signals for both NT-1 and NT-2 showed very similar distributions. They were observed in the ectoperitrophic space between the peritrophic membrane and the midgut wall throughout the mixed segment, and the strong NT-1 signals were detected especially in the anterior part of the mixed segment (Fig. 4A). The NT-2 signals were strong in the middle to the posterior part of the mixed segment (Fig. 4B). However, such signals were not observed in the gut lumen or in the ectoperitrophic space of the proctodeal side. Signals were not detected in control sections (Fig. 4C and D), which were hybridized with a mixture of 10 pmol of labeled probes per ml and an excess amount (100 pmol/ml) of nonlabeled probes.
FIG. 4.
In situ hybridization to 16S rRNA of the symbiotic bacteria in the mixed segment of N. takasagoensis. Arrowheads indicate the ectoperitrophic space of the mesenteric side, which is filled with bacteria. (A) Cross-section of the anterior part hybridized to NT-1. (B) The posterior part hybridized to NT-2. (C) Control section hybridized to NT-1. (D) Control section hybridized to NT-2. Bars, 50 μm.
Whole-cell hybridization.
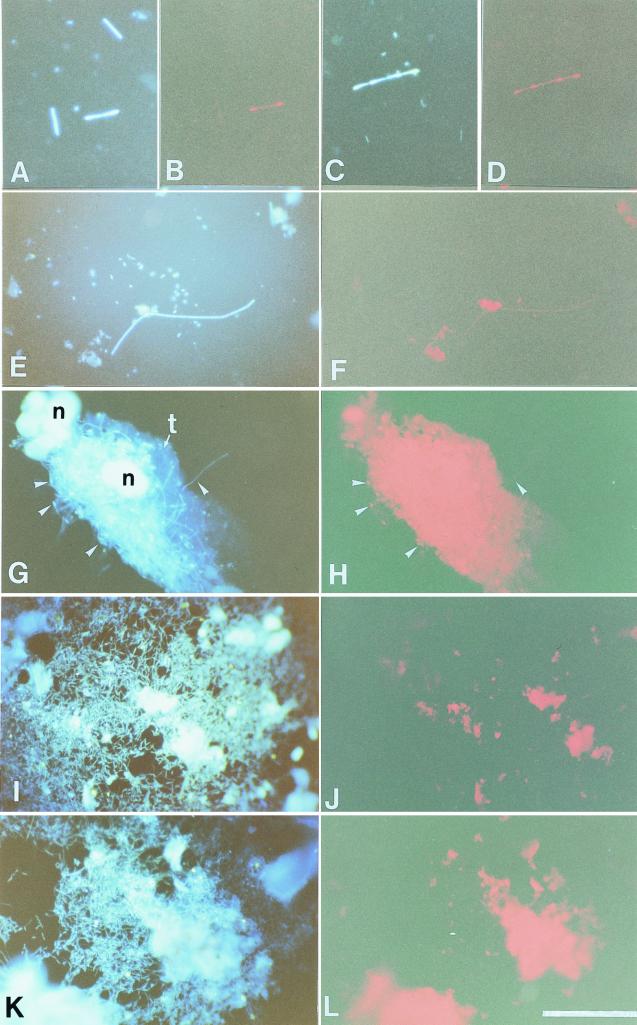
To observe morphologies of the symbiotic bacteria, we performed whole-cell hybridization to gut contents from the mixed segment using the rhodamine-labeled probes. Because the signals were too weak to recognize the individual bacteria by using only one probe, the aforementioned primers rSymA and rSymB were also labeled with rhodamine and applied to the gut contents with the former probes at the same time to strengthen the signals. The probes specific for NT-1 recognized short rods in chains, which have gnarl-like structures at the edge of each cell (Fig. 5A to D), while the probes specific for NT-2 detected not only bacteria similar to those recognized by NT-1 probes but also simple short rods in chains (Fig. 5E and F). Even after the gut contents were washed out, several chained short rods remained adhered to pieces of the mesenteric tissue (Fig. 5G). Although the strong red autofluorescence from the tissue interfered with the detection of signals from such bacteria, NT-2 probes could detect some signals from the bacteria attached to the midgut wall (Fig. 5H). It is not clear whether the NT-2 probes specifically detected NT-2 or whether they detected both NT-1 and NT-2 in this experiment because they stained two types of morphologies and it was difficult to perform the hybridization in more severe conditions due to very weak signals. However, in contrast, both probes did not hybridize to cocci in the mixed segment and bacteria present in the hindgut (Fig. 5I to L), suggesting that these probes are specific enough to detect the target bacteria NT-1 and NT-2.
FIG. 5.
Whole-cell hybridization to 16S rRNA of the symbiotic bacteria in the mixed segment of N. takasagoensis. Rhodamine-labeled probes specific for NT-1 were applied to the gut contents, and bacteria were viewed with the DAPI filter set (A and C) and the rhodamine filter set (B and D). Stained short rods in chains have gnarl-like structure at the edges of the cells. Bacteria hybridized with the NT-2 probes were viewed with the DAPI filter set (E) or the rhodamine filter set (F). Simple short rods in chains were weakly stained, and nonspecific autofluorescence from gut contents or contaminated tissues was detected throughout the experiments. Several bacteria (arrowheads) are adhered to a piece of the mesenteric tissue (G), and some of them were detected by the NT-2 probes (H), although the strong autofluorescence from the tissue interferes with the signals from most bacteria. (I and K) Numerous bacteria were observed in the hindgut by DAPI staining. However, these bacteria are not stained with NT-1 (J) or NT-2 (L) probes. t, mesenteric tissue; n, nucleus. Bar, 50 μm.
Electron microscopy.
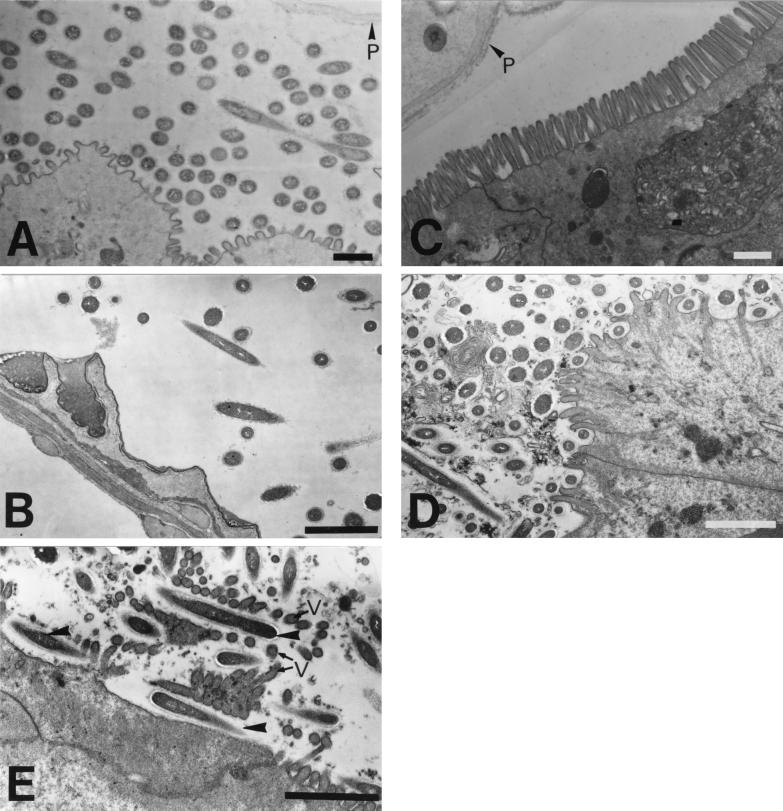
Transverse sections of the mixed segment were observed with an electron microscope to more precisely determine the distribution of the symbionts. Numerous bacteria were observed in the ectoperitrophic space of the mesenteric side (Fig. 6A), while some bacteria were sparsely distributed in the ectoperitrophic space of the proctodeal side (Fig. 6B). On the contrary, bacteria are very rare in the ectoperitrophic space of the actual midgut (Fig. 6C). Except for the most anterior part, several bacteria were situated between the microvilli, and some were attached to the midgut wall (Fig. 6D) throughout the mesenteric tissue of the mixed segment. The longitudinal section shows that several rods were situated between the microvilli (Fig. 6E), and this finding corresponds to the observations made by whole-cell hybridization. These results suggest close associations between these bacteria and the gut tissue.
FIG. 6.
Electron micrographs of the mixed segment in N. takasagoensis. (A) The most anterior part of the mixed segment. Numerous bacteria are present in the ectoperitrophic space between the mesenteric epithelium and peritrophic membrane. (B) Proctodeal epithelium and luminal bacteria. Some bacteria are sparsely distributed in the ectoperitrophic space. (C) Posterior part of the actual midgut. Bacteria are rare in the ectoperitrophic space, and microvilli are well developed. (D) Mesenteric side in the middle part of the mixed segment. Bacteria are observed not only in the ectoperitrophic space but also among the microvilli. (E) Longitudinal section of the apical surface of the mesenteric epithelium. Rods (arrowheads) are situated between microvilli (v). P, peritrophic membrane. Bars, 1 μm.
DISCUSSION
The role of symbiotic microbes is important for clarifying the digestive physiology in termites. However, precise information was not available about symbiotic bacteria in the mixed segment, except for their morphology. In the present study, we first clarified the phylogeny and distribution of symbiotic bacteria in the mixed segment.
Our phylogenetic study suggests that the symbiotic bacteria in the mixed segment are closely related to the Clostridium group, which is one of the largest bacterial genera. These symbionts differ from bacteria found in the wood-eating lower termite Reticulitermes speratus (36) and from the clostridia isolated from higher termites (21, 26) based on 16S rDNA sequences, and no low-G+C-content gram-positive bacteria have been discovered in the gut of the drywood-eating lower termite Cryptotermes domesticus (37). It is commonly known that Clostridium species are usually obligate anaerobes, and some are facultative anaerobes and appear as rods, short rods, or cocci (42). These characters correspond to the morphological observations in N. exitiosus (12) and N. takasagoensis. Since the microflora in the mixed segment consist mainly of two species in an aspect of 16S rDNA sequences, these bacteria probably contribute to the dramatic decrease of oxygen concentration observed in the mixed segment (7). Clostridia are usually known as soil bacteria and are also found in the rumens and intestines of animals and humans (11, 13, 19). Intestinal bacteria are important to the health of human beings (11), and some predicted roles of intestinal bacteria in termites (6) are similar to those in humans, such as producing short-chain fatty acids from carbohydrates or synthesizing amino acids (11). Because many clostridia degrade polysaccharides to produce acetone, alcohol, acetate, lactate, CO2, and hydrogen (9, 20, 24, 33, 41) and others can ferment nitrogenous or lipidic compounds (16, 25), it is expected that the symbiotic bacteria of N. takasagoensis play a role in the nutritional physiology of the termite; in fact, an acetogenic clostridial species has already been isolated from a soil-feeding termite (26). However, pathogenic activities are also well known in many anaerobic clostridia (4). The symbiotic bacteria form a clade with Tizzer's organism and C. colinum with high bootstrap percentages, suggesting that they share a common ancestor. Tizzer's organism, Bacillus piliformis is widely known as a pathogen for laboratory, domesticated, and wild animals and is characterized by multifocal necrotizing hepatitis, carditis, and/or severe hemorrhagic enteritis (18, 45). This organism has never been grown in a cell-free medium, although it has been cultured in mammalian cells and chicken eggs (45). B. piliformis was assigned to the genus Clostridium by 16S rRNA sequencing in 1993 and renamed Clostridium piliforme (15). Similarly, C. colinum is also known as a pathogenic anaerobe that causes ulcerative enteritis in avian species (4). They were both assigned to Cluster XIV of the clostridia, based on 16S rRNA gene sequences (10), which signifies that our bacteria are a new species belonging to this cluster. It is interesting that the symbionts of the termite are the most closely related to these pathogenic bacteria, which prefer the closed environment present in living organisms and demonstrate significant associations to gut tissues.
Our PCR experiments suggest that bacteria are generally present throughout the guts of termites. This fact in turn suggests that many of the bacteria in termite guts are just passing through or are digested by termites as a nutritional source. Such bacteria could also be not-yet-identified species as well as essential gut residents. Therefore, it is important to distinguish the facultative residents from the essential residents in termites to understand the roles of microorganisms in termite guts. We assume that microflora in the foregut are occasional residents, because the foregut is an organ used simply for physical digestion of wood particles and is not a fermentation chamber like the paunch. Moreover, the foregut is the part nearest to the mouth and completely aerobic, in contrast to the hindgut (7, 17). The symbionts in this study were not detected in the foregut and, surprisingly, these bacteria were also not detected in the paunch, which is inhabited by numerous bacteria. Both symbionts are likely to be confined to the mixed segment. It is known that a dramatic increase in the pH value occurs in the mixed segment (3, 7, 8), and alkaline fluid, predominantly including potassium ions, is thought to be secreted from the mesenteric tissue of the mixed segment (1). These symbiotic bacteria seem to be completely suited to this region, and it might be interesting to study how they adapt to this very strict alkaline environment. Na+ or K+-H+ antiporters are known to be involved in the pH homeostasis of bacteria in some alkaline bacillus species that are closely related to clostridia (27). A K+-H+ antiporter-like structure is probably developed in the symbiotic bacteria of the mixed segment.
The in situ hybridization and electron microscopy clearly showed the localization of the symbiotic bacteria to the mesenteric side of the mixed segment. Since there is a trace amount of cellulase activity in this region (51), since the peritrophic membrane keeps the bacteria from wood particles in the lumen, and since these bacteria do not distribute uniformly around the peritrophic membrane, we suspect that their nutritional source does not come from the lumen. Electron microscopy revealed the close relationship between the bacteria and mesenteric epithelium, suggesting that the symbiotic bacteria utilize some substances secreted from the mesenteric tissue in the mixed segment. It is known that midgut cells are replenished very quickly in cockroaches (14). Such phenomena have also been observed in the termite R. speratus, in which the midgut cells that have fulfilled their function are believed to be endocytosed by neighbor epithelial cells and digested in phagolysosomes (54). Similar phagosomes are observed throughout the midgut columnar cells, including the mixed segment region of N. takasagoensis (data not shown); the symbionts trapped among the microvilli of old cells could be endocytosed by young columnar cells and digested with the old cells.
The transmission mechanism of these symbionts from generation to generation remains unclear. Since usually bacteria cannot pass through the peritrophic membrane (49) and since they are not present in any other gut regions, it is unlikely that they infect the termites at the stage when the peritrophic membrane is intact. However, the bacteria are present not only in worker insects but also in soldier insects, which cannot feed directly on wood and ingest stomodeal and/or proctodeal foods from workers (32). This suggests that a trace of the bacteria might be ingested as the contamination of stomodeal and/or proctodeal foods and reaches the mixed segment immediately after molting, when the peritrophic membrane has not yet formed.
The results of phylogeny and distribution studies of the symbiotic bacteria in the mixed segment indicated that these bacteria might play a significant role in the gut physiology of termites. Since the mixed segments of soil-feeding termites exhibit microflora different from that of wood-eating termites (1), it is possible that symbionts in the mixed segment have a close relationship to the feeding habits, gut physiology, and/or phylogeny of the hosts. Clarification of the physiological function of the symbiotic clostridia will greatly contribute to our understanding of host-symbiont interaction and the role of the mixed segment in higher termites.
ACKNOWLEDGMENT
This work was supported by Program for Promotion of Basic Research Activities for Innovative Biosciences of Bio-oriented Technology Research Advancement Institution.
REFERENCES
- 1.Bignell D E, Oskarsson H, Anderson J M, Ineson P. Structure, microbial associations and function of the so-called “mixed segment” of the gut in two soil-feeding termites, Procubitermes aburiensis and Cubitermes severus (Termitidae, Termitinae) J Zool. 1983;201:445–480. [Google Scholar]
- 2.Bignell D E. Soil-feeding and gut morphology in higher termites. In: Hunt J H, Nalepa C A, editors. Nourishment and evolution in insect societies. Boulder, Colo: Westview Press, Inc.; 1994. pp. 131–158. [Google Scholar]
- 3.Bignell D E, Eggleton P. On the elevated intestinal pH of higher termites (Isoptera: Termitidae) Insectes Soc. 1995;42:57–69. [Google Scholar]
- 4.Borriello S P. Clostridial disease of the gut. Clin Infect Dis. 1995;20:S242–S250. doi: 10.1093/clinids/20.supplement_2.s242. [DOI] [PubMed] [Google Scholar]
- 5.Brauman A, Kane M D, Labat M, Breznak J A. Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science. 1992;257:1384–1387. doi: 10.1126/science.257.5075.1384. [DOI] [PubMed] [Google Scholar]
- 6.Breznak J A, Brune A. Role of microorganisms in the digestion of lignocellulose by termites. Annu Rev Entomol. 1994;39:453–487. [Google Scholar]
- 7.Brune A, Emerson D, Breznak J A. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl Environ Microbiol. 1995;61:2681–2687. doi: 10.1128/aem.61.7.2681-2687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune A, Kühl M. pH profiles of the extremely alkaline hindguts of soil-feeding termites (Isoptera: Termitidae) determined with microelectrodes. J Insect Physiol. 1996;42:1121–1127. [Google Scholar]
- 9.Chen J S. Alcohol dehydrogenase: multiplicity and relatedness in the solvent-producing clostridia. FEMS Microbiol Rev. 1995;17:263–273. doi: 10.1111/j.1574-6976.1995.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 10.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 11.Cummings J H, Macfarlane G T. Role of intestinal bacteria in nutrient metabolism. J Parenter Enteral Nutr. 1997;21:357–365. doi: 10.1177/0148607197021006357. [DOI] [PubMed] [Google Scholar]
- 12.Czolij R, Slaytor M, O'Brien R W. Bacterial flora of the mixed segment and the hindgut of the higher termite Nasutitermes exitiosus Hill (Termitidae, Nasutitermitinae) Appl Environ Microbiol. 1985;49:1226–1236. [Google Scholar]
- 13.Dehority B A. Cellulose degradation in ruminants. In: Haigler C H, Weimer P J, editors. Biosynthesis and biodegradation of cellulose. New York, N.Y: Marcel Dekker, Inc.; 1991. pp. 327–354. [Google Scholar]
- 14.Dow J A T. Insect midgut function. Adv Insect Physiol. 1986;19:188–328. [Google Scholar]
- 15.Duncan A J, Carman R J, Olsen G J, Wilson K H. Assignment of the agent of Tizzer's disease to Clostridium piliforme comb. nov. on the basis of 16S rRNA sequence analysis. Int J Syst Bacteriol. 1993;43:314–318. doi: 10.1099/00207713-43-2-314. [DOI] [PubMed] [Google Scholar]
- 16.Elsden S R, Hilton M G. Amino acid utilization patterns in clostridial taxonomy. Arch Microbiol. 1979;123:137–141. doi: 10.1007/BF00446812. [DOI] [PubMed] [Google Scholar]
- 17.Eutick M L, O'Brien R W, Slaytor M. Aerobic state of gut of Nasutitermes exitiosus and Coptotermes lacteus, high and low caste termites. J Insect Physiol. 1976;22:1377–1380. [Google Scholar]
- 18.Goto K, Itoh T. Detection of Bacillus piliformis by specific amplification of ribosomal sequences. Exp Anim. 1994;43:389–394. [PubMed] [Google Scholar]
- 19.Haagsma J. Pathogenic bacteria and the environment. Rev Sci Tech Off Int Epizoot. 1991;10:749–764. doi: 10.20506/rst.10.3.569. [DOI] [PubMed] [Google Scholar]
- 20.Hazlewood G P, Gilbert H J. Xylan and cellulose utilization by the clostridia. Bio/Technology. 1993;25:311–341. [PubMed] [Google Scholar]
- 21.Hethener P, Brauman A, Garcia J-L. Clostridium termitidis sp. nov., a cellulolytic bacterium from the gut of the wood-feeding termite Nasutitermes lujae. Syst Appl Microbiol. 1992;15:52–58. [Google Scholar]
- 22.Hirota S, Ito A, Morii E, Wanaka A, Tohyama M, Kitamura Y, Nomura S. Localization of mRNA for c-kit receptor and its ligand in the brain of adult rats: an analysis using in situ hybridization. Mol Brain Res. 1992;15:47–54. doi: 10.1016/0169-328x(92)90150-a. [DOI] [PubMed] [Google Scholar]
- 23.Hogan M E, Slaytor M, O'Brien R W. Transport of volatile fatty acids across the hindgut of the cockroach Panesthia ciribrata Saussure and the termite, Mastotermes darwiniensis Froggatt. J Insect Physiol. 1985;31:587–591. [Google Scholar]
- 24.Johnston N C, Goldfine H. Phospholipid aliphatic chain composition modulates lipid class composition, but not lipid asymmetry in Clostridium butyricum. Biochim Biophys Acta. 1985;813:10–18. doi: 10.1016/0005-2736(85)90339-6. [DOI] [PubMed] [Google Scholar]
- 25.Jones D T, Woods D R. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50:484–525. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane M D, Brauman A, Breznak J A. Clostridium mayombei sp. nov., an H2/CO2 acetogenic bacterium from the gut of the African soil-feeding termite Cubitermes speciosus. Arch Microbiol. 1991;156:99–104. [Google Scholar]
- 27.Krulwich T A, Ito M, Gilmour R, Hicks D B, Guffanti A A. Energetics of alkaliphilic Bacillus species: physiology and molecules. Adv Microb Physiol. 1998;40:401–438. doi: 10.1016/s0065-2911(08)60136-8. [DOI] [PubMed] [Google Scholar]
- 28.Kuhnigk T, Branke J, Krekeler D, Cypionka H, König H. A feasible role of sulfate-reducing bacteria in the termite gut. System Appl Microbiol. 1996;19:139–149. [Google Scholar]
- 29.Leadbetter J R, Schmidt T M, Graber J R, Breznak J A. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science. 1999;283:686–689. doi: 10.1126/science.283.5402.686. [DOI] [PubMed] [Google Scholar]
- 30.Lilburn T G, Schmidt T M, Breznak J A. Phylogenetic diversity of termite gut spirochetes. Environ Microbiol. 1999;1:331–345. doi: 10.1046/j.1462-2920.1999.00043.x. [DOI] [PubMed] [Google Scholar]
- 31.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mcmahan E A. Feeding relationships and radioisotope techniques. In: Krishna K, Weesner F M, editors. Biology of termites. Vol. 1. New York, N.Y: Academic Press; 1969. pp. 387–406. [Google Scholar]
- 33.Mitchell W J. Carbohydrate assimilation by saccharolytic clostridia. Res Microbiol. 1992;143:245–250. doi: 10.1016/0923-2508(92)90016-h. [DOI] [PubMed] [Google Scholar]
- 34.Odelson D A, Breznak J A. Volatile fatty acid production by the hindgut microbiota of xylophagous termites. Appl Environ Microbiol. 1983;45:1602–1613. doi: 10.1128/aem.45.5.1602-1613.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkuma M, Noda S, Horikoshi K, Kudo T. Phylogeny of symbiotic methanogens in the gut of the termite Reticulitermes speratus. FEMS Microbiol Lett. 1995;134:45–50. doi: 10.1111/j.1574-6968.1995.tb07912.x. [DOI] [PubMed] [Google Scholar]
- 36.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkuma M, Kudo T. Phylogenetic analysis of the symbiotic intestinal microflora of the termite Cryptotermes domesticus. FEMS Microbiol Lett. 1998;164:389–395. [Google Scholar]
- 38.Ohkuma M, Iida T, Kudo T. Phylogenetic relationships of symbiotic spirochetes in the gut of diverse termites. FEMS Microbiol Lett. 1999;181:123–129. doi: 10.1111/j.1574-6968.1999.tb08834.x. [DOI] [PubMed] [Google Scholar]
- 39.Ohkuma M, Noda S, Usami R, Horokoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasti M B, Belli M L. Cellulolytic activity of Actinomycetes isolated from termites (Termitidae) gut. FEMS Microbiol Lett. 1985;26:107–112. [Google Scholar]
- 41.Rainey F A, Stackebrandt E. 16S rDNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol Lett. 1993;113:125–128. doi: 10.1111/j.1574-6968.1993.tb06501.x. [DOI] [PubMed] [Google Scholar]
- 42.Rieu-Lesme F, Dauga C, Morvan B, Bouvet O M, Grimont P A, Dore J. Acetogenic coccoid spore-forming bacteria isolated from the rumen. Res Microbiol. 1996;147:753–764. doi: 10.1016/s0923-2508(97)85122-4. [DOI] [PubMed] [Google Scholar]
- 43.Schäfer A, Konrad R, Kuhnigk T, Kämpfer P, Hertei H, König K. Hemicellulose-degrading bacteria and yeasts from the termite gut. J Appl Bacteriol. 1996;80:471–478. doi: 10.1111/j.1365-2672.1996.tb03245.x. [DOI] [PubMed] [Google Scholar]
- 44.Simada H, Tada Y. Rapid isolation of a rice waxy sequence: a simple PCR method for the analysis of recombinant plasmids from intact Escherichia coli cells. Gene. 1991;98:243–248. doi: 10.1016/0378-1119(91)90180-j. [DOI] [PubMed] [Google Scholar]
- 45.Smith K J, Skelton H G, Hilyard E J, Hadfield C T, Moeller R S, Tuur S, Decker C, Wagner K F, Angritt P. Bacillus piliformis infection (Tizzer's disease) in a patient infected with HIV-1: confirmation with 16S ribosomal RNA sequence analysis. J Am Acad Dermatol. 1996;34:343–348. doi: 10.1016/s0190-9622(07)80005-3. [DOI] [PubMed] [Google Scholar]
- 46.Taguchi F, Chang J D, Mizukami N, Saito-Taki T, Hasegawa K, Morimoto M. Isolation of a hydrogen-producing bacterium, Clostridium beijerinckii strain AM21B, from termites. Can J Microbiol. 1993;39:726–730.93. [Google Scholar]
- 47.Taguchi F, Hasagawa K, Saito-Taki T, Hara K. Simultaneous production of xylanase and hydrogen using xylan in batch culture of Clostridium sp. strain X53. J Ferment Bioeng. 1996;81:178–180. [Google Scholar]
- 48.Tayasu I, Sugimoto A, Wada E, Abe T. Xylophagous termites depending on atmospheric nitrogen. Naturwissenschaften. 1994;81:229–231. [Google Scholar]
- 49.Tellam R L, Wijffels G, Willadsen P. Peritrophic matrix proteins. Insect Biochem Mol Biol. 1999;29:87–101. doi: 10.1016/s0965-1748(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 50.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tokuda G, Watanabe H, Matsumoto T, Noda H. Cellulose digestion in the wood-eating higher termite, Nasutitermes takasagoensis (Shiraki): distribution of cellulases and properties of endo-β-1,4-glucanase. Zool Sci. 1997;14:83–93. doi: 10.2108/zsj.14.83. [DOI] [PubMed] [Google Scholar]
- 52.Trager W. The cultivation of a cellulose-digesting flagellate Trichomonas termopsidis and of certain other termite protozoa. Biol Bull. 1934;66:182–190. [Google Scholar]
- 53.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaoka I, Nagatani Y. Phagocytic cells in the midgut epithelium of the termite, Reticulitermes speratus (Kolbe) Zool Mag. 1980;89:308–311. [Google Scholar]