Abstract
We report detailed functional analyses and genotype-phenotype correlations in 392 individuals carrying disease-causing variants in SCN8A, encoding the voltage-gated Na+ channel Nav1.6, with the aim of describing clinical phenotypes related to functional effects. Six different clinical subgroups were identified: Group 1, benign familial infantile epilepsy (n = 15, normal cognition, treatable seizures); Group 2, intermediate epilepsy (n = 33, mild intellectual disability, partially pharmaco-responsive); Group 3, developmental and epileptic encephalopathy (n = 177, severe intellectual disability, majority pharmaco-resistant); Group 4, generalized epilepsy (n = 20, mild to moderate intellectual disability, frequently with absence seizures); Group 5, unclassifiable epilepsy (n = 127); and Group 6, neurodevelopmental disorder without epilepsy (n = 20, mild to moderate intellectual disability). Those in Groups 1–3 presented with focal or multifocal seizures (median age of onset: 4 months) and focal epileptiform discharges, whereas the onset of seizures in patients with generalized epilepsy was later (median: 42 months) with generalized epileptiform discharges. We performed functional studies expressing missense variants in ND7/23 neuroblastoma cells and primary neuronal cultures using recombinant tetrodotoxin-insensitive human Nav1.6 channels and whole-cell patch-clamping.
Two variants causing developmental and epileptic encephalopathy showed a strong gain-of-function (hyperpolarizing shift of steady-state activation, strongly increased neuronal firing rate) and one variant causing benign familial infantile epilepsy or intermediate epilepsy showed a mild gain-of-function (defective fast inactivation, less increased firing). In contrast, all three variants causing generalized epilepsy induced a loss-of-function (reduced current amplitudes, depolarizing shift of steady-state activation, reduced neuronal firing). Functional effects were known for 170 individuals. All 136 individuals carrying a functionally tested gain-of-function variant had either focal (n = 97, Groups 1–3) or unclassifiable (n = 39) epilepsy, whereas 34 individuals with a loss-of-function variant had either generalized (n = 14), no (n = 11) or unclassifiable (n = 6) epilepsy; only three had developmental and epileptic encephalopathy. Computational modelling in the gain-of-function group revealed a significant correlation between the severity of the electrophysiological and clinical phenotypes. Gain-of-function variant carriers responded significantly better to sodium channel blockers than to other anti-seizure medications, and the same applied for all individuals in Groups 1–3.
In conclusion, our data reveal clear genotype-phenotype correlations between age at seizure onset, type of epilepsy and gain- or loss-of-function effects of SCN8A variants. Generalized epilepsy with absence seizures is the main epilepsy phenotype of loss-of-function variant carriers and the extent of the electrophysiological dysfunction of the gain-of-function variants is a main determinant of the severity of the clinical phenotype in focal epilepsies. Our pharmacological data indicate that sodium channel blockers present a treatment option in SCN8A-related focal epilepsy with onset in the first year of life.
Keywords: SCN8A, epilepsy, genetics, personalized medicine
Johannesen et al. report detailed functional analyses and genotype-phenotype correlations in almost 400 individuals with SCN8A-related disorders: individuals carrying gain-of-function variants have early-onset focal epilepsy, while individuals with loss-of-function variants have late-onset generalized epilepsy.
Introduction
Since the first pathogenic SCN8A variant was discovered in an individual with epilepsy,1 a wide clinical spectrum of neurodevelopmental phenotypes has been reported. The spectrum ranges from benign familial infantile epilepsy (BFIE) with self-limiting seizures and typical cognitive development,2-4 over an intermediate phenotype with variable seizure onset, treatable seizures and mild to moderate intellectual disability (ID)5 to developmental and epileptic encephalopathies (DEEs) with onset in the first year of life and moderate to severe ID,6–10 often with movement disorders, cortical visual impairment, severe gastrointestinal symptoms and increased risk of premature death.7,10–15 Furthermore, rare clinical presentations with ID, autism spectrum disorder (ASD) and movement disorders without epilepsy have been described.16–19
SCN8A encodes Nav1.6, which is one of four voltage-gated sodium channels expressed in the mammalian brain. Nav1.6 is found in the central and peripheral nervous system with predominant expression in excitatory, but also in inhibitory, neurons.20 Previous studies have revealed that BFIE and DEEs are caused by missense variants with gain-of-function (GOF) effects, whereas truncating variants, deletions and certain missense variants causing loss-of-function (LOF) or both LOF and GOF effects have been associated with ID, ASD and movement disorders with or without seizures.3,11,17,19,21
Treatment of SCN8A-related DEEs has resulted in frequent resistance to anti-seizure medications (ASMs), although treatment with sodium channel blockers (SCBs), especially high-dosage phenytoin, is beneficial in some affected individuals.22
Here, we combined a detailed clinical analysis of the largest cohort of individuals with SCN8A-related neurodevelopmental disorders investigated to date with functional studies of newly detected variants in mammalian cells and primary neurons and explored the genotype-phenotype correlations in functional studies, computational modelling and treatment response in SCN8A.
Materials and methods
Cohort ascertainment and phenotyping
Affected individuals were recruited through a network of collaborating clinicians, as well as GeneMatcher,23 by means of a standardized phenotyping sheet to assess clinical characteristics (medical history, seizure and physical characteristics, family history, neurodevelopment and cognition), EEG, neuroimaging and retrospective data on anti-epileptic treatment. Seizures and epilepsy syndromes were classified according to the latest ILAE guidelines.24,25
Based on the information available about the presence and severity of the epilepsy, seizure onset and cognitive status, the affected individuals were categorized into the following a priori defined subgroups: Group 1: BFIE, infantile onset focal seizures with onset during infancy and normal cognitive development3; Group : intermediate epilepsy, individuals with a focal epilepsy (seizure types and EEG) of intermediate severity, reflecting neither BFIE nor DEE5; Group 3: DEE,14 individuals with focal seizure types and EEG; Group 4: generalized epilepsy, individuals with a generalized epilepsy (seizure types and EEG), thus not fitting Groups 1, 2 or 3; Group 5: unclassifiable epilepsy, individuals with insufficient data to be classified into one of the above groups, or, rarely, both focal and generalized seizure types or epileptiform discharges; and Group 6: neurodevelopmental disorder without epilepsy (NDDwoE).
See Supplementary Fig. 2 for a flow chart describing the categorization of SCN8A-related phenotypes.
Treatment response was evaluated by the referring treatment providers as freedom from seizures (at least 6 months without seizures), seizure reduction (affected individuals still on ASMs as the treatment provider and parents consider there to be a beneficial effect), no change (ASMs terminated) or seizure aggravation noted by treatment providers and parents. Phenytoin, carbamazepine, oxcarbazepine, lacosamide, lamotrigine and zonisamide were all classified as SCBs.
Variant pathogenicity was assessed according to the ACMG guidelines.26 All variants were absent from gnomAD r2.1.1 (https://gnomad.broadinstitute.org/) and BRAVO (https://bravo.sph.umich.edu/freeze8/hg38/). Pathogenic or likely pathogenic variants were included. SCN8A transcript NM_014191.3 was used for coding variant nomenclature.
Previously published cases
A PubMed search for ‘SCN8A’ was performed, and all publications that included affected individuals were included in the present study. The latest search was performed on 15 May 2020. Papers not available in English were excluded. Only original cases and only probands were included. Pathogenic or likely pathogenic variants were included, and the variant locations were remapped on NM_014191.3 when necessary to harmonize their presentation in this article. Data on functional studies were also collected. If affected individuals were published more than once, all papers were included in the list. Duplications of affected individuals were avoided by follow-up with the referring clinician/corresponding author.
Ethics
The study was approved by the local ethical committees or followed other local guidelines. Previously unpublished individuals (or parents, in the case of minors) provided informed consent.
Frequency
The Danish Epilepsy Centre is the only tertiary hospital in Denmark that specializes in treating epilepsy, and the majority of affected individuals with non-acquired epilepsy are referred to the centre for genetic testing. Furthermore, inquiries were sent to all national clinical genetics departments. Thus, we were able to estimate the frequency of SCN8A variants in the Danish population by using the birth cohort from 2006 to 2017 in Statistics Denmark, the electronic population database of Danish national statistics.
Functional studies
Variants were chosen for functional studies according to the clinical phenotype of the variant carriers and the location of variants in the Nav1.6 channel protein to cover the phenotypic spectrum of SCN8A-related disorders (see the ‘Results’ section for details). All methods have been previously described19 and are summarized in the Supplementary material.
Neuronal simulations
To investigate how changes in Na+ current properties affect neuronal firing behaviour, we simulated a model neuron with sodium, potassium and leak currents (details provided in the Supplementary material). We simulated firing by injecting step currents of 0–0.75 nA for 2 s and analysed the voltage traces for the firing rate after the neuron had adapted to the injected current. The resulting input-output curve was then analysed for its area under the curve (AUC). We modified the following six parameters of theNa+ current. The half activation voltages V1/2 of the activation and inactivation curves were shifted within ±10 mV of the original values. The slopes k of the activation and inactivation curves as well as the maximum conductance gNa were multiplied with a scaling factor ranging from 0.5 to 2. The persistent current z of inactivation ranged from 0 to 5% of the maximum inactivation. Only a single parameter was modified at a time. The effect of alteration i on the AUC was quantified relative to the AUC of the unchanged model AUC0 by:
| (1) |
For each altered parameter, the slope mj of the regression line through the origin between the parameter changes x and its resulting AUC contrast was calculated. To compare the magnitudes of effects across different alteration scales, the parameter changes x were set to and log(scaling factori) for k and gNa.
These slopes were then used to score the biophysical changes j of ionic current variants to estimate their effect on the firing behaviour and the severity of their neurological impairments with:
| (2) |
Where scorej is the scoring for one alteration of a biophysical parameter. This resulted in the following equations for the biophysical scores of SCN8A variants:
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
Finally, these scores were summed to predict the severity of the respective variants:
| (9) |
Classification of phenotype severity
For the correlation analysis between electrophysiological score and severity of the disease in SCN8A variant carriers, we classified the individuals into four severity groups: (i) BFIE; (ii) intermediate epilepsy; (iii) DEE with mild or moderate ID; and (iv) DEE with severe ID. We assigned weights to the four grades of severity (BFIE = 1, intermediate epilepsy = 2, DEE with mild or moderate ID = 3 and DEE with severe ID = 4). In case the same variant was associated with different phenotypes, we calculated a weighted average score. As this analysis was performed for the GOF variants only, the generalized epilepsy, unclassifiable epilepsy and NDDwoE groups were not included.
Statistical analysis
Clinical data were analysed using Stata version 15.1 for Mac (StataCorp, College Station, Texas, USA). For categorical data, Fisher’s exact test was used, and for continuous data the Kruskal-Wallis test and Mann-Whitney test were used. Significance was evaluated using a two-tailed test of proportions and significance was reached if P < 0.05. The do-file used to perform the analyses is available upon request.
Electrophysiological data were analysed using Clampfit software of pClamp 10.6 (Axon Instruments), Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) or Igor Pro (Wavemetrics, Portland, OR, USA). Statistics were performed using one-way ANOVA with Dunnett’s post hoc test, ANOVA on ranks with Dunn’s post hoc test or Fisher’s exact test in GraphPad prism (GraphPad software, San Diego, CA, USA). For all statistical tests, significance is indicated in the figures (*P < 0.05, **P < 0.01, ***P < 0.001) or in the text.
Data availability
Data will be made available upon request.
Results
Phenotypes within the whole cohort
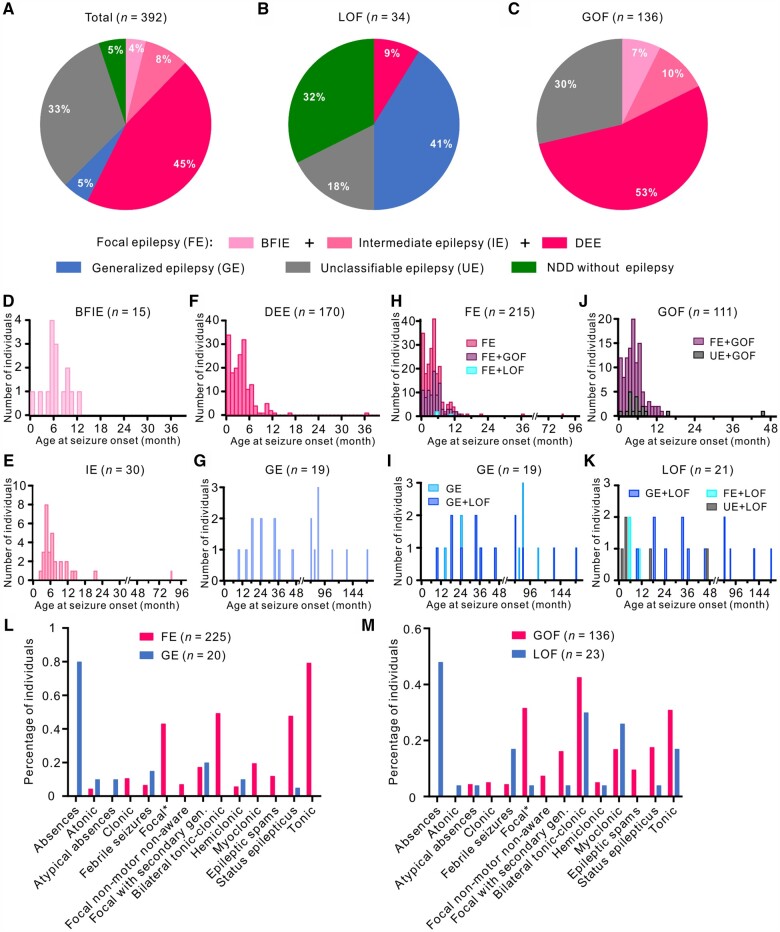
We assessed a cohort of 91 unpublished and 301 previously published affected individuals (392 affected individuals in total). One hundred and twenty-seven affected individuals had unclassifiable epilepsy due to a lack of information and have not been included in the analyses below. Age at follow-up/inclusion ranged from 2 months to 44 years (median 4.3 years). We differentiated the following phenotypes:
BFIE (n = 15): median age at seizure onset in this subgroup was 6 months (range: 2 weeks to 1 year). The most prevalent seizure types were focal (40%), focal to bilateral tonic-clonic (27%) and bilateral tonic-clonic without identifiable focal onset (27%). Cognition was normal in all affected individuals. Treatment response was known in 10 patients, all of whom were seizure free. Six patients were treated with SCBs, one with a non-SCB and three with a combination. Seven of 15 (47%) variants were inherited from an affected parent. Two variants were recurrent: p.(Glu1483Lys) and p.(Asn1877Ser).2,3
Intermediate epilepsy (n = 33): intermediate epilepsy was defined as a focal epilepsy fitting neither the BFIE nor the DEE categories (see Supplementary Fig. 2 for a flow chart and distinction criteria). Median age at seizure onset was 5 months (range: 2 months to 7 years). The most prevalent seizure types were focal (64%), tonic (33%) and bilateral tonic-clonic (55%). Cognition was normal in 33% of the patients, while mild ID was present in 52% and moderate ID in 15%. Additional features included speech delay (36%), behavioural disorders (attention-deficit hyperactivity disorder, autistic features, aggression; 19%) or ataxia (11%). Treatment response was known for 31 affected individuals. Fourteen of them were seizure free; eight individuals had been treated with an SCB, three with a non-SCB and three with a combination. Three variants were inherited from an affected parent, while four were unknown and the remainder were de novo. Two variants were recurrent: p.(Gly1475Arg) and p.(Asn1877Ser).5
DEE (n = 177): median age at seizure onset was 3 months (range: first day of life to 36 months). The most frequent seizure types were focal (70%), tonic (76%) and bilateral tonic-clonic (89%). Cognition ranged from moderate (22%) to severe (73%) ID and was unknown in the remainder (5%). Additional features included hypotonia in 50% and cortical vision impairment in 32%. Treatment response was known in 128 affected individuals. Twenty-six affected individuals (20%) were seizure free; 11 of them were treated with SCBs (42%), seven with non-SCBs (27%) and eight with a combination (31%). The vast majority of variants occurred de novo (90%), three were inherited from an affected parent, while inheritance was unknown in 11 individuals. Twenty-five variants were recurrent: p.(Arg850Gln/Gly) and p.(Arg1872Trp/Gln/Leu) were the most common10,14,27 (Supplementary Table 1).
Generalized epilepsy (n = 20): the median age at seizure onset was 42 months (range: 9 months to 14 years). The most prevalent seizure types were absence (80%), generalized tonic-clonic (20%) and febrile (15%). Cognition was normal in 20% of individuals and ranged from mild (30%) or moderate (35%) to severe ID (10%). Cognition was unknown in 5% of the patients. Additional features included ataxia (5%), behavioural disorders (autism, aggression, anxiety; 28%) and speech delay (19%). Treatment response was known in 16 affected individuals. Six were seizure free. One was treated with an SCB and five with a non-SCB. Ten variants occurred de novo, five were inherited from an affected parent, and five were of unknown inheritance. None of the variants were recurrent.
NDDwoE (n = 20): individuals with NDDwoE did not have epilepsy at time of inclusion (median age 9 years, range: 3–35 years). Cognition ranged from normal (10%) to mild (45%), moderate (20%) or severe ID (15%) and was unknown in 10%. Additional features included behavioural disorders (ASD, attention-deficit hyperactivity disorder; 43%), delayed speech (24%) and microcephaly (19%). Six variants occurred de novo, whereas nine were inherited from affected or mosaic parents and the remainder were unknown. Three variants were recurrent: p.(Gly384Arg), p.(Arg931Gln) and p.(Ala1622Asp).
Clinical details for all previously unpublished and published individuals (as data were available) are summarized in Supplementary Table 1.
Functional studies
We examined seven SCN8A variants functionally, chosen to represent the most important aspects of the clinical spectrum, particularly the newly identified phenotype with generalized epilepsy, for which functional studies have not been performed to date. Three variants [p.(Leu840Pro), p.(Phe846Ser) and p.(Asn1877Ser)] were seen in affected individuals with onset of seizures in the first year of life; the first two appeared in affected individuals with DEE who were resistant to all ASMs including high-dose SCBs,7 the third was identified in affected individuals with BFIE or intermediate epilepsy who were responsive to SCBs.2,5 Three variants [p.(Ile1654Asn), p.(Val1758Ala), p.(Thr1787Pro)] were seen in affected individuals with generalized epilepsy and late onset absence seizures in two cases preceded by febrile seizures.5,28 The last variant [p.(Asn374Lys)] was found in an affected individual with focal epilepsy and moderate ID with late onset at 7 years of age—an unusual phenotype for which the pathogenicity of the SCN8A variant had to be shown.5
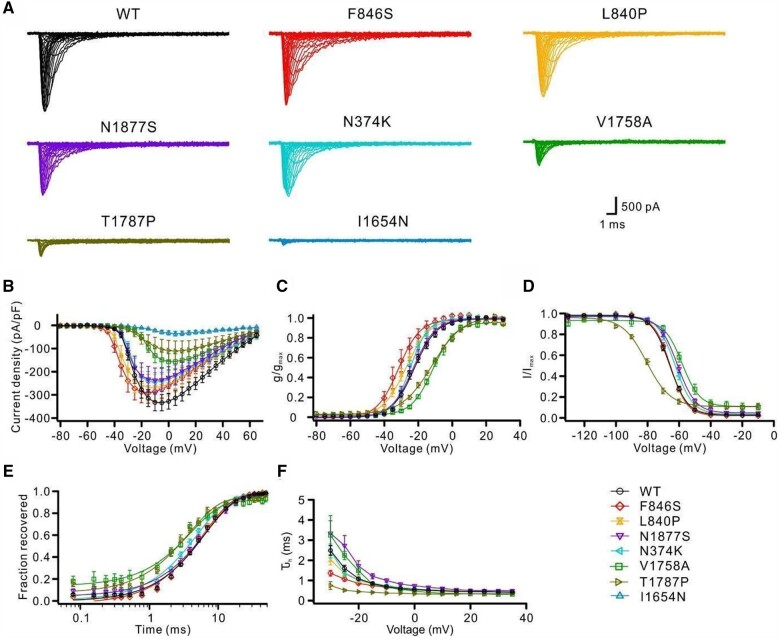
The biophysical consequences of these seven variants were first studied in ND7/23 cells. p.(Phe846Ser) and p.(Leu840Pro) variants induced hyperpolarizing shifts of the activation curves, indicating clear GOF effects (Fig. 1B and C and Supplementary Table 2). The p.(Asn1877Ser) variant caused a significant depolarizing shift of steady-state fast inactivation and slightly slowed the time course of fast inactivation (Fig. 1D and F and Supplementary Table 2). In contrast, p.(Ile1654Asn), p.(Val1758Ala) and p.(Thr1787Pro) variants dramatically reduced the peak current density. In particular, cells transfected with p.(Ile1654Asn) exhibited almost no Na+ current; hence no gating parameters were obtained for this variant (Fig. 2A and B and Supplementary Table 2). Furthermore, the p.(Val1758Ala) and p.(Thr1787Pro) variants shifted the activation curves toward more depolarized potentials, suggesting LOF effects, although the former variant impaired, while the latter enhanced, fast inactivation (Fig. 2B–D and F and Supplementary Table 2). Additionally, these two variants, as well as p.(Asn374Lys) to a minor extent, accelerated recovery from fast inactivation (Fig. 2E and Supplementary Table 2).
Figure 1.
Functional characterizations of SCN8A variants in the neuroblastoma cell line ND7/23. Wild-type or mutant Nav1.6 channels were transfected into ND7/23 cells and recorded in the presence of tetrodotoxin to block endogenous Na+ channels. (A) Representative Na+ current traces for transfected Nav1.6 wild-type (WT, black) or mutant channels (colour code in the bottom right). (B) Peak Na+ currents normalized by cell capacitance were plotted versus voltage. Both the p.(Phe846Ser) and p.(Leu840Pro) variants caused a hyperpolarizing shift of the current-voltage relationship, whereas the p.(Val1758Ala) and p.(Thr1787Pro) variants caused a depolarizing shift compared with the wild-type channels. The p.(Ile1654Asn), p.(Val1758Ala) and p.(Thr1787Pro) variants significantly decreased the current density in comparison with the wild-type. Wild-type: n = 30; mutants: n = 14–19. (C) Voltage-dependent steady-state activation curves. Lines represent Boltzmann functions fit to the data-points. (D) Voltage-dependent steady-state inactivation curves. Lines represent Boltzmann functions fit to the data-points. (E) Time course of recovery from fast inactivation at −100 mV. The p.(Val1758Ala), p.(Thr1787Pro) and p.(Asn374Lys) variants accelerated the recovery from fast inactivation compared with the wild-type. (F) Voltage-dependence of the major time constant of fast inactivation τh. Shown are means ± SEM (B–F). Numbers of recorded cells and statistical analysis for all experiments are provided in Supplementary Table 2.
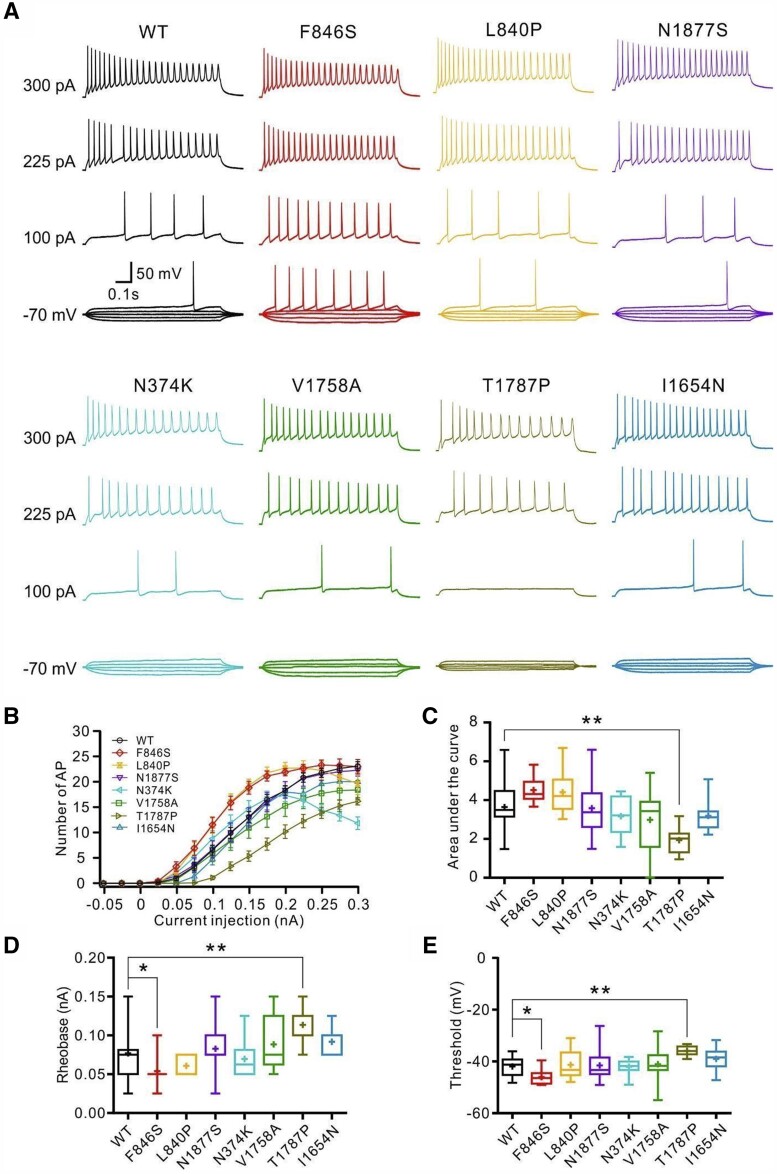
Figure 2.
Effects of SCN8A variants in primary cultured hippocampal mouse neurons in the absence of tetrodotoxin. Neurons were transfected with wild-type or mutant Nav1.6 channels and recorded in the absence of tetrodotoxin. (A) Representative voltage traces of evoked action potentials (APs) in the absence of tetrodotoxin from neurons transfected with wild-type (WT, black) or mutant neurons (colour code indicated in B). (B) Numbers of evoked action potentials plotted versus injected current in the absence of tetrodotoxin. Shown are means ± SEM. (C) Area under the curve for the input-output relationships. The p.(Thr1787Pro) variant shows a significantly decreased area under the curve compared with the wild-type channels. Rheobase (D) and threshold (E) of action potentials were decreased for neurons transfected with the p.(Phe846Ser) variant but increased for neurons transfected with the p.(Thr1787Pro) variant compared with the wild-type channels. Box-and-whisker plots (C–E) show means (plus sign), the 25th, 50th and 75th percentiles, minima and maxima; *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA with Dunnett’s post hoc test or ANOVA on ranks with Dunn’s post hoc test were performed. Numbers of recorded cells and statistical analysis are provided in Supplementary Table 3.
Next, intrinsic and firing properties were examined in transfected cultured hippocampal mouse neurons in the absence or presence of tetrodotoxin. In the absence of tetrodotoxin, action potential firing was jointly determined by endogenous and transfected Na+ channels. Only the p.(Thr1787Pro) variant significantly decreased the action potential firing rate compared to neurons transfected with wild-type channels, as revealed by the area under the curve of input-output relationships (Fig. 2B and C and Supplementary Table 3). The p.(Phe846Ser) variant decreased, whereas the p.(Thr1787Pro) variant increased both the rheobase and action potential threshold compared with the wild-type (Fig. 2D and E and Supplementary Table 3), indicating the former enhanced, whereas the latter impaired, neuronal excitability.
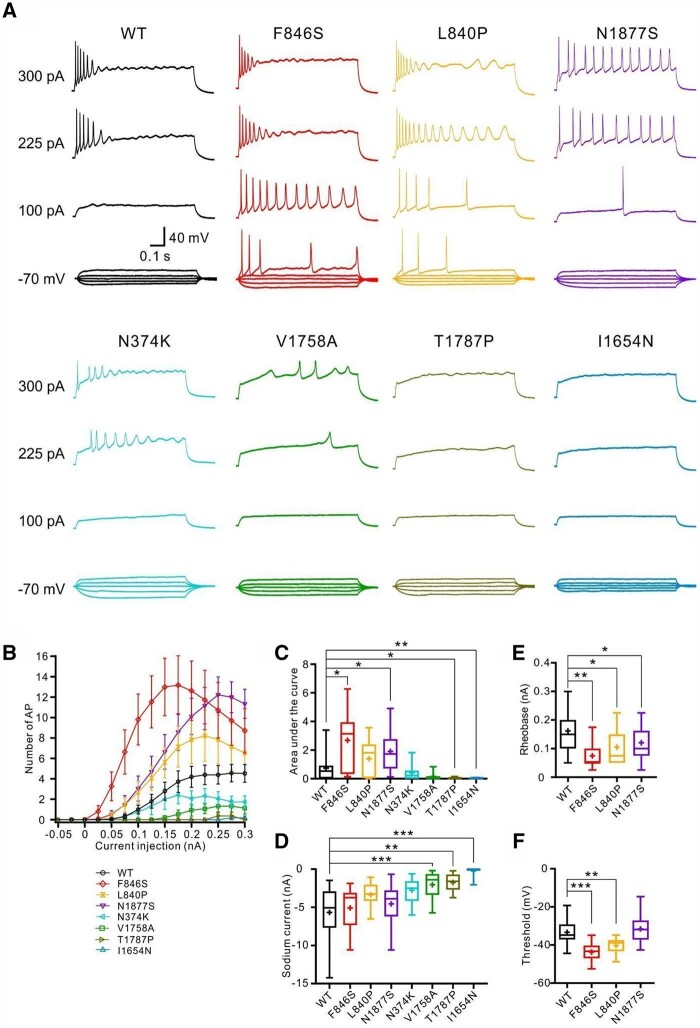
In the presence of tetrodotoxin, action potential firing was dependent on the tetrodotoxin-insensitive transfected Na+ channels. Under these conditions, p.(Phe846Ser) and p.(Asn1877Ser) variants significantly increased action potential firing, whereas p.(Thr1787Pro) and p.(Ile1654Asn) decreased action potential firing (Fig. 3B and C and Supplementary Table 4). Additionally, the p.(Phe846Ser), p.(Leu840Pro) and p.(Asn1877Ser) variants decreased the rheobase and the former two variants further reduced the threshold for action potential firing, indicating increased neuronal excitability. In contrast, neurons transfected with p.(Val1758Ala), p.(Thr1787Pro) or p.(Ile1654Asn) variants exhibited significantly decreased peak Na+ currents, resulting in very few action potentials firing in the presence of tetrodotoxin; hence, no parameters of single action potentials were obtained. Neurons expressing p.(Asn374Lys) showed comparable Na+ peak currents and action potential firing to wild-type channels (Fig. 3 and Supplementary Table 4).
Figure 3.
Neuronal properties carried only by transfected wild-type or mutant Nav1.6 channels. Hippocampal neurons transfected with wild-type or mutant Nav1.6 channels were recorded in the presence of tetrodotoxin to block endogenous Na+ channels. (A) Representative voltage traces of evoked action potentials (APs) from neurons transfected with wild-type (black) or mutant channels (colour code in the bottom left). (B) Numbers of evoked action potentials plotted versus injected current in the presence of tetrodotoxin. Shown are means ± SEM. (C) Area under the curve for the input-output relationships. (D) Peak Na+ current amplitudes of neurons transfected with wild-type or mutant Nav1.6 channels in the presence of tetrodotoxin. Rheobase (E) and threshold (F) were significantly decreased in neurons transfected with p.(Phe846Ser) or p.(Leu840Pro) variants compared with the wild-type channels. The p.(Asn1877Ser) variant also significantly decreased the rheobase. The rheobase or threshold could not be obtained in neurons transfected with p.(Asn374Lys), p.(Val1758Ala), p.(Thr1787Pro) and p.(Ile 1654Asn) mutant channels due to very few evoked action potentials. Box-and-whisker plots (C–F) show means (plus symbol), the 25th, 50th and 75th percentiles, minima and maxima; *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA with Dunnett’s post hoc test or ANOVA on ranks with Dunn’s post hoc test were performed. The numbers of recorded cells and statistical analysis are provided in Supplementary Table 4.
In summary, p.(Phe846Ser), p.(Leu840Pro) and p.(Asn1877Ser) showed a GOF effect or increased neuronal firing; p.(Asn374Lys) showed mild GOF effects and did not alter neuronal firing; and p.(Val1758Ala), p.(Thr1787Pro) and p.(Ile1654Asn) showed LOF effects or decreased neuronal firing.
Genotype-phenotype correlations in gain- or loss-of-function variant carriers and extrapolation to the cohort
Clinical data combined with functional results of this and earlier studies enabled us to explore genotype-phenotype correlations in 170 affected individuals carrying a known LOF (n = 34) or GOF (n = 136) variant. Detailed phenotypic analysis in those affected individuals and a comparison with the whole cohort may allow the extrapolation of genotype-phenotype correlations that could be valid for all individuals in which causative SCN8A variants have been detected. These phenotypic comparisons are provided below (Fig. 4).
Figure 4.
Distribution of affected individuals carrying SCN8A variants according to phenotype and age of seizure onset. Phenotypic subgroups of the total cohort (A) and individuals carrying LOF (B) or GOF (C) variants. (A) In the total cohort, 225 affected individuals (57.4%) had BFIE, intermediate epilepsy or DEE; 20 individuals (5.1%) had generalized epilepsy; and 20 individuals (5.1%) had a NDDwoE. (B) Twenty-five affected individuals had generalized epilepsy or an NDDwoE, accounting for 73.5% of LOF variant carriers, whereas 71.6% of GOF variant carriers had BFIE, intermediate epilepsy or DEE (C). Histogram of the age at seizure onset in affected individuals with BFIE (D), intermediate epilepsy (E), DEE (F) and generalized epilepsy (G). (H) Stacked histogram of age at seizure onset in affected individuals with focal epilepsy (FE) carrying GOF (FE+GOF), LOF (FE+LOF) or SCN8A variants which are not functionally characterized. (I) Stacked histogram of age at seizure onset in affected individuals with generalized epilepsy (GE) carrying LOF (GE+LOF) or SCN8A variants which are not functionally characterized. (J) Stacked histogram of age at seizure onset in affected individuals carrying GOF variants with FE (FE+GOF) or unclassifiable epilepsy (UE+GOF). (K) Stacked histogram of age at seizure onset in affected individuals carrying LOF variants with generalized epilepsy (GE+LOF), FE (FE+LOF) or unclassifiable epilepsy (UE+GOF). Affected individuals with BNIE, intermediate epilepsy or DEE exhibited a significant earlier age at seizure onset than individuals with generalized epilepsy (Kruskal-Wallis test, P < 0.001), which is also observed for GOF versus LOF variant carriers (Mann-Whitney test, P < 0.001). Histogram bin size = 1 month. Seizure type distributions of affected individuals with FE versus generalized epilepsy (L) and GOF versus LOF variant carriers (M).
Phenotypes of affected individuals with loss-of-function variants
Thirty-four affected individuals (12 previously unpublished) carried clear LOF variants that were deletions, splice-site, frameshift or stop-variants, or missense variants, with an LOF effect confirmed either in this or previous studies.17–19,21 Twenty-three individuals (68%) had epilepsy. The median age at seizure onset was 24 months (range: 1 month to 14 years). Seizure types included absence (11/23), bilateral tonic-clonic (7/23) and myoclonic (5/23). Additionally, 4/23 had febrile seizures within the first year of life, all with later onset (>2 years of age) of afebrile seizure types. A phenotype of generalized epilepsy was found in 14/34 individuals, 11/34 had a NDDwoE, 3/34 had DEE and the remaining 6/34 had an unclassifiable epilepsy. EEGs showed background slowing, generalized spike-wave activity, slow rhythmic activity or remained normal.
Of 23 affected individuals with epilepsy, nine were seizure-free: one with a SCB (lamotrigine), seven with non-SCBs (valproate monotherapy in four affected individuals) and one on a combination (lamotrigine and topiramate). Out of four additional individuals who tried SCBs, one experienced a worsening of seizures with zonisamide, two showed no effect and one exhibited a reduction of seizures with lamotrigine.
None of the individuals with LOF variants died prematurely.
When we compared this presentation of clear LOF variant carriers with the phenotypes of the whole cohort of affected individuals, there was a clear correlation to two groups: one with generalized epilepsy and one with NDDwoE. The only exceptions were three affected individuals with DEE in the LOF group. The main phenotypic features of these groups, including analyses of the age at seizure onset (except febrile seizures) and seizure types, are summarized and contrasted to the groups with GOF variants in Fig. 4 and Table 1.
Table 1.
Clinical characteristics of LOF and GOF variants and phenotypic subgroups
| BFIE | IE | DEE | GE | NDDwoE | LOF | GOF | |
|---|---|---|---|---|---|---|---|
| n | 15 | 33 | 177 | 20 | 20 | 34 | 136 |
| Percentage with epilepsy | 100% | 100% | 100% | 100% | 0% | 68% | 100% |
| Median age at seizure onset | 6 months | 5 months | 3 months | 42 months | – | 24 months | 4 months |
| Most common seizure types | Focal, focal to bilateral TC and bilateral TC | Focal, bilateral TC and tonic | Bilateral TC, focal and tonic | Absences, bilateral TC and febrile seizures | – | Absence seizures, bilateral TC and myoclonic seizures | Bilateral TC, tonic and focal seizures |
| Phenotype subgroups | – | – | – | – | – |
|
|
| Cognition | Normal 100% |
|
|
|
|
|
|
| Comorbidities | Paroxysmal kinesigenic dyskinesia | Language delay, behavioural issues | Hypotonia, CVI, ataxia | Language delay, behavioural issues | Behavioural disorders (ASD, ADHD), delayed speech, microcephaly | Language delay, ASD, behavioural issues, ataxia | Hypotonia, CVI, dyskinesia, ataxia |
| Mortality | 0% | 0% | 10.2% | 0% | 0% | 0% | 9.0% |
| Precision medicine | SCBs | SCBs | SCBs | SCBs |
ADHD = attention-deficit hyperactivity disorder; bilateral TCs = bilateral tonic-clonic seizures; CVI = cortical vision impairment; GE = generalized epilepsy; IE = intermediate epilepsy; UE = unclassifiable epilepsy.
Phenotypes of affected individuals with gain-of-function variants
One hundred and thirty-six affected individuals from the total cohort (29 unpublished) carried missense variants that were shown to cause GOF.1,6,17,19,29,30 All 136 patients suffered from epilepsy. The most prevalent seizure types were focal (32%), focal tonic (31%) or bilateral tonic-clonic seizures (43%). Febrile seizures were seen in 4% of the individuals. The median age at seizure onset was 4 months (range: first day of life to 45 months). Half of the affected individuals (54%) were diagnosed with DEE, followed by unclassifiable epilepsy in 29%, intermediate epilepsy in 10% and BFIE in 7%. Individuals with BFIE had either no interictal epileptiform abnormalities or rare diffuse spike and wave complexes. Individuals with intermediate epilepsy had heterogeneous EEG features with trains of beta and delta activity and focal spike and slow waves, bilaterally in the parieto‐occipital regions, with or without diffuse spreading. Most individuals with severe DEE had background slowing, polymorphic delta and beta activity and multifocal spike and slow waves, predominant in the posterior quadrants.
Treatment data were available for 84 affected individuals. Twenty-six of the 84 affected individuals were seizure-free; 16 with SCBs. Seizure reduction was seen in 47 affected individuals, 18 with SCBs. Two affected individuals showed worsening of seizures with SCBs; both affected individuals were resistant to several ASMs and had increased seizure frequency with oxcarbazepine and levetiracetam.31 Eight affected individuals did not try SCB treatment: two had a mild phenotype and were seizure free with valproate or valproate + levetiracetam,5,32 and one affected individual also had a mild phenotype and seizure reduction with valproate.33 The remaining five affected individuals all had pharmacoresistant seizures; it is unknown why SCBs were not tried.
Twelve affected individuals were prematurely deceased (9%), all with a DEE phenotype; 10 deaths were due to a general worsening in the patients’ overall and neurological condition followed by organ failure, and two were due to definite or probable sudden unexpected death in epilepsy (SUDEP) (1.5%).
As in the LOF group, there was a clear correlation in the GOF group to specific phenotypes in the whole cohort, namely BFIE, intermediate epilepsy and DEE. None of the GOF variant carriers had generalized epilepsy or NDDwoE.
A summary of these data is provided in Fig. 4, Table 1 and Supplementary Table 5. The obvious difference in the age of onset was statistically significant between the GOF and LOF variant carriers (Mann-Whitney test, P < 0.001) and between BFIE/intermediate epilepsy/DEE and generalized epilepsy (Kruskal-Wallis test, P < 0.001). Statistical analysis also confirmed that affected individuals with LOF variants mostly had a generalized epilepsy, whereas affected individuals with GOF had one of the focal epilepsy phenotypes (BFIE, intermediate epilepsy or DEE; Fisher’s exact test, P < 0.001).
Correlation of clinical and electrophysiological phenotypes in the gain-of-function group
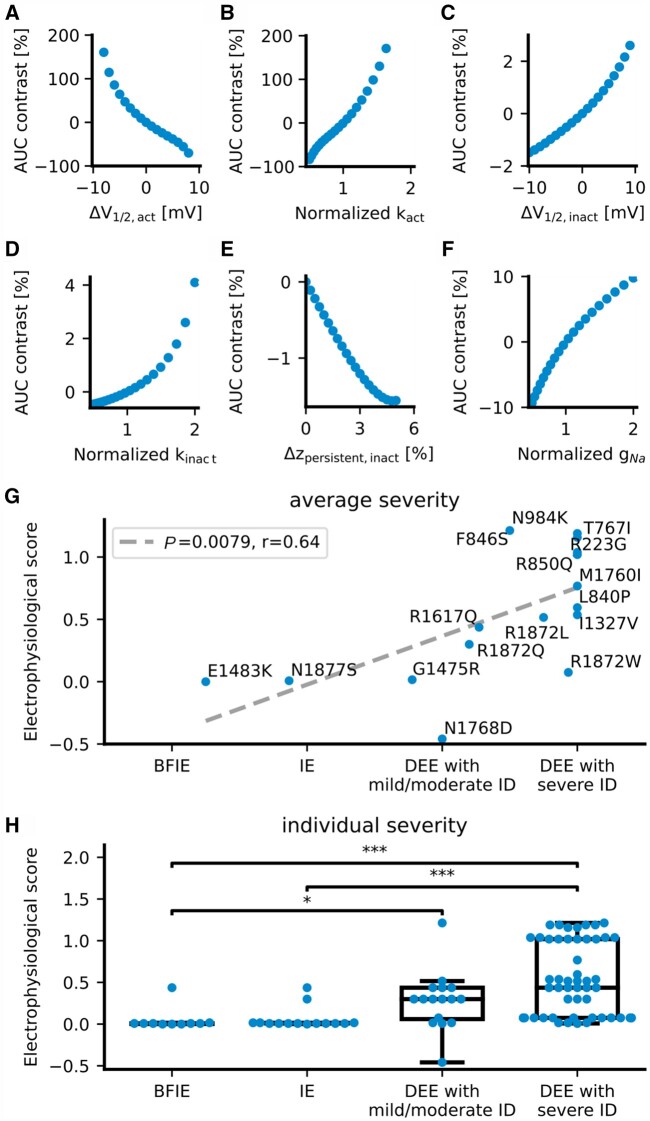
We scored all GOF variants according to their functional characterizations from this and previous studies, including a total of 16 variants. The LOF variants were not included because of a large proportion of deleterious variants, which lead to non-functional proteins that cannot be well differentiated. The electrophysiological score was determined by the effect of biophysical changes of variants on neuronal firing simulated by a single-compartment conductance-based model (see the ‘Materials and methods’ section and Supplementary material). This analysis revealed that (i) alterations of activation properties severely affect neuronal firing; (ii) changes in the sodium conductivity have a moderate effect; and (iii) changes of inactivation properties and the persistent current affect neuronal firing mildly (Fig. 5A–F and Supplementary Table 5).
Figure 5.
Correlation of a computed electrophysiological score with the clinical severity of SCN8A GOF variants. Electrophysiological scores of SCN8A GOF variants were obtained according to the effect of variants on action potential firing simulated by a single-compartment conductance-based model. (A–F) The contrast of the simulated area under the input-output curve (AUC, Equation 1) as a function of the changes in single Na+ current gating parameters, such as: (A) the V1/2 of the activation curve; (B) the slope factor k of the activation curve; (C) the V1/2 of the fast inactivation curve; (D) the slope factor k of the fast inactivation curve; (E) the persistent Na+ current; and (F) the Na+ conductivity (or current density). Changes in the V1/2 and the slope of the activation curve had a much stronger effect on the AUC than other parameters. (G) Correlation (dashed grey line) of the simulation-based score (Equation 9) with the severity of each SCN8A GOF variant averaged over affected individuals (blue dots with the respective one-amino acid code). (H) Distributions of simulation-based scores of each patient (blue dots) for each of the four categories of clinical severities. *P < 0.05; ***P < 0.001 (ANOVA on ranks with Dunn’s post hoc test).
The electrophysiological scores of GOF variants significantly correlated with the clinical severity of affected individuals carrying these variants (Fig. 5G; P = 0.0079, r = 0.64), which were previously classified into BFIE, intermediate epilepsy, DEE with mild/moderate ID and DEE with severe ID (scored as 1, 2, 3 and 4, respectively). Variants causing different phenotypes were averaged over this score (see the ‘Materials and methods’ section). In a separate analysis, we also compared the individual severity of each affected individual with the electrophysiological score of the variants. The scores in the groups with DEE are significantly higher than the ones in BFIE and intermediate epilepsy, but there is no significant difference between BFIE and intermediate epilepsy or DEE with mild/moderate ID and DEE with severe ID (Fig. 5H).
Genetic landscape of SCN8A variants
We analysed 256 different variants in 392 individuals. Missense variants (n = 233) accounted for the majority of disease-causing variants (91%). Nineteen variants were deleterious frameshift, stop-gain or canonical splice-site variants. Seven were inframe indels or deletions. Three patients had biallelic missense variants.34 Sixty-one recurring missense variants were found in 244 affected individuals. Two hundred and ninety-seven variants occurred de novo, 31 were inherited from an affected or unaffected mosaic parent, and for 64 segregation was unknown.
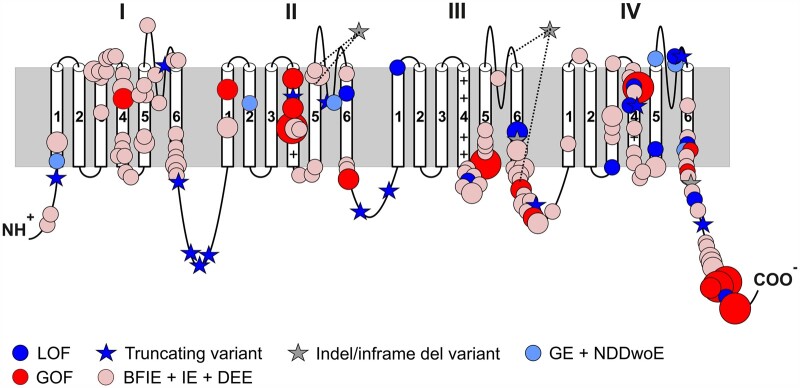
The distribution of variants across the Nav1.6 channel protein is shown in Fig. 6. Most GOF variants and also variants causing focal epilepsy (BFIE/intermediate epilepsy/DEE) without functional data are concentrated at the cytoplasmic side and in the voltage sensors (protein regions given as codon positions: 212–263, 398–418, 838–883, 966–986, 1283–1336, 1450–1523, 1602–1660 and 1753–1775), whereas most LOF variants were found in the pore region (protein regions: 274–408, 893–976, 1347–1460 and 1669–1765). The region-specific distribution of GOF and BFIE/intermediate epilepsy/DEE variants in comparison to the distribution of LOF and generalized epilepsy/NDDwoE variants is highly significant (Fisher’s exact P-value < 0.00001). This is also illustrated in a 3D structural model which additionally reveals that population variants taken from the gnomAD database are located in different regions. Most disease-causing variants were located in functionally important and evolutionary conserved regions of the channel, whereas variants from the gnomAD database are in less conserved regions [Supplementary Fig. 1 and Supplementary Video 1 (the video is available to view at Figshare: https://doi.org/10.6084/m9.figshare.15141018)].
Figure 6.
Location of SCN8A variants associated with neurodevelopmental disorders. Schematic 2D representation of the Nav1.6 channel displaying the location of pathogenic variants. A comparison of the location of missense variants with proven GOF or a BFIE/intermediate epilepsy/DEE phenotype (without functional analysis) on one hand, and variants with proven LOF or generalized epilepsy/NDDwoE phenotypes (without functional analysis) on the other, revealed a significant difference in the distribution of variants (see main text and Supplementary material). Recurring variants are indicated with larger symbol size relative to the number of patients and in frame indels or deletions are indicated showing the whole affected regions [p.(Ile888_Val892delinsMet), p.(Glu1774_Ala1777del) and p.(Pro1428_Lys1473del)].
Frequency of SCN8A-related disorders in the Danish population
From the year 2006 to 2017, an average of 60 934 children were born per year. During the same period, 13 affected individuals were diagnosed with an SCN8A-related disorder in Denmark, yielding an estimated frequency of 1/56 247.
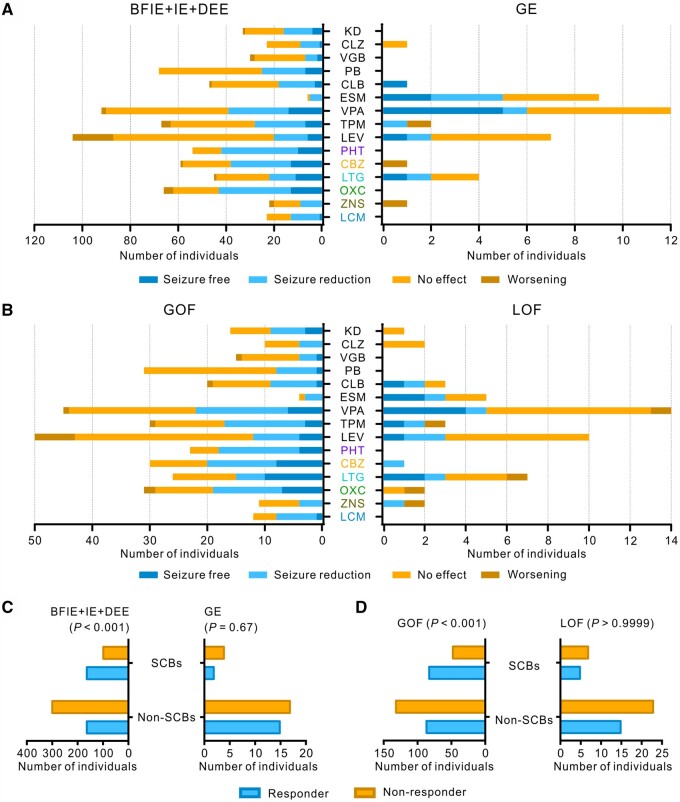
Evaluation of treatment effects
Treatment responses to specific ASMs were grouped into four categories—seizure-free, seizure reduction, no change and worsening—as described in the methods section. We differentiated between SCBs and non-SCBs, in LOF and GOF variant carriers, generalized epilepsy and focal epilepsies (BFIE/intermediate epilepsy/DEE). The results are shown in Fig. 7. GOF variant carriers responded better to SCBs than to non-SCBs (Fisher’s exact test, P < 0.001), whereas there was no difference between treatment with SCBs and non-SCBs for LOF variant carriers (numbers very small, Fisher’s exact test, P = 0.48). Similar results were obtained for the treatment with SCBs versus non-SCBs in affected individuals with focal epilepsy versus those with generalized epilepsy.
Figure 7.
Treatment responses to anti-seizure medications in affected individuals carrying SCN8A variants. Treatment effects of ASMs on seizures in affected individuals with BFIE, intermediate epilepsy or DEE versus those with generalized epilepsy (A) and those carrying SCN8A GOF versus LOF variants (B). Phenytoin, carbamazepine, lamotrigine, oxcarbazepine, zonisamide and lacosamide are SCBs. (C and D) Individuals with BFIE, intermediate epilepsy or DEE and those carrying SCN8A GOF variants responded significantly better to SCBs than non-SCBs, whereas treatment with SCBs or non-SCBs did not cause a different effect in individuals with generalized epilepsy and those carrying SCN8A LOF variants (please consider the very small numbers in these latter categories). P values derived from Fisher’s exact test are provided in the figure. Responders were defined as those who became seizure-free or experienced a seizure reduction while staying on the drug; non-responders were defined as those experiencing no effect or seizure worsening. CBZ = carbamazepine; CLB = clobazam; CLZ = clonazepam; ESM = ethosuximide; KD = ketogenic diet; LCM = lacosamide; LEV = levetiracetam; LTG = lamotrigine; OXC= oxcarbazepine; PB = phenobarbital; PHT = phenytoin; TPM = topiramate; VGB = vigabatrin; VPA = valproate; ZNS = zonisamide.
Discussion
Our study provides a detailed analysis of the correlation between clinical phenotypes, genotypes and electrophysiological characterizations, suggesting the following four main conclusions:
There are five main phenotypes in SCN8A-related disorders in this largest series of 392 individuals collected to date: BFIE, intermediate epilepsy, DEE, generalized epilepsy and NDDwoE.
There is a significant correlation between GOF variants causing focal epilepsy of different/increasing severity (BFIE, intermediate epilepsy, DEE) and LOF variants causing generalized epilepsy or NDDwoE, allowing an extrapolation from the clinical phenotypes of clear GOF and LOF variant carriers to the whole cohort. There was only one exception from this rule: three DEE affected individuals with onset between 5 and 10 months carrying stop-gain or canonical splice-site (hence, predicted LOF) variants (see below for exact phenotypes).
The clinical severity of epilepsies associated with GOF variants is at least partially determined by the degree of the electrophysiological dysfunction, as there was a significant correlation between the clinical and electrophysiological phenotypes using a new system to estimate the influence of different gating parameters on neuronal firing.
As previously suggested in smaller studies, seizures in individuals with disease-causing GOF variants respond better to SCBs than to other ASMs.
Functional studies
Altogether, we examined 14 variants (this study and previously19) covering the whole clinical spectrum of SCN8A-related phenotypes. Using both neuroblastoma cells to study the effects of the mutant channels on gating properties and murine primary neuronal cultures to study the effects on intrinsic neuronal properties and firing, we found mild GOF effects or an increase in neuronal firing to be associated with BFIE or intermediate epilepsy [p.(Gly1475Arg), p.(Glu1483Lys) and p.(Asn1877Ser)], whereas more severe gating defects, particularly strong hyperpolarizing shifts of steady-state activation that also on average led to a stronger increase in neuronal firing, were associated with DEE [p.(Arg223Gly), p.(Leu840Pro), p.(Phe846Ser), p.(Met1760Ile) and p.(Arg1872Trp)]. One previously published variant, p.(Arg223Gly), was initially suggested to cause a predominant LOF11 and had a phenotype of spasms with onset at 6 months, severe ID and an epileptic encephalopathy. However, there were also GOF features identified, and when we retested this variant in our system, GOF effects predominated and it increased neuronal firing (Liu et al., unpublished results). When we used a one-compartment neuronal model system, we were able to weight the gating defects of all GOF variants that were functionally investigated to date according to their effect on neuronal firing, revealing a significant correlation between the severity of the clinical and electrophysiological phenotypes. Such a correlation was not observed when we used a different scoring system (data not shown) based only on the degree of the electrophysiological dysfunction (i.e. without weighting the effects on neuronal firing), which, however, worked well previously to correlate clinical and electrophysiological phenotypic severity in SCN2A-related epilepsy.35
In contrast, the p.(Ile1654Asn), p.(Thr1787Pro) and p.(Val1758Ala) variants, all causing generalized epilepsy with absence seizures as a new recurring phenotype, showed an LOF in neuroblastoma cells or decreased neuronal firing. Also, variants not causing epilepsy either caused a clear LOF and decreased neuronal firing [p.(Gly964Arg) and p.(Arg1620Leu)] or a strong GOF on channel gating leading to a depolarization block, i.e. LOF on a neuronal level [p.(Ala1622Asp)].19 Additional variants characterized by other groups confirm these results. Many other variants causing GOF effects mainly on channel gating (only few studies with limited results on neuronal firing) were clearly associated with focal epilepsies (Supplementary Table 1).
The p.(Ala1319Thr) variant caused a depolarizing shift of the activation curve of Na+ current in X. laevis oocytes36,37 and decreased firing of cerebellar Purkinje cells,38,39 indicating an LOF. The Scn8a Medjo mouse carrying the corresponding variant in the mouse channel exhibited absence epilepsy revealed by slow-wave discharges in EEG recordings associated with absence-like seizures,40 reproducing the clinical phenotype found in the affected individual carrying the p.(Ala1319Thr) variant. Other epilepsy-related LOF variants were also associated with absence epilepsy, such as p.(Asn544fs*39) and p.(Glu587*).5,31 In another conditional mouse model, a complete Scn8a LOF restricted to inhibitory neurons of the thalamic reticular nucleus also caused absence-like epilepsy with generalized slow-wave discharges.40,41 It is therefore intriguing to speculate that SCN8A variants cause absences and other generalized seizures due to an LOF in inhibitory neurons, whereas the GOF effects are more important in excitatory neurons, as discussed in more detail below.
The p.(Asn374Lys) variant showed only very mild GOF gating defects but did not change parameters in neurons. The phenotype with late onset focal epilepsy at 7 years without other neuropsychiatric symptoms also does not fit the five main categories. Thus, while we cannot exclude a mild contribution of this variant to seizures in this affected individual, we consider it to be a variant of unknown significance.
Phenotype correlations in gain- versus loss-of-function variant carriers
When we compared the clinical phenotypes of the 136 confirmed GOF and 34 confirmed LOF variant carriers with the whole group of 392 individuals, there were strikingly congruent phenotypes, which strongly suggested that BFIE, intermediate epilepsy and DEE are generally caused by GOF, while generalized epilepsy and NDDwoE are caused by LOF variants. Not only did the seizure types resemble each other in those groups, but also an analysis of the age of onset of seizures strongly confirmed this hypothesis (Table 1 and Fig. 4). Only three cases with presumed DEE and an LOF variant did not fit into this pattern. One patient carried a p.(Met1481Ilefs*12) variant with onset of pharmaco-resistant tonic and bilateral tonic-clonic seizures and episodes of status epilepticus at 5 months of age, severe ID, no speech and poor eye contact. Two patients with seizure onset at 5 or 10 months carried a splice-site variant predicting exon skipping to cause a large in-frame deletion cutting out the whole pore region of domain III, p.(Pro1428_Lys1473del; Fig. 6). They had severe ID, no speech, poor eye contact and hypotonia. EEG showed multifocal epileptiform discharges. Thus, these single cases could not be differentiated from the DEE cohort caused by GOF variants. The epilepsy phenotype of two further patients with similar splice-site variants was unclassifiable. Alternative consequences on the Nav1.6 protein other than those predicted here [p.(Pro1428_Lys1473del) and p.(Met148Ilefs*12)], e.g. resulting from activation of cryptic splice sites, cannot be ruled out completely.
In a previous study on SCN2A,42 we demonstrated that different neonatal and infantile epilepsy syndromes were caused by GOF variants, whereas later onset seizures were due to an LOF of Nav1.2. In SCN2A, the boundary for the onset of seizures between GOF and LOF was very early—approximately 3 months of age.42 In the present study, the onset in both groups was later (median age at onset of 4 months in the GOF subgroup and 24 months in the LOF subgroup), and the ranges were more overlapping. This difference may reflect the differential developmental expression of SCN2A versus SCN8A, as SCN2A is expressed neonatally and earlier than SCN8A.43,44 An additional difference is that SCN2A variant carriers have better defined electroclinical epilepsy syndromes. Both groups share a phenotype of BFNIE/BFIE, with a later onset in SCN8A-related epilepsy. Some individuals carrying SCN8A variants develop paroxysmal kinesigenic dyskinesia,3 whereas SCN2A variant carriers may develop a form of later-onset episodic ataxia.45 There were also differences regarding the syndromes of LOF variant carriers in both genes. Generalized epilepsy was not generally observed in SCN2A, although absence epilepsy has been reported in mice with complete LOF of Scn2a when it has been knocked out in excitatory neurons.46 Instead, West syndrome was a common phenotype in human SCN2A, but phenotypes with generalized seizures and EEG abnormalities were also found in the LOF group (epilepsy with myoclonic-atonic seizures or Lennox-Gastaut syndrome). For SCN8A, the phenotypes were rather unspecific generalized epilepsy, mainly with absence seizures. However, we found that an LOF subgroup had a phenotype resembling SCN1A LOF, with febrile seizures preceding additional seizure types, resembling a generalized/genetic epilepsy with febrile seizure plus (GEFS+) phenotype. SCN8A is expressed in both excitatory and inhibitory neurons,47 which may well explain the phenotypic similarity between some of the SCN1A- and SCN8A-related phenotypes, since SCN1A is considered to be the main sodium channel in inhibitory GABAergic neurons.48,49 Additionally, it has been found that knockout of Scn8a in mice decreases cortical excitability, resulting in convulsive seizure protection.40 Furthermore, mice with global reduction of Scn8a have fewer seizures compared with mice that exhibit Scn8a deletion only in inhibitory neurons.41 In contrast, mice carrying a GOF Scn8a variant [p.(Arg1872Trp)] exhibited convulsive seizures and premature death; however, activation of this variant only in inhibitory neurons did not induce seizures or overt neurological dysfunctions.20 Altogether, SCN8A GOF variants may mainly cause hyperexcitation in excitatory neurons leading to tonic-clonic and focal seizures, whereas SCN8A LOF variants mainly induce hypoexcitation in inhibitory neurons, causing generalized and, particularly, absence seizures. Further studies may elucidate the distinct roles of SCN8A and the other sodium channel genes in different neuronal circuits.
Genetic landscape
The vast majority of the detected variants were missense, and we found several recurrent variants. Those detected in more than 10 affected individuals included: p.(Arg850Gln/Gly), seen in 12 affected individuals; p.(Gly1475Arg), seen in 19 affected individuals; p.(Arg1617Trp/Gln/Leu), seen in 18 affected individuals; p.(Arg1872Trp/Gln/Leu), seen in 41 affected individuals; and p.(Asn1877Ser), seen in 23 affected individuals. All of these variants cause a clear GOF. Why some of these variants, especially p.(Gly1475Arg) and p.(Arg1617Trp/Gln/Leu), have a high phenotypic variability and others not, remains to be elucidated. Other genetic or environmental factors might play an important role. Additionally, we found families with intra-familial variability, such as the family of Proband 377, whose mother and two older siblings had BFIE and were seizure-free with normal intellect, whereas the proband suffered from pharmacoresistant DEE.
Estimated frequency
We found the frequency of SCN8A-related disorders in the Danish population to be 1/56 247, which is higher than for SCN2A (1/78 608),42 but lower than for SCN1A (1/22 000).50 Since these three genes are of similar size and homology, we would assume a similar variant frequency. Large-scale genetic testing in isolated ASD cohorts have failed to detect large numbers of SCN8A-affected individuals,51 suggesting that a large number of undiagnosed SCN8A-ASD affected individuals does not exist and that this is not the cause behind the discrepancy in numbers. It could be that some individuals with truncating/LOF variants are too mildly affected (learning disabilities, mild ID etc.) to be candidates for genetic testing but are also not healthy enough to be part of control populations (such as blood donors), as reflected in the almost complete absence of truncating variants in gnomAD (pLI score: 1). Intrauterine death in SCN8A-related disease could be a third cause, which has not been investigated to date.
Treatment implications
In general, and as expected, affected individuals with GOF effects showed an overall positive response to treatment with SCBs, as has been hypothesized previously.22 However, many of the affected individuals with DEE still have pharmacoresistant epilepsy (81%), underlining the severity of SCN8A-related epilepsy. A recent study evaluated the effect of a novel SCB: GS967, which primarily targets the elevated persistent currents without affecting peak currents, as well as having increased potency compared with phenytoin.52 Long-term treatment with GS967 was shown to protect Scn8aN1768/+ mice against premature death and also alleviated seizure burden.52 GS967/PRAX-330 (https://adisinsight.springer.com/drugs/800050600) and another new drug—XEN901 (https://clinicaltrials.gov/ct2/show/NCT03467100) specifically targeting SCN8A with the goal of rectifying the effects of GOF variants—are currently in phase I clinical trials. Furthermore, antisense oligonucleotide therapy was recently shown to prolong survival in Scn8aR1872W/+ (GOF) mice.53 These novel compounds provide promising advances for precision medicine in SCN8A-affected individuals. As functional testing is not yet readily available for all affected individuals, it will be important to have clinical markers that may predict the underlying functional effect, when specific blockers of the Nav1.6 channel become available on the market. Our study provides an important contribution in this direction, as genotype-phenotype correlations revealed a clearly differential pattern for both GOF and LOF variants with very few exceptions.
Supplementary Material
Acknowledgements
We thank the affected individuals and their families for participating in this study.
The authors wish to acknowledge the resources of MSSNG (www.mss.ng), Autism Speaks and The Centre for Applied Genomics at The Hospital for Sick Children, Toronto, Canada. We thank the Epi25 Collaborative for providing the SCN8A variants in three individuals (see supporting grants in the ‘Funding’ section). We also thank the participating families for their time and contributions to this database, as well as the generosity of the donors who supported this program.
Funding
This work was supported by the German Research Foundation and the Fonds Nationale de la Recherche in Luxembourg (Research Unit FOR-2715, DFG grants Le1030/15–1 and/16–1 to H.L., He8155/1–1 to U.B.S.H., He5415/7–1 to I.H., FNR grant INTER/DFG/17/11583046 to P.M.), the German Federal Ministry for Education and Research (BMBF, Treat-ION, 01GM1907A to H.L., D.L. and P.M.), the foundation no epilep (to H.L. to partially support L.S.), the Medical Faculty of the University of Tuebingen (the Fortüne program 2430–00 to SL). K.S., P.L. and M.V. were funded by grant: MH CR AZV NU20-04–00279. Epi25 was supported by the National Human Genome Research Institute (NHGRI) grants UM1 HG008895 and 5U01HG009088-02 and the Stanley Center for Psychiatric Research at the Broad Institute. S.I. was employed by and received a salary from Ambry Genetics. S.S. hold the GlaxoSmithKline Endowed Chair in Genome Sciences at the Hospital for Sick Children and University of Toronto. P.S. has received speaker fees and participated at advisory boards for Biomarin, Zogenyx and GW Pharmaceuticals and has received research funding from ENECTA BV, GW Pharmaceuticals, Kolfarma Srl. and Eisai. E.M. was funded by a grant NIH NINDS K08 NS097633 and the Foerderer Award for Excellence from The Children’s Hospital of Philadelphia Research Institute. P.V. received a speaker fee from Nutricia GmbH, Dr Sch¨ar AG/SPA, Eisai, Nestlé, Pediatrica. F.Z. and P.S. developed this work within the framework of the DINOGMI Department of Excellence of MIUR 2018–2022 (legge 232 del 2016). I.H. was also supported by The Hartwell Foundation through an Individual Biomedical Research Award. This work was also supported by the National Institute for Neurological Disorders and Stroke (K02 NS112600), including support through the Center Without Walls on ion channel function in epilepsy (‘Channelopathy-associated Research Center’, U54 NS108874), the Eunice Kennedy Shriver National Institute of Child Health and Human Development through the Intellectual and Developmental Disabilities Research Center (IDDRC) at the Children’s Hospital of Philadelphia and the University of Pennsylvania (U54 HD086984) and by intramural funds from the Children’s Hospital of Philadelphia through the Epilepsy NeuroGenetics Initiative (ENGIN). Research reported in this publication was also supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001878. This project was also supported in part by the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics at the Perelman School of Medicine of the University of Pennsylvania. H.E.O. was funded by K23 NS107646-03, PI Olson.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- ASD
autism spectrum disorder
- ASM
anti-seizure medication
- BFIE
benign familial infantile epilepsy
- DEE
developmental and epileptic encephalopathy
- GOF
gain-of-function
- ID
intellectual disability
- LOF
loss-of-function
- NDDwoE
neurodevelopmental disorder without epilepsy
- SCB
sodium channel blocker
Contributor Information
Katrine M Johannesen, Department of Epilepsy Genetics and Personalized Treatment, The Danish Epilepsy Center, 4293 Dianalund, Denmark; Institute for Regional Health Services, University of Southern Denmark, 5230 Odense, Denmark.
Yuanyuan Liu, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Mahmoud Koko, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Cathrine E Gjerulfsen, Department of Epilepsy Genetics and Personalized Treatment, The Danish Epilepsy Center, 4293 Dianalund, Denmark.
Lukas Sonnenberg, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany; Institute for Neurobiology, University of Tuebingen, 72072 Tuebingen, Germany.
Julian Schubert, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Christina D Fenger, Department of Epilepsy Genetics and Personalized Treatment, The Danish Epilepsy Center, 4293 Dianalund, Denmark.
Ahmed Eltokhi, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Maert Rannap, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Nils A Koch, Institute for Neurobiology, University of Tuebingen, 72072 Tuebingen, Germany.
Stephan Lauxmann, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany; Institute for Neurobiology, University of Tuebingen, 72072 Tuebingen, Germany.
Johanna Krüger, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Josua Kegele, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Laura Canafoglia, Department of Diagnostics and Technology, Fondazione IRCCS Istituto Neurologio Carlo Besta, 20125 Milan, Italy.
Silvana Franceschetti, Department of Diagnostics and Technology, Fondazione IRCCS Istituto Neurologio Carlo Besta, 20125 Milan, Italy.
Thomas Mayer, Epilepsy Center Kleinwachau, 01454 Dresden-Radeberg, Germany.
Johannes Rebstock, Epilepsy Center Kleinwachau, 01454 Dresden-Radeberg, Germany.
Pia Zacher, Epilepsy Center Kleinwachau, 01454 Dresden-Radeberg, Germany.
Susanne Ruf, Department of Pediatric Neurology and Developmental Medicine, University Children’s Hospital, 72072 Tuebingen, Germany.
Michael Alber, Department of Pediatric Neurology and Developmental Medicine, University Children’s Hospital, 72072 Tuebingen, Germany.
Katalin Sterbova, Department of Child Neurology, 2nd Faculty of Medicine, Charles University and University Hospital Motol, 10000 Prague, Czech Republic.
Petra Lassuthová, Department of Child Neurology, 2nd Faculty of Medicine, Charles University and University Hospital Motol, 10000 Prague, Czech Republic.
Marketa Vlckova, Department of Child Neurology, 2nd Faculty of Medicine, Charles University and University Hospital Motol, 10000 Prague, Czech Republic.
Johannes R Lemke, Institute of Human Genetics, University of Leipzig Hospitals and Clinics, 4275 Leipzig, Germany.
Konrad Platzer, Institute of Human Genetics, University of Leipzig Hospitals and Clinics, 4275 Leipzig, Germany.
Ilona Krey, Institute of Human Genetics, University of Leipzig Hospitals and Clinics, 4275 Leipzig, Germany.
Constanze Heine, Institute of Human Genetics, University of Leipzig Hospitals and Clinics, 4275 Leipzig, Germany.
Dagmar Wieczorek, Institute of Human Genetics, University Clinic, Heinrich-Heine-University, 40210 Düsseldorf, Germany.
Judith Kroell-Seger, Children’s Department, Swiss Epilepsy Centre, Clinic Lengg, 8001 Zurich, Switzerland.
Caroline Lund, National Centre for Rare Epilepsy-Related Disorders, Oslo University Hospital, 0001 Oslo, Norway.
Karl Martin Klein, Departments of Clinical Neurosciences, Medical Genetics and Community Health Sciences, Hotchkiss Brain Institute and Alberta Children’s Hospital Research Institute, Cumming School of Medicine, University of Calgary, Calgary, AB T2P 0A1, Canada.
P Y Billie Au, Department of Medical Genetics, Alberta Children’s Hospital Research Institute, University of Calgary, AB T6G 2T4, Canada.
Jong M Rho, Section of Pediatric Neurology, Alberta Children’s Hospital, Cumming School of Medicine, University of Calgary, Calgary, AB T2P 0A1, Canada.
Alice W Ho, Section of Pediatric Neurology, Alberta Children’s Hospital, Cumming School of Medicine, University of Calgary, Calgary, AB T2P 0A1, Canada.
Silvia Masnada, Department of Child Neurology, V. Buzzi Children’s Hospital, 20125 Milan, Italy.
Pierangelo Veggiotti, Department of Child Neurology, V. Buzzi Children’s Hospital, 20125 Milan, Italy; ‘L. Sacco’ Department of Biomedical and Clinical Sciences, University of Milan, 20157 Milan, Italy.
Lucio Giordano, Child Neuropsychiatric Unit, Civilian Hospital, 25100 Brescia, Italy.
Patrizia Accorsi, Child Neuropsychiatric Unit, Civilian Hospital, 25100 Brescia, Italy.
Christina E Hoei-Hansen, Department of Pediatrics, Copenhagen University Hospital Rigshospitalet, 2200 Copenhagen, Denmark; Department of Clinical Medicine, University of Copenhagen, 2200 Copenhagen, Denmark.
Pasquale Striano, Department of Neurosciences, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health, University of Genoa, 16121 Genova, Italy; IRCCS ‘G. Gaslini’ Institute, 16121 Genoa, Italy.
Federico Zara, IRCCS ‘G. Gaslini’ Institute, 16121 Genoa, Italy.
Helene Verhelst, Department of Pediatrics, Division of Pediatric Neurology, Gent University Hospital, 9042 Gent, Belgium.
Judith S Verhoeven, Academic Center for Epileptology, Kempenhaeghe/Maastricht University Medical Center, 5591 Heeze, The Netherlands.
Hilde M H Braakman, Department of Pediatric Neurology, Amalia Children’s Hospital, Radboud University Medical Center, 6525 Nijmegen, The Netherlands.
Bert van der Zwaag, Department of Genetics, University Medical Center Utrecht, Utrecht University, 3553 Utrecht, The Netherlands.
Aster V E Harder, Department of Genetics, University Medical Center Utrecht, Utrecht University, 3553 Utrecht, The Netherlands.
Eva Brilstra, Department of Genetics, University Medical Center Utrecht, Utrecht University, 3553 Utrecht, The Netherlands.
Manuela Pendziwiat, Department of Neuropediatrics, Universitätsklinikum Schleswig Holstein Campus Kiel, 24106 Kiel, Germany.
Sebastian Lebon, Pediatric Neurology and Neurorehabilitation Unit, Woman Mother Child Department, Lausanne University Hospital (CHUV), 1000 Lausanne, Switzerland; University of Lausanne, 1000 Lausanne, Switzerland.
Maria Vaccarezza, Department of Pediatric Neurology, Hospital Italiano de Buenos Aires, C1428 Buenos Aires, Argentina.
Ngoc Minh Le, Center for Pediatric Neurology, Cleveland Clinic, Cleveland, OH 44102, USA.
Jakob Christensen, Department of Neurology, Aarhus University Hospital, 8000 Aarhus, Denmark.
Sabine Grønborg, Center for Rare Diseases, Department of Pediatrics and Department of Clinical Genetics, Copenhagen University Hospital Rigshospitalet, 2200 Copenhagen, Denmark.
Stephen W Scherer, McLaughlin Centre and Department of Molecular Genetics, University of Toronto, Toronto, ON 66777, Canada; The Centre for Applied Genomics and Department of Genetics and Genome Biology, The Hospital for Sick Children, Toronto, ON 66777, Canada.
Jennifer Howe, Department of Neuropediatrics, University Hospital Bonn, 53229 Bonn, Germany.
Walid Fazeli, Institute for Molecular and Behavioral Neuroscience, University of Cologne, 50667 Cologne, Germany; Neurology Department, The Royal Children’s Hospital Melbourne, 3002 Melbourne, Australia.
Katherine B Howell, Neurology Department, The Royal Children’s Hospital Melbourne, 3002 Melbourne, Australia; Murdoch Children’s Research Institute, 3052 Parkville, Australia; Department of Pediatrics, University of Melbourne, Royal Children’s Hospital, 3052 Parkville, Australia.
Richard Leventer, Neurology Department, The Royal Children’s Hospital Melbourne, 3002 Melbourne, Australia; Murdoch Children’s Research Institute, 3052 Parkville, Australia; Department of Pediatrics, University of Melbourne, Royal Children’s Hospital, 3052 Parkville, Australia.
Chloe Stutterd, Murdoch Children’s Research Institute, 3052 Parkville, Australia; Department of Pediatrics, University of Melbourne, Royal Children’s Hospital, 3052 Parkville, Australia.
Sonja Walsh, Department of Neuropediatrics, Children’s Hospital, University Hospital Carl Gustav Carus, Technical University, 1099 Dresden, Germany.
Marion Gerard, Genetics Department, CHU Côte de Nacre, 14118 Caen, France.
Bénédicte Gerard, Genetics Department, CHRU Strasbourg, 67000 Strasbourg, France.
Sara Matricardi, Child Neurology and Psychiatry Unit, Children’s Hospital G. Salesi, 60121 Ancona, Italy.
Claudia M Bonardi, Department of Woman’s and Child’s Health, Padova University Hospital, 35100 Padova, Italy.
Stefano Sartori, Child Neurology and Clinical Neurophysiology Unit, Padova University Hospital, 35100 Padova, Italy.
Andrea Berger, Department of Neuropediatrics, Klinikum Weiden, Kliniken Nordoberpfalz AG, 92637 Weiden, Germany.
Dorota Hoffman-Zacharska, Department of Medical Genetics, Institute of Mother and Child, 00-034 Warsaw, Poland.
Massimo Mastrangelo, Pediatric Neurology Unit, Vittore Buzzi Hospital, ASST Fatebenefratelli Sacco, 20100 Milan, Italy.
Francesca Darra, Department of Surgical Sciences, Dentistry, Gynecology and Pediatrics, University of Verona, 37121 Verona, Italy.
Arve Vøllo, Department of Pediatrics, Oestfold Hospital, 1712 Graalum, Norway.
M Mahdi Motazacker, Laboratory of Genome Diagnostics, Department of Clinical Genetics, Amsterdam UMC, University of Amsterdam, 1019 Amsterdam, Netherlands.
Phillis Lakeman, Department of Clinical Genetics, Amsterdam Reproduction and Development Research Institute, Amsterdam UMC, University of Amsterdam, 1019 Amsterdam, Netherlands.
Mathilde Nizon, Service de Génétique Médicale, CHU Nantes, 44093 Nantes, France.
Cornelia Betzler, Clinic for Neuropediatrics and Neurorehabilitation, Epilepsy Center for Children and Adolescents, Schön Klinik, 83569 Vogtareuth, Germany; Research Institute ‘Rehabilitation, Transition, Palliation’, PMU Salzburg, 5020 Salzburg, Austria.
Cecilia Altuzarra, Department of Pediatrics, St. Jacques Hospital, 25000 Besançon, France.
Roseline Caume, Clinique de Génétique Guy Fontaine, CHU Lille, 59000, Lille, France.
Agathe Roubertie, Département de Neuropédiatrie, INSERM, CHU Montpellier, 34000 Montpellier, France.
Philippe Gélisse, Département de Neuropédiatrie, INSERM, CHU Montpellier, 34000 Montpellier, France.
Carla Marini, Pediatric Neurology, Neurogenetics and Neurobiology Unit and Laboratories, Meyer Children’s Hospital, University of Florence, 50131 Florence, Italy.
Renzo Guerrini, IRCCS Stella Maris, 56121 Pisa, Italy.
Frederic Bilan, Service de Génétique, Centre Hospitalier Universitaire de Poitiers, 86021 Poitiers, France.
Daniel Tibussek, Child Neurology, Center for Pediatric and Teenage Health Care, 53757 Sankt Augustin, Germany.
Margarete Koch-Hogrebe, Vestische Kinder- und Jugendklinik, 45711 Datteln, Germany.
M Scott Perry, Justin Neurosciences Center, Cook Children’s Medical Center, Fort Worth, TX 76101, USA.
Shoji Ichikawa, Department of Clinical Diagnostics, Ambry Genetics, Aliso Viejo, CA 92637, USA.
Elena Dadali, Research Centre for Medical Genetics, 115522 Moscow, Russia; Veltischev Research and Clinical Institute for Pediatrics, Pirogov Russian National Research Medical University, 125412 Moscow, Russia.
Artem Sharkov, Veltischev Research and Clinical Institute for Pediatrics, Pirogov Russian National Research Medical University, 125412 Moscow, Russia; Genomed Ltd., 100000 Moscow, Russia.
Irina Mishina, Research Centre for Medical Genetics, 115522 Moscow, Russia.
Mikhail Abramov, Veltischev Research and Clinical Institute for Pediatrics, Pirogov Russian National Research Medical University, 125412 Moscow, Russia.
Ilya Kanivets, Svt. Luka’s Institute of Child Neurology & Epilepsy, 100000 Moscow, Russia; Russian Medical Academy of Continuous Professional Education, 100000 Moscow, Russia.
Sergey Korostelev, Svt. Luka’s Institute of Child Neurology & Epilepsy, 100000 Moscow, Russia; I.M. Sechenov First Moscow State Medical University, 100000 Moscow, Russia.
Sergey Kutsev, Research Centre for Medical Genetics, 115522 Moscow, Russia.
Karen E Wain, Geisinger Autism & Developmental Medicine Institute, Lewisburg, PA 17837, USA.
Nancy Eisenhauer, Geisinger Autism & Developmental Medicine Institute, Lewisburg, PA 17837, USA.
Monisa Wagner, Geisinger Autism & Developmental Medicine Institute, Lewisburg, PA 17837, USA.
Juliann M Savatt, Geisinger Autism & Developmental Medicine Institute, Lewisburg, PA 17837, USA.
Karen Müller-Schlüter, Epilepsy Center for Children, University Hospital Neuruppin, Brandenburg Medical School, 16816 Neuruppin, Germany.
Haim Bassan, Pediatric Neurology & Development Center, Shamir Medical Center (Assaf Harofe), Be'er Ya'akov, Israel; Sackler Faculty of Medicine, Tel Aviv University, 5296001 Tel Aviv, Israel.
Artem Borovikov, Research Centre for Medical Genetics, 115522 Moscow, Russia.
Marie Cecile Nassogne, Pediatric Neurology Unit, Cliniques Universitaires Saint-Luc, Université Catholique de Louvain, 1000 Brussels, Belgium.
Anne Destrée, Institute for Pathology and Genetics, 6040 Gosselies, Belgium.
An Sofie Schoonjans, Department of Pediatrics and Pediatric Neurology, Antwerp University Hospital, University of Antwerp, 2650 Edegem, Belgium.
Marije Meuwissen, Pediatric Neurology, Marie Curie Hospital—CHU Charleroi, 6032 Charleroi, Belgium.
Marga Buzatu, Pediatric Neurology, Marie Curie Hospital—CHU Charleroi, 6032 Charleroi, Belgium.
Anna Jansen, Pediatric Neurology Unit, Department of Pediatrics, Universitair Ziekenhuis Brussel, Vrije Universiteit Brussel, 1050 Brussels, Belgium.
Emmanuel Scalais, Pediatric Neurology Unit, Department of Pediatrics, Centre Hospitalier de Luxembourg, 1313 Luxembourg, Luxembourg.
Siddharth Srivastava, Department of Neurology, Boston Children’s Hospital and Harvard Medical School, Boston, MA 02108, USA.
Wen Hann Tan, Department of Genetics, Boston Children’s Hospital, Boston, MA 02108, USA.
Heather E Olson, Department of Neurology, Boston Children’s Hospital and Harvard Medical School, Boston, MA 02108, USA; Epilepsy Genetics Program, Boston Children’s Hospital, Boston, MA 02108, USA.
Tobias Loddenkemper, Department of Neurology, Boston Children’s Hospital and Harvard Medical School, Boston, MA 02108, USA.
Annapurna Poduri, Department of Neurology, Boston Children’s Hospital and Harvard Medical School, Boston, MA 02108, USA; Epilepsy Genetics Program, Boston Children’s Hospital, Boston, MA 02108, USA.
Katherine L Helbig, Division of Neurology, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; The Epilepsy Neurogenetics Initiative (ENGIN), Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Ingo Helbig, Division of Neurology, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; The Epilepsy Neurogenetics Initiative (ENGIN), Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Biomedical and Health Informatics (DBHi), Children’s Hospital of Philadelphia, Philadelphia, PA 19104 USA; Department of Neurology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104 USA; Institute of Clinical Molecular Biology, Kiel University, 24105 Kiel, Germany; Department of Neuropediatrics, Kiel University, 24105 Kiel, Germany.
Mark P Fitzgerald, Division of Neurology, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; The Epilepsy Neurogenetics Initiative (ENGIN), Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Biomedical and Health Informatics (DBHi), Children’s Hospital of Philadelphia, Philadelphia, PA 19104 USA; Institute of Clinical Molecular Biology, Kiel University, 24105 Kiel, Germany.
Ethan M Goldberg, Division of Neurology, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; The Epilepsy Neurogenetics Initiative (ENGIN), Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Timo Roser, Division of Pediatric Neurology, Developmental Medicine and Social Pediatrics, Department of Pediatrics, Dr. von Haunersches Children’s Hospital, Ludwig-Maximilian-University of Munich, 80331 Munich, Germany.
Ingo Borggraefe, Division of Pediatric Neurology, Developmental Medicine and Social Pediatrics, Department of Pediatrics, Dr. von Haunersches Children’s Hospital, Ludwig-Maximilian-University of Munich, 80331 Munich, Germany; Comprehensive Epilepsy Center, Ludwig-Maximilian- University of Munich, 80331 Munich, Germany.
Tobias Brünger, Luxembourg Centre for Systems Biomedicine (LCSB), University Luxembourg, L-4243 Esch-sur-Alzette, Luxembourg.
Patrick May, Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 44102, USA.
Dennis Lal, Luxembourg Centre for Systems Biomedicine (LCSB), University Luxembourg, L-4243 Esch-sur-Alzette, Luxembourg; Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, OH 44102, USA; Stanley Center for Psychiatric Research, Broad Institute of Harvard and M.I.T ., Cambridge, MA 02138, USA; Cologne Center for Genomics (CCG), University of Cologne, 50667 Cologne, Germany.
Damien Lederer, Institute for Pathology and Genetics, 6040 Gosselies, Belgium.
Guido Rubboli, Department of Epilepsy Genetics and Personalized Treatment, The Danish Epilepsy Center, 4293 Dianalund, Denmark; University of Copenhagen, 2200 Copenhagen, Denmark.
Henrike O Heyne, Institute of Human Genetics, University of Leipzig Hospitals and Clinics, 4275 Leipzig, Germany; Finnish Institute for Molecular Medicine (FIMM), University of Helsinki, 320 Helsinki, Finland; Program for Medical and Population Genetics/Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02138, USA; Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA 02108, USA.
Gaetan Lesca, Department of Medical Genetics, Groupement Hospitalier Est and ERN EpiCARE, University Hospitals of Lyon (HCL), 69001 Lyon, France; Institut Neuromyogène, CNRS UMR 5310 - INSERM U1217, Université de Lyon, Université Claude Bernard Lyon 1, 69001 Lyon, France.
Ulrike B S Hedrich, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Jan Benda, Institute for Neurobiology, University of Tuebingen, 72072 Tuebingen, Germany.
Elena Gardella, Department of Epilepsy Genetics and Personalized Treatment, The Danish Epilepsy Center, 4293 Dianalund, Denmark; Institute for Regional Health Services, University of Southern Denmark, 5230 Odense, Denmark.
Holger Lerche, Department of Neurology and Epileptology, Hertie Institute for Clinical Brain Research, University of Tuebingen, 72072 Tuebingen, Germany.
Rikke S Møller, Department of Epilepsy Genetics and Personalized Treatment, The Danish Epilepsy Center, 4293 Dianalund, Denmark; Institute for Regional Health Services, University of Southern Denmark, 5230 Odense, Denmark.
References
- 1. Veeramah KR, O'Brien JE, Meisler MH, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90(3):502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anand G, Collett-White F, Orsini A, et al. Autosomal dominant SCN8A mutation with an unusually mild phenotype. Eur J Paediatr Neurol. 2016;20(5):761–765. [DOI] [PubMed] [Google Scholar]
- 3. Gardella E, Becker F, Moller RS, et al. Benign infantile seizures and paroxysmal dyskinesia caused by an SCN8A mutation. Ann Neurol. 2016;79(3):428–436. [DOI] [PubMed] [Google Scholar]
- 4. Han JY, Jang JH, Lee IG, Shin S, Park J. A novel inherited mutation of SCN8A in a Korean family with benign familial infantile epilepsy using diagnostic exome sequencing. Ann Clin Lab Sci. 2017;47:747–753. [PubMed] [Google Scholar]
- 5. Johannesen KM, Gardella E, Encinas AC, et al. The spectrum of intermediate SCN8A-related epilepsy. Epilepsia. 2019;60(5):830–844. [DOI] [PubMed] [Google Scholar]
- 6. Estacion M, O'Brien JE, Conravey A, et al. A novel de novo mutation of SCN8A (Nav1.6) with enhanced channel activation in a child with epileptic encephalopathy. Neurobiol Dis. 2014;69:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohba C, Kato M, Takahashi S, et al. Early onset epileptic encephalopathy caused by de novo SCN8A mutations. Epilepsia. 2014;55(7):994–1000. [DOI] [PubMed] [Google Scholar]
- 8. Vaher U, Noukas M, Nikopensius T, et al. De novo SCN8A mutation identified by whole-exome sequencing in a boy with neonatal epileptic encephalopathy, multiple congenital anomalies, and movement disorders. J Child Neurol. 2014;29(12):Np202–206. [DOI] [PubMed] [Google Scholar]
- 9. Fung LW, Kwok SL, Tsui KW. SCN8A mutations in Chinese children with early onset epilepsy and intellectual disability. Epilepsia. 2015;56(8):1319–1320. [DOI] [PubMed] [Google Scholar]
- 10. Larsen J, Carvill GL, Gardella E, et al. ; EuroEPINOMICS RES Consortium CRP . The phenotypic spectrum of SCN8A encephalopathy. Neurology. 2015;84(5):480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Kovel CG, Meisler MH, Brilstra EH, et al. Characterization of a de novo SCN8A mutation in a patient with epileptic encephalopathy. Epilepsy Res. 2014;108(9):1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rolvien T, Butscheidt S, Jeschke A, et al. Severe bone loss and multiple fractures in SCN8A-related epileptic encephalopathy. Bone. 2017;103:136–143. [DOI] [PubMed] [Google Scholar]
- 13. Wang J, Gao H, Bao X, et al. SCN8A mutations in Chinese patients with early onset epileptic encephalopathy and benign infantile seizures. BMC Med Genet. 2017;18(1):104- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gardella E, Marini C, Trivisano M, et al. The phenotype of SCN8A developmental and epileptic encephalopathy. Neurology. 2018;91(12):e1112–e1124. [DOI] [PubMed] [Google Scholar]
- 15. Johannesen KM, Gardella E, Scheffer I, et al. Early mortality in SCN8A-related epilepsies. Epilepsy Res. 2018;143:79–81. [DOI] [PubMed] [Google Scholar]
- 16. Trudeau MM, Dalton JC, Day JW, Ranum LP, Meisler MH. Heterozygosity for a protein truncation mutation of sodium channel SCN8A in a patient with cerebellar atrophy, ataxia, and mental retardation. J Med Genet. 2006;43(6):527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blanchard MG, Willemsen MH, Walker JB, et al. De novo gain-of-function and loss-of-function mutations of SCN8A in patients with intellectual disabilities and epilepsy. J Med Genet. 2015;52(5):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagnon JL, Barker BS, Ottolini M, et al. Loss-of-function variants of SCN8A in intellectual disability without seizures. Neurol Genet. 2017;3(4):e170- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y, Schubert J, Sonnenberg L, et al. Neuronal mechanisms of mutations in SCN8A causing epilepsy or intellectual disability. Brain. 2019;142(2):376–390. [DOI] [PubMed] [Google Scholar]
- 20. Bunton-Stasyshyn RKA, Wagnon JL, Wengert ER, et al. Prominent role of forebrain excitatory neurons in SCN8A encephalopathy. 2019;142(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagnon JL, Mencacci NE, Barker BS, et al. Partial loss-of-function of sodium channel SCN8A in familial isolated myoclonus. Hum Mutat. 2018;39(7):965–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boerma RS, Braun KP, van de Broek MP, et al. Remarkable phenytoin sensitivity in 4 children with SCN8A-related Epilepsy: A molecular neuropharmacological approach. Neurotherapeutics. 2016;13(1):192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fisher RS, Cross JH, French JA, et al. Operational classification of seizure types by the International League Against Epilepsy: Position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58(4):522–530. [DOI] [PubMed] [Google Scholar]
- 26. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Denis J, Villeneuve N, Cacciagli P, et al. Clinical study of 19 patients with SCN8A-related epilepsy: Two modes of onset regarding EEG and seizures. Epilepsia. 2019;60(5):845–856. [DOI] [PubMed] [Google Scholar]
- 28. Epi25 . Ultra-rare genetic variation in the epilepsies: A whole-exome sequencing study of 17,606 individuals. Am J Hum Genet. 2019;105(2):267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagnon JL, Meisler MH. Recurrent and non-recurrent mutations of SCN8A in epileptic encephalopathy. Front Neurol. 2015;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pan Y, Cummins TR. Distinct functional alterations in SCN8A epilepsy mutant channels. J Physiol. 2020;598(2):381–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schreiber JM, Tochen L, Brown M, et al. A multi-disciplinary clinic for SCN8A-related epilepsy. Epilepsy Res. 2020;159:106261. [DOI] [PubMed] [Google Scholar]
- 32. Epifanio R, Zanotta N, Giorda R, Bardoni A, Zucca C. Novel epilepsy phenotype associated to a known SCN8A mutation. Seizure. 2019;67:15–17. [DOI] [PubMed] [Google Scholar]
- 33. Ranza E, Z'Graggen W, Lidgren M, et al. SCN8A heterozygous variants are associated with anoxic-epileptic seizures. Am J Med Genet Part A. 2020;182(5):1209–1216. [DOI] [PubMed] [Google Scholar]
- 34. Wengert ER, Tronhjem CE, Wagnon JL et al. Biallelic inherited SCN8A variants, a rare cause of SCN8A-related developmental and epileptic encephalopathy. Epilepsia. 2019;60(11):2277–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lauxmann S, Verbeek NE, Liu Y, et al. Relationship of electrophysiological dysfunction and clinical severity in SCN2A-related epilepsies. Hum Mutat. 2018;39(12):1942–1956. [DOI] [PubMed] [Google Scholar]
- 36. Kohrman DC, Smith MR, Goldin AL, Harris J, Meisler MH. A missense mutation in the sodium channel Scn8a is responsible for cerebellar ataxia in the mouse mutant jolting. J Neurosci. 1996;16(19):5993–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith MR, Goldin AL. A mutation that causes ataxia shifts the voltage-dependence of the Scn8a sodium channel. Neuroreport. 1999;10(14):3027–3031. [DOI] [PubMed] [Google Scholar]
- 38. Dick DJ, Boakes RJ, Harris JB. A cerebellar abnormality in the mouse with motor end-plate disease. Neuropathol Appl Neurobiol. 1985;11(2):141–147. [DOI] [PubMed] [Google Scholar]
- 39. Harris JB, Boakes RJ, Court JA. Physiological and biochemical studies on the cerebellar cortex of the murine mutants "jolting" and "motor end-plate disease". J Neurol Sci. 1992;110(1-2):186–194. [DOI] [PubMed] [Google Scholar]
- 40. Papale LA, Beyer B, Jones JM, et al. Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum Mol Genet. 2009;18(9):1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Makinson CD, Tanaka BS, Sorokin JM, et al. Regulation of Thalamic and Cortical Network Synchrony by Scn8a. Neuron. 2017;93(5):1165–1179.e1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolff M, Johannesen KM, Hedrich UBS, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140(5):1316–1336. [DOI] [PubMed] [Google Scholar]
- 43. Liao Y, Deprez L, Maljevic S, et al. Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain. 2010;133(Pt 5):1403–1414. [DOI] [PubMed] [Google Scholar]
- 44. Brunklaus A, Du J, Steckler F, et al. Biological concepts in human sodium channel epilepsies and their relevance in clinical practice. Epilepsia. 2020;61(3):387–399. [DOI] [PubMed] [Google Scholar]
- 45. Schwarz N, Bast T, Gaily E, et al. Clinical and genetic spectrum of <em>SCN2A</em>-associated episodic ataxia. Eur J Paediatr Neurol. 2019. [DOI] [PubMed] [Google Scholar]
- 46. Ogiwara I, Miyamoto H, Tatsukawa T, et al. Nav1.2 haplodeficiency in excitatory neurons causes absence-like seizures in mice. Commun Biol. 2018;1:96-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88(4):1407–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu FH, Mantegazza M, Westenbroek RE, et al. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9(9):1142–1149. [DOI] [PubMed] [Google Scholar]
- 49. Ogiwara I, Miyamoto H, Morita N, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: A circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27(22):5903–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bayat A, Hjalgrim H, Moller RS. The incidence of SCN1A-related Dravet syndrome in Denmark is 1:22,000: A population-based study from 2004 to 2009. Epilepsia. 2015;56(4):e36–e39. [DOI] [PubMed] [Google Scholar]
- 51. Werling DM, Brand H, An JY, et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet. 2018;50(5):727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baker EM, Thompson CH, Hawkins NA, et al. The novel sodium channel modulator GS-458967 (GS967) is an effective treatment in a mouse model of SCN8A encephalopathy. Epilepsia. 2018;59(6):1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lenk GM, Jafar-Nejad P, Hill SF, et al. Scn8a antisense oligonucleotide is protective in mouse models of SCN8A encephalopathy and Dravet syndrome. Ann Neurol. 2020;87(3):339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon request.